Abstract
Survivin promotes cell division and suppresses apoptosis in many human cancers, and increased abundance correlates with metastasis and poor prognosis. Here, we showed that a pool of survivin that localized to the mitochondria of certain tumor cell lines enhanced the stability of oxidative phosphorylation Complex II, which promoted cellular respiration. Survivin also supported the subcellular trafficking of mitochondria to the cortical cytoskeleton of tumor cells, which was associated with increased membrane ruffling, increased focal adhesion complex turnover, and increased tumor cell migration and invasion in cultured cells, and enhanced metastatic dissemination in vivo. Therefore, we found that mitochondrial respiration enhanced by survivin contributes to cancer metabolism, and relocalized mitochondria may provide a “regional” energy source to fuel tumor cell invasion and metastasis.
INTRODUCTION
The Inhibitor-of-Apoptosis (IAP) family member survivin functions in multiple mechanisms, including chromosomal segregation, microtubule dynamics, apoptosis resistance, and cellular stress responses (1). The transcription of the gene encoding survivin is greater in tumors than in normal tissues, and the presence of survivin in cancer has been linked to metastatic disease (2), but the underlying mechanism(s) has not been clearly defined. In tumors, a pool of survivin localizes to mitochondria (3), where it promotes resistance to apoptosis (4) and influences organelle bioenergetics (5), thus acting as a potential cancer driver.
Although indispensable for normal tissue and organ bioenergetics, the role of mitochondria in cancer has been debated (6). Most tumors rewire their energy sources towards aerobic glycolysis at the expense of mitochondrial respiration (7), the so-called “Warburg effect” (8), a process that is important for disease progression (9). Further, mutations in oxidative phosphorylation genes produce “oncometabolites” (10) or stabilize oncogenes, such as HIF1α (11), suggesting that mitochondrial respiration may have limited roles in cancer bioenergetics (12), and, at least in some cases, actually function as a “tumor suppressor” (13).
Conversely, there is evidence that oxidative phosphorylation remains an important source of ATP for many tumors (14) and may affect important cancer traits, such as “stemness” (15), tumor repopulation after oncogene ablation (16), and resistance to therapy (17). Whether there are cancer-specific regulators of mitochondrial respiration is presently unknown, but protein folding quality control within the unique anatomy of mitochondria (18) is required to buffer the risk of proteotoxic stress (19), and provides a key requirement for oxidative phosphorylation in tumors (20). Mechanistically, this involves the chaperone activity of Heat Shock Protein-90 (Hsp90) family proteins, which accumulate in tumor mitochondria compared to normal tissues (21), and maintain the stability and folding of multiple bioenergetics effectors, including succinate dehydrogenase (SDH), an iron- and sulfur-containing subunit of oxidative phosphorylation Complex II (22).
In this study, we explored a link between mitochondrial survivin, tumor metabolic reprogramming and metastatic competency.
RESULTS
Survivin-mediated regulation of tumor bioenergetics
We began this study by examining the distribution of mitochondrial survivin in androgen-independent prostate cancer PC3 cells (3). Analysis of sub-mitochondrial fractions revealed that survivin localized to the inner membrane and matrix, but not to outer membrane or inter-membrane space (fig. S1A). With this topography, survivin co-localized with effectors of mitochondrial protein folding, including the AAA+ matrix protease CLPP and the molecular chaperones Hsp90 and TRAP-1 (TNFR-associated protein-1) (23) (fig. S1A).
Transfection of PC3 cells with a previously characterized small interfering RNA (siRNA) directed against survivin (24) efficiently depleted the mitochondrial pool of survivin (fig. S1B). In addition, treatment of PC3 cells with YM155, a small molecule survivin “suppressant” currently in clinical trials (2), also abrogated the mitochondrial pool of survivin (fig. S1C). At the low concentrations of YM155 used and short incubation times, survivin targeting did not affect mitochondrial membrane potential (fig. S1D), nuclear morphology (fig. S1E) or cell cycle transitions (fig. S1F), and only modestly reduced cell proliferation (fig. S1G).
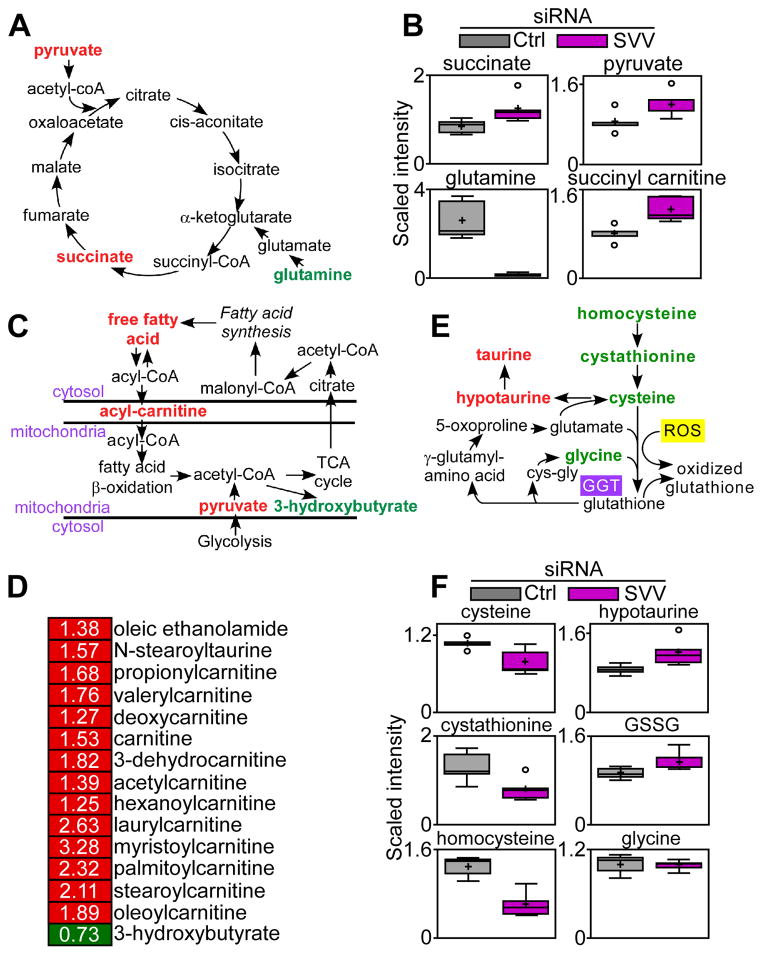
To determine whether mitochondrial survivin affected bioenergetics, we next profiled the metabolome of PC3 cells transfected with control or survivin-directed siRNA. Survivin silencing induced defective mitochondrial bioenergetics, with increased concentrations of various oxidative phosphorylation metabolites, including succinate and pyruvate, and a trend of increased α-ketoglutarate concentrations, associated with a substantial depletion of glutamine (Fig. 1A, B, Table S1), suggestive of compensatory glutaminolysis. Although increased succinate has been linked to a tumorigenic, “pseudo-hypoxic” state (6), survivin knockdown did not affect HIF1α protein abundance (fig. S1H), which mediates transcriptional responses to hypoxia (11). Consistent with impaired mitochondrial bioenergetics, survivin-silenced cells accumulated certain species of long chain fatty acids (Fig, 1C, Table S1), with increased concentrations of several carnitine-conjugated lipids involved in fatty acid import into mitochondria, and reduced concentrations of the ketone body 3-hydroxybutyrate (Fig. 1D, Table S1). In addition, survivin depletion in PC3 cells decreased the concentrations of homocysteine, cystathionine and glycine, all of which are implicated in redox mechanisms (Fig. 1E and F, Table S1); reduced isoleucine and leucine, which are involved in the metabolism of branched chain amino acids (fig. S1I and table S1), and reduced aspartate, ornithine, and putrescine concentrations and a trend of decreased concentrations of proline, which are implicated in arginine metabolism (fig. S1J and table S1).
Fig. 1. Survivin targeting impairs mitochondrial metabolism.
(A–F) Global metabolomics screening of PC3 cells transfected with control or survivin-directed (SVV) siRNA (n=5 biological replicates; see also table S1). Changes in the concentrations of metabolites implicated in oxidative phosphorylation (A and B), fatty acid β oxidation (C and D), or glutathione metabolism (E and F) are shown. For the heatmap (D), red indicates increased concentration; green indicates decreased concentration. Only significant changes (p<0.05) are shown. For box plots (B and F), relative metabolite abundance is represented. The limit of upper and lower quartiles, median values (straight line), and maximum and minimum distribution are shown. Cross, mean value; circle, extreme data point.
Modulation of oxidative phosphorylation by mitochondrial survivin
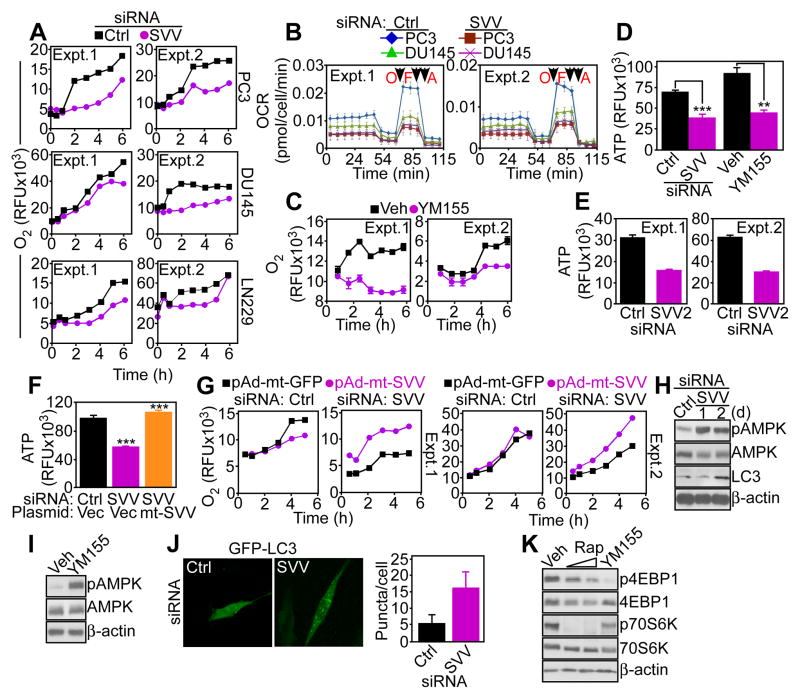
Consistent with the metabolomics data, siRNA-mediated silencing of survivin in prostate adenocarcinoma PC3 or DU145, or glioblastoma LN229 cells decreased oxygen (O2) consumption (Fig. 2A), and oxygen consumption rates (OCR) as assessed by real-time analysis of cellular respiration (Fig. 2B, fig. S2A). Targeting survivin with YM155 comparably suppressed O2 consumption in PC3 cells (Fig. 2C). Impaired bioenergetics in survivin-targeted PC3 cells correlated with reduced production of ATP, compared to control cultures (Fig. 2D). Silencing of survivin with an independent, previously characterized siRNA sequence (24) (fig. S2C) also inhibited ATP production (Fig. 2E).
Fig. 2. Survivin regulation of mitochondrial bioenergetics.
(A) Prostate cancer cell lines PC3 or DU145 or glioblastoma LN229 cells transfected with control (Ctrl) or survivin-directed (SVV) siRNA were analyzed for oxygen (O2) consumption. RFU, relative fluorescence units. The results of two independent experiments are shown. (B) siRNA silenced cells as in (A) were analyzed for O2 consumption rates (OCR) under basal conditions or in response to oligomicyin (O), FCCP (F), or antimycin (A). Arrows, time of drug addition. Extra-mitochondrial respiration after addition of antimycin was subtracted as background. The profiles of two independent experiments (Expt.) are shown. (C) PC3 cells were treated with vehicle (Veh) or small molecule survivin suppressant YM155 and analyzed for O2 consumption. RFU, relative fluorescence units. Two independent experiments (Expt.) are shown. (D) PC3 cells transfected with the indicated siRNA or treated with vehicle (Veh) or YM155 were analyzed for ATP production. Graph shows means±SEM from three independent experiments. ***, p<0.0001; **, p=0.0011. (E) PC3 cells transfected with control siRNA (Ctrl) or a second independent siRNA to survivin (SVV2) were analyzed for ATP production. Two independent experiments (Expt.) are shown. (F) PC3 cells transfected with control (Ctrl) or survivin-directed siRNA were reconstituted with vector or mt-SVV cDNA and analyzed for ATP production. Graph shows means±SEM from three independent experiments. ***, p<0.0001. (G) PC3 cells were transfected with control siRNA (Ctrl) or survivin (SVV)-directed siRNA, reconstituted with pAd-mitochondrial-targeted GFP (pAd-mt-GFP) or pAd-mitochondrial-targeted survivin (pAd-mt-SVV), and analyzed for O2 consumption. RFU, relative fluorescence units. Two independent experiments (Expt.) are shown. (H and I) PC3 cells transfected with control or survivin-directed siRNA (H) or treated with vehicle or YM155 (I) were analyzed by Western blotting. d, days. p, phosphorylated. Blots are representative of two independent experiments. (J) PC3 cells transfected with control (Ctrl) or survivin-directed siRNA (SVV) plus GFP-LC3 were analyzed by fluorescence microscopy. Right, the number of GFP-LC3 puncta (autophagosomes) per cell was quantified (n=50 cells imaged/transfection condition in two independent experiments). Scale bar, 10 μm. (K) PC3 cells treated with vehicle (Veh), Rapamycin (Rap) or YM155 were analyzed by Western blotting after 24 h. p, phosphorylated. Blots are representative of two independent experiments.
To examine the specificity of these findings, we next reconstituted PC3 cells silenced for endogenous survivin by siRNA with a mitochondrial-targeted survivin variant (mt-SVV) (3) that accumulates in mitochondria (fig. S2D), and localizes to the inner membrane and matrix in a similar manner to endogenous survivin (fig. S2E). Reconstitution of survivin-silenced PC3 cells with mt-SVV (fig. S2F) restored ATP production to that of control cultures (Fig. 2F). Similarly, transfection of mt-SVV increased ATP production in breast adenocarcinoma MCF-7 cells (fig. S2G) as well as rat insulinoma INS-1 cells (fig. S2H) that lack mitochondrial survivin (3). Furthermore, reconstitution of survivin-depleted PC3 cells with adenovirus (pAd) encoding mt-SVV (3) (pAd-mt-SVV) stimulated O2 consumption (Fig. 2G). In contrast, PC3 cells transfected with non-targeting siRNA and reconstituted with pAd-mt-SVV or pAd-mt-GFP had comparable O2 consumption (Fig. 2G). Accordingly, recombinant survivin accumulated to a greater extent in mitochondrial fractions compared to cytosol of PC3 cells (fig. S2I). Functionally, accumulation of mitochondrial survivin in MCF-7 or INS-1 cells did not change cell cycle transitions as assessed by propidium iodide staining (fig. S2J) or cell proliferation as assessed by BrdU incorporation (fig. S2K and L).
Consistent with impaired mitochondrial bioenergetics, siRNA (Fig. 2H) or YM155 (Fig. 2I) targeting of survivin induced hallmarks of cellular starvation, including increased phosphorylation of the energy sensor AMP-activated kinase (AMPK), induction of autophagy as assessed by microtubule-associated LC3 conversion (Fig. 2H) and the appearance of punctate GFP-LC3 fluorescence corresponding to autophagosome formation (Fig. 2J). The response to YM155 was accompanied by decreased mammalian target of rapamycin (mTOR) signaling, as indicated by reduced phosphorylation of the downstream effectors 4EBP1 and p70S6 kinase (Fig. 2K).
Survivin-mediated regulation of mitochondrial oxidative phosphorylation Complex II
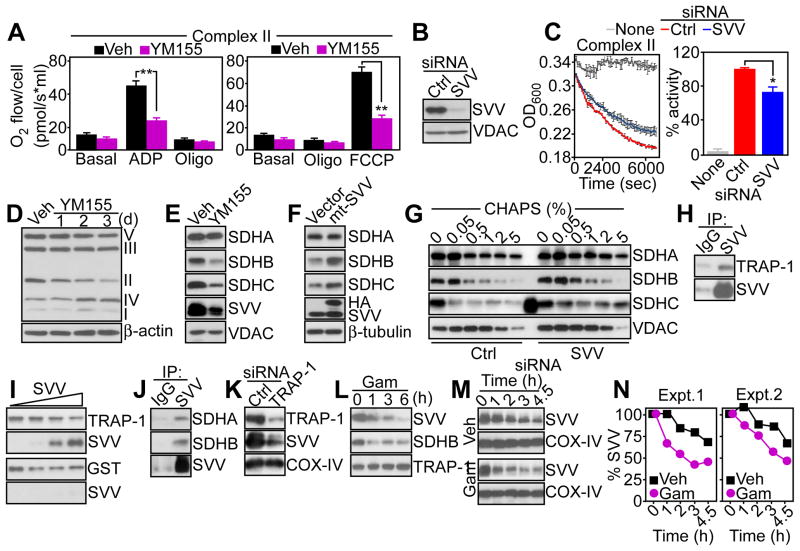
We next examined the role of survivin on the function of individual oxidative phosphorylation complexes. High-resolution respirometry of permeabilized PC3 cells showed that targeting survivin with YM155 inhibited the activity of Complex II (Fig. 3A) and Complex I activity (fig. S3A) but not that of Complex III (fig. S3B). Similarly, immuno-captured Complex II from PC3 cells silenced for endogenous survivin by siRNA (Fig. 3B) showed reduced SDH activity (Fig. 3C), whereas citrate synthase or Complex I activity was not affected (fig. S3C).
Fig. 3. Survivin regulation of mitochondrial Complex II.
(A) PC3 cells treated with vehicle (Veh) or YM155 were permeabilized with digitonin and analyzed for Complex II activity in the presence of succinate and rotenone. Oligo, oligomycin. Graphs shows means±SEM from three independent experiments. **, p<0.01. (B) PC3 cells transfected with control (Ctrl) or survivin-directed (SVV) siRNA were analyzed by Western blotting. Blots are representative of two independent experiments. (C) Mitochondrial Complex II was immunoprecipitated from siRNA-transfected PC3 cells as in (B) and analyzed for enzymatic activity. Right, quantification. Graph shows means±SEM from three independent experiments. *, p=0.037. (D) Mitochondria from PC3 cells treated with vehicle (Veh) or YM155 were analyzed by Western blotting. The position of oxidative phosphorylation complex subunits is indicated. d, days. Blots are representative of two independent experiments. (E and F) PC3 cells treated with vehicle (Veh) or YM155 (E), or MCF-7 cells transfected with vector or mt-SVV cDNA (F) were analyzed by Western blotting. In (F), the position of endogenous (SVV) or transfected (HA) survivin is indicated. Blots in (E) and (F) are representative of two independent experiments. (G) PC3 cells were transfected with the indicated siRNAs and proteins remaining insoluble at increasing detergent concentrations (CHAPS) were analyzed by Western blotting. The extra band in the SDHC lane corresponds to non-specific reactivity with a molecular weight marker. Blots are representative of two independent experiments. (H) Mitochondrial extracts from PC3 cells were immunoprecipitated (IP) with IgG or an antibody to survivin, and pellets were analyzed by Western blotting. Blots are representative of two independent experiments. (I) Aliquots of GST-TRAP-1 (top) or GST (bottom) were incubated with recombinant survivin (SVV), and bound proteins were analyzed by Western blotting. The molar ratio of survivin to TRAP-1 was 0, 0.1, 0.5 and 1. Blots are representative of two independent experiments. (J) PC3 mitochondrial extracts were immunoprecipitated (IP) with IgG or an antibody to survivin, and pellets were analyzed by Western blotting. Blots are representative of two independent experiments. (K and L) PC3 cells were transfected with control (Ctrl) or TRAP-1-directed siRNA (K), or treated with mitochondrial-targeted small molecule Hsp90 inhibitor, Gamitrinib (Gam) (L), and extracts were analyzed after 48 h (K) or at the indicated time intervals (L), by Western blotting. Blots are representative of two independent experiments for (K) and (L). (M and N) PC3 cells were treated with vehicle (Veh) or Gamitrinib (Gam), then cycloheximide, and analyzed by Western blotting (M), with quantification of survivin half-life by densitometry (N). The quantification from two independent experiments (Expt.) is shown.
Next, we asked how survivin affected mitochondrial Complex II activity. PC3 cells exposed to YM155 exhibited time-dependent degradation of Complex II-associated proteins, but not of other mitochondrial phosphorylation complexes (Fig. 3D), a response that involved SDHB and SDHC subunits of Complex II, but not the SDHA subunit (Fig. 3E). Conversely, transfection of mt-SVV in survivin-silenced PC3 cells was sufficient to increase the abundance of SDHB and SDHC subunits, but not that of SDHA subunit (Fig. 3F). To determine whether changes in SDH abundance reflected protein (mis)folding, we next quantified the amount of Complex II proteins that remained insoluble over a broad range of detergent concentrations. Survivin silencing in PC3 cells increased the amount of detergent-insoluble Complex II proteins, compared to control transfectants (fig. S3D), suggestive of protein misfolding. Complex V abundance was also modestly reduced, whereas the abundance of Complex I, III or IV was not affected (fig. S3D). In terms of individual subunits, loss of survivin increased the detergent insolubility of SDHB and SDHC, but not that of SDHA (Fig. 3G, fig. S3D).
Complex II-SDHB protein folding requires the mitochondrial chaperone Hsp90 and its homolog TRAP-1 (22). In co-immunoprecipitation experiments from mitochondrial extracts, survivin formed a complex with TRAP-1 (Fig. 3H). Similarly, recombinant survivin bound to GST-TRAP-1, but not to GST, in pull down experiments in vitro, indicating that this interaction was direct (Fig. 3I). Survivin immune complexes also contained the Complex II subunits SDHA and SDHB (Fig. 3J). Silencing of TRAP-1 by siRNA (Fig. 3K) or inhibiting its chaperone activity with the mitochondrial-targeted small molecule ATPase antagonist Gamitrinib (20) (Fig. 3L), induced time-dependent degradation of mitochondrial survivin. Accordingly, Gamitrinib treatment shortened the half-life of mitochondrial survivin from >4.5 h to approximately 2.4 h (Fig. 3M and N), suggesting that survivin is a “client protein” of TRAP-1 in mitochondria. Consistent with chaperone-dependent SDHB folding (22), Gamitrinib decreased SDHB abundance in PC3 cells (Fig. 3L). In contrast, recombinant survivin did not affect TRAP-1 ATPase activity in vitro (fig. S3F).
Association between survivin-mediated regulation of bioenergetics and subcellular mitochondrial trafficking
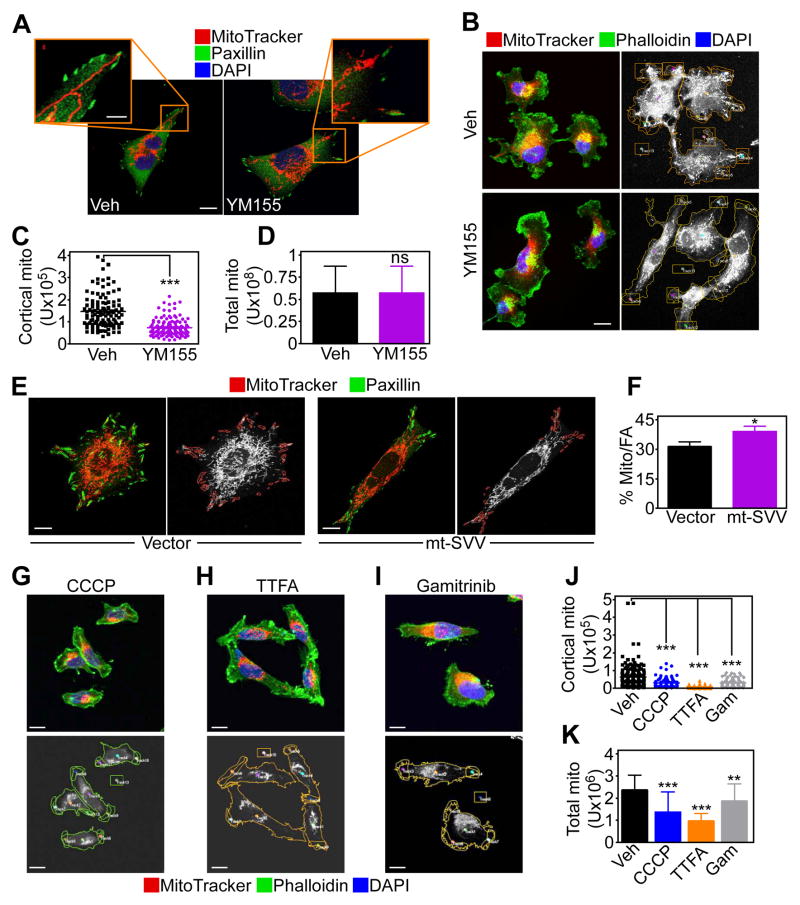
Next, we asked whether survivin-directed bioenergetics affected tumor cell behavior. We found that chemotactic stimuli changed the morphology of mitochondria in PC3 cells, with the appearance of elongated organelles that infiltrated regions of the cortical cytoskeleton, close to membrane protrusions that participate in cell motility (Fig. 4A and fig. S4A). In contrast, YM155 inhibited the subcellular trafficking of mitochondria (Fig. 4A and fig. S4A), and depleted the number of mitochondria associated with the cortical cytoskeleton after chemoattractant stimulation (Fig. 4C) without affecting total mitochondrial content (Fig. 4D). Reciprocally, transfection of MCF-7 cells with mt-SVV cDNA increased the localization of elongated mitochondria to paxillin-positive regions of the cortical cytoskeleton (Fig. 4E and F) corresponding to focal adhesion complexes (25), compared to control transfectants.
Fig. 4. Survivin-mediated regulation of subcellular mitochondrial trafficking.
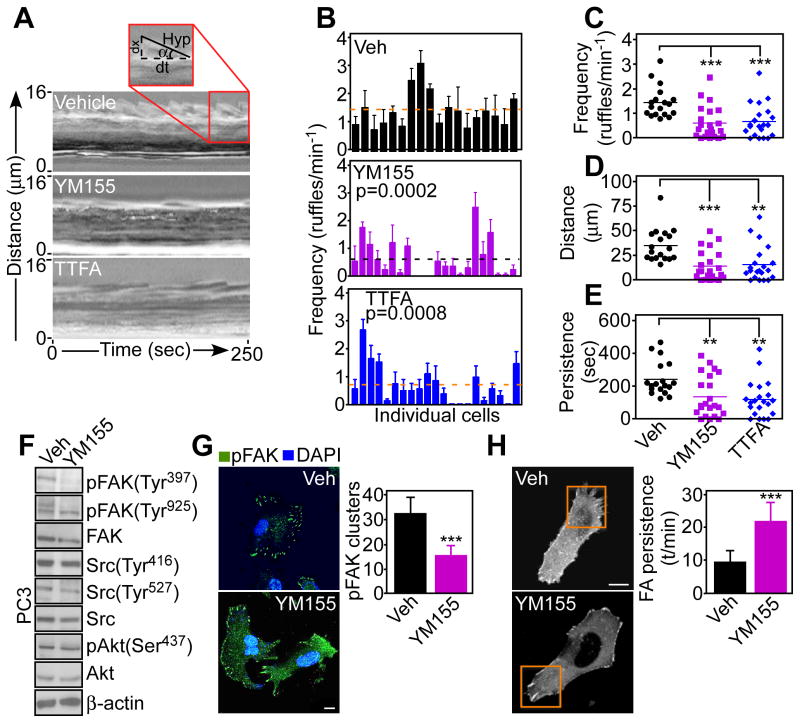
(A) PC3 cells were treated with vehicle (Veh) or YM155, labeled for mitochondria (MitoTracker, red), focal adhesion (FA) complexes (Paxillin, green) and DNA (DAPI, blue), and analyzed by confocal microscopy. 20 cells/treatment condition imaged in two independent experiments. Scale bar, 10 μm; insets, 5 μm. (B) PC3 cells treated as in (A) were labeled for mitochondria (MitoTracker, red), actin (phalloidin, green) and DNA (DAPI, blue), and analyzed by confocal microscopy (left). Right, masked images used to quantify mitochondrial localization to phalloidin-positive protrusions. 20 cells/treatment condition imaged in two independent experiments. Scale bar, 10 μm. (C and D) The conditions are as in (B), and the amount of cortical mitochondria (C) or total mitochondria (D) was quantified in the presence of vehicle or YM155. Each point corresponds to an individual measurement. 45 cells/treatment condition imaged in two independent experiments. ***, p<0.0001; n.s., not significant. (E and F) MCF-7 cells were transduced with vector or mt-SVV, labeled for mitochondria (MitoTracker, red) and paxillin (green) and analyzed by fluorescence microscopy (E). Right, masked (right panels) images were used to quantify mitochondrial localization to paxillin-containing regions (F). Data are expressed as percent of mitochondria localization to paxillin-containing focal adhesion (FA) complexes per cell. Mean±SEM. Vector, n=26 cells; mt-SVV, n=27; imaged in two independent experiments. *, p=0.0013. Scale bar, 10 μm. (G–I) The condition are as in (B) except that PC3 cells were treated with the mitochondrial uncoupler CCCP (G), the small molecule Complex II inhibitor TTFA (H), or Gamitrinib (I) and analyzed by confocal microscopy (top). Bottom, masked images used to quantify mitochondria localization to phalloidin-positive cellular protrusions. 45 cells/treatment condition imaged in two independent experiments. Scale bar, 10 μm. (J and K) Treated PC3 cells were analyzed by confocal microscopy as in (G–I), and the amount of cortical mitochondria (J) or total mitochondria (K) was quantified. Each point corresponds to an individual measurement. ***, p<0.0001; **, p=0.0013. 45 cells/treatment condition imaged in two independent experiments.
Next, we looked at the requirements of mitochondrial trafficking to the cortical cytoskeleton in PC3 cells. Dissipation of mitochondrial membrane potential with CCCP (Fig. 4G, inhibition of Complex II activity with the small molecule SDHB antagonist thenoyltrifluoroacetone (TTFA) (22) (Fig. 4H) or inhibition of SDHB degradation by Gamitrinib (22) (Fig. 4I) all suppressed mitochondrial trafficking to the cortical cytoskeleton (Fig. 4H–J). These responses were associated with a reduction in mitochondrial content (Fig. 4K), potentially reflecting organelle fragmentation (fig. S4B). At the concentrations used, TTFA did not affect PC3 cell proliferation (fig. S4C).
Regulation of membrane dynamics by cortical mitochondria
We reasoned that redistribution of mitochondria to the cortical cytoskeleton near focal adhesion complexes (Fig. 4E and F) could fuel the energy-intensive process of tumor cell movements. Consistent with this possibility, targeting survivin with YM155 or inhibiting SDHB with TTFA in PC3 cells suppressed membrane lamellipodia dynamics (Fig. 5A, B and fig. S5A), which are required, but not sufficient for cell motility (25). Single-cell stroboscopic microscopy demonstrated that either treatment decreased the formation (Fig. 5C), total distance traveled (Fig. 5D), and total persistence of membrane ruffles (Fig. 5E). As a result, individual ruffles that formed in the presence of YM155 or TTFA extended for shorter distances (fig. S5B, C) with slower retraction speed (fig. S5D, E). In this analysis, average ruffling persistence did not change (fig. S5F, G). Instead, a post-analysis of cumulative ruffling dynamics that takes into account all ruffling events (20–60 ruffles measured per cell in a 5 min time interval) showed that YM155 or TTFA treatment reduced the overall distance and persistence of cell dynamics (Fig. 5D, E). In contrast, control cells exposed to conditioned media used as a chemotactic stimulus exhibited vigorous membrane lamellipodia dynamics (Fig. 5A and fig. S5A).
Fig. 5. Mitochondrial bioenergetics requirements of membrane dynamics.
(A) Representative stroboscopic images of FBS-stimulated PC3 cells treated with vehicle, YM155 or the Complex II inhibitor TTFA. Inset, SACED parameters of cell protrusion dynamics. (B) Quantification of membrane ruffle frequency in PC3 cells treated with vehicle (Veh), YM155 or TTFA. Each bar corresponds to an individual cell. Cells were imaged in two independent experiments. Broken lines, average values. The p values in YM155- or TTFA-treated cultures compared to Veh are indicated. (C–E) PC3 cells treated with vehicle (Veh), YM155 or TTFA were analyzed by SACED stroboscopic microscopy for average frequency of membrane ruffling (C), ruffle distance traveled (D), or ruffle persistence time (E). The quantified values are as follows (number of cells examined in parentheses): frequency of membrane ruffling (C), Veh, 1.44±0.14 (n=18); YM155, 0.61±0.14 (n=22), p=0.0002; TTFA, 0.68±0.14 (n=21), p=0.0008; ruffle distance traveled (D), Veh, 35±3.9 (n=18); YM155, 14.2±3.2 (n=22), p=0.0002; TTFA, 16±3.89 (n=21), p=0.0016; time of ruffle persistence (E), Veh, 243.3±24.3 (n=18); YM155, 136.4±26.9 (n=22), p=0.006; TTFA, 120.3±24.1 (n=21), p=0.001. Cells were imaged in two independent experiments. (F) PC3 cells treated with vehicle (Veh) or YM155 were analyzed by Western blotting. p, phosphorylated. Blots are representative of two independent experiments. (G) PC3 cells were treated with vehicle (Veh) or YM155, labeled with an antibody to phosphorylated FAK (Tyr397, pFAK) and analyzed by confocal microscopy. DNA was stained with DAPI. Right, quantification of phosphorylated FAK clusters. Mean±SEM. ***, p<0.0001. 20 cells/treatment condition imaged in two independent experiments. Scale bar, 10 μm. (H) PC3 cells treated as in (G) were transfected with α-actinin-GFP cDNA and analyzed by confocal microscopy. 10 cells (10 movies)/treatment condition imaged in two independent experiments. ***, p<0.0001. Scale bar, 10 μm. Right, FA dynamics quantified by time-lapse videomicroscopy. FA complexes examined: Veh, n=42; YM155, n=47.
Focal adhesion kinase (FAK) is a key effector of membrane dynamics, focal adhesion complexes and cell motility that becomes activated through autophosphorylation at Tyr397 and Src-mediated phosphorylation at Tyr925in response to growth factor stimulation and cell-extracellular matrix contact (26). Treatment with YM155 (Fig. 5F) or TTFA (fig. S5H) suppressed the autophosphorylation of FAK (26) at Tyr397 without affecting phosphorylation of Tyr925 (Fig. 5F). In contrast, YM155 did not affect the phosphorylation of Src or Akt, kinases that have also been implicated in cell motility, and total FAK abundance was unchanged (Fig. 5F). Consistent with these results, YM155 reduced the recruitment of phosphorylated FAK to focal adhesion complexes (Fig. 5G), while increasing the persistence of mature complexes (Fig. 5H and fig. S5I). (25). In contrast, vehicle-treated cells assembled focal adhesion complexes at cellular protrusions that contained Tyr397-phosphorylated FAK (Fig. 5G).
Control of tumor cell invasion and metastasis by mitochondrial survivin
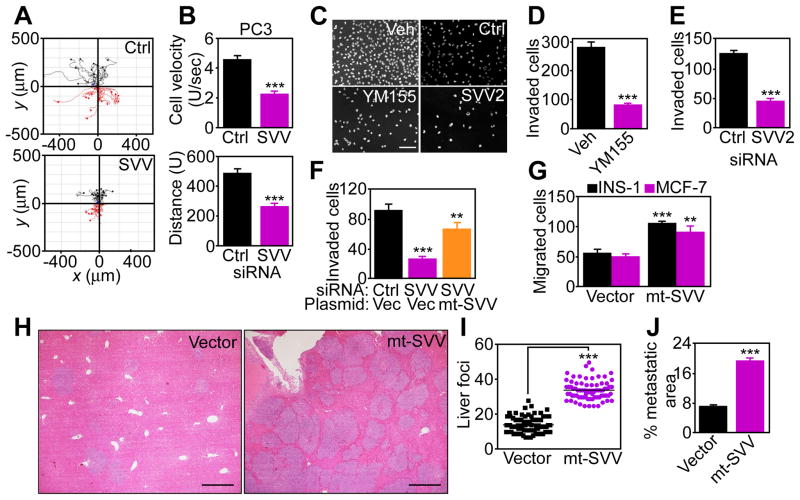
Next, we asked whether survivin bioenergetics and regulation of membrane dynamics affected PC3 cell movements. When analyzed for 2D chemotaxis, survivin targeting with siRNA (Fig. 6A) or YM155 (fig. S6A) decreased PC3 cell velocity and distance travelled, as assessed by time lapse videomicroscopy in scratch closure assays (Fig. 6B, fig. S6B), which mimic wound healing in vitro. In addition, YM155 (Fig. 6C) or survivin silencing by siRNA (fig. S6C, Fig. 6D and E) inhibited PC3 cell invasion across Matrigel-coated inserts, whereas reconstitution of survivin-knockdown cells with mt-SVV partially restored PC3 cell invasion (Fig. 6F). Furthermore, stable expression of mt-SVV in poorly migratory INS-1 or MCF-7 cells was sufficient to increase their invasive ability (Fig. 6G). In contrast, expression of a mitochondrial-targeted survivin Cys84→Ala (C84A) dominant negative mutant, which accumulated in the mitochondria, but not the cytosol of MCF-7 cells (fig. S6C), did not increase invasion (fig. S6D). To determine whether survivin regulation of tumor cell invasion was important in vivo, we measured liver metastasis in immunocompromised mice injected with MCF-7 cells in the spleen. Mice injected with mt-SVV-expressing cells had more liver metastases with greater surface area than those injected with vector-expressing cells (Fig. 6H, I, J).
Fig. 6. Regulation of tumor cell motility, invasion, and metastasis by mitochondrial survivin.
(A) siRNA-transfected PC3 cells were analyzed in a scratch closure assay and individual cell movements were tracked by time-lapse videomicroscopy. Ctrl siRNA, number of cells examined (n) =29; SVV siRNA, n=30; imaged in two independent determinations. (B) Average cell velocity (top) or total distance traveled (bottom) of the cells in (A) was quantified per each condition. Mean±SEM. p<0.0001. (C–E) PC3 cells treated with vehicle (Veh) or YM155 or transfected with survivin-directed siRNA (SVV2) were analyzed for Matrigel invasion. DAPI-stained nuclei of invaded cells (C) after YM155 treatment (D) or survivin knockdown (E) were quantified. Mean±SEM. ***, p<0.0001. 100–250 cells/treatment or transfection condition imaged in two independent experiments. Scale bar, 200 μm. (F) PC3 cells silenced for survivin by siRNA were reconstituted with vector or mt-SVV cDNA and analyzed for Matrigel invasion. Mean±SEM. **, p=0.0026; ***, p<0.0001. 100–250 cells/treatment or transfection condition imaged in two independent experiments. (G) INS-1 or MCF-7 cells transfected with vector or mt-SVV cDNA were analyzed for Matrigel invasion. Mean±SEM. **, p=0.003; ***, p<0.0001. 100–250 cells/treatment or transfection condition imaged in two independent experiments. (H) MCF-7 cells transfected with vector or mt-SVV cDNA were injected in the spleen of immunocompetent mice (three mice per transfection condition). Representative images of hematoxylin-eosin stained liver sections (n=15 per each transfection condition). Scale bar, 500 μm. (I and J) Morphometric quantification of total number of metastatic foci (I) and metastatic surface area (J) in serial liver sections (n=15 per each transfection condition). Mean±SEM. ***, p<0.0001.
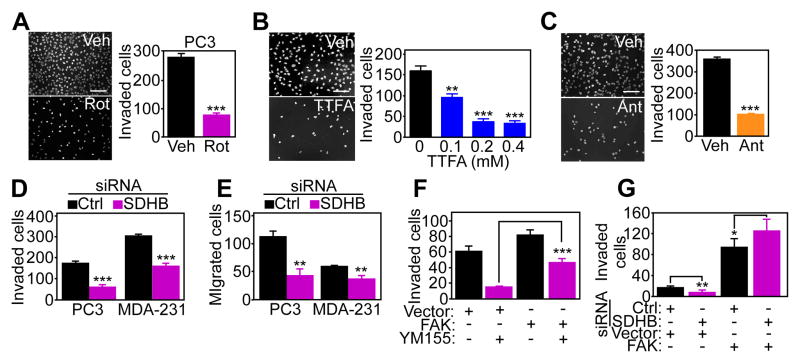
Finally, we asked whether oxidative phosphorylation was required for tumor cell invasion mediated by mitochondrial survivin. As expected, treatment of PC3 cells with the Complex I inhibitor Rotenone or the Complex III inhibitor Antimycin A suppressed ATP production (fig. S7A). Inhibition of the respiratory chain with Rotenone (Fig. 7A), TTFA (which inhibits the SDHB subunit of Complex II) (Fig. 7B), or Antimycin A (Fig. 7C) reduced PC3 cell invasion. Furthermore, siRNA-directed silencing of SDHB (fig. S7B) increased the phosphorylation of AMPK in PC3 cells (fig. S7B), which occurs under conditions of nutrient deprivation, and inhibited the invasion (Fig. 7D and fig. S7C) and migration (Fig. 7E) of PC3 and breast adenocarcinoma MDA-231 cells. Consistent with a requirement of FAK in this response (Fig. 5E), transfection of PC3 cells with a cDNA encoding wild-type FAK that could be phosphorylated on Tyr397 (fig. S7D) partially or fully reversed the inhibitory effect of YM155 (Fig. 7F) or SDHB knockdown (Fig. 7G) on tumor cell invasion. In contrast, siRNA-mediated silencing of FIP200 (fig. S7E), an upstream autophagy initiator and endogenous inhibitor of FAK (27) did not rescue the inhibition of tumor cell invasion mediated by YM155 (fig. S7F). Similarly, inhibition of NF-κB signaling, which has been implicated in regulation of cell motility by cytosolic survivin (28), by transfection of an IκBα super-repressor mutant, did not rescue the inhibition of tumor cell invasion mediated by YM155 (fig. S7G).
Fig. 7. Requirements of mitochondrial regulation in tumor cell invasion.
(A–C) PC3 cells were incubated with vehicle or the Complex I inhibitor Rotenone (Rot) (A), the Complex II inhibitor TTFA (B), or the Complex III inhibitor antimycin A (C) and analyzed for Matrigel invasion. Right panels, quantification of DAPI-stained nuclei of invaded cells in the presence of the various inhibitors. Mean±SEM. **, p=0.0053; ***, p<0.0001. 100–250 cells/treatment condition imaged in two independent experiments. Scale bar, 200 μm. (D and E) PC3 or MDA-231 cells were transfected with control (Ctrl) or SDHB-directed siRNA, and analyzed for Matrigel invasion (D) or cell migration (E). Mean±SEM. **, p=0.001–0.0028; ***, p<0.0001. 100–250 cells/treatment condition imaged in two independent experiments. (F and G) PC3 cells transfected with vector or FAK cDNA were treated with vehicle (Veh) or YM155 (F), or silenced for SDHB by siRNA (G) and quantified for Matrigel invasion. Mean±SEM. *, p=0.03; **, p=0.0032; ***, p<0.0001. 100–250 cells/treatment condition imaged in two independent experiments.
DISCUSSION
In this study, we showed that a mitochondrial pool of survivin (3) cooperates with Hsp90 chaperones in the folding of the oxidative phosphorylation Complex II subunits SDHB and SDHC. Genetic, molecular or pharmacologic interference with this pathway impaired mitochondrial respiration, lowered ATP production, and produced a phenotype of cellular starvation characterized by phosphorylation of AMPK and suppression of mTOR signaling. Conversely, oxidative phosphorylation supported by survivin enabled the trafficking of mitochondria to the cortical cytoskeleton of tumor cells, which may provide a “regional” energy source to fuel membrane lamellipodia dynamics, disassembly of FAK-containing focal adhesion complexes, increased tumor cell migration and invasion, and heightened metastatic dissemination in vivo.
IAP proteins, including survivin (28), have been implicated in cell motility (29). The mechanistic underpinnings of this pathway have not been completely elucidated, because at least under certain conditions, IAPs can inhibit cell migration, potentially through ubiquitination of Rac1 (30), or stimulation of RAF destruction (31). However, most data point to IAPs (32, 33), including survivin (28), as evolutionarily conserved (34) activators of cell migration, invasion and metastatic dissemination (29). In the case of survivin, this response has been linked to the formation of a survivin-XIAP complex (35), promoting NFκB-dependent activation of process that promote cell motility, including increased production and deposition of fibronectin (28).
The data here uncover a mechanism by which survivin promotes tumor cell invasion through mitochondrial bioenergetics. Accordingly, a pool of survivin localized to tumor mitochondria (3), and previously implicated in apoptosis resistance upon discharge in the cytosol (4), emerged here as an intrinsic regulator of oxidative phosphorylation in tumors. We showed that mitochondrial survivin cooperated with the chaperone TRAP-1 (23) to maintain SDH folding and Complex II activity in tumors, consistent with a role of protein quality control in preserving organelle bioenergetics (22). This finding conflicts with other data showing that mitochondrial survivin promotes mitochondrial fission, inhibits Complex I-dependent respiration, and enhances aerobic glycolysis (5).
How survivin affects a TRAP-1-SDH interaction (22) in maintaining mitochondrial protein folding quality control remains to be elucidated. Several chaperones, including Hsp90 (36), the aryl-hydrocarbon receptor-interacting protein (AIP) (37), and Hsp60 (38) associate with survivin, regulating protein stability and subcellular trafficking. In the case of TRAP-1, the recruitment of survivin to this complex may protect co-associated SDH subunits (22) from the mitochondrial protein degradation machinery. This scenario is reminiscent of how cytosolic survivin prevents access of the ubiquitin-conjugating enzyme UbcH5 to XIAP, maintaining XIAP stability and increasing apoptosis resistance (35).
A role of mitochondrial respiration in cancer metabolism has been debated (6) against the backdrop of the Warburg effect (8). Whether this process contributes to tumor cell motility, and potentially, metastasis, has also been controversial (39), with findings suggesting that oxidative phosphorylation is important (40), not important (41), or must be dysfunctional (42) in order to support tumor cell invasion. In addition, a prevailing model is that metabolites that accumulate in a highly glycolytic (43) and hypoxic (44) microenvironment suppress mitochondrial respiration (7), and enhance metastatic competency in vivo. The data here suggest a complementary scenario, in which oxidative phosphorylation maintained by survivin-TRAP-1 protein folding is integral to cancer metabolism (39), and enables the repositioning of energetically active mitochondria to the cortical cytoskeleton of tumor cells, where they provide a concentrated, “regional” energy source to support cell motility (25). Targeting mitochondrial survivin, destabilizing Complex II, or inhibiting different steps in oxidative phosphorylation interfered with this pathway, prevented the accumulation of cortical mitochondria, suppressed the formation of membrane lamellipodia (45) and focal adhesion dynamics (26), and inhibited tumor cell migration and invasion. There is precedent for a model of “regional” mitochondrial bioenergetics to support highly energy-intensive cellular tasks. For instance, neurons reposition mitochondria (46) to subcellular sites of high ATP demands, providing an efficient energy source to power synapses, growth cones, and dendritic branching (47). Whether the same machinery controlling mitochondrial trafficking in neurons is exploited for metastatic competency in tumors is unknown. However, mitochondrial dynamics has been implicated in tumor cell migration (48), and asymmetric mitochondrial distribution towards the leading edge may contribute to the directionality of tumor cell movements (49).
These observations thus identify survivin as a potential cancer therapeutic target (1). Accordingly, expression of mitochondrial survivin was sufficient to convert poorly migratory MCF-7 cells, into cells with an invasive and metastatic phenotype in vivo, in line with clinical data that links survivin abundance to metastatic dissemination in humans (2). Although the specificity of YM155 as a survivin antagonist (50) remains to be fully elucidated (51), our data show that this agent disrupted mitochondrial bioenergetics, suppressed cell invasion, and activated tumor suppressor mechanisms, namely AMPK activation or mTOR inhibition.
In summary, we have demonstrated that survivin controls mitochondrial respiration, enabling organelle trafficking and potentially “regional” bioenergetics to fuel tumor cell motility and metastasis. These observations reinforce a key role of mitochondrial bioenergetics as a driver of tumor progression (22), and open new prospects for targeting oxidative phosphorylation as a cancer therapeutic strategy (52).
MATERIALS AND METHODS
Antibodies and reagents
The following antibodies to survivin (NOVUS Biologicals), succinate dehydrogenase complex subunit B (SDHB, Abcam), SDHA (Abcam), SDHC (Abcam), Cox-IV (Cell Signaling), Thr172-phosphorylated AMPKα (Cell Signaling), AMPKα (Cell Signaling), Tyr397-phosphorylated FAK (Invitrogen), Tyr925-phosphorylated FAK (Cell Signaling), FAK (Cell Signaling), Tyr416-phosphorylated Src (Cell Signaling), Tyr527-phosphorylated Src (Cell Signaling), Src (Cell Signaling), Ser473-phosphorylated Akt (Cell Signaling), Akt (Cell Signaling), FIP200 (Novus Biologicals), LC-3B (Cell Signaling), TRAP-1 (BD Biosciences), Hsp90 (BD Biosciences), Thr37/46-phosphorylated 4EBP1 (Cell Signaling), 4EBP1 (Cell Signaling), Thr389-phosphorylated p70S6 kinase (Cell Signaling), 70S6 kinase (Cell Signaling), VDAC (Cell Signaling), Cox-IV (Cell Signaling), CypD (Millipore), CLPP (Santa Cruz Biotechnology), β-actin (Sigma-Aldrich), and β-tubulin (Sigma-Aldrich) were used. An oxidative phosphorylation antibody cocktail (Mitoscience) directed against the 20-kD subunit of Complex I (20 kD), cytochrome C oxidase subunit II of Complex IV (22 kD), SDHB subunit of Complex II (30 kD), core 2 of complex III (~50 kD), and F1α (ATP synthase) of Complex V (~60 kD) was used.
The complete chemical synthesis, HPLC profile, and mass spectrometry of mitochondrial-targeted small molecule Hsp90 antagonist, Gamitrinib (GA mitochondrial matrix inhibitors) has been previously reported (53). The Gamitrinib variant containing triphenylphosphonium as a mitochondrial-targeting moiety was used in this study (53). Oligomycin, carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP), carbonyl cyanide m-chlorophenylhydrazone (CCCP), antimycin A, and thenoyltrifluoroacetone (TTFA) were obtained from Sigma-Aldrich. Antimycin A (Ant) and Rotenone (Rot) were obtained from Abcam Biochemicals. A small molecule survivin suppressant, YM155, which inhibits Sp1-dependent transcription of the survivin locus (50), was from Selleckem. In all experiments, TTFA was used at concentrations of 100–400 μM and YM155 was used at 10 nM. All chemicals were of the highest purity commercially available.
Cell culture
Human glioblastoma LN229, prostate adenocarcinoma PC3 and DU145, breast adenocarcinoma MCF-7, MCF-10A and MDA-MB231 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA), and maintained in culture according to the supplier’s specifications. Rat insulinoma INS-1 cells stably transfected with vector or mitochondrial-targeted survivin (mt-SVV) have been previously described (3).
Transfections
For gene knockdown experiments, tumor cells were transfected using control, non-targeting small interfering RNA (siRNA) pool (Dharmacon, cat. no. D-001810), two independent custom-prepared survivin-directed siRNA with the sequence GAGCCAAGAACAAAAUUGC (SVV) or GGACCACCGCAUCUCUACA (SVV2) (Thermo Scientific) characterized in previous studies (24), or siRNA pools targeting TRAP-1 (Dharmacon, cat. no. L-010104) or FIP200 (Dharmacon, cat. no. L-021117), as described (54). The various siRNAs were transfected at 10–30 nM in the presence of Lipofectamine RNAiMAX in a 1:1 ratio (Invitrogen). Cells were incubated for 48 h, validated for target protein knockdown by Western blotting, and processed for subsequent experiments. Plasmid DNA transfections were carried out using X-tremeGENE HP DNA transfection reagent (Roche). A replication-deficient adenovirus (pAd) encoding mitochondrial-targeted GFP (pAd-GFP) or mitochondrial-targeted GFP-survivin (pAd-mt-SVV) has been previously described (3). For reconstitution experiments, PC3 cells were silenced for endogenous survivin by siRNA and transfected with plasmid encoding mitochondrial-targeted survivin (mt-SVV), or, alternatively pAd-mt-SVV. In some experiments, MCF-7 or MCF-10A cells were transfected with vector, or mitochondrial-targeted wild type (WT) survivin or mitochondrially-targeted Cys84→Ala (C84A) survivin dominant negative mutant, characterized in previous studies (55). For rescue experiments, PC3 cells treated with vehicle or YM155, or, alternatively, silenced for endogenous SDHB expression by siRNA, were transfected with vector or cDNA encoding FAK, FIP200 (54) or IκBα super-repressor mutant (28) characterized in previous studies, before analysis of Matrigel invasion.
Quantification of autophagy
PC3 cells were co-transfected with siRNA against survivin and plasmid encoding green fluorescent protein (GFP) fused to human dynein light chain 3 (LC3) cDNA in the presence of Lipofectamine 2000 Transfection Reagent (Invitrogen). After 48 h, transfected cells were fixed in 4% paraformaldehyde for 15 min at 37°C, washed, and examined by confocal microscopy (Leica, SP5). The numbers of GFP-LC3 punctate dots per cell was determined from two independent experiments. A minimum of 50 GFP-LC3-positive cells assessed from at least ten random fields per sample were counted in triplicate per each condition, as previously described (20).
Cell proliferation, cell cycle analysis and mitochondrial membrane potential
Potential changes in cell proliferation in various tumor cell types (2x104–1.25x105 cells) were evaluated after 48–72 h by direct cell counting. Alternatively, tumor cells were labeled with 1:1000 dilution bromodeoxyuridine (BrdU) (Amersham Pharmacia Biotech) in cultured medium for 1 h, and analyzed by multiparametric flow cytometry with quantification of BrdU+ cells. In some experiments, tumor cell types (1x106) were fixed in glacial 70% ethanol for 24 h, followed by incubation with propidium iodide (2.5 μg/ml) in the presence of RNAse A for 10 min at room temperature. Twenty thousand events were acquired on a Calibur flow cytometer, with quantification of individual cell cycle transitions using Cell Quest Pro software (Becton Dickinson), as previously described (20).
In some experiments, PC3 cells grown at low confluency (1–2x104/well) on optical grade coverslips were transfected with control or survivin-directed siRNA or treated with vehicle or YM155 (10 nM) for 24 h, fixed in 4% formaldehyde for 15 min at 37°C, washed in PBS, pH 7.4, and permeabilized with 0.1% Triton X-100 for 5 min at 22°C. Slides were washed in PBS, pH 7.4, blocked in 1% BSA/PBS for 30 min, and analyzed for changes in nuclear morphology by DAPI staining on a Leica TCS SP5 fluorescence microscope with a 100X oil objective. To test the impact of survivin targeting on mitochondrial inner membrane potential, PC3 cells were treated with 10 nM YM155 for 16 h or transfected with siRNA against survivin for 48 h, incubated with 0.1 μM tetramethylrhodamine methyl ester (TMRE), and analyzed for changes in fluorescence emission by flow cytometry.
Subcellular fractionation
Mitochondrial fractions were isolated from treated cells as previously described (20). Briefly, the various tumor cell types were mechanically disrupted by 70 strokes with a Dounce homogenizer in isolation buffer containing protease inhibitor cocktail. Cell debris was removed by centrifugation at 700 g for 10 min. The supernatant was further centrifuged at 3,000 g for 15 min, and supernatants or mitochondrial pellets were processed for further analysis.
Protein analysis
For Western blotting, protein lysates were prepared from the different cell types in RIPA buffer (150 mM NaCl, 1.0% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) containing EDTA-free Protease Inhibitor Cocktail (Roche) and Phosphatase Inhibitor Cocktail 2 and 3 (Sigma-Aldrich). Equal amounts of protein lysates were separated by SDS gel electrophoresis, transferred to PVDF membranes and incubated with primary antibodies of various specificities. Protein bands were visualized by chemiluminescence. In some experiments, mitochondria were isolated from PC3 cells, and lysed in 0.5% CHAPS buffer, containing 1% n-dodecyl-β-D-maltopyranoside plus protease inhibitors (Roche) for 30 min at 4°C under constant agitation. For immunoprecipitation experiments, aliquots (500 μg) of isolated mitochondrial extracts were incubated with non-binding IgG or an antibody to survivin for 16 h at 4°C. Immune complexes were coupled with Protein A-sepharose beads (Calbiochem) for 2 h at 4°C. After washes in TBST, the immune complexes were separated by SDS gel electrophoresis, and analyzed by Western blotting.
Mitochondrial protein folding
Mitochondrial protein folding assays were performed as previously described (20, 22). Briefly, mitochondrial fractions were isolated after 24–48 h from PC3 cells transfected with control non-targeting or survivin-directed siRNA and suspended in equal volume of mitochondrial fractionation buffer containing increasing concentrations of CHAPS (0, 0.05, 0.5, 1, 2, or 5%) or NP-40 (0, 0.05, 0.2, 0.5 or 2%). Samples were incubated for 25 min on ice with vortexing every 5 min and detergent-insoluble protein aggregates were isolated by centrifugation (20,000 g) for 20 min, separated on SDS polyacrylamide gels and analyzed by Western blotting. In some experiments, PC3 cells were treated with 5 μM Gamitrinib for 6 h, incubated with 100 μg/ml of the protein synthesis inhibitor cycloheximide and aliquots of mitochondrial extracts were collected at T0 and after 1, 2, 3 and 4.5 h, followed by Western blotting. Protein bands at the indicated time intervals were quantified by densitometry and survivin half-life was calculated.
Submitochondrial fractionation
Purified mitochondrial pellets isolated by sucrose step gradient were suspended in swelling buffer (10 mM KH2PO4, pH 7.4, plus protease inhibitors) and incubated for 20 min at 0°C with gentle mixing, as previously described (21). Mitochondria were mixed with equal volume of shrinking buffer (10 mM KH2PO4, pH 7.4, 32% sucrose, 30% glycerol, 10 mM MgCl2, and protease inhibitors) for 20 min at 0°C. After centrifugation at 10,000 × g for 10 min, the supernatant was collected as containing outer membrane and inter-membrane space fractions. Pellets were washed three times with 1:1 mixture of swelling-shrinking buffer, suspended in swelling buffer, and sonicated to disrupt the inner membrane, which was collected as containing inner membrane and matrix fractions. Aliquots containing outer membrane and inter-membrane space fractions and inner membrane and matrix fractions were further fractionated by centrifugation at 150,000 g for 1 h at 4°C. The pellets were collected as outer membrane and inter-membrane space fractions, respectively. Supernatants were concentrated using Centricon 10K and Microcon 10K centrifugal filters (Millipore) and collected as inner membrane and matrix fractions, respectively.
Metabolomics screen
A global metabolomics profiling to examine changes in expression of 301 individual metabolites in PC3 cells transfected with control non-targeting siRNA or survivin-directed siRNA was performed by Metabolon, as previously described (22).
Tumor bioenergetics
Various tumor cell types treated with 10 nM YM155 for 16 h or, alternatively, transfected with control non-targeting siRNA or survivin-directed siRNA for 48 h were analyzed for oxygen consumption (ENZO Lifesciences cat. No. ENZ-51045-1) or ATP generation (BioChain cat. No. Z5030041), as previously described (20, 22). Aliquots of cultured medium were collected for analysis of glucose consumption (Enzyme Cat. No. CA-G005), or lactate production (Abcam cat No. ab65331), as previously described (20, 22).
Cellular respiration
Oxygen consumption rates (OCR) were assayed in intact cells using an Extracellular Flux System 24 Instrument (Seahorse Bioscience, Billerica, MD), as previously described (22). Briefly, PC3 and DU145 cells in complete medium were transfected with the various siRNAs for 24–36 h. After trypsinization and re-suspension in growth medium, 2.5x104 cells were plated in each well of a Seahorse XF24 cell culture plate (100 μl) in complete medium. After 4-h incubation to allow cells to adhere to plates, an additional 150 μl of medium was added to each well, and cells were grown in 5% CO2 for 24 h at 37°C. The medium was then exchanged with unbuffered DMEM XF assay medium (Seahorse Bioscience) supplemented with 2 mM glutaMAX, 1 mM sodium pyruvate and 5 mM glucose (pH 7.4 at 37°C), and equilibrated for 30 min at 37°C and ~0.04% CO2 before the experiment. Where indicated, the unbuffered DMEM XF assay medium was supplemented with high (10 mM) or low (1 mM) glucose concentrations. Cellular oxygen consumption was monitored at basal conditions (before any addition) and after addition of oligomycin (1.25 μM), FCCP (0.4 μM), and antimycin (1.8 μM), all dissolved in DMSO. The three drugs were injected sequentially, and the OCR measured with three cycles of mixing (150 seconds), waiting (120 seconds), and measuring (210 seconds). This cycle was repeated following each injection.
For quantification of cellular respiration in permeabilized cultures, PC3 cells were treated with vehicle (DMSO) or 10 nM YM155 in complete medium for 16 h. After trypsinization, PC3 cells were suspended in MiR06 incubation medium (Oroboros Instruments) at a density of 1.5x106 cells/ml (3x106 cells total), permeabilized with digitonin (10 μg/1x106 cells), and oxygen consumption was measured at 37°C with an Oroboros Oxygraph-2k (Oroboros Instruments), 2 chamber high-resolution respirometer, Clark-type oxygen electrode, in closed 2-ml chambers equipped with magnetic stirring. The respiration medium contained 10 mM glutamate and 2 mM malate (complex I assay), 10 mM succinate and 0.5 μM rotenone (complex II assay), 5 mM malonic acid and 10 mM glycerol phosphate (complex III assay). During the experiment, the following agents were added: ADP (200 μM), oligomycin (Oligo, 2.5 μM), FCCP (1 μM) and antimycin (2 μM).
Mitochondrial respiration complex activity
PC3 cells were analyzed for changes in oxidative phosphorylation complex activity with Abcam reagents (Cat. no. ab109721 – Complex I and ab109908 – Complex II) using lysed isolated mitochondria, as described previously. Briefly, PC3 cells were transfected with control non-targeting or survivin-directed siRNA, validated for protein knockdown by Western blotting, and two μg of mitochondrial extracts from each condition were assayed for Citrate Synthase activity (ScienCell). Aliquots of mitochondrial lysates with comparable citrate synthase activity were applied to mitochondrial complex-specific microplates for immunocapture, with continuous quantification of enzymatic activity in a microplate reader for either an increase in absorbance at 450 nm (Complex I) or a decrease in absorbance at 600 nm (Complex II). NADH or ubiquinone was used as substrate for Complex I or Complex II activity, respectively. Relative complex activities were calculated by determining the change in absorbance over time in the linear range of the measurements.
Immunofluorescence
To quantify mitochondrial subcellular trafficking, PC3 cells were incubated with MitroTracker Red CM-H2XRos (Invitrogen) diluted in conditioned media for 1 h at 37°C. Cells were fixed with 3.7% paraformaldehyde for 20 min, permeabilized in 0.2% Triton X-100 for 15 min, washed with 100 mM glycine, blocked with 1%BSA/0.05% Triton X-100 for 30 min, and further incubated with Alexa-Fluor 488-Phalloidin (Life Technologies) in blocking buffer for 20 min. After three washes in PBS, coverslips were mounted in Prolong Gold mounting media with DAPI (Invitrogen). In some experiments, cells were treated with the mitochondrial uncoupler CCCP (12.5 μM), the SDHB inhibitor thenoyltrifluoroacetone (TTFA, 200 μM), or the mitochondrially-targeted small molecule Hsp90 inhibitor Gamitrinib (5 μM) for 16 h before immunofluorescence staining. For analysis of phosphorylated FAK clusters, PC3 cells were seeded on fibronectin (10 μg/ml) for 2 h in the presence of vehicle (DMSO) or YM155 (10 nm). Cells were fixed with 3.7% paraformaldehyde for 20 min, permeabilized in 0.2% Triton X-100 for 15 min, washed with 100 mM glycine, blocked with 1% BSA/0.05% Triton X-100 for 30 min and incubated with an antibody to phosphorylated FAK (pFAK, Tyr397) for 1 h (1:100) in blocking buffer. After 3 washes in PBS, cells were incubates with a FITC-conjugated secondary antibody (1:1000) for 1 h, washed, and coverslips were mounted in Prolong Gold mounting media containing DAPI (Invitrogen) for analysis by confocal microscopy. For quantification of mitochondria-containing focal adhesion complexes, MCF7 cells were transfected with vector or mitochondrial-targeted WT survivin cDNA for 48 h. Cells were then seeded on fibronectin-covered coverslips, incubated with MitroTracker Red for 1 h at 37°C and stained with an antibody to Paxillin (1:100) as a marker for focal adhesions before analysis by confocal fluorescence microscopy.
For quantification of microscopy data, a minimum of 50 cells per experiment was imaged using a Leica TCS SP5 II Scanning Laser Confocal Microscope system with an HCX PL APO CS 63X 1.40NA oil UV objective using the same laser intensity and exposure time. A full cell Z-stack of a minimum of 11 sequential steps of 0.5 μm size were collected. Total fluorescence intensity of mitochondria for whole cell and within the lamellipodia regions were obtained by creating masks for each cell and areas of interests (AOIs) for lamellipodia, determined by co-staining with phalloidin. For scoring of pFAK-containing clusters per cell, or focal adhesions per cell, an automated counting was applied by thresholding pFAK-containing clusters or focal adhesions based on fluorescence intensity using ImagePro software. Outlines of focal adhesions were created and overlayed on images showing mitochondrial staining, and intensity measurements of MitoTracker in the outlines of FAs were obtained to quantify positive staining in focal adhesion regions. Control images were additionally taken for subtraction of background signal.
Stroboscopic Analysis of Cell Dynamics (SACED)
Quantification of membrane ruffle dynamics in live cells was carried out as described previously (54). Briefly, 3–5 x104 cells were grown on high optical quality 96 well μ-plates (Ibidi) and imaged with a 40X objective on a Nikon TE300 inverted time-lapse microscope equipped with a video system containing an Evolution QEi camera and a time-lapse video cassette recorder. The atmosphere was equilibrated to 37°C and 5% CO2 in an incubation chamber. Phase contrast images were captured at 0.5s intervals for 250 seconds (500 frames) and merged into sequence files using ImagePro Plus 7. To monitor dynamics of a particular region by Stroboscopic Analysis of Cell Dynamics (SACED) (54), the sequence files were imported into Image J, and a particular region of 16.2 mm x 0.162 mm (“SACED line”) was selected, duplicated and montaged in sequence to display the region over time in a stroboscopic image. This process was repeated to obtain 4 SACED lines and therefore 4 stroboscopic images per cell, and structures such as protruding lamellipodia and ruffles were manually labeled. For each cell, the frequency of ruffles per min, the ruffling retraction speed (μm/min), the ruffle migration distance (nm) and time of ruffle persistence (msec) were calculated. Mean values were calculated from at least 18 cells from 2 separate wells. All experiments were repeated at least twice.
Time-lapse analysis of focal adhesion complex dynamics
PC3 cells were transfected with a GFP-α-actinin cDNA for 48 h. Cells were seeded on glass plates coated with 10 μg/ml fibronectin, treated with 10 nM YM155 or vehicle (DMSO) for 4 h and kept at 37°C and 5% CO2 during imaging. Time-lapse videomicroscopy was done using a Leica TCS SP5 II Scanning Laser Confocal Microscope system with an HCX PL APO CS 63X 1.40NA oil UV objective. Acquisition of live cells using an integrated Leica LAS software was performed every 1 min for a total interval of 30 min using the Argon laser at 476 nm for detection of GFP-α-actinin. Actinin-positive focal adhesion complexes were identified based on fluorescent intensity, thresholded and masked using FIJI software. Identified focal adhesion complexes for every cell were tracked using generated masks through every image taken at each min of the experiment, and individual focal adhesion life spans and newly formed focal adhesion complexes were recorded throughout the 30 min time lapse interval. The analysis was carried out in 10 cells per condition in 10 independent time-lapse experiments using a FIJI software package.
Cell migration and invasion
Tumor cell lines were treated as indicated in each experiment, suspended in 0.1% BSA/DMEM and seeded (1.6–3.2x103 cells/mm2, depending on the cell line) in the upper compartments of 8 μM pore diameter BD Transwells (BD). NIH3T3 conditioned medium was placed in the lower compartment as a chemoattractant, as previously described (54). Except for the Complex I inhibitor, Rotenone (4 μM), which was preincubated with PC3 cells, YM155 (10 nM) or inhibitors of Complex II-SDHB, TTFA (100–400 μM) or Complex III (Antimycin A, 30 μM) were added to the top and bottom chambers. After 6–18 h incubation at 37°C, Transwell membranes were recovered and cells on the upper side (non-migratory) were wiped off the surface. Cells on the lower side of the membrane were fixed in methanol, rinsed in water and mounted on glass slides with Vectashield medium containing DAPI (Vector Laboratories). Migrated cells on each membrane were counted by fluorescence microscopy in 5 different fields. For cell invasion assays, transwell membranes were coated with Matrigel and processed as described above (54). For cell migration using a scratch closure assay, PC3 cells were transfected with control non-targeting siRNA or survivin-directed siRNA, or, alternatively treated with vehicle or YM155, and seeded in 24 well plates (5x105 per well). Scratch “wounds” were made with a 10 μl pipet tip, cell debris was washed off and cultures were maintained in complete medium containing 10% FBS at 37°C and 5% CO2 for 24 h. Time-lapse imaging of migrating cells was performed using a TE300 Inverted Microscope (Nikon) equipped with an incubator set at 37°C, 5% CO2 and 95% relative humidity. Each image was acquired utilizing a 10x objective of the same fields every 10 min time interval for a total 24 h. Velocity of cell migration and distance traveled were quantified under the different conditions using ImageJ’s Chemotaxis and Migration Tool, Version 1.01. A minimum of 30 cells per condition were individually tracked between frames generating a live track for each individual cell. Each track was then uploaded to the Migration Tool yielding Average Velocity and Distance Traveled.
Liver metastasis model
All in vivo experiments were carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH). Protocols were approved by an Institutional Animal Care and Use Committee (IACUC) at the Wistar Institute. A liver metastasis model was carried out generally as previously described (28). Briefly, six- to eight-weeks old female SCID/beige mice (3 mice per experimental conditions) were anesthetized with ketamine hydrochloride, the abdominal cavity was exposed by laparotomy, and 1.5×106 MCF-7 cells transfected with vector or mitochondrial-targeted survivin (mt-SVV) cDNA injected into the spleen. Spleens were removed the first day after injection to minimize potentially confounding effects on metastasis due to variable growth of primary tumors. Animals were sacrificed 11 days after injection, and their livers were resected, fixed in formalin and paraffin embedded. Serial liver sections 500 μm apart (n=15 per each condition) were stained with hematoxilin and eosin and analyzed histologically. Metastatic foci were quantified by morphometry and expressed as number of lesions and surface areas of tumor growth compared to total surface area, as previously described (28).
Statistical analysis
Data were analyzed using the two-sided unpaired t tests or ANOVA using a GraphPad software package (Prism 4.0) for Windows. Data are expressed as mean±SD or mean±SEM of at least three independent experiments. A p value of 0.05 was considered as statistically significant. For a metabolomics screening, missing values (if any) were assumed to be below the level of detection. However, biochemicals that were detected in all samples from one or more groups, but not in samples from other groups were assumed to be near the lower limit of detection in the groups in which they were not detected. In this case, the lowest detected level of these biochemicals was imputed for samples in which that biochemical was not detected. Following log transformation and imputation with minimum observed values for each compound, Welch’s two-sample t-test was used to identify biochemicals that differed significantly between experimental groups. The false discovery rate (FDR) in the metabolomics screen was estimated using the q value per each compound detected. Pathways were assigned for each metabolite, allowing examination of overrepresented pathways. For classification studies, random forest analyses were performed. Statistical analyses were performed with the program “R” (http://cran.r-project.org/).
Supplementary Material
Fig. S1. Mitochondrial survivin.
Fig. S2. Survivin regulation of mitochondrial bioenergetics.
Fig. S3. Modulation of oxidative phosphorylation by survivin.
Fig. S4. Survivin regulation of mitochondrial trafficking.
Fig. S5. Regulation of membrane dynamics by survivin.
Fig. S6. Expression of mitochondrial-targeted survivin dominant negative mutant.
Fig. S7. Mitochondrial survivin-Complex II regulation of tumor cell motility.
Acknowledgments
We thank James Hayden and Frederick Keeney of the Wistar Imaging Shared Resource for excellent assistance.
FUNDING: This work was supported by the National Institutes of Health (NIH) grants P01 CA140043 (DCA and LRL), R01 CA78810 and CA190027 (DCA), R01 CA089720 (L.R.L.), F32 CA177018 (M.C.C.), and the Office of the Assistant Secretary of Defense for Health Affairs through the Prostate Cancer Research Program under Award No. W81XWH-13-1-0193 (DCA). Support for Core Facilities utilized in this study was provided by Cancer Center Support Grant (CCSG) CA010815 to The Wistar Institute.
Footnotes
AUTHOR CONTRIBUTIONS: D.R., M.C.C., J.H.S., and D.C.A. designed research; D.R., M.C.C., J.H.S., and A.A. performed research; D.R., M.C.C., J.H.S., A.A., D.C.W., L.R.L. and D.C.A. analyzed data, and D.R., M.C.C., J.H.S., and D.C.A. wrote the paper.
COMPETING INTERESTS: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Altieri DC. Survivin - The inconvenient IAP. Semin Cell Dev Biol. 2015 doi: 10.1016/j.semcdb.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Discov. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 3.Dohi T, Beltrami E, Wall NR, Plescia J, Altieri DC. Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J Clin Invest. 2004;114:1117–1127. doi: 10.1172/JCI22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dohi T, Xia F, Altieri DC. Compartmentalized phosphorylation of IAP by protein kinase A regulates cytoprotection. Mol Cell. 2007;27:17–28. doi: 10.1016/j.molcel.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagenbuchner J, Kuznetsov AV, Obexer P, Ausserlechner MJ. BIRC5/Survivin enhances aerobic glycolysis and drug resistance by altered regulation of the mitochondrial fusion/fission machinery. Oncogene. 2013;32:4748–4757. doi: 10.1038/onc.2012.500. [DOI] [PubMed] [Google Scholar]
- 6.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 8.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 12.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 14.Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor’s dilemma? Biochim Biophys Acta. 2011;1807:552–561. doi: 10.1016/j.bbabio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Janiszewska M, Suva ML, Riggi N, Houtkooper RH, Auwerx J, Clement-Schatlo V, Radovanovic I, Rheinbay E, Provero P, Stamenkovic I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26:1926–1944. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, Kost-Alimova M, Muller F, Colla S, Nezi L, Genovese G, Deem AK, Kapoor A, Yao W, Brunetto E, Kang Y, Yuan M, Asara JM, Wang YA, Heffernan TP, Kimmelman AC, Wang H, Fleming JB, Cantley LC, DePinho RA, Draetta GF. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, Korbel C, Laschke MW, Gimotty PA, Philipp SE, Krause E, Patzold S, Villanueva J, Krepler C, Fukunaga-Kalabis M, Hoth M, Bastian BC, Vogt T, Herlyn M. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 19.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 20.Chae YC, Caino MC, Lisanti S, Ghosh JC, Dohi T, Danial NN, Villanueva J, Ferrero S, Vaira V, Santambrogio L, Bosari S, Languino LR, Herlyn M, Altieri DC. Control of tumor bioenergetics and survival stress signaling by mitochondrial HSP90s. Cancer Cell. 2012;22:331–344. doi: 10.1016/j.ccr.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang BH, Plescia J, Dohi T, Rosa J, Doxsey SJ, Altieri DC. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell. 2007;131:257–270. doi: 10.1016/j.cell.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 22.Chae YC, Angelin A, Lisanti S, Kossenkov AV, Speicher KD, Wang H, Powers JF, Tischler AS, Pacak K, Fliedner S, Michalek RD, Karoly ED, Wallace DC, Languino LR, Speicher DW, Altieri DC. Landscape of the mitochondrial Hsp90 metabolome in tumours. Nat Commun. 2013;4:2139. doi: 10.1038/ncomms3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavery LA, Partridge JR, Ramelot TA, Elnatan D, Kennedy MA, Agard DA. Structural Asymmetry in the Closed State of Mitochondrial Hsp90 (TRAP1) Supports a Two-Step ATP Hydrolysis Mechanism. Mol Cell. 2014;53:330–343. doi: 10.1016/j.molcel.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beltrami E, Plescia J, Wilkinson JC, Duckett CS, Altieri DC. Acute ablation of survivin uncovers p53-dependent mitotic checkpoint functions and control of mitochondrial apoptosis. J Biol Chem. 2004;279:2077–2084. doi: 10.1074/jbc.M309479200. [DOI] [PubMed] [Google Scholar]
- 25.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14:598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehrotra S, Languino LR, Raskett CM, Mercurio AM, Dohi T, Altieri DC. IAP regulation of metastasis. Cancer Cell. 2010;17:53–64. doi: 10.1016/j.ccr.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fulda S. Regulation of cell migration, invasion and metastasis by IAP proteins and their antagonists. Oncogene. 2014;33:671–676. doi: 10.1038/onc.2013.63. [DOI] [PubMed] [Google Scholar]
- 30.Oberoi TK, Dogan T, Hocking JC, Scholz RP, Mooz J, Anderson CL, Karreman C, Meyer zu Heringdorf D, Schmidt G, Ruonala M, Namikawa K, Harms GS, Carpy A, Macek B, Koster RW, Rajalingam K. IAPs regulate the plasticity of cell migration by directly targeting Rac1 for degradation. EMBO J. 2012;31:14–28. doi: 10.1038/emboj.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dogan T, Harms GS, Hekman M, Karreman C, Oberoi TK, Alnemri ES, Rapp UR, Rajalingam K. X-linked and cellular IAPs modulate the stability of C-RAF kinase and cell motility. Nat Cell Biol. 2008;10:1447–1455. doi: 10.1038/ncb1804. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Zhang D, Luo W, Yu Y, Yu J, Li J, Zhang X, Zhang B, Chen J, Wu XR, Rosas-Acosta G, Huang C. X-linked inhibitor of apoptosis protein (XIAP) mediates cancer cell motility via Rho GDP dissociation inhibitor (RhoGDI)-dependent regulation of the cytoskeleton. J Biol Chem. 2011;286:15630–15640. doi: 10.1074/jbc.M110.176982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Zhang D, Liu J, Li J, Yu Y, Wu XR, Huang C. RhoGDI SUMOylation at Lys-138 increases its binding activity to Rho GTPase and its inhibiting cancer cell motility. J Biol Chem. 2012;287:13752–13760. doi: 10.1074/jbc.M111.337469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geisbrecht ER, Montell DJ. A role for Drosophila IAP1-mediated caspase inhibition in Rac-dependent cell migration. Cell. 2004;118:111–125. doi: 10.1016/j.cell.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 35.Dohi T, Okada K, Xia F, Wilford CE, Samuel T, Welsh K, Marusawa H, Zou H, Armstrong R, Matsuzawa S, Salvesen GS, Reed JC, Altieri DC. An IAP-IAP complex inhibits apoptosis. J Biol Chem. 2004;279:34087–34090. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- 36.Fortugno P, Beltrami E, Plescia J, Fontana J, Pradhan D, Marchisio PC, Sessa WC, Altieri DC. Regulation of survivin function by Hsp90. Proc Natl Acad Sci U S A. 2003;100:13791–13796. doi: 10.1073/pnas.2434345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang BH, Xia F, Pop R, Dohi T, Socolovsky M, Altieri DC. Developmental control of apoptosis by the immunophilin aryl hydrocarbon receptor-interacting protein (AIP) involves mitochondrial import of the survivin protein. J Biol Chem. 2011;286:16758–16767. doi: 10.1074/jbc.M110.210120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh JC, Dohi T, Kang BH, Altieri DC. Hsp60 regulation of tumor cell apoptosis. J Biol Chem. 2008;283:5188–5194. doi: 10.1074/jbc.M705904200. [DOI] [PubMed] [Google Scholar]
- 39.Tan AS, Baty JW, Berridge MV. The role of mitochondrial electron transport in tumorigenesis and metastasis. Biochim Biophys Acta. 2014;1840:1454–1463. doi: 10.1016/j.bbagen.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 40.LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, Asara JM, Kalluri R. PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shiraishi T, Verdone JE, Huang J, Kahlert UD, Hernandez JR, Torga G, Zarif JC, Epstein T, Gatenby R, McCartney A, Elisseeff JH, Mooney SM, An SS, Pienta KJ. Glycolysis is the primary bioenergetic pathway for cell motility and cytoskeletal remodeling in human prostate and breast cancer cells. Oncotarget. 2014 doi: 10.18632/oncotarget.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porporato PE, Payen VL, Perez-Escuredo J, De Saedeleer CJ, Danhier P, Copetti T, Dhup S, Tardy M, Vazeille T, Bouzin C, Feron O, Michiels C, Gallez B, Sonveaux P. A mitochondrial switch promotes tumor metastasis. Cell Rep. 2014;8:754–766. doi: 10.1016/j.celrep.2014.06.043. [DOI] [PubMed] [Google Scholar]
- 43.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 44.Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest. 2013;123:3664–3671. doi: 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boldogh IR, Pon LA. Mitochondria on the move. Trends Cell Biol. 2007;17:502–510. doi: 10.1016/j.tcb.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Saxton WM, Hollenbeck PJ. The axonal transport of mitochondria. J Cell Sci. 2012;125:2095–2104. doi: 10.1242/jcs.053850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW, Abel PW, Tu Y. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desai SP, Bhatia SN, Toner M, Irimia D. Mitochondrial localization and the persistent migration of epithelial cancer cells. Biophys J. 2013;104:2077–2088. doi: 10.1016/j.bpj.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Church DN, Talbot DC. Survivin in solid tumors: rationale for development of inhibitors. Curr Oncol Rep. 2012;14:120–128. doi: 10.1007/s11912-012-0215-2. [DOI] [PubMed] [Google Scholar]
- 51.Rauch A, Hennig D, Schafer C, Wirth M, Marx C, Heinzel T, Schneider G, Kramer OH. Survivin and YM155: how faithful is the liaison? Biochim Biophys Acta. 2014;1845:202–220. doi: 10.1016/j.bbcan.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9:447–464. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 53.Kang BH, Plescia J, Song HY, Meli M, Colombo G, Beebe K, Scroggins B, Neckers L, Altieri DC. Combinatorial drug design targeting multiple cancer signaling networks controlled by mitochondrial Hsp90. J Clin Invest. 2009;119:454–464. doi: 10.1172/JCI37613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caino MC, Chae YC, Vaira V, Ferrero S, Nosotti M, Martin NM, Weeraratna A, O’Connell M, Jernigan D, Fatatis A, Languino LR, Bosari S, Altieri DC. Metabolic stress regulates cytoskeletal dynamics and metastasis of cancer cells. J Clin Invest. 2013;123:2907–2920. doi: 10.1172/JCI67841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li F, Ackermann EJ, Bennett CF, Rothermel AL, Plescia J, Tognin S, Villa A, Marchisio PC, Altieri DC. Pleiotropic cell-division defects and apoptosis induced by interference with survivin function. Nat Cell Biol. 1999;1:461–466. doi: 10.1038/70242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Mitochondrial survivin.
Fig. S2. Survivin regulation of mitochondrial bioenergetics.
Fig. S3. Modulation of oxidative phosphorylation by survivin.
Fig. S4. Survivin regulation of mitochondrial trafficking.
Fig. S5. Regulation of membrane dynamics by survivin.
Fig. S6. Expression of mitochondrial-targeted survivin dominant negative mutant.
Fig. S7. Mitochondrial survivin-Complex II regulation of tumor cell motility.