Abstract
Herd immunity can potentially induce a change of circulating viruses. However, it remains largely unknown that how bacterial pathogens adapt to vaccination. In this study, Bordetella pertussis, the causative agent of whooping cough, was selected as an example to explore possible effect of vaccination on the bacterial pathogen. We sequenced and analysed the complete genomes of 40 B. pertussis strains from Finland and China, as well as 11 previously sequenced strains from the Netherlands, where different vaccination strategies have been used over the past 50 years. The results showed that the molecular clock moved at different rates in these countries and in distinct periods, which suggested that evolution of the B. pertussis population was closely associated with the country vaccination coverage. Comparative whole-genome analyses indicated that evolution in this human-restricted pathogen was mainly characterised by ongoing genetic shift and gene loss. Furthermore, 116 SNPs were specifically detected in currently circulating ptxP3-containing strains. The finding might explain the successful emergence of this lineage and its spread worldwide. Collectively, our results suggest that the immune pressure of vaccination is one major driving force for the evolution of B. pertussis, which facilitates further exploration of the pathogenicity of B. pertussis.
The 20th century witnessed enormous successes in vaccination campaigns against infectious diseases, drastically reducing burden of these diseases. However, certain recent studies have reported that the effectiveness of viral vaccines, such as the influenza vaccine, has been affected by mismatches between vaccine components and the circulating virus due to evolutionary drift1,2. In the case of viral pathogens, herd immunity is considered to potentially contribute to evolutionary drift1,3, but it is unclear that how the bacterial pathogens adapt to vaccination due to bacteria’s slower rate of evolution compared with that of viruses.
Bordetella pertussis could serve as a favourable example for illustrating the adaptation of bacterial pathogens. Before the introduction of mass vaccination, pertussis was one of the primary causes of infant mortality, but since the introduction of whole-cell pertussis vaccines (WCVs) in many countries in the 1940s–1960s, the morbidity and mortality of pertussis have been declined dramatically4. However, since 1990s, pertussis resurgence has been observed in developed countries that have attained high vaccination coverage such as the Netherland, the United Kingdom and the United States4,5,6.
Although B. pertussis is a monomorphic pathogen, the most common antigenic alleles among current circulating strains, such as pertussis toxin A subunit (ptxA) 1 and pertactin (prn) 2, have been shown to differ from the alleles of the vaccine strains (ptxA2 or ptxA3 and prn1) used in several countries in which vaccination has been implemented since the 1940s–1960s7,8,9,10, suggesting that pathogen adaptation may play a role in the emergence of pertussis5,10,11,12,13,14,15. Furthermore, a novel allele of pertussis toxin promoter (ptxP) 3 emerged in the 1990s, and has become prevalent in developed countries15,16,17,18. Compared with ptxP1 strain, the ptxP3 can produce more Ptx, which results in increased bacterial virulence19; this suggests that the expansion of clones carrying ptxP3 might be associated with pertussis resurgence in the Netherlands and Australia16,19.
Contrasting the observations from developed countries with long histories of high vaccine coverage, in China, the prn 2 allele was first detected in 200011, and its frequency was found to be only 16% during the 1997–2005 period. Furthermore, many of the pulsed-filed gel electrophoresis (PFGE) profiles of the examined Chinese strains belonged to the three lineage clusters (group I, II and III) that resembled most of the strains circulating in Europe before the 1990s11. In Senegal, where large-scale pertussis vaccinations with WCVs were started in the late 1980s and the coverage was generally low, the predominant strains also belong to these 3 groups20. These observations indicate that distinct immunization strategies may be linked to the microevolution of B. pertussis.
Recently, a comparative genomic study based on a global collection of 343 B. pertussis isolates revealed that the worldwide B. pertussis population underwent major changes during last 60 years; the phylo-geographic analysis of these strains suggests that adaptive evolution of this pathogen is closely associated with vaccine introduction and that new emerged strains spread rapidly between countries21. However, several questions remain unanswered, including these: (1) Why do the types of isolates prevalent before the introduction of vaccination still predominate in areas of low vaccination coverage? (2) Do the evolution rates of B. pertussis population in certain countries with high vaccination coverage differ from the rates in countries in which the coverage is low? To further understand how vaccination affects B. pertussis evolution and obtain insight into how this pathogen has adapted to immune pressure from vaccination, we performed whole-genome sequencing to characterise 40 isolates from 1956 to 2008 in Finland, a country with high vaccination coverage and China, a country with relatively low coverage. The results were also compared with the sequences of 11 previously published strains from the Netherlands7,22, where pertussis vaccination was introduced in 1953.
Results
SNPs and phylogenetic analyses
The sequence reads obtained for each isolate were assembled in the genomes, which resulted on average in 105-fold coverage per genome (Supplementary Table S1). These sequences, combined with 11 previously published whole-genome sequences of Dutch isolates, were mapped against the complete chromosome of the B. pertussis strain CS. Our analysis focused on the non-repetitive components of each genome. Repetitive sequences, including variable-number tandem-repeat sequences and 3 types of insertion sequences (IS), IS 481 (239 copies), IS 1002 (6 copies) and IS 1663 (17 copies), accounted for 6.3% of genome of the strain CS23. After excluding these repetitive sequences, 825 SNP sites were identified in the non-repetitive genome (Supplementary Tables 2 and 3). Further analysis revealed that 120 of these SNPs appeared in high-density clusters, indicating that they might have originated from recent recombinant events24; 101 SNPs in B. pertussis isolates from recent periods (since 1990s) (Supplementary Table S3). Moreover, although 50.8% (61/120) of the SNPs were located at non-coding regions and genes of unknown function, it was noted that certain recombination events had occurred and included genetic determinants for recognised surface antigens and two-component regulatory systems, such as genes encoding Ptx, filamentous hemagglutinin (FHA) and two-component histidine kinase, which modulated host-cell interactions.
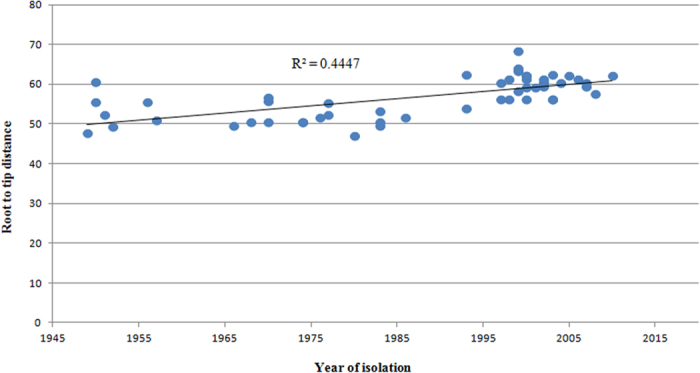
We excluded the aforementioned SNPs from the phylogenetic analyses because the recombination masked the true phylogenetic signal. Using B. bronchiseptica as the outgroup, a maximum likelihood phylogenetic tree that also included the published genome of B. pertussis Tohama I25 was constructed based on the remaining 705 SNPs (Fig. 1). Detailed information regarding these SNPs was provided in Supplementary Table S2. We also constructed a phylogenetic tree with roots by using the B. pertussis typing strain 18323 (Supplementary Fig. S1). The two trees exhibited the same topology, further supporting the hypothesis that strain 18323 is highly phylogenetically distinct from other B. pertussis strains, and might have been derived through a unique evolutionary process from the most recent common ancestor (MRCA)21,26. The resulting phylogeny recapitulated the 4 major groups previously as identified by the PFGE analyses11,12 but provided more accurate estimates of branch lengths and substantially increased the resolution. The strains from different countries within the same period (pre- or post- vaccination era) were clustered closely in the phylogenetic tree. Furthermore, the strains within each group were separated by up to 300 SNPs, and the strains of clade IV appeared to be the most diverse. 35 SNPs shared by all members of group III and IV discriminated them from the isolates in other groups. Lastly, clade IV contained most of the strains (88.6%, 31/35) from 1977 to 2008, and this clade was divided into 2 subgroups, IVa and IVb, separated by 121 SNPs. The results indicated that the number of SNPs from the root varied between the different clades and that the root-to-tip distances of individual isolates correlated with their date of isolation (R2 = 0.44; Fig. 2), suggesting that B. pertussis adapted through successive SNP accumulation over time7,21. Furthermore, the correlation between the year of isolation and the root-to-tip distances of individual Chinese strains (R2 = 0.58) differed from those of Finnish and Dutch strains (R2 = 0.35; Supplementary Fig. S2), indicating that SNP accumulation rates over time might vary between different countries.
Figure 1. Maximum likelihood phylogenetic tree for B. pertussis based on SNPs across the entire core genome, excluding probable recombination events.
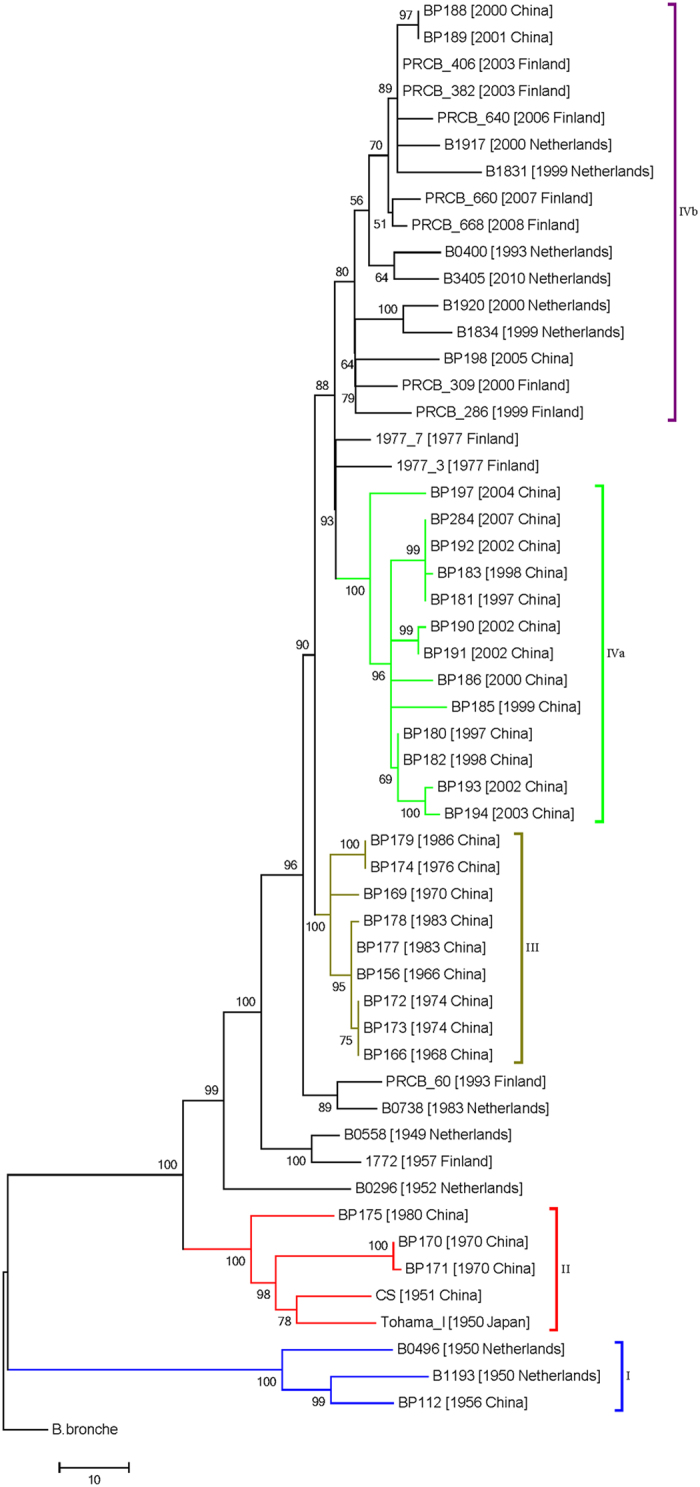
B. bronchiseptica was used as the outgroup to root the tree. The evolutionary relationships were inferred using the neighbor-joining method. The percentages of replicate trees in which the associated taxa clustered together based on the bootstrap test (100 replicates) are presented next to the branches.
Figure 2. Linear regression plot of B. pertussis displaying the correlation (R2) between the root-to-tip distance (y-axis) and the date of isolation (x-axis).
Variations in mutation rate
Single-copy genes are conserved genetic markers and are widely used for phylogenetic analyses of numerous species27,28,29. To investigate the potential cause of the aforementioned heterogeneity in genomic diversity, we performed a BEAST analysis on the single-copy genes in the genomes of isolates from different periods and countries. Our results demonstrated that the substitution rates of the isolates from period 2 were 6.0-fold higher than those of isolates from period 1 [substitutions per site per year: 4.60 × 10−5, 95% confidence interval (CI): 2.03–7.78 × 10−5 versus 7.64 × 10−6, 95% CI: 3.03 × 10−10 to 1.63 × 10−5] (Supplementary Fig. S3). Furthermore, the substitution rates of the Finnish and Dutch B. pertussis populations were higher than those of the Chinese population (substitutions per site per year: 1.59 × 10−5, 95% CI: 3.98 × 10−6 to 2.87 × 10−5 versus 3.06 × 10−6, 95% CI: 1.25–5.18 × 10−6) (Supplementary Fig. S3). Similar results were also obtained in the BEAST analysis of SNPs in the B. pertussis population (Supplementary Fig. S4). These results suggested that the substitution mutation rate differed substantially between isolates from different countries or periods.
Potential signals of selection
Our results identified 307 nonsynonymous SNPs, which represented 43.5% of all SNPs found here (Supplementary Table S2 and Supplementary Fig. S5). Further analysis showed that the nonsynonymous SNP densities of isolates from clades IVa and IVb were higher than those of isolates from clade I and II (P < 0.01). It is generally accepted that homoplasies are traits that species share due to convergent evolution that constitute a dependable signal of selective pressure. Here, 42 homoplastic SNPs were identified, of which 27 occurred within the clade III and IV lineages (Supplementary Table S4); 16 of these 27 SNPs were located in coding regions, 11 were intergenic. Intriguingly, one SNP were present at between the bvgA-encoded virulence factors transcription regulator and the fhaB encoded adhesion protein FHA. Furthermore, 11 of the homoplastic SNPs associated with coding genes were nonsynonymous SNPs. Although these coding genes containing homoplasies are poorly characterised, a few of the representative genes are of interest, such as sphB1 and bscl, which are under the control of the Bordetella virulence regulon (bvg). The product of sphB1 was reported to be an outer membrane protein, associated with host-cell adhesion and invasion of B. pertussis30,31. The gene bscl identified in B. bronchiseptica and B. pertussis encodes a type III secretion protein that is recognised to be essential for bacterial pathogenicity23,25,32. Another bvg-regulated virulence gene, ptxc which encodes Ptx subunit 3, also contained a homoplastic SNP. Collectively, these data indicated that adaptive bacterial evolution might contribute to the substitution-rate differences.
Genomic deletions and ongoing gene loss
To further investigate the evolution of distinct lineages, we analysed the nucleotide changes that induced gene disruption and genome size reduction. The clade II strain CS, which was isolated in Beijing, China, in 1951, was used as a reference to compare the genome sequences of the examined isolates. In addition to base substitutions, genomic deletions were common in the sequenced isolates, but insertions were infrequent. The mean deletion size largely exceeded the mean insertion size: only 732 bp were gained, but 19.4 kb were lost (Supplementary Fig. S6). The deletions ranged in size from 1 bp to 6.4 kb, and most of the deleted regions included protein-coding sequences, resulting in partial or complete deletion of B. pertussis genes (Supplementary Fig. S6). Deletions, insertions and nucleotide substitutions affect the corresponding gene. However, their effects on protein function cannot be readily to predict; thus we focused on genes that were completely deleted. Of the 150 deleted genes found in various lineages (Supplementary Table S5), a few corresponded to variant regions previously identified based on DNA microarray analyses33. Several of these genes are involved in replication, recombination and repair, transcription and unknown functions. When the deletions were mapped to the phylogenetic tree, these lineages were found to display gene loss over time (Supplementary Figs S6 and S7), further supporting the ongoing reductive evolution in the global B. pertussis population21.
In addition to identifying the aforementioned genes affected by deletion events, we detected 9 nonsense SNPs that have occurred since the MRCA. These SNPs introduce stop codons into protein-coding genes and terminate translation (Supplementary Table S2). The published strains Tohama and CS both contain more than 300 pseudogenes23,25. Pseudogenes constitute 9.4% of all B. pertussis genes, a considerably higher proportion than that in other Bordetella species, such as B. bronchiseptica (0.4%), B. parapertussis (5.0%), B. avium (2.0%) and B. petrii (2.5%)25,34,35.
Emergence of the ptxP3 lineage
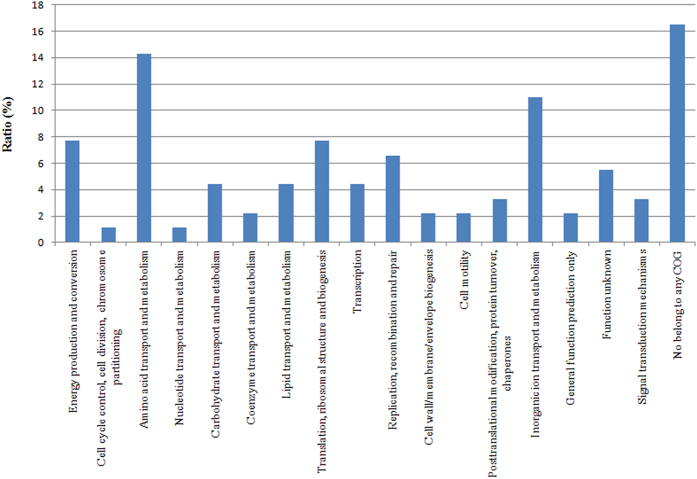
Almost all subgroup IVb isolates contained the novel ptxP3 allele, whereas subgroup IVa isolates contained ptxP1. The phylogenetic tree clearly revealed that these 2 subgroups shared a common ancestor. The results of Bayesian phylogenetic analysis suggested that these lineages diverged more than 20 years ago, which agrees with the earliest reports of ptxP3 strains in the 1990s19,36. We also examined the distribution of SNPs in the ptxP3 lineage, and identified 116 specific SNPs, of which 54 and 10 SNPs were distributed in our Finnish and Chinese isolates, respectively (Supplementary Table S2). Besides the above 3 previously mentioned SNPs in coding regions (ptxc, sphb1 and bscl), 85 other SNPs were located in coding regions and 28 SNPs were intergenic (located in non-coding regions). The SNPs in the coding regions corresponded to 17 different clusters of orthologous genes (COGs). The highest COG frequencies were related to amino acid transport and metabolism (14.4%), and inorganic ion transport and metabolism (11.1%) (Fig. 3). Furthermore, one nonsynonymous SNP was located in fim3 and caused an alanine (A) to glutamic acid (E) substitution. Fimbriae 3 (Fim3), also known as pili or agglutinogen, is expressed on the surface of B. pertussis and is considered to be involved in the bacterial attachment to the respiratory tract37.
Figure 3. Clusters of orthologous genes associated with the specific SNPs in the ptxP3 strains.
Discussion
Isolate selection is critical for comparative analyses because it uncovers mutations that differ between the examined isolates. The 29 Chinese and 11 Finnish isolates studies here were sampled from reliable phylogenies based on the molecular genotyping of >500 strains from Finland and >100 strains from China11,12,38 to provide a satisfactory representation of the genetic diversity of B. pertussis in these two countries. To capture most of the variation in B. pertussis isolates from different countries at the whole-genome level and to further understand how vaccination affects bacterial evolution, 11 previously published genomes of B. pertussis isolates obtained between 1949 and 2010 from the Netherlands7,22 were also included in this study.
Vaccinations against pertussis have been introduced since 1950s in many countries. The initial WCVs were replaced by less reactogenic acellular pertussis vaccines (ACVs) in the 1990s in developed countries4. Many studies have demonstrated the marked changes in the B. pertussis populations in these countries following the introduction of ACVs11,17,18,21. B. pertussis strains from different countries with similar vaccination histories are genetically similar22,39,40,41. Our studies also revealed that Finnish and Chinese B. pertussis isolates from similar period were phylogenetically closely related, further supporting the hypothesis that B. pertussis strains do not exhibit geographic specificity21,33. Although the exact cause for this is unknown, it might be associated with recent population migration, human admixture, and strain transmission.
B. pertussis is an obligate human pathogen, and evolutionary pressure is likely dominantly mediated by the host immune response9. Comparative genomic analysis has provided evidence that the world-wide B. pertussis population is evolving in response to vaccine introduction21. Extending previous findings11,21, our results showed that the molecular clock moved at a dissimilar rate in different countries and periods, which might have resulted in the large heterogeneity in the number of SNPs that accumulated over time. According to WHO estimates, the rate of vaccination coverage was only 58% in 1983 in China, as compared with 94% and 97% in Finland and the Netherlands, respectively42. A variation in vaccination coverage might exert different selective pressures on B. pertussis populations11,20. In Finland and the Netherlands, ACVs were introduced in 2005, whereas in China, WCVs were not completely replaced by ACVs until 2012. For understanding the evolution of B. pertussis, evaluating the relative impact of ACVs and WCVs is critical. However, we could not analyse this impact because our study included only 3 Finnish strains collected after ACV introduction. Recently, Sealey et al have demonstrated that in the ACV era, genes encoding ACV antigens are evolving more rapidly than other cell surface proteins of B. pertussis isolates43. Collectively, these results provide direct evidence that B. pertussis evolution has accelerated since the introduction of vaccination. This evolution might also explain why isolates belonging to the PFGE group III, which predominated before the introduction of ACVs in industrialised countries, are still prevalent in areas with low vaccination coverage, and thus contribute to the differences observed in the frequency of prn2 alleles between these countries11,20.
In agreement with previous studies21,43, an over abundance of nonsynonymous and homoplastic SNPs was identified in B. pertussis isolates from both China and Finland, which provided additional evidence for adaptive selection in B. pertussis strains from the post-vaccination era. However, most of the detected homoplasies were located in modern isolates, which were primarily categorised into clades III and IV, indicating that convergent evolution might occur over short periods. Although the precise effects of these homoplasies in B. pertussis genes are unknown, we found 3 homoplastic SNPs in 3 genes (ptxc, sphB1 and bscl) likely associated with bacterial virulence under the control of bvg, which consisted with 33% of the homoplastic SNPs located in bvg-activated genes in global phylogenic analysis of B. pertussis21. Notably, one homoplasic SNP in bscl was identified in B. pertussis strains collected in different countries21. The bscl encodes a component of the Bordetella type III secretion system32, which subverts innate and adaptive immune responses during B. pertussis infection and promotes bacterial persistence44. Recently, Han et al have reported that the type III effector BteA is expressed at higher levels in B. pertussis non-vaccine type strains than in vaccine-type strains, which suggests that increased BteA protein expression might play a key role in the evolution of B. pertussis45. Intergenic SNPs have been suggested to be capable of altering gene expression level46. Furthermore, an abundance of homoplasic SNPs in intergenic region might affect the transcription of related genes. Although the functions of most of the identified genes are unknown, we suspect that the intergenic SNP located at between bvgA and fhaB might be associated with pathogen adaptation. Taken together, the adaptive mutations detected in surface antigenic and bvg-controlled genes further emphasize the strong selective pressure on the B. pertussis genome due to herd immunity21.
Our whole-genome comparison provided insights into the spectrum of genetic variation in the B. pertussis population. In contrast to the accumulation of SNP mutations over time, numerous deletions were identified in different lineage and continuous gene loss was observed. Although a high coverage of bacterial genomes was obtained using the Illumina sequencing platform and de novo assemblies were performed, a few gaps might remain between contigs due to the limitation of unfinished genomes47. Therefore, to reduce error by increasing alignment accuracy, we only analysed insertions or deletion flanked by at least 20 bp of gapless matches. Similar to other genetically monomorphic bacterial pathogens, such as Yersinia pestis48, a remarkably high number of transposed sequences, IS elements and pseudogenes are present in the B. pertussis genomes compared with other Bordetella species25,34,35. Indeed, IS elements have been reported to play a major role on causing gene loss in B. pertussis population, which benefitted from the increased fitness21,33. Furthermore, our whole-genome analysis also revealed an ongoing formation of pseudogenes in B. pertussis. High pseudogene frequencies are associated with host restriction in a variety of bacterial species49. This phenomenon is potentially caused by high rates of mutational fixation resulting from the accelerated genetic shift due to evolutionary bottlenecks associated with host adaptation and a reduction in the effective population size. Collectively, these data further support the hypothesis that evolution in this host-restricted pathogen might be dominated by genetic shift and gene loss rather than by the acquisition of new genes21,33,43. On the other hands, previous studies have also reported that rearrangements mediated by IS elements contribute to the genome reduction in B. pertussis when it evolved from a B. bronchiseptica like ancestor25. Chromosomal rearrangements can even occur during subculturing of B. pertussis in vitro50. Altogether, rearrangement might also serve as a key mechanism in Bordetella evolution.
Recent studies also demonstrated that ptxP3 is the dominant allele in clinical isolates from many countries with high vaccine coverage15,16,36. In this study, we showed that ptxP3 was associated with the emergence of sub-branch that contained the currently circulating isolates from different countries. Mooi et al reported that the ptxP3 strains can lead to the increased production of Ptx than the ptxP1, resulting in increased virulence and immune suppression19. Aside from the SNP mutations in the ptxP promoter, our whole-genome analysis identified 116 specific SNPs in the ptxP3 lineage. These polymorphisms might facilitate the adaptability of the ptxP3 strains. Notably, 3 SNPs specific to British ptxP3 strains43 were also identified in this present study. Collectively, these results highlight that the SNPs identified among ptxP3 strains will serve as potential genotypic markers in phylogenetic genotyping studies, particularly in the monitoring of ptxP3 strains worldwide. A recent genome-wide association study have also demonstrated that compared to ptxP1 strains, the ptxP3 strains express higher levels of a series of genes belonging to amino acid biosynthesis, energy metabolism, regulation and central/intermediary metabolism categories, and that wild-type ptxP3 strains are relatively more effective colonizers in an animal infection model51. These characteristics might have caused the recent emergence of the ptxP3 lineage and its worldwide spread.
In conclusion, our whole-genome comparative analysis supports the hypothesis that the evolution of B. pertussis has been dominated by ongoing genetic shift and gene loss21,33,43. Furthermore, numerous SNPs specific to the ptxP3-containing strains were identified here, which might explain the recent emergence and worldwide spread of this successful lineage. More importantly, we found that the molecular clock moved at different rates in different countries and periods, suggesting that the evolution of the B. pertussis population is closely associated with the extent of vaccination coverage. These results provide new and crucial evidence that the immune pressure from vaccination is one major driving force for the evolution of B. pertussis. We expect these data to enrich the understanding of evolutionary mechanisms in pathogens in the presence of different vaccination coverage levels. The genomic data presented here will also facilitate further exploration of the pathogenicity of B. pertussis.
Methods
Bacterial strains
We selected 29 Chinese and 11 Finnish isolates from 1956 to 2008 based on molecular genotyping in accordance with their occurrence frequencies11,38. The selected strains were representative of the most prevalent PFGE profiles and genotypes during each period (Table 1 and Supplementary Table S1). Furthermore, B. pertussis strain CS, which isolated in China in 1951 and is widely used as a vaccine strain for ACV production in China, was included in the analysis23. All isolates were cultured on Bordet-Gengou agar supplemented with 15% sheep blood for 72 h at 37 °C, and genomic DNA was isolated using the Promega Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Table 1. Pertussis vaccination program of the three countries.
| Country | Start | Vaccination schedule |
|---|---|---|
| Finland | 1952 | Primary: DTwP at 3,4,5 months;Boosters: DTwP at 20–24 months |
| 2003 | Primary: DTwP at 3,4,5 months;Boosters: DTwP at 20–24 months and dTap at 6 years | |
| 2005 | Primary: DTaP at 3,5,12 months;Boosters: dTap at 4 and 14 years | |
| China* | 1960s | Primary: DTwP at 3,4,5 months; |
| 1978 | Primary: DTwP at 3,4,5 months;Boosters: DTwP at 18–24 months | |
| 2007 | Primary: DTwP or DTaP at 3,4,5 months;Boosters: DTwP or DTaP at 18–24 months | |
| Netherlands | 1953 | Primary: DTwP at 3,4,5 months;Boosters: DTwP at 4 years |
| 1962 | Primary: DTwP-IPV at 3,4,5, 11 months | |
| 1993 | Primary: DTwP-IPV-Hib at 3,4,5, 11 months | |
| 1999 | Primary: DTwP-IPV-Hib at 2,3,4, 11 months | |
| 2001 | Primary: DTwP-IPV-Hib at 2,3,4, 11 months;Booster: DTaP at 4 years | |
| 2005 | Primary: DTaP-IPV-Hib at 2,3,4, 11 months;Booster: DTaP at 4 years | |
| 2006 | Primary: DTaP-IPV-Hib at 2,3,4, 11 months;Booster: dTap at 4 years |
Notes: *Although DTaP was phased in from the late 1990s, DTwP have been completely replaced by DTaP until 2012 in China. DTwP, diphtheria-tetanus-whole cell pertussis; DTaP, diphtheria-tetanus-acellular pertussis; dTap, diphtheria-tetanus-whole cell pertussis (with reduced content of pertussis antigens); IPV, inactivated polio virus; Hib, haemophilus influenza type b
To comprehensively investigate how vaccination affects the evolution of bacterial genomes, 11 published genomes of Dutch isolates from 1949 to 2010 were included in the phylogenetic analysis of B. pertussis. These isolates were separated into 2 periods based on the time of WCV introduction and the vaccine coverage in each country (Table 1). Period 1 comprised the period before the onset of mass vaccination using WCVs and the early post-vaccination period that resulted in a low coverage rate, including 10 Chinese isolates from 1951 to 1977 and 5 European isolates (1 Finnish, 4 Dutch) from 1949 to 1957. Period 2 covered the post-vaccination period, including 20 Chinese isolates from 1980 to 2007 and 17 European isolates (10 Finnish, 7 Dutch) from 1977 to 2010 (Supplementary Table S1).
Genome sequencing and comparative genomics
Whole-genome sequencing of 40 isolates was performed using the Solexa Genome Analyzer IIx 75-bp paired-end platform (Illumina, Little Chesterford, UK). The Illumina data were de novo assembled using Velvet, and multi-contig draft genomes were generated for each isolate. Details for each genomic assembly were summarized in Supplementary Table S1. Open reading frames of at least 30 amino acids in length were predicted using Glimmer 3.0 and were manually verified according to the annotation of B. pertussis CS23. The transfer RNA and ribosomal RNA genes were predicted using tRNAscan-SE. Artemis was used to collate the data and facilitate the annotation52. Functional predictions were based on BLASTp similarity searches using the UniProtKB, GenBank, and Swiss-Prot protein databases. Genome comparisons were performed to search for insertion and deletion events. Blocks of sequences that were substantially shared by B. pertussis CS and each isolate were identified using BLAST, and the alignment within each block was based on the Mauve method53 using a seed length of 15. In order to reduce error by improving alignment accuracy, only insertions or deletions, flanked by at least 20 bp of gapless matches were considered to be indel events54.
Detection of SNPs and phylogenetic analysis
The generated Solexa reads were called from duplicates containing identical reads that were presumably derived from replicates during the PCR step. Then, the reads were mapped to the genome of B. pertussis CS (accession number NC_017223) using BWA55 under the default parameters, which allow for 3% mismatches. All reads of extremely large or small insert size (<50% or >200% of normal) were remapped by using BLASTn at an e-value of 0.0001 and using the -F F flag. We only considered read pairs that mapped at an appropriate separation and contained at least one mapped end in a non-repeat region. The SAMtools program was used to calculate the per-position coverage and the base calls for each position56. The SNPs in the sequenced strain were called if the position was covered by at least 10 reads and at least 80% of the covering reads displayed the same mismatch57. SNP clusters within the recombination region were excluded as described previously24, and SNPs from regions that were not present in all genomes were also excluded. A SNP was considered homoplastic if the allele pattern did not agree with the tree topology. A phylogeny was constructed based on the SNPs using RAxML, which was based on the root of B. bronchiseptica or B. pertussis strain 18323. A regression analysis was conducted on the root-to-tip genetic distances against the sampling date according to a neighbor-joining tree using the program Path-O-Gen V1.4 (http://tree.bio.ed.ac.uk/software/pathogen/). The SNP densities were calculated by determining the number of SNPs per bp of each gene, and the differences between distinct clades were analysed using the nonparametric Wilcoxon test of SPSS 20.0 software (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered statistically significant.
To comprehensively investigate the evolution sites in B. pertussis strains from different countries and periods, the SNPs and single-copy orthologous clusters identified in this study were utilized to estimate the nucleotide substitution rate of each site (substitutions per site per year) using the BEAST package58. BEAST analysis can incorporate the date of isolates into the model and provide a source of information on the overall rate of evolutionary change when the differences in the dates are associated with the tips of the phylogenetic tree. The single-copy orthologous gene families were identified as previously described28,29. Briefly, we compared and clustered the complete genomes of all B. pertussis strains in this study using the software OrthoMCL29, identified 179 single-copy orthologous families shared among all genomes. We used the jModelTest program59 to select the best-fitting substitution model for our sequences according to the Bayesian information criterion, and the date of isolation of each isolate was used to calibrate the tree, and finally these analyses were performed using a log-normal relaxed molecular clock, which allows for different mutation rates on different branches. Each run (4 in total) spanned 100 million generations, and the model parameter values and trees were sampled every 10,000 generations. The results were visualised using Tracer v1.5 to assess convergence.
Additional Information
How to cite this article: Xu, Y. et al. Whole-genome sequencing reveals the effect of vaccination on the evolution of Bordetella pertussis. Sci. Rep. 5, 12888; doi: 10.1038/srep12888 (2015).
Supplementary Material
Acknowledgments
This work was supported by the National Major Project from the Ministry of Science and Technology, People’s Republic of China (No. 2013ZX09304101, 2013ZX10004216-001-001 and 2013ZX10004221), the National 973 Program of China Grant (No.2011CB504900) and The National Natural Science Fund (No. 81201249).
Footnotes
Author Contributions S.Z., L.W. and Q.H. conceived and designed the study. Y.X., B.L., K.G., Y.T., L.F., T.K., L.W. and D.P. performed the experiments. Y.X., B.L. and K.G. conducted the bioinformatic analyses of the data. Y.X., H.Z., B.L., S.Z., L.W. and Q.H. wrote the paper. All authors reviewed and approved the final manuscript.
References
- Carrat F. & Flahault A. Influenza vaccine: the challenge of antigenic drift. Vaccine 25, 6852–6862 (2007). [DOI] [PubMed] [Google Scholar]
- Belongia E. A. et al. Effectiveness of inactivated influenza vaccines varied substantially with antigenic match from the 2004-2005 season to the 2006-2007 season. J Infect Dis 199, 159–167 (2009). [DOI] [PubMed] [Google Scholar]
- Boni M. F., Gog J. R., Andreasen V. & Christiansen F. B. Influenza drift and epidemic size: the race between generating and escaping immunity. Theor Popul Biol 65, 179–191 (2004). [DOI] [PubMed] [Google Scholar]
- Galanis E., King A. S., Varughese P. & Halperin S. A. Changing epidemiology and emerging risk groups for pertussis. CMAJ 174, 451–452 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Melker H. E. et al. Pertussis in The Netherlands: an outbreak despite high levels of immunization with whole-cell vaccine. Emerg Infect Dis 3, 175–178 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guris D. et al. Changing epidemiology of pertussis in the United States: increasing reported incidence among adolescents and adults, 1990-1996. Clin Infect Dis 28, 1230–1237 (1999). [DOI] [PubMed] [Google Scholar]
- Bart M. J. et al. Comparative genomics of prevaccination and modern Bordetella pertussis strains. BMC Genomics 11, 627 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez V., Caro V., Levillain E., Guigon G. & Guiso N. Genomic content of Bordetella pertussis clinical isolates circulating in areas of intensive children vaccination. PLoS One 3, e2437 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi F. R. Bordetella pertussis and vaccination: the persistence of a genetically monomorphic pathogen. Infect Genet Evol 10, 36–49 (2010). [DOI] [PubMed] [Google Scholar]
- Mooi F. R., Van Loo I. H. & King A. J. Adaptation of Bordetella pertussis to vaccination: a cause for its reemergence? Emerg Infect Dis 7, 526–528 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. Effect of vaccination on Bordetella pertussis strains, China. Emerg Infect Dis 16, 1695–1701 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elomaa A. et al. Strain variation among Bordetella pertussis isolates in finland, where the whole-cell pertussis vaccine has been used for 50 years. J Clin Microbiol 43, 3681–3687 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingermann M. et al. Differences of circulating Bordetella pertussis population in Argentina from the strain used in vaccine production. Vaccine 24, 3513–3521 (2006). [DOI] [PubMed] [Google Scholar]
- Hallander H. O., Advani A., Donnelly D., Gustafsson L. & Carlsson R. M. Shifts of Bordetella pertussis variants in Sweden from 1970 to 2003, during three periods marked by different vaccination programs. J Clin Microbiol 43, 2856–2865 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Advani A., Gustafsson L., Ahren C., Mooi F. R. & Hallander H. O. Appearance of Fim3 and ptxP3-Bordetella pertussis strains, in two regions of Sweden with different vaccination programs. Vaccine 29, 3438–3442 (2011). [DOI] [PubMed] [Google Scholar]
- Octavia S. et al. Newly emerging clones of Bordetella pertussis carrying prn2 and ptxP3 alleles implicated in Australian pertussis epidemic in 2008-2010. J Infect Dis 205, 1220–1224 (2012). [DOI] [PubMed] [Google Scholar]
- Petersen R. F., Dalby T., Dragsted D. M., Mooi F. & Lambertsen L. Temporal trends in Bordetella pertussis populations, Denmark, 1949-2010. Emerg Infect Dis 18, 767–774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Advani A. et al. Pulsed-field gel electrophoresis analysis of Bordetella pertussis isolates circulating in Europe from 1998 to 2009. J Clin Microbiol 51, 422–428 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi F. R. et al. Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis 15, 1206–1213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njamkepo E., Cantinelli T., Guigon G. & Guiso N. Genomic analysis and comparison of Bordetella pertussis isolates circulating in low and high vaccine coverage areas. Microbes and infection/Institut Pasteur 10, 1582–1586 (2008). [DOI] [PubMed] [Google Scholar]
- Bart M. J. et al. Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. MBio 5, e01074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gent M., Bart M. J., Van Der Heide H. G., Heuvelman K. J. & Mooi F. R. Small Mutations in Bordetella pertussis Are Associated with Selective Sweeps. PLoS One 7, e46407 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. et al. Complete genome sequence of Bordetella pertussis CS, a Chinese pertussis vaccine strain. J Bacteriol 193, 4017–4018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L. et al. A recalibrated molecular clock and independent origins for the cholera pandemic clones. PLoS One 3, e4053 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J. et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 35, 32–40 (2003). [DOI] [PubMed] [Google Scholar]
- Park J. et al. Comparative genomics of the classical Bordetella subspecies: the evolution and exchange of virulence-associated diversity amongst closely related pathogens. BMC Genomics 13, 545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amselem J. et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet 7, e1002230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Mueller L. A., Crouzillat D., Petiard V. & Tanksley S. D. Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics 174, 1407–1420 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J. Jr. & Roos D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deora R., Bootsma H. J., Miller J. F. & Cotter P. A. Diversity in the Bordetella virulence regulon: transcriptional control of a Bvg-intermediate phase gene. Mol Microbiol 40, 669–683 (2001). [DOI] [PubMed] [Google Scholar]
- Coutte L., Antoine R., Drobecq H., Locht C. & Jacob-Dubuisson F. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J 20, 5040–5048 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo S., Yuk M. H., Huang L. L. & Miller J. F. Regulation of type III secretion in Bordetella. Mol Microbiol 52, 1201–1214 (2004). [DOI] [PubMed] [Google Scholar]
- King A. J., Van Gorkom T., Van Der Heide H. G., Advani A. & Van Der Lee S. Changes in the genomic content of circulating Bordetella pertussis strains isolated from the Netherlands, Sweden, Japan and Australia: adaptive evolution or drift? BMC Genomics 11, 64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross R. et al. The missing link: Bordetella petrii is endowed with both the metabolic versatility of environmental bacteria and virulence traits of pathogenic Bordetellae. BMC Genomics 9, 449 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebaihia M. et al. Comparison of the genome sequence of the poultry pathogen Bordetella avium with those of B. bronchiseptica, B. pertussis, and B. parapertussis reveals extensive diversity in surface structures associated with host interaction. J Bacteriol 188, 6002–6015 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallonen T., Mertsola J., Mooi F. R. & He Q. Rapid detection of the recently emerged Bordetella pertussis strains with the ptxP3 pertussis toxin promoter allele by real-time PCR. Clin Microbiol Infect 18, E377–379 (2012). [DOI] [PubMed] [Google Scholar]
- Irons L. I., Ashworth L. A. & Robinson A. Release and purification of fimbriae from Bordetella pertussis. Dev Biol Stand 61, 153–163 (1985). [PubMed] [Google Scholar]
- Kallonen T. et al. Differences in the genomic content of Bordetella pertussis isolates before and after introduction of pertussis vaccines in four European countries. Infect Genet Evol 11, 2034–2042 (2011). [DOI] [PubMed] [Google Scholar]
- Mooi F. R. et al. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect Immun 66, 670–675 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrantonio P. et al. Antigenic variants in Bordetella pertussis strains isolated from vaccinated and unvaccinated children. Microbiology 145, (Pt 8) 2069–2075 (1999). [DOI] [PubMed] [Google Scholar]
- Van Loo I. H., Heuvelman K. J., King A. J. & Mooi F. R. Multilocus sequence typing of Bordetella pertussis based on surface protein genes. J Clin Microbiol 40, 1994–2001 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. WHO-UNICEF estimates of DTP3 coverage. Available at : http://apps.who.int/immunization_monitoring/globalsummary/timeseries/tswucoveragedtp3.html (Accessed: 5th November 2014).
- Sealey K. L. et al. Genomic Analysis of Isolates From the United Kingdom 2012 Pertussis Outbreak Reveals That Vaccine Antigen Genes Are Unusually Fast Evolving. J Infect Dis 212, 294–301(2015). [DOI] [PubMed] [Google Scholar]
- Fennelly N. K. et al. Bordetella pertussis expresses a functional type III secretion system that subverts protective innate and adaptive immune responses. Infect Immun 76, 1257–1266 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H. J., Kuwae A., Abe A., Arakawa Y. & Kamachi K. Differential expression of type III effector BteA protein due to IS481 insertion in Bordetella pertussis. PLoS One 6, e17797 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villaruz A. E. et al. A point mutation in the agr locus rather than expression of the Panton-Valentine leukocidin caused previously reported phenotypes in Staphylococcus aureus pneumonia and gene regulation. J Infect Dis 200, 724–734 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. R., Koren S. & Sutton G. Assembly algorithms for next-generation sequencing data. Genomics 95, 315–327 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M. et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci USA 101, 17837–17842 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E. & Ochman H. Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res 33, 3125–3132 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stibitz S. & Yang M. S. Genomic fluidity of Bordetella pertussis assessed by a new method for chromosomal mapping. J Bacteriol 179, 5820–5826 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. J. et al. Genome-wide gene expression analysis of Bordetella pertussis isolates associated with a resurgence in pertussis: elucidation of factors involved in the increased fitness of epidemic strains. PLoS One 8, e66150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T. J. et al. ACT: the Artemis Comparison Tool. Bioinformatics 21, 3422–3423 (2005). [DOI] [PubMed] [Google Scholar]
- Darling A. E., Treangen T. J., Messeguer X. & Perna N. T. Analyzing patterns of microbial evolution using the mauve genome alignment system. Methods Mol Biol 396, 135–152 (2007). [DOI] [PubMed] [Google Scholar]
- Tanay A. & Siggia E. D. Sequence context affects the rate of short insertions and deletions in flies and primates. Genome Biol 9, R37 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan R. P. et al. The multiplicity of divergence mechanisms in a single evolving population. Genome Biol 13, R41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J. & Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7, 214 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol 25, 1253–1256 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.