Abstract
We conducted a genome wide SNP association study on 1,803 Urinary Bladder Cancer (UBC) cases and 34,336 controls from Iceland and the Netherlands and follow up studies in seven additional case control groups (2,165 cases and 3,800 controls). The strongest association was observed with allele T of rs9642880 on chromosome 8q24, 30kb upstream of the c-Myc gene (allele specific OR=1.22; P=9.34×10−12). Approximately 20% of individuals of European ancestry are homozygous for rs9642880 (T) and their estimated risk of developing UBC is 1.49 times that of non-carriers with population attributable risk (PAR) of 17%. No association was observed between UBC and the four 8q24 variants previously associated with prostate, colorectal and breast cancers, nor did rs9642880 associate with any of these three cancers. A weaker signal, but nonetheless of genome wide significance, was captured by rs710521 (A) located near the TP63 gene on chromosome 3q28 (allele specific OR=1.19; P=1. 15× 10−7).
Urinary bladder cancer (UBC) is the 6th most common type of cancer in the United States with approximately 67,000 new cases and 14,000 deaths from the disease in 2007 1. Cigarette smoking and occupational exposure to specific carcinogens are the strongest known risk factors for UBC. Familial clustering of UBC cases suggests that there is a genetic component to the risk of the disease 2–4. Segregation analyses have suggested that this component consists of many genes, each conferring a small risk 5. Epidemiological studies have evaluated potential associations between sequence variants in candidate genes and UBC but the results have in many cases been difficult to replicate. Recently, genome wide association (GWA) studies have led to discoveries of variants in the sequence of the human genome that confer risk of common diseases, including cancers 6–10. To search for loci associated with risk of UBC, we carried out a genome wide association analysis on Icelandic and Dutch UBC cases and controls, using the Illumina Infinium Whole-Genome Genotyping microarray technology.
We genotyped 525 cases and 32,504 controls from Iceland as well as 1,278 cases and 1,832 controls from the Netherlands on the HumanHap300/HumanCNV370-duo BeadChips (Table 1). After removing SNPs failing quality control checks, 302,140 SNPs were tested for association with UBC. The results were adjusted for relatedness between individuals and for potential population stratification using the method of genomic control. With this sample size the combined Iceland/Netherlands GWA study has a 88% power to achieve a genome-wide significant result for a variant with an OR of 1.3 and a minor allele frequency of 50%. The power drops to 22% for an OR of 1.2 and the same frequency.
Table 1.
Description of the case control groups used in the studies
| Study Group | #cases | #controls | Average age at diagnosis (range) | % males (cases) | Study type |
|---|---|---|---|---|---|
| Discovery groups (GWA) | |||||
| Iceland | 525 | 32.504 | 67 (22–94) | 76 | Population based |
| The Netherlands | 1278 | 1,832 | 62 (25–93) | 81 | Population based |
| Follow up groups | |||||
| UK | 724 | 530 | 73 (30–101) | 71 | Hospital-based |
| Italy-Torino | 328 | 389 | 63 (40–75) | 100 | Hospital-based |
| Italy-Brescia | 181 | 192 | 63 (22–80) | 100 | Hospital-based |
| Belgium | 197 | 379 | 68 (40–93) | 86 | Population based |
| Eastern Europe - Hungary, Romania, Slovakia | 213 | 521 | 65 (36–90) | 83 | Hospital-based |
| Sweden | 349 | 930 | 69 (32–97) | 67 | Population based |
| Spain | 173 | 859 | 65 (27–94) | 87 | Hospital-based |
| Total | 3,968 | 38,136 | |||
No single SNP reached our genome wide significance threshold (P<1.6×10−7; corresponding to 0.05/302,140) either in the combined or individual analysis of the Icelandic or Dutch GWA sample sets. The 10 most significant SNPs (all P<5×10−5) were genotyped using Centaurus single track assays in additional 2,165 UBC cases and 3,800 controls from seven follow up groups, all of European ancestry (Table 1). The associations of each of these SNPs in the initial GWA, the joint follow up groups and the combined analysis of all groups are listed in Supplementary Table 1. The strongest association with UBC, reaching genome wide significance in the overall analysis of the discovery and follow up groups, was observed for the T allele of rs9642880 at 8q24.21 (combined odds ratio, OR=1.22 (95% confidence interval 1.15–1.29), P=9.34 × 10−12). This was followed by rs710521 (A) on 3q28 (combined OR=1.19 (95% CI 1.12–1.27), P=1.15 × 10−7) (Table 2). Of the 10 SNPs analyzed, only rs9642880 and rs710521 were nominally significant (P<0.05) in the combined analysis of the follow up groups (Supplementary Table 1), both reaching genome wide significance in the combined analysis and showing no heterogeneity between the ORs of the 9 study groups (Phet=0.69 and 0.77, respectively) (Table 2).
Table 2.
Association of rs9642880 (T) on 8q24.21 and rs710521 (A) on 3q28 with UBC
| Study population (N cases/N controls) | rs9642880 T
|
rs710521 A
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Frequency
|
OR | 95% CI | P value | Frequency
|
OR | 95% CI | P value | |||
| Cases | Controls | Cases | Controls | |||||||
|
|
|
|||||||||
| Discovery groups (GWA) | ||||||||||
| Iceland (523/32,458) a | 0.53 | 0.48 | 1.21 | 1.07–1.37 | 3.10x10−3 | 0.76 | 0.72 | 1.23 | 1.07–1.42 | 4.32x10−3 |
| The Netherlands (1,269/1,824) a | 0.53 | 0.48 | 1.22 | 1.10–1.36 | 2.53x10−4 | 0.76 | 0.72 | 1.24 | 1.10–1.40 | 4.87x10−4 |
| Follow up groups | ||||||||||
| UK (695/507) | 0.49 | 0.45 | 1.21 | 1.03–1.42 | 0.021 | 0.75 | 0.72 | 1.14 | 0.95–1.37 | 0.147 |
| Italy-Torino (323/381) | 0.50 | 0.41 | 1.43 | 1.16–1.77 | 8.69x10−4 | 0.76 | 0.75 | 1.04 | 0.82–1.33 | 0.747 |
| Italy-Brescia (174/183) | 0.44 | 0.41 | 1.13 | 0.84–1.52 | 0.419 | 0.74 | 0.69 | 1.23 | 0.89–1.69 | 0.207 |
| Belgium (195/372) | 0.49 | 0.45 | 1.15 | 0.90–1.46 | 0.254 | 0.79 | 0.73 | 1.34 | 1.00–1.79 | 0.048 |
| Eastern Europe (186/515) | 0.52 | 0.44 | 1.40 | 1.10–1.78 | 5.72x10−3 | 0.78 | 0.75 | 1.13 | 0.85–1.49 | 0.401 |
| Sweden (319/909) | 0.51 | 0.48 | 1.16 | 0.96–1.40 | 0.116 | 0.79 | 0.75 | 1.27 | 1.03–1.57 | 0.026 |
| Spain (171/836) | 0.48 | 0.45 | 1.11 | 0.88–1.40 | 0.375 | 0.73 | 0.74 | 0.96 | 0.73–1.27 | 0.773 |
| GWA (1,792/34,282) b | 0.48 | 1.21 | 1.12–1.31 | 2.72x10−6 | 0.72 | 1.23 | 1.13–1.35 | 7.10x10−6 | ||
| Follow up groups (2,063/3,703) b | 0.44 | 1.22 | 1.13–1.33 | 7.98x10−7 | 0.73 | 1.15 | 1.05–1.26 | 2.80x10−3 | ||
| All combined (3,855/37,985) b | 0.45 | 1.22 | 1.15–1.29 | 9.34x10−12 | 0.73 | 1.19 | 1.12–1.27 | 1.15x10−7 | ||
All P values shown are two-sided. Shown are the corresponding numbers of cases and controls (N), allelic frequencies of variants in affected and control individuals, the allelic odds-ratio (OR) with P values based on the multiplicative model.
Results presented for Iceland and the Netherlands were individually adjusted by the method of genomic control (see Supplementary Methods).
For the combined study populations, the reported control frequency was the average, unweighted control frequency of the individual populations, while the OR and the P value were estimated using the Mantel-Haenszel model.
From the initial genome wide association scan, 2 markers (rs12547643 and rs4733677) close to rs9642880 on chromosome 8q24.21 were nominally associated with UBC (P<0.01). When results for these markers were analyzed in the 9 combined groups, only rs4733677 (T) was significantly associated to UBC (P=7.15 x 10−6) while rs12547643 (A) was not (P=0.078). After adjusting for the effect of rs9642880, rs4733677 (T) was no longer significant (P =0.15). To investigate the mode of inheritance more carefully, we computed the genotype-specific OR for rs9642880. Results from all groups combined demonstrated that the association of rs9642880 (T) to UBC did not deviate from the multiplicative model (P=0.76). Relative to the non-carriers, the ORs for heterozygous and homozygous carriers of the risk allele T were 1.22 and 1.49, respectively. Assuming that the frequency of the allele is 45%, i.e. the average of the frequency of all the populations studied (Table 2), then individuals homozygous for rs9642880 (TT) represent ~20% of the population. The estimated population attributable risk (PAR) of rs9642880 (T) is 17%.
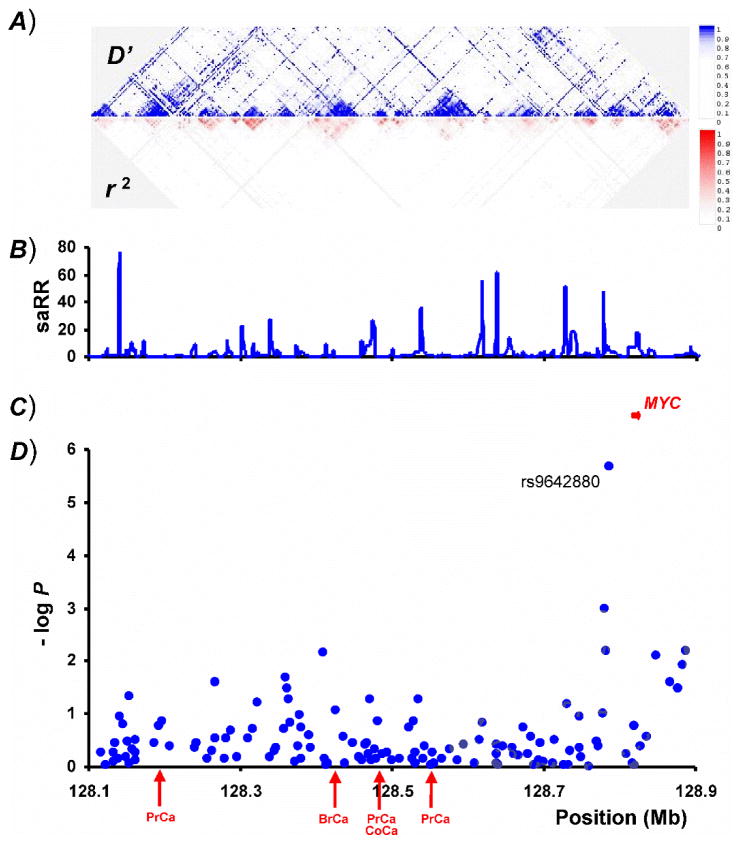
The SNP rs9642880 is located in the LD block immediately adjacent to the c-Myc oncogene and only 30kb upstream of it (Figure 1). c-Myc is the only known gene close to rs9642880, but a predicted gene, BC042052, is also in the same region (Supplementary Figure 2). We genotyped samples from 7 of the 9 study groups (all populations except Sweden and Spain) for two known missense mutations in the c-Myc gene (G175C/rs4645960 and N26S/rs4645959) but found no association with UBC. rs4645960 (T) was very rare, with only two cases in the combined sample sets carrying the allele. rs4645959 (G) was weakly correlated with rs9642880 (T) (D′=1, r2=0.04) with a frequency of 3.3% in cases and 3.7% in controls (OR=0.91, P=0.29). c-Myc is a nuclear phosphoprotein involved in transcriptional regulation. The normal function of c-Myc involves growth regulation, cell differentiation and apoptosis and its deregulation is associated with malignant growth 11. Altered expression, methylation and/or copy number variations at 8q24 have been found in bladder tumors, suggesting that c-Myc may have a role in tumor formation 12. The LD block of rs9642880 has been shown to contain an evolutionary conserved 1.6kb insulator element (~ 2.5 kb upstream of the c-Myc transcription initiation site) 13 as well as predicted transcription factor binding sites 14. We have previously measured the expression of 23,720 transcripts using an Agilent microarray and RNA from both adipose tissue and blood for 674 and 1,002 Icelandic individuals, respectively 15. Of those individuals, 602 with adipose tissue and 744 with blood have been genotyped with the Illumina 317 chip. We used this dataset to investigate the correlation between the rs9642880 (T) and c-Myc expression. No significant correlation was observed between c-Myc mRNA expression and the number of copies of the T allele of rs9642880 carried, either in whole blood or in adipose tissue (P=0.86 for whole blood and P=0.74 for adipose tissue). However, it should be noted that these data are not conclusive as other regulatory mechanism for c-Myc expression might be present in urothelial tissue.
Figure 1.
The second strongest signal in the combined analysis was observed for rs710521 (A) on chromosome 3q28 (OR=1.19, P=1.15 × 10−7) (Table 2). The association of rs710521 (A) to UBC did not deviate from the multiplicative model (P=0.70) and the estimated population attributable risk (PAR) of rs710521 (A) was 23%. The ORs for heterozygous and homozygous carriers of the risk allele T were 1.19 and 1.41, respectively, relative to the risk of non-carriers. With the average frequency of the risk allele in all the study populations at 73% (Table 2), 53% of the population may be homozygous carriers of rs710521 (A). No significant interaction was observed between the effects of rs9642880 and rs710521 (P=0.23). The rs710521 SNP is located in an LD block that overlaps the TP63 gene (encoding tumor protein p63) and no other gene is found in this block. The TP63 gene has strong homology with the tumor suppressor gene TP53 and the TP73 gene that encodes the p53-related protein p73. Like TP53, the TP63 gene regulates cell-cycle arrest and is involved in apoptosis. It is involved in the normal development of stratified epithelia including urothelium and is possibly associated with urothelial differentiation 16. The expression of the gene is often lost in urothelial tumors and it has been suggested that TP63 may play a critical role in the progression of urothelial neoplasia 17. No significant correlation was observed between TP63 mRNA expression and the number of copies of the A allele of rs710521 carried, in whole blood from 741 individuals and adipose tissue from 601 individuals (P=0.58 for whole blood and P=0.40 for adipose tissue).
Information on stage and grade was available for 7 of the 9 groups. Based on this information the UBC cases were classified into patients with a good prognosis (‘low risk’: tumor confined to the bladder mucosa and not poorly differentiated) or patients with a considerable risk of tumor progression (‘high risk’: tumor invasion in or beyond the lamina propria or poorly differentiated) (See Methods). Comparing the frequency of rs9642880 (T) between the 1,318 patients with low risk and 1,635 patients with high risk tumors showed some heterogeneity across the study groups (Phet=0.025). Overall patients with low risk tumors had a higher frequency of rs9642880 (T) than patients with high risk tumors (combined OR=1.15, P=0.011; Supplementary Table 3). This potential association with risk of progression needs to be investigated further. No difference was detected in frequency of rs710521 (A) between patients with low vs. high risk of progression.
Genome wide association studies have repeatedly reported cancer-associated variants on 8q24, 200–700 kb proximal to rs9642880 and c-Myc. We and others found SNPs at 8q24.21 to be strongly associated with cancer of the prostate (rs1447295, rs6983267 and rs16901979) 6,18,19. Subsequently, rs6983267 was also shown to associate with colorectal cancer 10 and most recently rs13281615 with breast cancer 9. These 4 variants are dispersed over a 500kb region (Figure 1) and are in weak LD with each other and with rs9642880 (Table 3). We found no association between these 4 SNPs and UBC in the combined study groups (Table 3). Moreover, we found no association between rs9642880 and prostate, breast, colorectal or lung cancer in Icelandic case control samples (Supplementary Table 2). Presently, it is not known how these variants at 8q24 affect cancer risk but the proximity of the c-Myc gene suggests that it may be implicated in this process. The UBC variant described here is closer to the c-Myc gene than any of the other known cancer-associated variants as rs9642880 is located in the LD block adjacent to c-Myc and only 30kb upstream of it (Figure 1).
Table 3.
Markers at 8q24.21 previously reported for cancer association, their LD to rs9642880, average control frequency and association with UBC
| Marker | Allelea | Position (build 36) | # UBC cases | # controls | Allele Freq | Association to UBC | LD to rs9642880 | Published Cancer Association | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||
| OR | 95% CI | P value | D′ | r2 | |||||||
| rs16901979 | A | 128,194,098 | 3,115 | 12,020b | 0.03 | 0.98 | 0.81–1.19 | 0.84 | 0.259 | 0.001 | Prostate |
| rs672888c | C | 128,424,800 | 3,383 | 36,212 | 0.40 | 1.02 | 0.96–1.08 | 0.51 | 0.156 | 0.017 | Breast |
| rs6983267 | G | 128,482,487 | 3,420 | 36,241 | 0.51 | 1.03 | 0.97–1.10 | 0.36 | 0.209 | 0.042 | Prostate, Colorectal |
| rs1447295 | A | 128,554,220 | 3,437 | 36,280 | 0.09 | 1.06 | 0.95–1.18 | 0.30 | 0.230 | 0.0031 | Prostate |
The SNPs rs16901979, rs672888, rs6983267 and rs1447295 were typed in UBC cases and controls from Iceland, The Netherlands, UK, Italy (Torino and Brescia), Belgium and Eastern Europe
Shown are the reported at risk alleles corresponding to published cancer association
The rs16901979 marker is not on the Illumina 317/370 chip and was typed by a single track assay for all UBC cases and all control samples, except in Iceland, where it was typed in 8,828 of the 32,504 control individuals.
Tagging G allele of rs13281615; r2 = 0.97
We and others have recently found an association between a variant in the nicotinic acetylcholine receptor gene cluster on 15q24 and nicotine addiction, smoking behavior, lung cancer and peripheral arterial disease 20–23. Since smoking is a strong risk factor for UBC, we tested the reported smoking-associated variant, rs1051730, in all the 9 UBC case control groups but found no association between the risk allele and disease (combined OR=1.03, P=0.26). No difference in the frequencies of rs9642880 (T) on chromosome 8 or rs710521 (A) on chromosome 3 was observed between ever smoking and never smoking cases (P=0.47 and P=0.55 respectively). Similarly, analysis of the Icelandic and Dutch controls showed that the results observed for rs9642880 and rs710521 cannot be explained by an association of these SNPs to smoking initiation or smoking quantity. rs9642880 (T) and rs710521 (A) did not correlate with age at diagnosis (data not shown). A nominally significant difference in the frequency of rs710521 (A) on chromosome 3 was observed between male and female cases (combined OR=1.14, P=0.049) with frequency in males being greater than females, no difference was observed for rs9642880 (P=0.13).
To summarize, we describe two sequence variants, on chromosomes 8q24 and 3q28, that associate with UBC in 9 study groups of European descent. The strongest variant is on chromosome 8q24.21, only 200 kb downstream of a 500kb region previously reported to associate with three other cancer types. Clustering of independent cancer-associated variants in this region suggests the existence of a common molecular mechanism of susceptibility. Unlike the variants reported in prostate, colorectal and breast cancer, the rs9642880 variant is located in the LD block adjacent to and only 30kb upstream of the c-Myc gene. Functional studies of this region are needed to understand the mechanism by which these variants affect carcinogenesis.
METHODS
Subjects
Nine study populations were used in this work, two discovery populations (Iceland and the Netherlands) and 7 follow up sample sets. The Icelandic sample set consists of 525 patients and 32,504 controls and the Dutch sample set consists of 1,278 cases and 1,832 controls. Follow up genotyping was performed in sample sets from Leeds, UK (724 cases and 530 controls), Torino, Italy (329 cases and 389 controls), Brescia, Italy (182 cases and 192 controls), Leuven, Belgium (195 cases and 382 controls), Stockholm, Sweden (349 cases and 930 controls), Zaragoza, Spain (173 cases and 859 controls) and an Eastern European sample set obtained from the German Cancer Research Center (DKFZ) in Heidelberg (213 cases and 521 controls). All participants gave informed consent and the studies were approved by the appropriate institutional review boards and/or ethics committees. Detailed description of the study populations is in the Supplementary Material.
Genotyping
Samples from Iceland and the Netherlands were used for the genome-wide association study (GWA) and were assayed with either the Infinium humanHap300 or humanCNV370 SNP chips (Illumina). The analysis was restricted to 302,140 SNPs that passed quality filters and were deemed usable due to yield, Hardy-Weinberg equilibrium and consistency in genotype frequencies between the two arrays. All samples had call rates above 98%. All follow up genotyping was carried out applying single track Centaurus assays (Nanogen). The quality of the Centaurus SNP assays was evaluated by genotyping each assay in the CEU HapMap samples and comparing the results with the publicly released HapMap data. Assays with >1.5% mismatch rate were not used and a linkage disequilibrium (LD) test was used for SNPs known to be in LD. The concordance rate of genotypes derived from the two genotyping platforms (Illumina and Centaurus) was >99.5%.
Statistical analysis
Association analysis
A likelihood procedure described in a previous publication 24 and implemented in the NEMO software was used for the association analyses. An attempt was made to genotype all individuals and all SNPs reported had a yield that was higher than 95% in every study group. The SNPs rs4645960 and rs16901979 are not part of the Human Hap300/HumanCNV370-duo chips. For these SNPs, a subset of the large Icelandic control set as well as all Icelandic cases and all individuals from the other study groups were genotyped by single track assays. We tested the association of an allele to UBC using a standard likelihood ratio statistic that, if the subjects were unrelated, would have asymptotically a χ2 distribution with one degree of freedom under the null hypothesis. Allelic frequencies rather than carrier frequencies are presented for the markers in the main text. Allele-specific ORs and associated P values were calculated assuming a multiplicative model for the two chromosomes of an individual 25. Results from multiple case-control groups were combined using a Mantel-Haenszel model 26 in which the groups were allowed to have different population frequencies for alleles and genotypes but were assumed to have common relative risks.
Correction of the GWA studies by genomic control
To adjust for possible population stratification and the relatedness amongst individuals, we divided the χ2 test statistics from the individual scans using the method of genomic control 27, i.e. the 302,140 χ2 test statistics were divided by their means, which were 1.04 and 1.075 for Iceland and the Netherlands, respectively. Supplementary Figure 1 is a quantile-quantile (Q-Q) plot of the chi-square statistics, before and after adjustment, against the chi-square distribution.
Correlation between genotype and expression of c-Myc in whole blood and adipose tissue
c-Myc expression was analyzed in whole blood and adipose tissue from 744 and 602 Icelandic individuals respectively and correlated with rs9642880 genotype status. Collection of whole blood and adipose tissue samples, mRNA isolation and expression profiling was described previously 15. Expression changes between two samples were quantified as mean logarithm (log10) expression ratio (MLR), i.e. expression ratios compared to background corrected intensity values for the two channels for each spot on the arrays 28. The hybridizations went through the standard QC process, i.e. signal to noise ratio, reproducibility and accuracy at spike-in compounds. The correlation between MLR for c-Myc and the genotypes of the SNP rs9642880 was tested by regressing the MLR’s on the number of copies of the at-risk T allele of rs9642880, adjusting for age, sex and, for whole blood, the differential blood cell count. All P-values were adjusted for relatedness of the individuals by simulating genotypes through the Icelandic genealogy as previously described 29. The probes used to test the expression of c-Myc and TP63 were NM_002467 and NM_003372 respectively.
Classification of “low risk” and “high risk” patients
Based on stage and grade information, all patients were classified with regards to risk of progression. Patients with low risk of progression were defined as having TNM stage pTa in combination with WHO 1973 differentiation grade 1 or 2 or WHO/ISUP 2004 low grade 30. All other tumors were classified as high risk of progression (stage pTis or ≥ pT1 or WHO 1973 grade 3 or WHO/ISUP 2004 high grade).
Supplementary Material
Acknowledgments
We thank the individuals that participated in the study and whose contribution made this work possible. We also thank the nurses at Noatun (deCODE’s participant recruitment center) and the personnel at the deCODE core facilities. We acknowledge the Icelandic Cancer Registry for assistance in the ascertainment of the Icelandic UBC patients. Collection of samples and data in Iceland and The Netherlands was funded in part by the European Commission (POLYGENE: LSHC-CT-2005-018827 and GENADDICT: LSHM-CT-2004-005166), the National Institute of Health (R01-DA017932) and a research investment grant of the Radboud University Nijmegen Medical Centre. The Leeds Bladder Cancer Study was funded by Cancer Research UK and Yorkshire Cancer Research. Torino Bladder Cancer Case Control Study was supported by a grant to ECNIS (Environmental Cancer Risk, Nutrition and Individual Susceptibility), a network of excellence operating within the European Union 6th Framework Program, Priority 5: “Food Quality and Safety” (Contract No 513943); by a grant of the compagnia di San Paolo, the Italian Association for Cancer Research, Italy and the Piedmont Region Progetti di Ricerca Sanitaria Finalizzata.
Footnotes
AUTHOR CONTRIBUTIONS: The study was designed and results were interpreted by LAK, ST, PS, FG, SNS, JG, JRG, UT, TR and KS. Statistical analysis was carried out by PS, FG, DG, GT and AK. Patient ascertainment, recruitment, biological material collection and collection of clinical and lifestyle information was organized and carried out by KKHA, JAW, SHV, CAH, DWS, MP, EBC, HV, TET, DP, MC, CA, KK, EG, PR, EK, SP, SG, CS, MS, BS, GV, PdV, CR, AL, KG, DTB, MAK, SN, VP, EJ, GG, BK, JIM, GS, SP, FB, MPZ, TF, RK, GM, PV and AEK. Principal collaborators for the UBC follow up populations were AEK (UK), GM and PV (Torino), SP (Brescia), MPZ and FB (Belgium), RK and TF (Eastern Europe), JIM (Spain) and GS (Sweden). Genotyping and laboratory experiments were carried out by SNS, JG, MJ, AS, TB, KTK, MM and SS. Expression analysis was carried out by JTB, TB, AS, GT and PS. Bioinformatic analysis was carried out by ST, PS, AS, TB and SAG. Authors LAK, ST, PS, FG, UT, TR and KS drafted the manuscript. All authors contributed to the final version of the paper.
References
- 1.Surveillance, Epidemiology, and End Results Program, 1975–2004. Division of Cancer Control and Population Sciences, National Cancer Institute; 2007. Available at: http://seer.cancer.gov/csr/1975_2004/results_merged/sect_27_urinary_bladder.pdf. [Google Scholar]
- 2.Aben KK, et al. Familial aggregation of urothelial cell carcinoma. Int J Cancer. 2002;98:274–8. doi: 10.1002/ijc.10191. [DOI] [PubMed] [Google Scholar]
- 3.Amundadottir LT, et al. Cancer as a Complex Phenotype: Pattern of Cancer Distribution within and beyond the Nuclear Family. PLoS Med. 2004;1:e65. doi: 10.1371/journal.pmed.0010065. Epub 2004 Dec 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murta-Nascimento C, et al. Risk of bladder cancer associated with family history of cancer: do low-penetrance polymorphisms account for the increase in risk? Cancer Epidemiol Biomarkers Prev. 2007;16:1595–600. doi: 10.1158/1055-9965.EPI-06-0743. [DOI] [PubMed] [Google Scholar]
- 5.Aben KK, et al. Segregation analysis of urothelial cell carcinoma. Eur J Cancer. 2006;42:1428–33. doi: 10.1016/j.ejca.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 6.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 7.Gudmundsson J, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–83. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 8.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 9.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 11.DePinho RA, Schreiber-Agus N, Alt FW. myc family oncogenes in the development of normal and neoplastic cells. Adv Cancer Res. 1991;57:1–46. doi: 10.1016/s0065-230x(08)60994-x. [DOI] [PubMed] [Google Scholar]
- 12.Mhawech-Fauceglia P, Cheney RT, Schwaller J. Genetic alterations in urothelial bladder carcinoma: an updated review. Cancer. 2006;106:1205–16. doi: 10.1002/cncr.21743. [DOI] [PubMed] [Google Scholar]
- 13.Gombert WM, et al. The c-myc insulator element and matrix attachment regions define the c-myc chromosomal domain. Mol Cell Biol. 2003;23:9338–48. doi: 10.1128/MCB.23.24.9338-9348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallikas O, et al. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 15.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 16.Urist MJ, et al. Loss of p63 expression is associated with tumor progression in bladder cancer. Am J Pathol. 2002;161:1199–206. doi: 10.1016/S0002-9440(10)64396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koga F, et al. Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clin Cancer Res. 2003;9:5501–7. [PubMed] [Google Scholar]
- 18.Amundadottir LT, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;7:7. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 19.Yeager M, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–9. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 20.Saccone SF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008 doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 24.Gretarsdottir S, et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat Genet. 2003;35:131–8. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 25.Falk CT, Rubinstein P. Haplotype relative risks: an easy reliable way to construct a proper control sample for risk calculations. Ann Hum Genet. 1987;51 (Pt 3):227–33. doi: 10.1111/j.1469-1809.1987.tb00875.x. [DOI] [PubMed] [Google Scholar]
- 26.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 27.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 28.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 29.Stefansson H, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 30.Epstein JI, Amin MB, Reuter VR, Mostofi FK. The World Health Organization/International Society of Urological Pathology consensus classification of urothelial (transitional cell) neoplasms of the urinary bladder. Bladder Consensus Conference Committee. Am J Surg Pathol. 1998;22:1435–48. doi: 10.1097/00000478-199812000-00001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.