Abstract
Automated image analysis of unperturbed cells reveals a new sequence of events underlying protrusion of the cell membrane.
Many vital cellular behaviors require physical contact between the multi-component cytoskeleton and super-molecular structures like the plasma membrane. Each of these interfaces can comprise hundreds of proteins, many of which have overlapping or redundant functions. Such complexity confounds assigning specific roles to individual proteins, especially given the cell's ability to adapt to perturbations on-the-fly and the contrasting, method-specific phenotypes seen after perturbations.
In this issue of Cell Systems, Lee et al. (2015) introduce an image analysis strategy that avoids loss-of-function manipulations and rather relies upon harnessing information within the naturally occurring fluctuations present in unperturbed cells. Their technique allows the authors to define the intrinsic spatial and temporal sequence or “hierarchy” of molecular events involved in the dynamic behavior of a cellular superstructure—in this case, the leading edge—in the absence of the confounding compensatory mechanisms underlying cellular adaptation.
The filamentous actin (F-actin) cytoskeleton is one of the major structural and force-generating systems in eukaryotic cells. Actin dynamics promote cell migration by fueling plasma membrane protrusion and retraction events at the cell's leading edge. However, inherent stochastic switching between protrusion and retraction phases has further complicated assigning molecular functions to the plethora of regulatory proteins that facilitate polymerization, remodeling, and adhesion of the highly branched lamellipodial F-actin network. One clearly critical component in the formation of this network is the Arp2/3 complex, which is the only identified nucleator of F-actin branches.
Recent studies have shown that cells proceed to migrate in the absence of Arp2/3 function (Suraneni et al., 2012; Wu et al., 2012). These findings may highlight compensatory mechanisms provided by other F-actin nucleating and polymerizing proteins. Further, the contrasting phenotypes observed by Sunareni et al. and Wu et al. may suggest something deeper: context-dependent crosstalk or feedback between F-actin nucleators and cytoskeletal modulators may adjust the cell's molecular strategy for actin-based protrusion when Arp2/3 function is compromised.
The robust, adaptive strategies that cells employ when Arp2/3 is perturbed may be far removed from the native system's function. Lee et al. have developed a local image sampling and registration approach to study Arp2/3 and other cellular machinery underlying leading edge dynamics in unperturbed PtK1 cells, a rat kangaroo cell line widely used to study the cytoskeleton. By analyzing spontaneous cellular (leading edge) and molecular (cytoskeletal/adhesion components) fluctuations present within microscopy data acquired with high spatial and temporal resolution, the authors have dissected the hierarchy of molecular events in actin-driven membrane protrusion.
In an elegant, non-invasive combination of imaging and computer-vision analysis, near resolution-limit-sized regions of interest along the leading edge of migrating epithelial cells were tracked and categorized based upon defined activities: initiation of leading edge retraction, initiation of protrusion, and the time of maximal protrusion velocity. All of the windows containing one category of activity were then grouped, and the time-series data were temporally aligned, using the defined activity as a common reference point. This enhanced statistical power and revealed previously hidden, dynamic correlations between the localization of F-actin, actin regulatory proteins, adhesion components, and the generation of traction against the extracellular substrate.
For example, although Arp2/3 has been considered the foundation of the lamellipodial F-actin network, the authors demonstrate that the formin actin nucleator, mDia1, is recruited to the leading edge along with nascent adhesions prior to protrusion onset (Figure 1). This suggests that mDia1 initiates protrusion. In contrast, accumulation of the Arp2/3 complex occurs subsequent to protrusion onset. Lee et al. hypothesize that Arp2/3 reinforces lamellipodial F-actin against the increasing membrane tension generated in newly adhered mDia1-initiated protrusions. They go on to propose a heretofore unexpected idea: the Arp2/3 complex is enlisted at the leading edge by a mechano-sensitive feedback mechanism. In such a mechanism, F-actin polymerization is coupled to substrate adhesions while simultaneously pushing against the plasma membrane. This increases membrane tension, which would be sensed by an unknown factor that recruits or activates the Arp2/3 complex.
Figure 1.
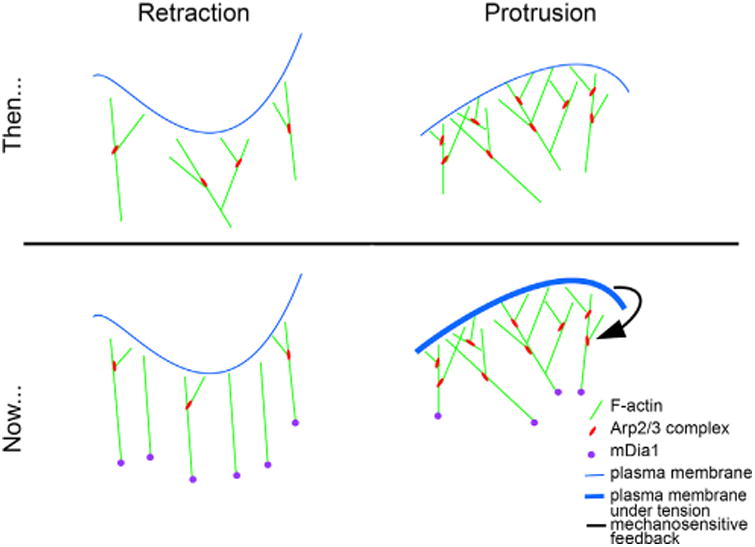
Models of Leading Edge Retraction and Protrusion, Before and After the Work of Lee Et Al. Presented in This Issue
Such mechano-sensitive feedback is plausible. For example, mechanically sensitive integrins could modulate the activity of Rho GTPases upstream of nucleating promoting factors, which in turn activate Arp2/3. Alternatively, mechanical strain originating from substrate adhesion and actin polymerization may confer a preferred binding site along mDia1-nucleated actin filaments for the Arp2/3 complex, and thus may explain why mDia1 accumulation correlated with subsequent Arp2/3 recruitment.
This work by Lee et al. highlights the impact of exploiting computer vision analysis to increase statistical power and isolate discrete, molecular functions in a redundant and adaptive cellular system. As such, it underscores the importance of directly observing F-actin and actin modulators at the leading edge while the cell is engaged in its native behavior in vitro. Importantly, this strategy may provide the key to understanding the adaptive responses consequent to genetic or pharmacological perturbations. Coupling this approach with more specific pharmacological tools or synthetic biology approaches that provide spatiotemporal control of protein function or membrane tension—by optogenetics or the light-oxygen-voltage (LOV) domain technology, for example—may allow visualization and analysis of system adaptation in real time (Weitzman and Hahn, 2014, Govorunova et al., 2015).
This approach is also broadly applicable. Other multi-component stochastic cellular events, such as filopodia formation and dynamics, focal adhesion maturation and disassembly, chromosome alignment and segregation, vesicle endocytosis or exocytosis, would benefit from this rigorous strategy of coupling image registration and statistical analysis. For example, such analysis may further reveal how mDia2 and Ena/VASP cooperate in filopodia formation (Barzik et al., 2014, Nowotarski et. al., 2014), how the multitude of proteins that promote disassembly or depolymerization of F-actin, such as cofilin and GMFbeta are involved in cell edge retraction (Haynes et al., 2015, Ghosh et al., 2004), as well as help alleviate the controversies of how actin dynamics are involved in the progression of vesicle scission and fusion during endocytosis and exocytosis.
Acknowledgments
This work was also supported by NIH intramural funds from the National Institute on Deafness and Other Communication Disorders (NIDCD) 1 Z01 DC000039-17 to T. Friedman (Laboratory of Molecular Genetics, NIDCD, NIH).
References
- Barzik M, McClain LM, Gupton SL, Gertler FB. Mol Biol Cell. 2014;25:2604–2619. doi: 10.1091/mbc.E14-02-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL. Science. 2015:aaa7484. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes EM, Asokan SB, King SJ, Johnson HE, Haugh JM, Bear JE. J Cell Biol. 2015;209:803–812. doi: 10.1083/jcb.201501094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Elliott HL, Oak Y, Zee CT, Groisman A, Tytell JD, Danuser G. Cell Systems. 2015;1 doi: 10.1016/j.cels.2015.07.001. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotarski SH, McKeon N, Moser RJ, Peifer M. Mol Biol Cell. 2014;25:3147–3165. doi: 10.1091/mbc.E14-05-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. J Cell Biol. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman M, Hahn KM. Curr Opin Cell Biol. 2014;30:112–120. doi: 10.1016/j.ceb.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]


