Abstract
Ammonia- and nitrite-oxidizers are collectively responsible for the aerobic oxidation of ammonia via nitrite to nitrate and play essential roles for the global biogeochemical nitrogen cycle. The physiology of these nitrifying microbes has been intensively studied since the first experiments of Sergei Winogradsky more than a century ago. Urea and ammonia are the only recognized energy sources that promote the aerobic growth of ammonia-oxidizing bacteria and archaea. Here we report the aerobic growth of a pure culture of the ammonia-oxidizing thaumarchaeote Nitrososphaera gargensis1 on cyanate as the sole source of energy and reductant, the first organism known to do so. Cyanate, which is a potentially important source of reduced nitrogen in aquatic and terrestrial ecosystems2, is converted to ammonium and CO2 by this archaeon using a cyanase that is induced upon addition of this compound. Within the cyanase gene family, this cyanase is a member of a distinct clade that also contains cyanases of nitrite-oxidizing bacteria of the genus Nitrospira. We demonstrate by co-culture experiments that these nitrite-oxidizers supply ammonia-oxidizers lacking cyanase with ammonium from cyanate, which is fully nitrified by this consortium through reciprocal feeding. Screening of a comprehensive set of more than 3,000 publically available metagenomes from environmental samples revealed that cyanase-encoding genes clustering with the cyanases of these nitrifiers are widespread in the environment. Our results demonstrate an unexpected metabolic versatility of nitrifying microbes and suggest a previously unrecognized importance of cyanate for N-cycling in the environment.
Cyanate is a small molecule containing carbon, nitrogen, and oxygen atoms. It is formed spontaneously within cells from urea and carbamoylphosphate3,4, but also occurs in the environment where it may be produced from the (physico)chemical decomposition of urea or cyanide5,6. Until recently, environmental cyanate concentrations were virtually unavailable as analytical methods were inadequate for sub-micromolar detection. Furthermore, cyanate is not chemically stable and decomposes relatively slowly to ammonium and carbon dioxide. This decomposition rate is linearly related to the concentration of cyanate and thus cyanate is rather stable at low concentrations (Extended Data Figure 1). A more sensitive chromatographic method for the detection of cyanate in aquatic samples was very recently developed and revealed nanomolar-range cyanate concentrations in seawater6. These cyanate levels are in the same order of magnitude as ammonium concentrations typically found in oligotrophic marine environments7. Consistently, cyanate has been postulated to serve as a nitrogen source for the growth of certain marine cyanobacteria under nitrogen limitation2,8. For the assimilation of cyanate, these phototrophic bacteria convert it to ammonium (and CO2) by a dedicated enzyme called cyanate lyase or cyanase. Cyanases are also found in a variety of other bacteria and archaea where they have been reported to play a role in nitrogen assimilation or detoxification as cyanate chemically modifies proteins via carbamylation9,10. However, no microbe has been described that can grow on cyanate as source of energy and reductant.
Nitrifying microbes are generally considered to be highly specialized chemolithoautotrophs that oxidize either ammonia or nitrite for generating energy and reductant for growth and use CO2 as carbon source. Over the last decades, this perception has been challenged by several studies11-13. For example it was reported that uncultured thaumarchaeota closely related to the described ammonia-oxidizer N. gargensis thrive in wastewater treatment plants by using other (unknown) sources of energy and reductant than ammonium or urea14 and that nitrite-oxidizers of the genus Nitrospira can derive energy for growth by aerobic hydrogen oxidation15. Furthermore, the growth of some thaumarchaeotal ammonia-oxidizers is stimulated by the addition of organic compounds16 and others may be obligate mixotrophs17. However, aerobic growth of ammonia-oxidizing microbes has still only been demonstrated in the presence of urea or ammonium.
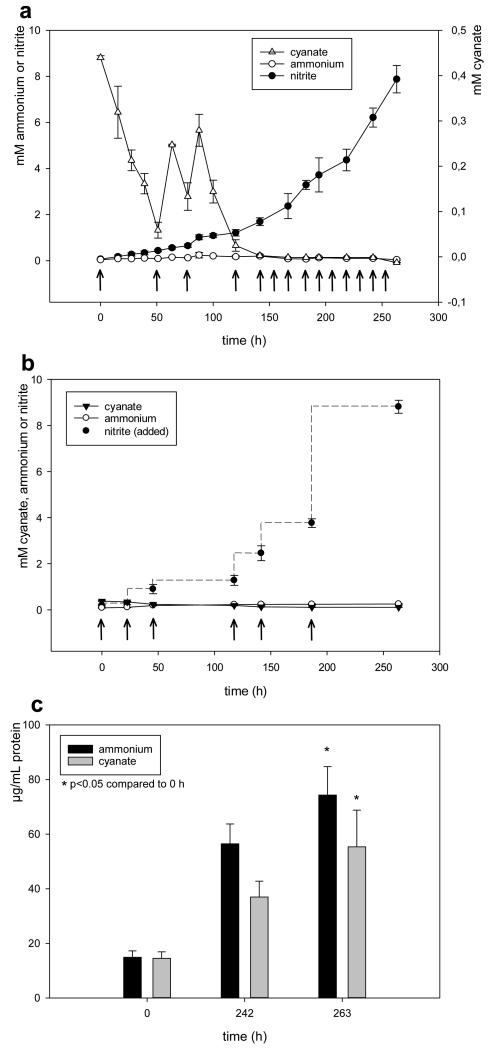
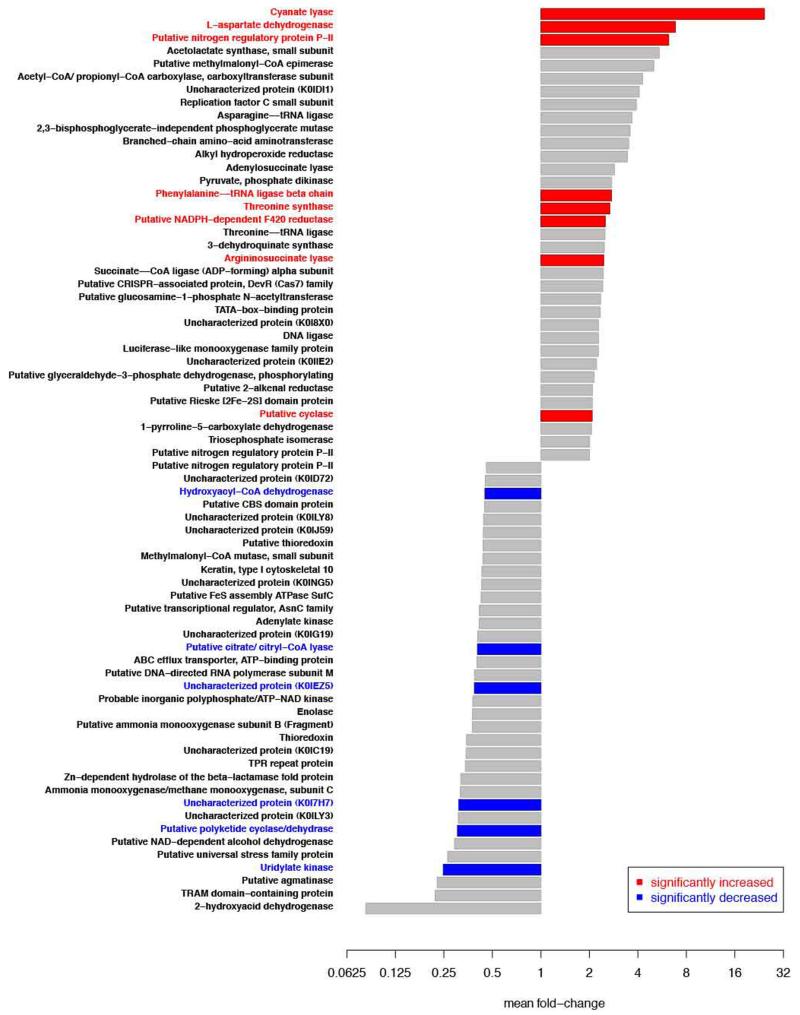
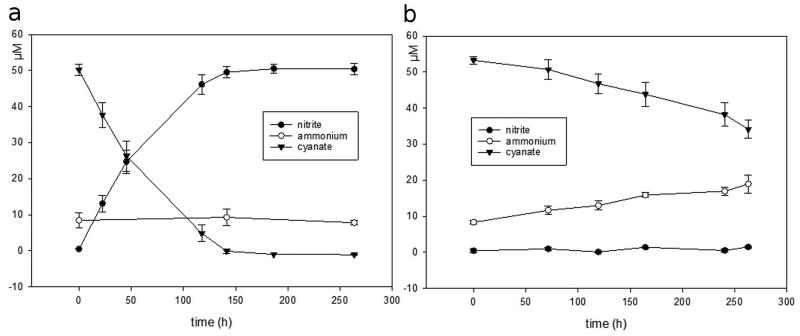
Recently we sequenced the genome of the thaumarchaeotal ammonia-oxidizer N. gargensis that was enriched from a thermal spring sample1. Unexpectedly, a gene encoding a putative cyanase was detected in this genome close to the gene of a putative cyanate/nitrite/formate transporter18. In contrast, all other sequenced genomes of archaeal or bacterial ammonia-oxidizers including its closest relative N. viennensis19 do not contain a cyanase-encoding gene. As N. gargensis shares most central metabolic pathways with other thaumarchaeotes it is very unlikely that its cyanase is required for detoxification of internally produced cyanate. We therefore hypothesized that N. gargensis might use cyanate as a source of energy and reductant for growth. Prior to testing our hypothesis a pure culture of N. gargensis was obtained by repeated serial dilutions over a period of 16 months (Supplementary Information 1). The pure culture of N. gargensis grew well in the presence of 2 mM ammonium and growth was not inhibited by addition of 0.5 mM cyanate. After a few days of growth in the presence of both compounds, biomass of N. gargensis was transferred to a medium containing cyanate as the only source of energy, reductant and nitrogen. In this medium, N. gargensis stochiometrically converted cyanate via ammonium to nitrite (Figure 1) and cyanate degradation was the rate-limiting step of the overall process (Extended Data Figure 1). A much slower cyanate conversion to ammonium reflecting chemical decay was observed in control experiments with equal amounts of dead biomass of N. gargensis (Figure 1). Importantly, growth of N. gargensis in the medium containing cyanate as the sole source of energy and reductant was demonstrated by total protein measurements (Figure 1) and by a qPCR assay targeting its 16S rRNA gene (Extended Data Figure 2). During growth on 0.5 mM cyanate, N. gargensis showed according to total protein measurements a mean generation time of 136.3 h (+/− 11.4 SD), which is slightly higher than the mean generation time observed during growth on 0.5 mM ammonium, which was determined to be 113.4 h (+/− 6.1 SD). This difference might reflect toxicity of cyanate despite the presence of a cyanase or the additional energy demand for the synthesis of cyanase during growth on this compound. Proteomic analyses revealed that upon exposure of N. gargensis (that had not been exposed to cyanate) to 0.5 mM cyanate for 48h, the cyanase of N. gargensis was the most strongly induced protein (Extended Data Figure 3; 32× fold change; mean from triplicates), confirming its key role for growth on this source of energy and reductant. Interestingly however, the putative cyanate/nitrite/formate transporter encoded in the same genomic region was not detected, despite the fact that a protocol optimized for extraction of membrane proteins was applied. This is likely caused by the fact that cyanate at mM concentration diffuses through biological membranes20. Interestingly, cyanate conversion by N. gargensis was also observed without a previous growth period in the presence of ammonium and cyanate. In addition, cyanate conversion to nitrite by N. gargensis could also be detected at a 10x lower concentration of the compound (0.05 mM) (Extended Data Figure 4).
Figure 1. N. gargensis grows on cyanate as the only source of energy and reductant.
a, Concentration changes of cyanate, ammonium, and nitrite during the growth of N. gargensis in a mineral medium containing 0.5 mM cyanate as the sole source of energy and reductant. Arrows indicate additions of 0.5 mM cyanate. b, Control experiment with identical amounts of dead biomass of N. gargensis. In this experiment nitrite was added at different time points as indicated by the arrows to mimic the conditions in the experiment with living biomass. All experiments shown in panels a and b were performed in four replicates and the chemical measurements were done in three technical replicates (averaged). Data points are mean values of four biological replicates, error bars show 1 SD. c, Total protein concentration of N. gargensis during the experiment. For comparison, the respective protein concentrations of N. gargensis after growth in medium with 0.5 mM ammonium are presented. Protein concentration increased 4.99 fold during growth on ammonium and 3.81 fold during growth on cyanate over 263 hours. Significance of difference was calculated by a paired t-test. Biomass increase was independently confirmed by qPCR (Extended Data Figure 2).
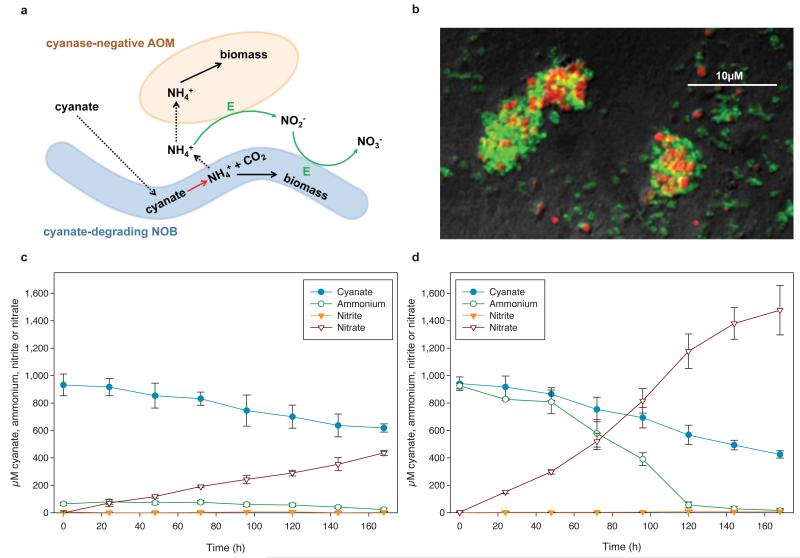
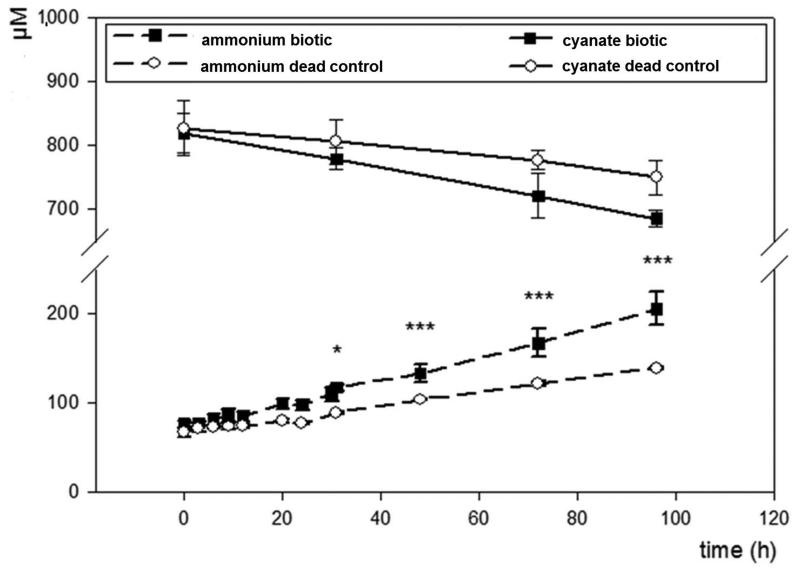
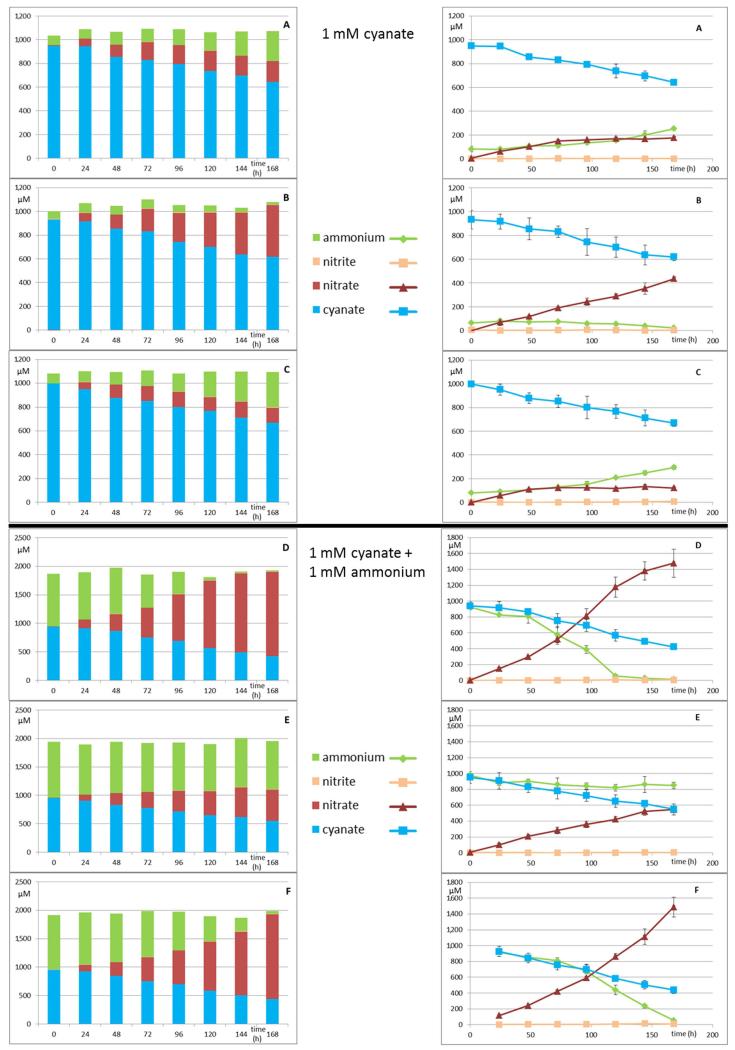
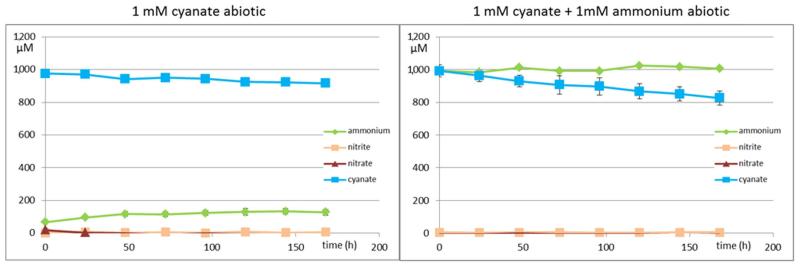
While N. gargensis is the only ammonia-oxidizing microbe with a sequenced genome in which a cyanase is present that was likely acquired from a Nitrospira strain via lateral gene transfer18, all nitrite-oxidizers for which a genome sequence is available contain a gene annotated as cyanase (Extended Data Table 1). To test whether these genes are functional, experiments were performed with a pure culture of the nitrite-oxidizer Nitrospira moscoviensis that possesses a cyanase closely related to the respective enzyme of N. gargensis. After 96 h of incubation in the presence of around 1 mM cyanate N. moscoviensis degraded significantly more cyanate leading to ammonium released from the cells than a negative control that included an identical amount of dead biomass of this strain (Extended Data Figure 5). Consequently, N. moscoviensis is capable of cyanate degradation. In a separate experiment addition of 1 mM cyanate only slightly decreased nitrite oxidation rates in N. moscoviensis, while higher concentrations showed a stronger effect (Extended Data Figure 6). The presence of a cyanase in the genomes of all nitrite-oxidizers might reflect that these nitrifiers make more cyanate as a side product of their metabolism than ammonia-oxidizing microbes. Cyanate is produced from both carbamoylphosphate metabolism and urea formation, and while the enzymatic repertoire involved in these processes is highly similar between ammonia- and nitrite-oxidizers, many members of the latter group (but also some thaumarchaeotes) do not contain enzymes for degradation of internally produced urea (Extended Data Table 1). In addition, nitrite-oxidizers might continuously import cyanate from the environment as some of their transporters for the uptake of nitrite from the environment also transport cyanate21. In both scenarios the presence of a cyanase is beneficial for nitrite-oxidizers as it allows them to detoxify cyanate and as the formed ammonium is not only available for assimilation, but after secretion (Extended Data Figure 5) might also serve as source of energy and reductant for ammonia-oxidizers which typically grow in close vicinity to nitrite-oxidizers22,23. The activity of the ammonia-oxidizers will lead to nitrite formation that can then be consumed by the nitrite-oxidizers (Figure 2a). This reciprocal feeding would enable nitrite-oxidizers as well as ammonia-oxidizers without a cyanase to convert cyanate for energy and reductant generation. We experimentally tested this hypothesis by establishing a co-culture of the ammonia-oxidizing bacterium Nitrosomonas nitrosa Nm9024 that has no cyanase activity but is not inhibited in its activity by 1 mM cyanate (Extended Data Figure 7) with the cyanase-encoding nitrite-oxidizer N. moscoviensis. Consistent with the reciprocal feeding hypothesis, this co-culture stoichiometrically converted cyanate to nitrate (Figure 2c; Extended Data Figure 8) and fluorescence in situ hybridization with specific 16S rRNA-targeted probes revealed that dense clusters containing both nitrifiers had formed (Figure 2b). Cyanate conversion rates to nitrate by this consortium could be accelerated by adding ammonium at the start of the experiment in order to allow consortium members to gain energy and reductant before interspecies cyanate degradation was fully established (Figure 2d). In contrast no nitrate formation was observed in abiotic control experiments using the same medium (Extended Data Figure 9).
Figure 2. Reciprocal feeding of ammonia- and nitrite-oxidizers during cyanate conversion.
a, Schematic illustration of the interaction between cyanate-degrading nitrite-oxidizing bacteria (NOB) and cyanase-negative ammonia-oxidizing microbes (AOM). Solid arrows represent conversions of compounds, dashed arrows the uptake or release of compounds. Green arrows represent conversions used for energy (E) and reductant generation. b, Co-aggregation of Nitrosomonas nitrosa (red) and N. moscoviensis (green) in the co-culture experiment shown in panel d, after 168 h, as revealed by FISH. c and d, Concentration changes of cyanate, ammonium, nitrite, and nitrate during the growth of the cyanase-negative ammonia-oxidizing bacterium N. nitrosa and the cyanase-positive nitrite-oxidizer N. moscoviensis in a mineral medium containing 1 mM cyanate (c) or 1 mM cyanate and 1mM ammonium (d). Error bars show 1 SD of three technical replicates. For each experiment three biological replicates were performed (one replicate displayed in panels c and d, all replicates including mass balances shown in Extended Data Figure 8). It should be noted that N. nitrosa did not grow equally well in all replicates.
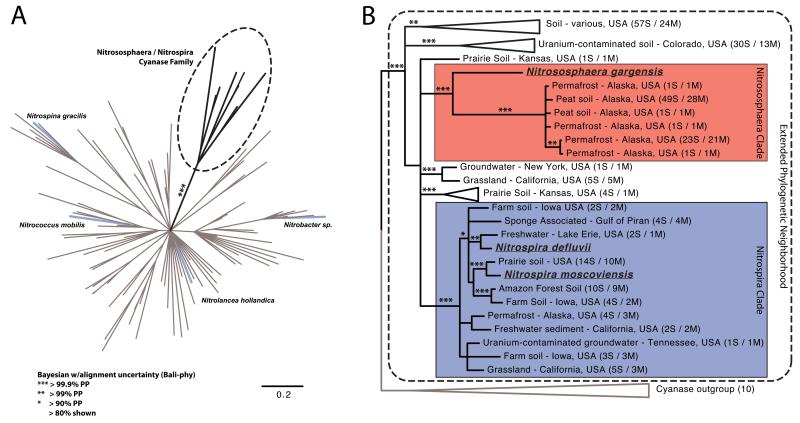
The cyanases found in N. gargensis and members of the genus Nitrospira form a deep-branching clade to the exclusion of other cultured organisms18. We searched a collection of 3,000 metagenomic datasets available from IMG25 and identified 225 additional metagenomic cyanase gene (fragments) that are related to the cyanases of these known nitrifiers (Figure 3). These findings show that the novel cyanase family is widespread in the environment. Most of these cyanases were located on very small contigs preventing an independent phylogenetic classification of the organisms carrying these genes. The metagenomic cyanase fragments most closely related to N. gargensis (47-55 % amino acid similarity) were retrieved from three different peat and permafrost soils in Alaska, while the sequences most closely affiliated with Nitrospira cyanases (67-80 % amino acid similarity) were mostly found in temperate forest and agricultural soil from lower latitudes as well as in lakes, freshwater sediment and groundwater, matching the known distribution of Nitrospira in a broad range of different ecosystems26 (Figure 3).
Figure 3. N. gargensis and Nitrospira cyanases form a distinct family containing sequences from various metagenomes.
a, Bayesian 80% consensus amino acid tree calculated with Bali-phy28. For clarity, posterior support is shown only for the branch separating this cyanase family from other cyanases. Cyanases from nitrite-oxidizing bacteria (NOB) are indicated by blue branches. b, Bayesian 80% consensus amino acid tree of the Nitrosophaera/Nitrospira cyanase family that contains separate well supported Nitrosophaera-related (red), and Nitrospira-related (blue) clades, respectively. Metagenomic cyanase sequences that showed more than 99% amino acid similarity were clustered using Usearch29. Behind each metagenomic sequence the total number of clustered sequences (S) and the number of metagenomic datasets (M) from which they were retrieved is displayed.
Our findings show that an archaeal ammonia-oxidizer can grow on cyanate as the sole source of energy, reductant and nitrogen. Furthermore, nitrite-oxidizers of the genus Nitrospira (and likely all nitrite-oxidizers) convert cyanate to ammonium and are capable of fully nitrifying it by a new type of reciprocal feeding with cyanase-negative ammonia-oxidizers. This metabolic capability potentially provides them with a selective advantage in environments where cyanate is present, in particular if ammonium concentrations are low, and thus might be an important facet of the ecology of nitrifiers. Cyanate forms spontaneously by isomerization of urea in aqueous solution. The high concentration of urea in many ecosystems (ranging from polar seawater and sea ice27 to the huge areas of urea-fertilized soils in global agriculture) combined with the wide distribution of nitrifier-related cyanase genes underscores the potential environmental ubiquity of this unique physiology.
Methods
Purification and standard cultivation of Nitrososphaera gargensis
A pure culture of the ammonia-oxidizing archaeon Nitrososphaera gargensis1 was obtained through a series of antibiotics treatments (50 mg/L kanamycin; 50 mg/L penicillin-G; 100 mg/L streptomycin; 100 mg/L carbenicillin; 50 mg/L ampicillin; 20 mg/L erythromycin; 20 mg/L doxycyclin) and repeated serial dilutions in the ammonia-oxidizer medium described below. Purity of the culture was confirmed by phase contrast microscopy and by using a specific CARD-FISH assay1 as well as by PCR targeting the 16S rRNA gene, using various universal eubacterial and archaeal primer combinations (27f 5′-AGAGTTTGATYMTGGCTCAG-3′; Arch21f 5′-TTCCGGTTGATCCYGCCGGA-3′; 907f 5′-AAACTCAAAKGAATTGACGG-3′; 909r 5′-CCGTCWATTCMTTTGAGT-3′; 1390r 5′-GACGGGCGGTGTGTACAA-3′; 1492r 5′-GGYTACCTTGTTACGACTT-3′) on DNA extracted by three different DNA isolation methods (Bead beating with Phenol-Chloroform extraction; MoBio UltraClean Soil DNA kit; FastDNA SPIN Kit for Soil). Any PCR product obtained was cloned and sequenced, retrieving only N. gargensis 16S rRNA gene sequences. In addition, no growth was observed if the N. gargensis culture was inoculated into various rich media like Lysogeny broth, Nutrient agar and Tryptic-Soy agar. Subsequently, N. gargensis was grown at 46 °C in a modified AOA medium30 containing (per l): 50 mg KH2PO4; 75 mg KCl; 50 mg MgSO4×7H2O; 584 mg NaCl; 4 g CaCO3 (mostly undissolved, acting as a solid buffering system and growth surface), 1 ml of specific trace element solution (AOA-TES), and 1 ml of selenium-wolfram solution31 (SWS). The composition of TES and SWS is described below. Both solutions were added to the autoclaved medium by sterile filtration using 0.2 μm pore size cellulose acetate filters (Thermo Scientific). The pH of the medium was around 8.4 after autoclaving and was kept around 8.2 during growth of N. gargensis by the CaCO3 buffering system. AOA-TES contained (per l): 34.4 mg MnSO4×1H2O; 50 mg H3BO3; 70 mg ZnCl2; 72,6 mg Na2MoO4×2H2O; 20 mg CuCl2×2H2O; 24 mg NiCl2×6H2O; 80 mg CoCl2×6H2O; 1 g FeSO4×7H2O. All salts except the FeSO4×7H2O were dissolved in 997.5 ml MQ water and 2.5 ml 37% (smoking) HCl was added before dissolving the FeSO4×7H2O salt. SWS contained (per l): 0.5 g NaOH; 3 mg Na2SeO3×5H2O; 4 mg Na2WO4×2H2O. After completing the medium, ammonium chloride (from an autoclaved 0.2 M stock solution) or potassium cyanate (filter sterilized, Sigma Aldrich) was added to the medium based on the experimental setups. All cultures were grown in the dark in screw-cap Schott bottles (Schott AG, Mainz, Germany) without shaking at 46°C.
Growth of N. gargensis on 0.5 mM cyanate
Cultures were induced with 0.5 mM (final concentration) potassium cyanate (KOCN) and 0.5 mM NH4Cl two days before the actual experiment. After 48 h cyanase-induced cultures were harvested by centrifugation (10,000×g for 30 min, room temperature), washed in AOA medium, centrifuged again, and inoculated into 20 ml fresh AOA medium in 50 ml CELLSTAR plastic suspension culture flasks (#690190, Greiner Bio-One), containing no ammonium but 0.5 mM KOCN final concentration. Biomass protein concentrations used for inoculation were 14.51±2.3 μg/ml. Cultures were incubated without shaking at 46°C in the dark, for 11 days (264 h). All incubations were done in 4 replicates. Samples for chemical-, protein- and qPCR-analysis were taken every 12 hours for the first 4 days, with daily sampling thereafter. After the experiment, the cells were harvested and washed as described above and then transferred into 20 ml of fresh AOA medium without ammonium and autoclaved. After cooling to room temperature, 20 μl TES, 20 μl SWS, and 0,5 mM of filter sterilized KOCN was added, and the dead biomass was incubated for 46°C in the dark for 264 h. To mimic the production of nitrite in these control experiments with dead biomass, NaNO2 was added at each sampling time point, according to the respective levels of nitrite in the biotic experiments at the next time point, resulting in a nitrite concentration, which is always at least as high as in the biotic parallels. In both experiments (either living or dead biomass) the pH stayed constant around 8.2±0.3 during the incubation time.
Growth of N. gargensis on 0.05 mM cyanate
Cultures were induced with 0.05 mM (final concentration) potassium cyanate (KOCN) and 0.5 mM NH4Cl two days before the actual experiment. After 48 h, cyanase-induced cultures were harvested by centrifugation (10,000×g for 30 min, room temperature), washed in AOA medium, centrifuged again, and inoculated into 20 ml fresh AOA medium in 50 ml CELLSTAR plastic suspension culture flasks (#690190, Greiner Bio-One), containing no ammonium but 0.05 mM KOCN final concentration. Cultures were incubated without shaking at 46°C in the dark for 264 h. In parallel, abiotic controls were started with similar parameters, without biomass. All incubations were done in 4 replicates. Samples for chemical analysis were taken at 7 time points during the 11 days. In both experiments (either biotic or abiotic) the pH stayed constant around 8.2±0.3 during the incubation time.
Cultivation of N. moscoviensis
The nitrite-oxidizing bacterium Nitrospira moscoviensis was pre-grown in mineral NOB medium32 containing (per 1 liter): 1,000ml distilled water; 10 mg CaCO3; 500 mg NaCl; 50mg MgSO4×7H20; 150 mg KH2PO4 as well as 1 ml filter sterilized NOB-specific trace elements solution (NOB-TES) added after autoclaving. The pH was initially adjusted to 8.6 which changed during autoclaving to 7.6. NOB-TES contained (per 1 liter): 34.4 mg MnSO4×1H2O; 50 mg H3BO3; 70 mg ZnCl2, 72.6 mg Na2MoO4×2H2O; 20 mg CuCl2×2H2O; 24 mg NiCL2×6H2O, 80 mg CoCL2×6H2O; 1g FeSO4×7H2O. All salts, except FeSO4×7H2O were dissolved in 997.6 ml distilled water and 2.5 ml of 37% (smoking) HCL was added before dissolving the FeSO4×7H2O salt. After autoclaving 1mM (final concentration) filter sterilized NaNO2 (if not stated otherwise) was added to the medium. All cultures were grown in the dark without shaking at 37°C. If all nitrite was consumed, it was re-added to a final concentration of 1 mM.
Cyanate degradation by N. moscoviensis
Nitrite-oxidizing cultures of N. moscoviensis were supplied with 0.5 mM (final concentration) KOCN and incubated for 48 hours at 37°C to induce the expression of cyanase. Biomass was harvested (8,500 rpm for 15 min at room temperature) and washed 2 times with fresh NOB medium without nitrite. Cells were then transferred into 50 ml NOB medium, which either contained 1 mM NaNO2 or 1 mM KOCN. Biomass concentrations were inferred from total protein concentrations, which were 27.6±3.9 μg/ml as measured by the Pierce BCA Protein Assay Kit (Thermo Scientific). In addition, abiotic experiments were performed by adding 1 mM mM KOCN to the NOB medium in the absence of nitrite. Dead biomass controls were performed by treating similar amounts of N. moscoviensis biomass fixed with paraformaldehyde (4%) as described above. The dead biomass was incubated in nitrite-free NOB medium containing 1 mM KOCN. All incubations were amended by filter sterilized 1.5 mM NaHCO3 (final concentration). All incubations were done in 250 ml Schott bottles closed by rubber stoppers without shaking at 37°C in the dark for 96 hours. All experiments were performed in triplicate.
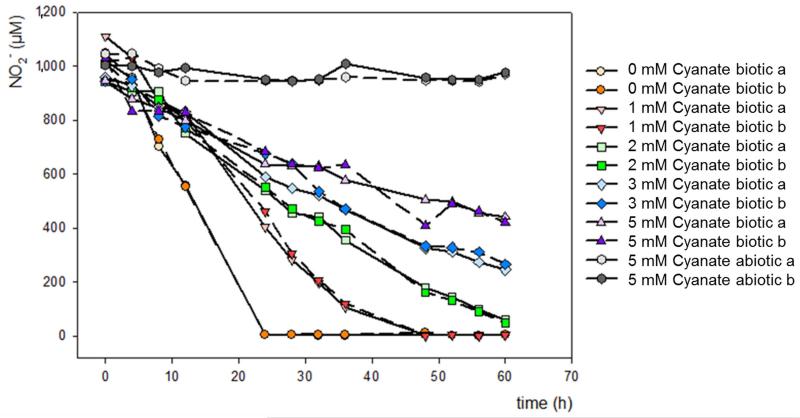
In order to evaluate the effect of increasing cyanate concentrations on nitrite oxidation by N. moscoviensis, biomass was harvested (9,300×g for 15 min at room temperature) and washed 2 times with fresh NOB medium without nitrite. Cells were then transferred into 100 ml NOB medium. Incubations were performed with 1 mM NaNO2 and 0 mM, 1 mM, 2 mM, 3 mM, 4 mM, and 5 mM of KOCN, respectively. In addition, medium containing 5 mM KOCN and 1 mM NaNO2 was incubated without addition of biomass as abiotic control. All incubations were done in 250 ml Schott bottles closed by rubber stoppers without shaking at 37°C in the dark for 60 hours. All experiments were performed in duplicate.
Response of Nitrosomonas nitrosa Nm90 to cyanate
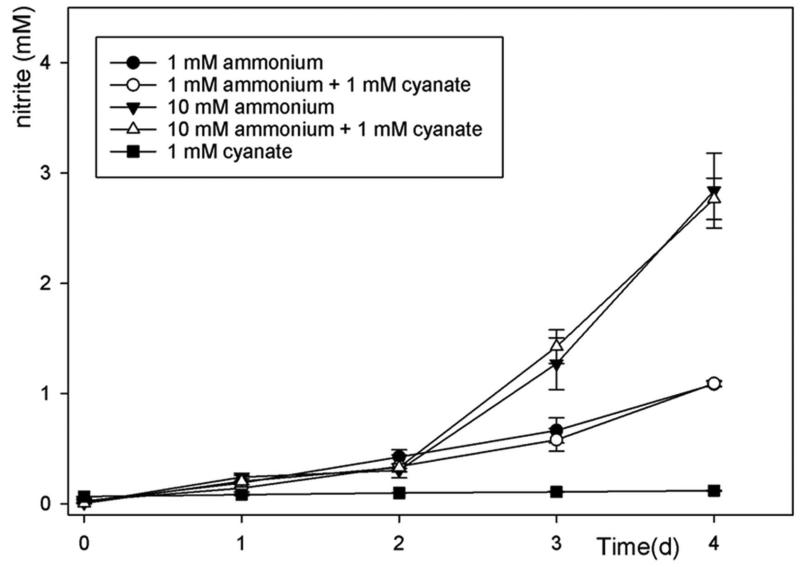
The ammonia-oxidizing bacterium N. nitrosa Nm90 (strain collection of the University of Hamburg, Germany) was grown in AOA medium amended with 10 mM NH4Cl at 37 °C. Biomass was harvested (8,500 rpm for 15 min at room temperature) and washed 2 times with fresh AOA medium without ammonium. Cells were inoculated into 25 ml batches of AOA medium containing either 1 mM KOCN alone or 1 mM NH4Cl plus 1 mM KOCN or 10 mM NH4Cl plus 1 mM KOCN. Cultures were incubated in 50 ml CELLSTAR plastic suspension culture flasks (#690190, Greiner Bio-One), at 37°C, in the dark and shaken at 150 RPM.
Co-culture experiments with N. nitrosa Nm90 and N. moscoviensis
Nitrite-oxidizing cultures of N. moscoviensis were supplied with 0.5 mM (final concentration) KOCN and incubated for 48 hours to induce the expression of cyanase. Biomass was harvested (8,500 rpm for 15 min at room temperature) and washed 2 times with fresh AOA medium without nitrite. N. nitrosa Nm90 was grown in AOA medium supplied with 10 mM NH4Cl. Biomass was harvested (8,500 rpm for 15 min at room temperature) and washed 2 times with fresh AOA medium without ammonium. Biomass concentrations were measured separately for N. moscoviensis and N. nitrosa Nm90 inferred from total protein concentrations, which were 446.5 and 164 μg/ml respectively in 50 ml final volumes for each culture-stock, measured by the Pierce BCA Protein Assay Kit (Thermo Scientific). All biomass were combined and diluted up to 1 liter serving as a master-mix, which was aliquoted to 100 ml batches for the experimental setups resulting a starting protein concentration 20 times less than in the separate stocks measured. Subsequently, either 1 mM KOCN or 1 mM KOCN plus 1mM NH4Cl or only 1 mM NH4Cl was added to the experiments (final concentrations). In addition, abiotic experiments were performed by adding either 1 mM KOCN or 1 mM KOCN plus 1mM NH4Cl to 100 ml AOA medium. All incubations were amended by filter sterilized 1.5 mM NaHCO3 (final concentration). All experiments were done in 250 ml Schott bottles closed by rubber stoppers, incubated without shaking at 37°C in the dark for 168 hours. All experiments were performed in triplicate.
Chemical analysis
Nitrite levels were measured by photometry with the sulfanilamide N-1-napthylethylenediamine dihydrochloride (NED) reagent method33. Ammonium levels were measured photometrically as described previously34. Cyanate was measured fluorimetrically after derivatization with 2-aminobenzoic acid to quinazoline-2,4-dione35, with the modification using fluorescence readout (excitation: 312 nm, emission: 370 nm). All photometric and fluorimetric reads were done by an Infinite 200 Pro spectrophotometer (Tecan Group AG, Männedorf, Switzerland).
qPCR quantification of N. gargensis
A qPCR assay was developed using the newly designed N.gargensis 16S rRNA gene-specific primers NG1052 5′-TAGTTGCTACCTCTGTTC-3′ and NG1436R 5′-ACCTTGTTACGACTTCTC-3′. The qPCR reactions were run with three technical replicates in a Bio-Rad C1000-CFX96 Real-Time PCR system, using the Bio-Rad iQ SYBR Green Supermix kit (Bio-Rad, Hercules, Ca, USA).
Fluorescence In Situ Hybridization (FISH)
Prior to FISH calcium carbonate containing formaldehyde fixed samples were treated with 0.1 M HCl for 3 min. After the calcium carbonate was dissolved the cells were centrifuged down (3 min, 10,000×g) and the supernatant discarded. The pellet was resuspended in 50 μl EtOH/1×PBS (50/50) and the cell suspension was spotted on slides.. The FISH procedure was performed according to the standard protocol with 16S rRNA-targeted probes Ntspa712 specific for the phylum Nitrospira26 and Nso1225 specific for betaproteobacterial ammonia-oxidizing bacteria36. Images were acquired with a Leica SP8 confocal laser scanning microscope (Leica, Wetzlar, Germany).
Total protein quantification
Protein concentrations were measured by using the Pierce BCA Protein Assay Kit (Thermo Scientific).
Replication of physiological experiments
Number of replications are detailed in the subsections for each specific experiment, and mostly were determined by the amount of biomass available for the different nitrifier cultures. In all experiments a minimum of three biological replications were used, except one auxiliary experiment: decelerating effect of increasing cyanate concentrations on nitrite oxidation by N. moscoviensis (Extended Data Figure 6).
Proteomic analysis
Concentrated N. gargensis biomass was inoculated in 140 ml modified AOA medium (amended with 1 mM ammonium final concentration, no cyanate) in three replicates. After a pre-incubation for 24 h, 40 ml samples were taken for proteomic analysis (time point 1) and the remaining cultures were amended with 0.5 mM KOCN and 0.1 mM NH4Cl (final concentrations) and further incubated. Cultures were regularly fed afterwards with 0.5 mM KOCN (final concentration), keeping the concentration between 0.1 and 0.6 mM based on residual KOCN levels calculated from the produced nitrite levels, which were measured every 12 hours. Forty-eight hours after switching to cyanate feeding, 40 ml samples were taken again for proteomic analyses. Cells in the samples from the two different time points were harvested by centrifugation (9,000×g, 30 min, 4°C) and stored at −80°C.
The harvested cell pellets were dissolved in 500 μL UT buffer (8 M urea; 2 M thiourea) and sonicated twice on ice for 1 min (amplitude 0.7; power 70%, UP50H, Hielscher Ultrasound technology). Subsequently, the samples were ultracentrifuged (100,000×g, 1 h, 4°C), and the supernatant was transferred into a fresh reaction tube. Pellets were dissolved in 200 μL preparation buffer (100 mM Tris-HCL; pH 7.5; 300 mM NaCl; 1% digitonin) and incubated overnight at 16°C with 1,200 rpm shaking. After centrifugation (12,000×g, 10 min, 4°C) the supernatant was combined with the supernatant of the previous preparation step. This combined lysate was precipitated with acetone (5× volume, ice-cold) by incubation for 1 h at −20°C, centrifuged (12,000×g, 15 min), and the protein pellet was air-dried. Protein concentrations of all extracts were determined photometrically using a Bradford Assay (Bio-Rad Laboratories, Hercules, CA, USA). SDS-PAGE preparation, reduction, alkylation, and proteolytic digestion by trypsin with subsequent C18-purification were performed as described previously37. Mass spectrometry was performed by a Orbitrap Fusion MS (Thermo Fisher Scientific, Waltham, MA, USA) coupled to a TriVersa NanoMate (Advion, Ltd., Harlow, UK). Five μL of the peptide lysates were separated with a Dionex Ultimate 3000 nano-LC system (Dionex/Thermo Fisher Scientific, Idstein, Germany).
MS raw files were processed using Proteome Discoverer (v1.4, Thermo Scientific). MS spectra were searched against a N. gargensis database (Uniprot/Swiss-Prot, containing 3,786 unreviewed sequence entries) and a common Repository of Adventitious Proteins (cRAP) database using the Sequest HT algorithm. Enzyme specificity was selected as trypsin with up to two missed cleavages allowed using 10 ppm peptide ion tolerance and 0.1 Da MS/MS tolerances. Oxidation (methionine) and carbamylation (lysine and arginine) were selected as variable modifications and carbamidomethylation (cysteine) as a static modification. Only peptides with a false discovery rate (FDR) <1% calculated by Percolator38 and peptide rank =1 were considered as identified.
Modeling biotic and abiotic cyanate degradation kinetics
For cyanate utilization experiments with N. gargensis, the chemical reaction kinetics for the species cyanate, ammonia, and nitrite were modeled as two consecutive first-order reactions. Reaction rates were then estimated using ordinary least squares optimization for a system of nonlinear equations39, as implemented by the nlsystemfit algorithm in the package systemfit40 in R41. For calculation of the abiotic degradation of cyanate/isocyanic acid as a function of temperature and pH, established reactions and values from the literature were used. Degradation was modeled as three first-order reactions (1) hydronium ion catalyzed hydrolysis of isocyanic acid, (2) direct hydrolysis of isocyanic acid, and (3) and direct hydrolysis of cyanate, as described previously42. Published values were used for rate constants and their temperature dependence42. A value of 3.7 was used for the acid dissociation constant for isocyanic acid (variously reported to be 3.29 to 3.92), which has no detectable temperature dependence range of 0°C to 80°C43.
Phylogenetics of cyanase genes in published metagenomes
Amino acid sequences for all members of the “Cyanase Family” (2425 entries) were downloaded from Uniprot44 and all predicted amino acid sequences annotated as cyanase, cyanate hydratase or cyanate lyase were downloaded from the IMG-ER (3028 sequences) and IMG-MER (5476 sequences) databases25 on August 8, 2014. Cyanase sequences from IMG were filtered according to inferred distance (<1.25 replacements per position) and bit scores (>56) from Uniprot references using alignment/distance calculation in Mafft45 and blastp46 (word_size 2, BLOSUM45) respectively. In addition, the predicted amino acid sequence of the N. gargensis cynS was used as a query in a tblastn search against publicly available metagenomes of the JGI IMG/M database. Hits were filtered by e-value (e-values<10−10), at least 50% length coverage of the query sequence, and assignment to the cyanase superfamily (e-value <10−10) of the Conserved Domain Database Database47 (CDD). All putative cyanase sequences were filtered for length (100 residues) and clustered at 99% identity using USEARCH29. The resulting 3340 Cyanase sequences were aligned in mafft to produce a distance matrix and clustered into 100 sequence clusters using the hclust(method=“complete”) and cutree(k=100) commands in R41. Clusters were examined manually and three singleton sequences that aligned poorly were discarded. Cyanase from N. gargensis and N. moscoviensis were added into the dataset. Alignment and phylogeny for the set of 99 representative cyanase genes was calculated using Bali-Phy28 with an initial alignment randomization and the number of iterations in each run set to 1100 with a burnin of 600. Posterior tree pools from three independent runs were combined to assess bipartition support. The 225 environmental cyanase sequences identified in a Nitrososphaera/Nitrospira clade were clustered into 61 representative sequences using USEARCH at 99% minimum identity. Alignment and phylogenetic reconstruction for these representative sequences and ten broadly sampled outgroup cyanases was carried out in Bali-Phy (randomize alignment, iterations=1100, burnin=600). Posterior tree pools from 4 independent runs were combined to generate an 80% PP consensus tree assess bipartition support.
Extended Data
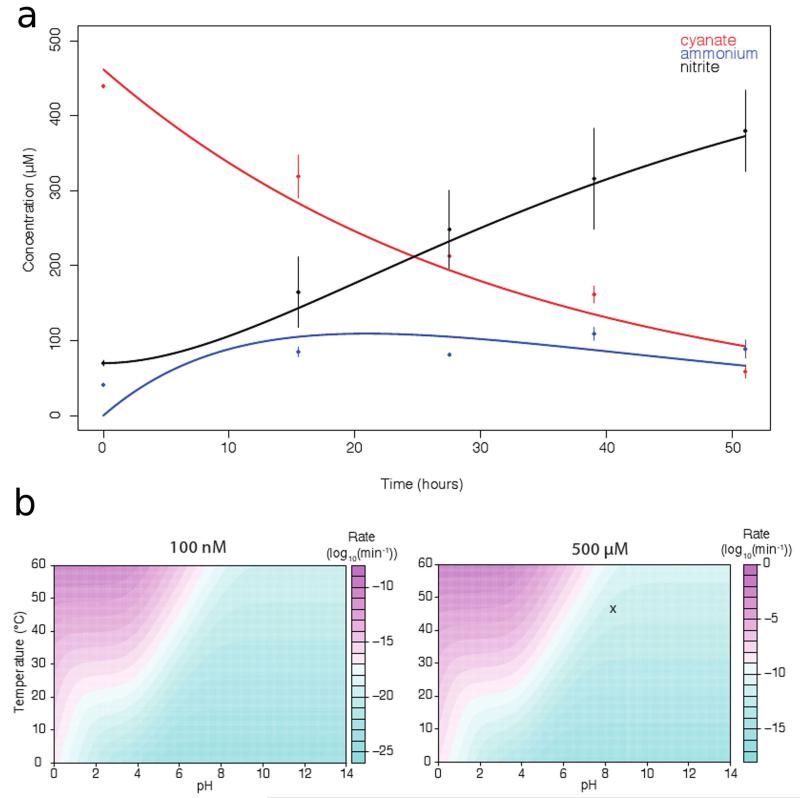
Extended Data Figure 1. Biotic and abiotic cyanate degradation kinetics.
a, Degradation of 500 μM cyanate and utilization of ammonium by N. gargensis modeled as two consecutive first order reactions (cyanate->ammonium->nitrite). Measured data are shown as dots and error bars (mean and s.e.m.) and model predictions with estimated rate parameters are shown as solid lines. Estimated rate constants were kcyanate-ammonium = 4.872 *10−4 min−1 and kammonium-nitrite= 1.064 *10−3 min−1. The abiotic hydrolysis of 500 μM cyanate in this medium was measured to be much slower than enzymatic degradation (kcyanate-hydrolysis = 8.71 *10−5 min−1.). b, The abiotic degradation of high (500 μM) and low (100 nM) concentrations of isocyanic acid/ cyanate across a range of temperatures and pH. Degradation was modeled using a well-established model of three first-order reactions (1) hydronium ion catalyzed hydrolysis of isocyanic acid (k1=e25.97*e−7201.29/T), (2) direct hydrolysis of isocyanic acid (k2=e72.30*e−21646.66/T)), and (3) direct hydrolysis of cyanate42 (k3=e22.23*e−8725/T). The log-transformed degradation rates are shown (as min−1). The conditions that were used to test cyanate degradation by N. gargensis are marked with an X.
Extended Data Figure 2. N. gargensis grows on cyanate.
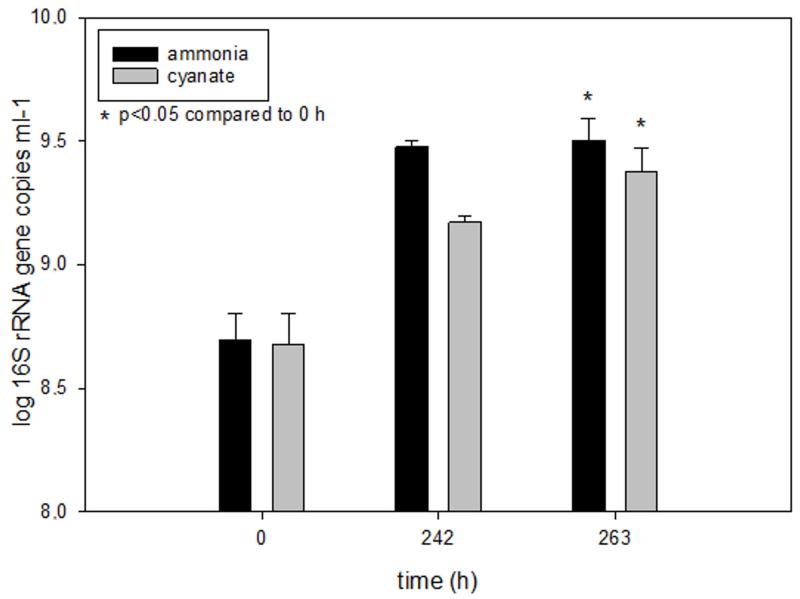
16S rRNA gene copy numbers of N. gargensis as determined by qPCR at three different time points during the experiment displayed in Figure 1. For comparison, the respective gene copy numbers after growth in medium with 0.5 mM ammonium as the sole source of energy and reductant are displayed. The gene copy numbers increased 6.49 fold during growth on ammonium and 4.98 fold during growth on cyanate over 263 hours. Columns show means, error bars show 1 SD of four biological replicates. Significance of difference was calculated by a paired t-test.
Extended Data Figure 3. Cyanase increase upon exposure of N. gargensis to cyanate.
Fold increase and decrease of the 35 most affected proteins after 48h of exposure of N. gargensis to 0.5 mM cyanate (in comparison to t=0 of N. gargensis biomass that has not been exposed to cyanate). Experiments were performed in biological triplicates. Proteins with a significant differential expression are color coded. Significance of difference was calculated by a one sample t-test on log-fold induction, with the Benjamini-Hochberg false discovery rate set to 0.05. For the proteomic analyses 10 μg protein and 500 ng peptide lysate per sample was used. Protein abundances within a sample were normalized by dividing the peak area for a given protein to median peak area for all detected proteins. It should be noted that during growth on cyanate N. gargensis experiences much lower concentrations of ammonium that during growth on ammonium in batch culture. This effect likely influences expression patterns of some of the listed proteins.
Extended Data Figure 4. Conversion of 0.05 mM cyanate by N. gargensis.
a, Concentration changes of cyanate, ammonium, and nitrite by N. gargensis in a mineral medium containing 0.05 mM cyanate as the only source of energy and reductant. b, Abiotic control experiment. All experiments were performed in four replicates and the chemical measurements were done in three technical replicates (averaged). Data points are mean values of four biological replicates, error bars show 1 SD.
Extended Data Figure 5. N. moscoviensis has a functional cyanase.
Concentration changes of cyanate and ammonium during incubation of N. moscoviensis (27.6±3.9 μg/ml protein) in a mineral medium containing cyanate, but no nitrite. Results from a control experiment with identical amounts of dead biomass of N. moscoviensis are also displayed. All experiments were performed in triplicates and the chemical measurements from each replicate were done in three replicates. Data points are mean values, error bars show 1 SD. Asterisks indicate statistical significance at P values of <0.05 (*) and <0.001 (***) between N. moscoviensis and dead biomass. Significance was tested by using two-way analysis of variance (ANOVA) including Tukey’s honest significant difference (HSD) test.
Extended Data Figure 6. Decelerating effect of increasing cyanate concentrations on nitrite oxidation by N. moscoviensis.
Biomass was incubated for 60 h in medium containing 1mM nitrite and increasing cyanate concentrations ranging from 0 mM to 5 mM and nitrite oxidation was monitored. Incubations were performed in duplicates.
Extended Data Figure 7. Nitrosomonas nitrosa Nm90 has no cyanase activity and is not inhibited by 1 mM cyanate.
Concentration changes of nitrite during incubation of N. nitrosa in a mineral medium containing (i) 1 mM ammonium, (ii) 1mM cyanate, (iii) 1mM cyanate and 1 mM ammonium, (iv) 10 mM ammonium, and (v) 1mM cyanate and 10 mM ammonium. All experiments were performed in three biological replicates, data points are mean values, error bars show 1 SD of biological replicates.
Extended Data Figure 8. Reciprocal feeding of ammonia- and nitrite-oxidizers during cyanate conversion.
As activities differed between biological replicates (as often observed for nitrifying strains that are very sensitive to rubber stoppers, contaminants on glass material etc) data are displayed for each replicate individually. Concentration changes of cyanate, ammonium, nitrite, and nitrate displayed as bar (left) and line charts (right) during the growth of the cyanase-negative ammonium-oxidizing bacterium Nitrosomonas nitrosa Nm90 and the cyanase-positive nitrite-oxidizer N. moscoviensis in a mineral medium containing 1 mM cyanate (upper panel) or 1 mM cyanate and 1mM ammonium as source of energy and reductant (lower panel). Data points are mean values, error bars show 1 SD of three technical replicates.
Extended Data Figure 9. Abiotic controls for the reciprocal-feeding experiment (Figure 2, Extended Data Figure 8).
Concentration changes of cyanate, ammonium, nitrite, and nitrate during 168 hours incubation under similar conditions to the biotic experiments: mineral medium containing 1 mM cyanate (left) or 1 mM cyanate and 1mM ammonium (right). Cyanate decay rate is much slower than in biotic setups (see Extended Data Figure 8). Cyanate decay in the presence of ammonium led to formation of a product other than ammonium (e.g. carbamylate). No nitrite or nitrate was formed abiotically. Data points are mean values, error bars show 1 SD of three technical replicates.
Extended Data Table 1. Presence of cyanase, nitrite/nitrate transporters and enzymes related to urea metabolism in ammonia- and nitrite-oxidizing microbes with fully sequenced genome.
| Nitrite/nitrate tansporter | Urea metabolism | ||||||
|---|---|---|---|---|---|---|---|
| Organism | Cyanase |
*FNT family |
†NNP Family |
‡ABC transporter |
§Urea transporter |
∥Urea production |
¶Urea degradation |
| Ammonia oxidizing archaea | |||||||
| Nitrososphaera gargensis | + | + | − | + | + | + | + |
| Nitrosophaera viennensis EN76 | − | − | − | + | + | + | + |
| Ca. Nitrosophaera evergladensis SR1 | − | − | − | + | + | + | + |
| Nitrosopumilus maritimus SCM1 | − | − | − | + | − | + | − |
| Nitrosopumilus sp. AR | − | − | − | + | + | + | + |
| Nitrosopumilus sp. SJ | − | − | − | + | − | + | − |
| Ca. Nitrosopumilus koreensis AR1 | − | − | − | + | − | + | − |
| Ca. Nitrosopumilus salaria BD31 | − | − | − | + | − | + | − |
| Ca. Nitrosopumilus sp. AR2 | − | − | − | − | + | + | + |
| Ca. Nitrosoarchaeum koreensis MY1 | − | − | − | + | − | + | − |
| Ca. Nitrosoarchaeum limnia BG20 | − | − | − | + | − | + | − |
| Ca. Nitrosoarchaeum limnia SFB1 | − | − | − | + | − | + | − |
| Ca. Nitrosotenuis uzonensis N4 | − | − | − | + | − | + | − |
| Cenarchaeum symbiosum A | − | − | − | + | + | + | + |
| Ammonia oxidizing bacteria | |||||||
| Nitrosococcus halophilus Nc4 | − | + | − | − | − | − | − |
| Nitrosococcus oceani AFC27 | − | + | − | − | + | + | + |
| Nitrosococcus oceani ATCC 19707 | − | + | − | − | + | + | + |
| Nitrosococcus watsonii C-113 | − | + | − | − | + | + | + |
| Nitrosomonas europaea ATCC 19718 | − | + | − | + | + | + | − |
| Nitrosomonas eutropha C91 | − | + | − | + | − | + | − |
| Nitrosomonas sp. AL212 | − | + | − | − | + | + | + |
| Nitrosomonas sp. ls79A3 | − | + | − | − | − | + | + |
| Nitrosospira multiformis ATCC 25196 | − | + | − | − | + | − | + |
| Nitrite oxidizing bacteria | |||||||
| Nitrospira moscoviensis | + | + | + | + | + | + | + |
| Ca. Nitrospira defluvii | + | + | + | − | − | + | − |
| Nitrobacter sp. Nb-311A | + | + | + | + | − | + | − |
| Nitrobacter winogradskyi Nb-255 | + | + | + | + | − | + | − |
| Nitrobacter hamburgensis X14 | + | + | + | + | − | + | − |
| Nitrococcus mobilis Nb-231 | + | + | + | − | − | + | − |
| Nitrolancea hollandica | + | + | + | − | + | + | − |
| Nitrospina gracilis | + | + | − | − | − | + | − |
FNT transporter family: Formate-nitrite transporter, encoded by focA/nirC – has been postulated to transport cyanate18
NNP transporter family: Nitrate/nitrite transporter, encoded by nark – might also transport cyanate due to its chemical similarity to nitrite
ABC transporter nitrate/sulfonate/bicarbonate – this transporter family has been shown to transport cyanate as well20
Urea transporter: ABC transporter, or urea channel
Urea produced internally by the enzymes agmatine amidinohydrolase or arginine amidinohydrolase
Urea consumed internally by the enzymes urease or urea carboxylase and allophanate hydrolase
Supplementary Material
Acknowledgements
We thank Elena Lebedeva for efforts in purifying N. gargensis. Maria Mooshammer for help with cyanate analytics, Kathleen Eismann and Benjamin Scheer for help with the proteomic analysis, and Markus Schmid for performing the FISH experiment. Andreas Pommerening-Röser is acknowledged for providing strain Nitrosomonas nitrosa Nm 90. M.Pa., M.Po., A.G., P.H., and M.W. were supported by the European Research Council via the Advanced Grant project “NITRICARE 294343” to M.W.. H.D., H.K., and D.B. were supported by the Austrian Science Fund (FWF, grants P25231-B21 and P26127-B20). The authors are grateful for using the analytical facilities of the Centre for Chemical Microscopy (ProVIS) at the Helmholtz Centre for Environmental Research, which is supported by European Regional Development Funds (EFRE–Europe funds Saxony) and the Helmholtz Association. M.vB. was partially funded by the Collaborative Research Centre AquaDiva of the German Research Foundation.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Hatzenpichler R, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamennaya NA, Post AF. Distribution and expression of the cyanate acquisition potential among cyanobacterial populations in oligotrophic marine waters. Limnol Oceanogr. 2013;58:1959–1971. [Google Scholar]
- 3.Qian M, Eaton JW, Wolff SP. Cyanate-mediated inhibition of neutrophil myeloperoxidase activity. Biochem J. 1997;326:159–166. doi: 10.1042/bj3260159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purcarea C, et al. Aquifex aeolicus aspartate transcarbamoylase, an enzyme specialized for the efficient utilization of unstable carbamoyl phosphate at elevated temperature. J Biol Chem. 2003;278:52924–52934. doi: 10.1074/jbc.M309383200. [DOI] [PubMed] [Google Scholar]
- 5.Ubalua AO. Cyanogenic glycosides and the fate of cyanide in soil. Aust J Crop Sci. 2010;4:223–237. [Google Scholar]
- 6.Widner B, Mulholland MR, Mopper K. Chromatographic determination of nanomolar cyanate concentrations in estuarine and sea waters by precolumn fluorescence derivatization. Anal Chem. 2013;85:6661–6666. doi: 10.1021/ac400351c. [DOI] [PubMed] [Google Scholar]
- 7.Rees AP, Woodward EMS, Joint I. Concentrations and uptake of nitrate and ammonium in the Atlantic Ocean between 60 degrees N and 50 degrees S. Deep-Sea Res Pt Ii. 2006;53:1649–1665. [Google Scholar]
- 8.Rocap G, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 9.Luque-Almagro VM, et al. Characterization of the Pseudomonas pseudoalcaligenes CECT5344 cyanase, an enzyme that is not essential for cyanide assimilation. Appl Environ Microb. 2008;74:6280–6288. doi: 10.1128/AEM.00916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamennaya NA, Chernihovsky M, Post AF. The cyanate utilization capacity of marine unicellular Cyanobacteria. Limnol Oceanogr. 2008;53:2485–2494. [Google Scholar]
- 11.Bock E. Growth of nitrobacter in the presence of organic matter. II. Chemoorganotrophic growth of Nitrobacter agilis. Arch Microbiol. 1976;108:305–312. doi: 10.1007/BF00454857. [DOI] [PubMed] [Google Scholar]
- 12.Bock E, Schmidt I, Stuven R, Zart D. Nitrogen loss caused by denitrifying Nitrosomonas cells using ammonium or hydrogen as electron-donors and nitrite as electron-acceptor. Arch Microbiol. 1995;163:16–20. [Google Scholar]
- 13.Hommes NG, Sayavedra-Soto LA, Arp DJ. Chemolithoorganotrophic growth of Nitrosomonas europaea on fructose. J Bacteriol. 2003;185:6809–6814. doi: 10.1128/JB.185.23.6809-6814.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mussmann M, et al. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci U S A. 2011;108:16771–16776. doi: 10.1073/pnas.1106427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch H, et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science. 2014;345:1052–1054. doi: 10.1126/science.1256985. [DOI] [PubMed] [Google Scholar]
- 16.Tourna M, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin W, et al. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci U S A. 2014;111:12504–12509. doi: 10.1073/pnas.1324115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spang A, et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012;14:3122–3145. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- 19.Stieglmeier M, et al. Nitrososphaera viennensis gen. nov., sp nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Micr. 2014;64:2738–2752. doi: 10.1099/ijs.0.063172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda S, Omata T. Nitrite transport activity of the ABC-type cyanate transporter of the cyanobacterium Synechococcus elongatus. J Bacteriol. 2009;191:3265–3272. doi: 10.1128/JB.00013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MunozCenteno MC, Paneque A, Cejudo FJ. Cyanate is transported by the nitrate permease in Azotobacter chroococcum. FEMS Microbiol Lett. 1996;137:91–94. [Google Scholar]
- 22.Schramm A, de Beer D, van den Heuvel JC, Ottengraf S, Amann R. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: Quantification by in situ hybridization and the use of microsensors. Appl Environ Microb. 1999;65:3690–3696. doi: 10.1128/aem.65.8.3690-3696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maixner F, et al. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol. 2006;8:1487–1495. doi: 10.1111/j.1462-2920.2006.01033.x. [DOI] [PubMed] [Google Scholar]
- 24.Koops HP, Bottcher B, Moller UC, Pommereningroser A, Stehr G. Classification of eight new species of ammonia-oxidizing bacteria: Nitrosomonas communis sp. nov., Nitrosomonas ureae sp. nov., Nitrosomonas aestuarii sp. nov., Nitrosomonas marina sp. nov., Nitrosomonas nitrosa sp. nov., Nitrosomonas eutropha sp. nov., Nitrosomonas oligotropha sp. nov. and Nitrosomonas halophila sp. nov. J Gen Microbiol. 1991;137:1689–1699. [Google Scholar]
- 25.Markowitz VM, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M. In situ characterization of Nitrospira-like nitrite oxidizing bacteria active in wastewater treatment plants. Appl Environ Microb. 2001;67:5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonso-Saez L, et al. Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci U S A. 2012;109:17989–17994. doi: 10.1073/pnas.1201914109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suchard MA, Redelings BD. BAli-Phy: simultaneous Bayesian inference of alignment and phylogeny. Bioinformatics. 2006;22:2047–2048. doi: 10.1093/bioinformatics/btl175. [DOI] [PubMed] [Google Scholar]
- 29.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
Additional references
- 30.Lebedeva EV, et al. Enrichment and genome sequence of the group I.1a ammonia-oxidizing Archaeon “Ca. Nitrosotenuis uzonensis” representing a clade globally distributed in thermal habitats. Plos One. 2013;8 doi: 10.1371/journal.pone.0080835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Widdel F. Anaerober Abbau von Fettsäuren und Benzoesäure durch neu isolierte Arten Sulfat-reduzierender Bakterien. Universität Göttingen; 1980. Dissertation. [Google Scholar]
- 32.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 33.Strickland JDH, Parsons TR. A Practical Handbook of Seawater Analysis. Fisheries Research Board of Canada; 1972. [Google Scholar]
- 34.Kandeler E, Gerber H. Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol Fert Soils. 1988;6:68–72. [Google Scholar]
- 35.Guilloton M, Karst F. A spectrophotometric determination of cyanate using reaction with 2-aminobenzoic acid. Anal Biochem. 1985;149:291–295. doi: 10.1016/0003-2697(85)90572-x. [DOI] [PubMed] [Google Scholar]
- 36.Mobarry BK, Wagner M, Urbain V, Rittmann BE, Stahl DA. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microb. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiffmann CL, et al. Proteome profile and proteogenomics of the organohalide-respiring bacterium Dehalococcoides mccartyi strain CBDB1 grown on hexachlorobenzene as electron acceptor. J Proteomics. 2014;98:59–64. doi: 10.1016/j.jprot.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 39.Gallant R. Nonlinear equation estimation. John Wiley and Sons; 1987. p. 610. [Google Scholar]
- 40.Henningsen A, Hamann JD. systemfit: A package for estimating systems of simultaneous equations in R. J Stat Softw. 2007;23 [Google Scholar]
- 41.Team, R. D. C. R: A language and environment for statistical computing. Proc Natl Acad Sci U S A. 2011 http://www.R-project.org/ [Google Scholar]
- 42.DeMartini N, Murzin DY, Forssen M, Hupa M. Kinetics of cyanate decomposition in alkaline solutions of high ionic strength: The catalytic effect of bicarbonate. Ind Eng Chem Res. 2004;43:4815–4821. [Google Scholar]
- 43.Taillades J, et al. A pH-dependent cyanate reactivity model: application to preparative N-carbamoylation of amino acids. J Chem Soc Perk T 2. 2001:1247–1254. [Google Scholar]
- 44.Apweiler R, et al. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local Alignment Search Tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Marchler-Bauer A, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.