Abstract
Different assembly processes drive the spatial structure of meta-communities (β-diversity). Recently, functional and phylogenetic diversities have been suggested as indicators of these assembly processes. Assuming that diversity is a good proxy for niche overlap, high β-diversity along environmental gradients should be the result of environmental filtering while low β-diversity should stem from competitive interactions. So far studies trying to disentangle the relative importance of these assembly processes provided mixed results. One reason for this may be that these studies often rely on a single measure of diversity and thus implicitly make a choice on how they account for species relative abundances and how species similarities are captured by functional traits or phylogeny.
Here, we tested the effect of gradually scaling the importance of dominance (the weight given to dominant vs. rare species) and species similarity (the weight given to small vs. large similarities) on resulting β-diversity patterns of an alpine plant meta-community. To this end, we combined recent extensions of the Hill numbers framework with Pagel’s phylogenetic tree transformation approach. We included functional (based on the Leaf-Height-Seed spectrum) and phylogenetic facets of β-diversity in our analysis and explicitly accounted for effects of environmental and spatial covariates.
We found that functional β-diversity was high when the same weight was given to dominant vs. rare species and to large vs. small species’ similarities. In contrast, phylogenetic β-diversity was low when greater weight was given to dominant species and small species’ similarities. Those results suggested that different environments along the gradients filtered different species according to their functional traits, while, the same competitive lineages dominated communities across the gradients.
Our results highlight that functional vs. phylogenetic facets, presence-absence vs. abundance structure and different weights of species’ dissimilarity provide complementary and important information on the drivers of meta-community structure. By utilizing the full extent of information provided by the flexible frameworks of Hill numbers and Pagel’s tree transformation, we propose a new approach to disentangle the patterns resulting from different assembly processes.
Keywords: Community assembly, β-diversity, Functional diversity, Phylogenetic diversity, Alpine communities, Hill numbers
Introduction
The spatial structure of meta-community diversity (β-diversity) is a key feature for understanding how the environment shapes biodiversity patterns (Kraft et al. 2011, Myers et al. 2013). While evaluating the change in species identities and relative abundances across communities has a long tradition in community ecology (Cody and Diamond 1975), recent work has highlighted the value of studying the change in species ecological similarities instead, in order to identify the spatial patterns that emerge from different historical, ecological and evolutionary processes (Graham and Fine 2008). In that perspective, distance measures applied to functional traits and phylogenetic trees have been increasingly used to estimate species ecological similarities (Pavoine and Bonsall 2011). Functional distances are based on species’ functional traits, i.e. measurable morphological, physiological or phenological features that impact their fitness via their effects on growth, reproduction and survival (Violle et al. 2007) and thus are directly connected to species’ niches (Thuiller et al. 2004). Pairwise species’ phylogenetic distances measure divergence times during evolutionary history and are often argued to be a good synthetic measure of species ecological differentiation as they do not require the identification and measurement of relevant traits (Faith 1992, Webb 2000, Mouquet et al. 2012).
However, functional and phylogenetic diversity do not necessarily provide similar information and patterns (Cadotte et al. 2013, Thuiller et al. 2014b). How strongly their patterns overlap depends on the strength of phylogenetic signal in the functional traits (i.e. the trend for closely related species to be more similar than distantly related species), which in turn depends on the underlying processes of niche evolution and species diversification (Losos 2008, Burns and Strauss 2011). The joint analysis of functional and phylogenetic facets of diversity can thus provide simultaneous hypotheses on the impacts of past evolutionary history (phylogenetic diversity) and specific phenotypic traits (functional diversity) on current ecological processes (Devictor et al. 2010, Safi et al. 2011, Cadotte et al. 2013).
If functional or phylogenetic similarities are suitable proxies for niche overlap then the observed patterns of β-diversity can shed light on the underlying ecological and evolutionary processes. High β-diversity along steep ecological gradients would identify a strong effect of ecological processes that foster the local co-occurrence of similar species and the regional differentiation of communities, suggesting either strong environmental filtering or dispersal limitation. Otherwise, very low β-diversity along steep ecological gradients reveals a stability of community structure, suggesting either an absence of environmental filtering on the species feature studied, unlimited dispersion or widespread local coexistence of competitive species (Spasojevic and al. 2014). These ecological processes are not exclusive and rather act simultaneously producing a complex pattern of diversity. We propose here that this complexity can be disentangled by gradually varying the effects of (1) species similarity and (2) species dominance in the functional and phylogenetic diversity patterns.
(1) A pervasive but never challenged assumption in studies of community assembly is that species ecological similarity varies linearly with interspecific functional or phylogenetic distance (e.g. Webb 2000, Mason et al. 2005). However, this is done for reasons of simplicity and with little theoretical foundation as the scenario of trait evolution that would result in this linear relationship is unlikely (see e.g. Fig. 3 in Thuiller et al. 2010). Studies on phylogenetic diversity patterns have shown that contrasted assembly processes can be detected when considering, for instance, all lineages of a meta-community or only a specific lineage (Cavender-Bares et al. 2006, Münkemüller et al. 2014). It has been also suggested that competitive interactions could often be restricted to specific lineages or functional groups (Cavender-Bares et al. 2006, Slingsby and Verboom 2006), while environmental filtering could be predominant when considering the assembly of distantly related lineages (Vamosi et al. 2009), due to broad climatic adaptations being conserved in angiosperms lineages (Crisp et al. 2009). Modeling species ecological similarities by assuming that all parts of the phylogeny or functional tree (i.e. a dendrogram based on species trait dissimilarities) are equally relevant may thus hinder the detection of assembly processes operating between closely related species or functionally close species vs. highly dissimilar ones. Instead, varying the importance given to small compared to large species similarities (i.e. to branches close to the root vs. to branches close to the tips of the phylogenetic or functional tree) in the diversity patterns analysis may allow to uncover the different patterns at different similarity scales (called “similarity effect” hereafter).
(2) Communities often exhibit an uneven species abundance distribution (Volkov et al. 2003). Usually a few species make most contribution to community biomass, vegetation cover or number of individuals while the majority of species are locally rare. Different ecological processes are responsible for this dominance pattern commonly observed (de Bello et al. 2012). Hierarchical scaling of community assembly rules (Lortie et al. 2004) and recent modeling developments have indeed hypothesized that while occurrence patterns may be primarily driven by environmental filtering, the local abundance of species mostly results from the interplay between biotic interactions and dispersal limitations (Boulangeat et al. 2012). We therefore expect that diversity patterns analyses yield contrasting results according to the importance given to dominant vs. rare species (called “dominance effect” hereafter).
The similarity and the dominance effects can impact the identification of patterns and the interpretation of underlying ecological processes and may thus be highly informative for our ecological understanding. However, in most diversity studies these effects are not explicitly considered. Instead, implicit weights are given to species similarities and abundance differences through the selection of an a priori diversity index (Tuomisto 2010a, Pavoine and Bonsall 2011). This lack of explicit consideration may be partly explained by the fact that comprehensive methods were unavailable so far. However, recent extensions of the Hill numbers (Hill 1973) now allow computing diversity indices with varying strength of the dominance effect, while at the same time considering species ecological dissimilarities (Pavoine et al. 2009, Chao et al. 2010, Leinster and Cobbold 2012). Additionally, studies of trait evolution have long used transformed trees to explicitly parameterize the importance of the phylogenetic similarity effect. One common tree transformation is the delta transformation (Pagel 1997). The rationale of this transformation is that a phylogenetic tree stretched close to the root puts more weight on large phylogenetic distances while a tree stretched close to the tips puts more weight on small phylogenetic distances. We use this approach to include and parameterize the strength of the similarity effect in our β-diversity analysis.
Here, we build on a multiplicative α,β,γ decomposition framework (Whittaker 1960, Jost 2007) in which we explicitly integrate the dominance and similarity effects into the study of functional and phylogenetic diversity patterns. Our study system is a plant meta-community composed of 120 community plots in a valley of the French Alps. Our hypothesis is that interacting environmental filters and competition drive the diversity patterns in these plant communities (Boulangeat et al. 2012). We ask whether the integration of the similarity and dominance effects allows us to identify diversity patterns that would have been hidden in a classical diversity analysis. More specifically, we test whether environmental filters can be detected based on β-diversity patterns that build on low dominance weights and strong weights on large species similarities and whether competition can be detected based on β-diversity patterns that build on high dominance weights and low weights on large species similarities. In addition we ask, whether trait diversity and phylogenetic diversity capture the expected patterns equally well.
Materials and Methods
Data
Study area
The study area was the 25 km long Guisane Valley located in the center of the French Alps (c. 260 km2; 44.9° N, 6.5° E). The valley is characterized by contrasted climatic conditions, with mean annual temperatures ranging from −8.1 °C to 7.7 °C. As in other valleys of the central Alps, the landscape is a mosaic of coniferous and deciduous forests, shrub heaths, subalpine grasslands and alpine meadows. All these habitats were represented in our dataset.
Environmental data
We used climatic variables (mean temperature of the coldest month of the year, relative summer wetness and sum of winter precipitations) and topographic variables (bedrock carbon content, topographic wetness index and topographic position, i.e. topographic convexity or concavity). The climatic variables were originally extracted from the AURELHY database (Benichou and Le Breton 1987), downscaled to a 100m resolution (Zimmermann et al. 2007), while the topographic variables came from a 50m resolution Digital Elevation Model.
Community plots
We worked with a meta-community of 120 community plots that have been sampled in the Guisane valley from 2009 onwards by the Alpine National Botanic Conservatory. Sites were representative of the heterogeneity of the valley’s climatic conditions. They were on average separated by 10 km (only 0.4% of site pairwise geographic distances fell below the threshold of 100 m set by the climatic variables resolution). The herbaceous strata of the community-plots were surveyed within an approximate area of 100 m2 of homogeneous vegetation by expert botanists. The abundance estimates were based on an abundance-dominance scale using six cover classes (Braun-Blanquet et al. 1946). Our meta-community dataset included initially a total of 531 species.
Functional tree
We chose three functional traits that describe species’ ecological strategies according to the Leaf-Height-Seed spectrum: specific leaf area, height and seed mass (LHS; Westoby 1998). These traits are strongly related to the fundamental processes of plant life, i.e. dispersal, establishment, and persistence (Weiher et al. 1999), and their combination has been useful to capture the existing variation in plant ecological strategies (Lavergne et al. 2003, Slingsby and Verboom 2006). Specific leaf area (SLA, i.e. light intercepting area per leaf dry mass) reflects the trade-off between resource acquisition and conservation in plants. Height at maturity is related to competitive ability and avoidance of environmental stress (Körner 2003). Seed mass strongly influences dispersal and is related to establishment (Pakeman et al. 2008). The trait information from each species was retrieved from the Alpine functional trait database (ANDROSACE, Thuiller et al. unpublished). The database includes trait information for Alpine plants from several in-house projects and freely available databases (see Appendix A for details). We excluded species for which less than two traits were available. The remaining 400 species still accounted for more than 80% of the total abundance of each studied community (Pakeman and Quested 2007). We then calculated the relative abundance of each species by dividing the abundance estimates by the total abundance of the remaining species in each community. Finally, we estimated the functional distance matrix from the trait-by-species matrix. Each trait was previously log-transformed to conform to normality and scaled between 0 and 1. We then constructed a functional tree as a prerequisite for performing the tree transformation detailed below. We used a hierarchical clustering approach to build an ultrametric functional dendrogram (functional tree, Mouchet et al. 2008) of all species, employing an average agglomeration method (UPGMA, function hclust in R).
Phylogenetic tree
We used an ultrametric genus-level phylogeny of alpine plants extracted from Thuiller et al. (2014a) that followed the workflow proposed in Roquet et al. (2013) with DNA sequences downloaded from Genbank (see Appendix A for details). The tips of the phylogenetic tree were resolved with polytomies to obtain a species-level phylogeny. The 400 species were vascular plants, mostly angiosperms (397 species) but also included six ferns species and a spike moss species.
Analysis
We performed our analyses in three steps. First, we calculated how strongly trait values relate to the phylogenetic tree, for each single trait but also for all traits together (phylogenetic signal). Second, we tested the effects of similarity and dominance on the estimation of meta-community β-diversities (functional and phylogenetic). Finally, we tested the strength of the influence of space and environment on inter-community pairwise diversities as a function of the similarity and dominance effects.
Phylogenetic signal in functional traits
We used Pagel’s λ (1997) to measure the strength of the phylogenetic signal of each functional trait. λ is a scaling parameter for the phylogeny. Its value is fitted so that the resulting transformation of the phylogeny ensures the best fit of trait data to the Brownian Motion model. If λ is not significantly different from 0, the trait distribution is independent from the phylogeny. We estimated λ with the function fitContinuous (Harmon et al. 2008, R-package geiger) and tested it against the hypothesis that the trait distribution was independent from the phylogeny (λ=0) using a log-likelihood ratio test (Münkemüller et al. 2012).
To test the phylogenetic signal of the LHS scheme (the three traits together), we performed a Mantel test between the matrix of the functional tree distances and the matrix of the square root phylogenetic distances, as recommended by Hardy and Pavoine (2012).
Diversity decomposition and meta-community β-diversity
We used the generalization of Chao et al. (2010) of Hill numbers (Hill 1973) to estimate the phylogenetic or functional α-diversity of each community and the γ-diversity of the whole meta-community. Following this generalization implies calculating a diversity index, which takes into account species similarities based on the branch lengths of either a phylogenetic or functional tree.
The index is a function of a parameter q, which varied between 0 and +∞ and reflects the effect of dominance on the diversity estimation. The more q increases, the more qD is influenced by dominant species and the less by rare species (Eq. 1).
| (Eq.1) |
where the summation is over all branches of an ultrametric phylogenetic or functional tree of tips-to-root distance T, Li is the length of branch i, and p = {pi} denotes the vector containing for each branch i, the summed relative abundance of all its descendent species.
To calculate the α-diversity for each community, p was calculated from the vector of the relative abundance of the N species occurring in the community, while to calculate the γ-diversity of the meta-community, p was calculated from the vector of the average relative abundance of the species over all communities (i.e. the entire meta-community). To improve the computational efficiency of our analysis, we used the mathematical formulation of qD given in the appendices of Leinster and Cobbold (2012, R-function available in Supplement 1).
Additionally, in Appendix B, we adapted the inclusion of species’ similarity to an alternative generalization of Hill’s number proposed by Leinster and Cobbold (2012) that relies on slightly different calculations (i.e. based on similarity matrices instead of trees). We compared the output of the two approaches and showed that they revealed largely similar results (see Appendix B for these analyses and discussion of relative advantages of both approaches).
Characteristics of the applied diversity measure
The diversity measure we used here is strongly related to other well-known measures. If species are considered to be equally similar (i.e. they are linked by a star-like tree), then qD (i) is equal to the number of species for q=0, (ii) tends toward the Shannon entropy exponential for q tending toward 1 and (iii) is the inverse of Simpson for q=2.
If species are not considered equally similar, then qD is equal (i) to Faith index for q=0 (Faith 1992), (ii) to the exponential of Allen’s index for q tending towards 1 (Allen et al. 2009) and (iii) equal to a monotonic transformation of Rao’s quadratic entropy, for q=2 (Rao 1986).
The effects of similarity were taken into account using a transformation of the functional and phylogenetic trees of the entire Guisane meta-community prior to calculating diversity indices. We influenced the effect of similarity using the delta transformation of trees proposed by Pagel (1997) in a phylogenetic context (Figure B1). The delta transformation raises the depth of the tree nodes to the power of δ. In concrete terms, it inflates (respectively deflates) the length of close-to-root branches compared to close-to-tips branches when the parameter δ is lower (respectively higher) than 1. When δ tends toward +∞, i.e. the transformed tree tends toward a star-like tree, all species are considered equally similar, and the diversity index approaches a measure of taxonomic diversity. In contrast, when δ̣ tends toward 0, the transformed tree is reduced to the two branches descending from the root and species are fused together according to this branching.
In species similarity terms, the delta transformation allows playing with the effect of similarity between species and shifts the scope of the analysis from large species cophenetic distances (weak similarity effect, δ <1) to small species cophenetic distances (strong similarity effect, δ>1, Figure B1).
All together, we thus computed γ-diversity and α-diversity using a function that depended on both the similarity (δ) and the dominance (q) effects.
Meta-community β-diversity standardized effect sizes
β-diversity was calculated as the ratio of γ-diversity and the average α-diversity of the meta-community estimated as the generalized mean of degree 1-q of the α-diversities of all communities (Jost 2007, Tuomisto 2010a, Chiu et al. 2014).
| (Eq.2) |
community j and S the number of communities.
β has a minimal possible value of 1 if all communities are identical in species abundances and species identities. Furthermore, qD obeys the replication principle. This means that if the studied communities had all an equal α-diversity, if they did not have any species in common, and if the species belonging to different communities are descending from different functional or phylogenetic tree branches, then the β(q,δ) of the study area would be maximized and equal to S (Chiu et al. 2014). The replication principle is a necessary condition for the independence of α and β and is thus essential to obtain meaningful measures of β-diversity (sensu Jost 2007). The value of β(q,δ) was then tested against a null model of tip-shuffling to access whether the observed β-diversity was higher or smaller than expected from a model of random assembly of the species from the meta-community pool. We then calculated the standardized effect size (SES) of the β-diversity as the mean of the distribution minus the observed β-diversity divided by the standard deviation of the null distribution. If the β-diversity was higher than expected (SES < 0) then the communities differed more than expected under a random assembly model; if the β-diversity was lower than expected (SES > 0), then the communities differed less than expected under a random assembly model.
Influence of environment and space on inter-community pairwise functional and phylogenetic diversities
Similarly to strong environmental filtering, dispersal limitation and ecological drift can also result in high β-diversity among communities (Ricklefs 2008). In this case, we can expect the inter-community pairwise functional and phylogenetic diversities pattern to be spatially auto-correlated. In order to avoid any misinterpretation of diversity patterns, which may be partially influenced by these confounding processes, and to fully characterize the fingerprint of environmental filtering, we explicitly linked the inter-community pairwise functional and phylogenetic diversities to environmental variables and space following a procedure based on Dray et al. (2012).
We then developed an approach to disentangle the relative effects (and their interaction) of space and environment on the structure of communities. To do so, we first defined space with Moran’s eigenvector maps (MEM) based on a Gabriel graph obtained from the sites geographical coordinates. We retained only the MEMs with significant Moran’s I (p < 0.05). To define environment, we performed a principal component analysis (PCA) on environmental variables and extract sites scores along all the PCA axes. Second, for both phylogenetic and functional information, we generated inter-community pairwise diversities matrices in function of the q and δ parameters by calculating the functional and phylogenetic β-diversities (Eq. 2) between all pairs of communities. We subtracted 1 (the minimal possible value) to each diversity value to build a matrix of inter-community pairwise diversities corresponding to the Whittaker’s effective species turnover (Whittaker 1960, Tuomisto 2010b). Third, we performed a principal coordinates analysis (PCOA) to separate the communities in a multivariate space and extract community scores along the PCOA axes. We then applied a forward selection procedure to the MEM spatial predictors to retain the most relevant spatial predictors for each inter-community pairwise diversities matrix (phylogenetic and functional, for each pair of q and δ; Blanchet et al. 2008). Finally, to partition the importance of space and environment to explain patterns of intercommunity pairwise diversities, we performed a variance partitioning procedure on the matrices of site scores deduced from the PCOA, with the matrix of relevant MEM and the matrix of site scores along the axes of the PCA on environmental variables as cofactors (Borcard et al. 1992). We therefore obtained for each pair of q and δ parameters and each diversity facet (functional and phylogenetic), the variance explained by environment after controlling for space (E\S), the variance explained by space after controlling for environment (S\E) and the variance explained by the interaction of space and environment (SxE). These explained variances were defined as adjusted R2.
All analyses were carried out using the software R 3.0.1. (R Development Core Team 2013) with the following packages: ade4, ape, geiger, packfor, snowfall, spacemakeR, spdep, vegan.
Results
Phylogenetic signal of functional traits
All the individual traits exhibited a significant phylogenetic signal. SLA and Height had moderate values of λ (Height: λ=0.52, χ2=70.54, df=1, p<0.001; SLA: λ=0.56, χ2=35.99, df=1, p<0.001). Seed mass had the strongest phylogenetic signal (Seed mass: λ=0.97, χ2=249.95, df=1, p<0.001). The phylogenetic signal of the species LHS scheme was significant but very low (Mantel-test, R2 =0.06; p<0.001).
Meta-community β-diversity standardized effect sizes
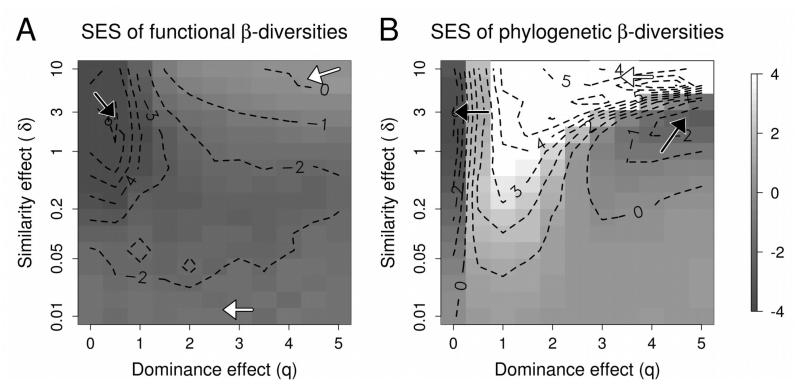
The standardized effect sizes (SES) of functional β-diversity were overall low, but more specifically (SES<−3) for low value of q (q<1) or for low to intermediate value of δ (0.02<δ<4, intermediate similarity effect, Figure 1A). For more extreme values of δ (δ<0.2 or δ>2) and high values of q (q>5), SES increased and the functional pattern of β-diversity became not discernible from the random expectation (SES > −2 and SES < 2). Overall this suggested a predominant influence of environmental filtering on functional diversity both when the dominance effect was weak (i.e. all species present have equal weight) and when the similarity effect was moderate (i.e. approximately unchanged functional tree branch lengths).
Figure 1.
Standard Effect Sizes (SES) of the functional (left panel) and phylogenetic (right panel) beta-diversity of the meta-community against a tip-shuffling null model, as a function of the strength of the dominance effect (q) and the strength of the similarity effect (δ). A low q value indicates that rare and dominant species were given about the same weight while a high q value indicates that more weight was given to dominant species. A low (respectively high) δ value indicates that small (respectively large) species’ similarities were given more weight (Figure 1). A low SES value indicates a higher than expected β-diversity, hence a predominant influence of environmental filtering, while a high SES value indicates a lower-than-expected β-diversity, hence a predominant influence of competition resulting in a limiting similarity pattern. Black and white arrows points towards respectively local minima and maxima.
When focusing on phylogenetic diversity, the pattern was radically different and more complex (Figure 1B). In general, SES of the meta-community phylogenetic β-diversity was lower than for functional diversity. Like the functional β-diversity, the phylogenetic β-diversity was noticeably low (SES < −3) for low value of q (q < 0.5). However, the β-diversity was noticeably high (SES >3) when q was between 0.8 and 2 or when δ was higher than 3. Otherwise the β-diversity did not differ strongly from the null expectation (SES > −2 and SES <2). Overall, this suggested that communities had a similar phylogenetic structure under a moderate dominance effect and when small phylogenetic distances were emphasized (δ > 3). When the dominance effect was reduced or ignored (q< 0.5), we detected environmental filtering, while we detected a random assembly process when considering a strong dominance effect.
Effects of environment and space on inter-community pairwise diversities
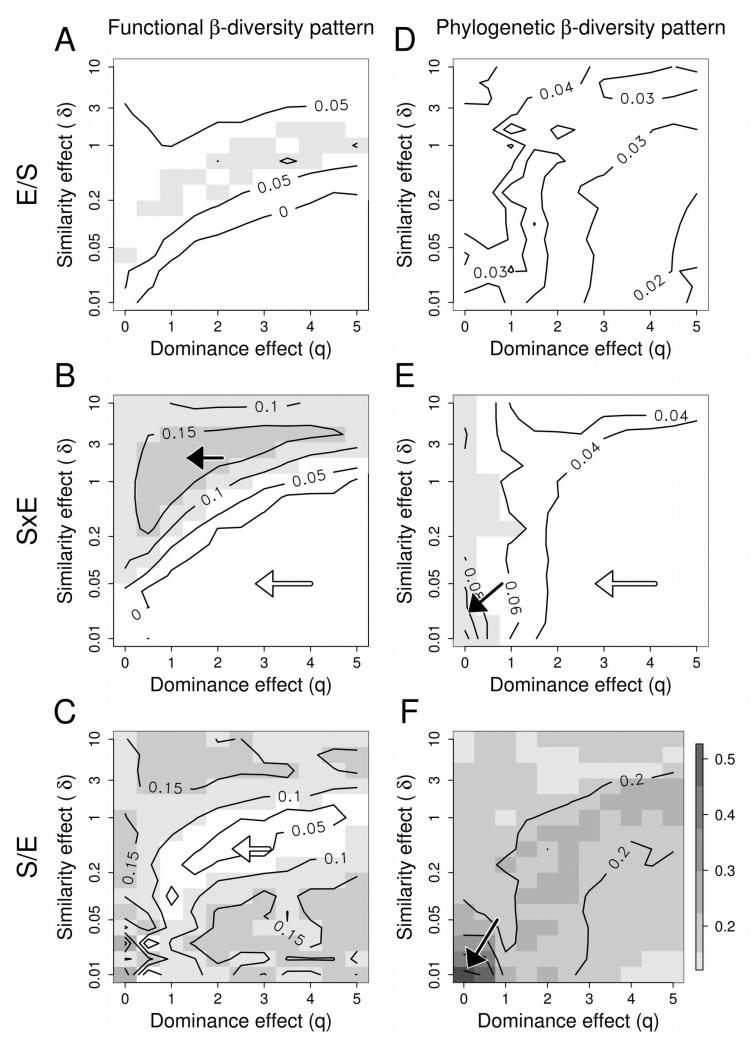
The second analysis yielded similar results as the first one and mainly confirmed that the functional β-diversity pattern was indeed driven by the strong environmental gradients of the Guisane valley. The purely environmental component (E/S) explained only a small portion of the variance of the functional and phylogenetic inter-community pairwise diversities regardless of the dual effects of dominance and similarity (R2 < 0.10; Figure 2A & 2D).
Figure 2.
Influence of environment and space on the inter-community pairwise functional and phylogenetic distances, as a function of the strength of the dominance effect (q) and the strength of the similarity effect (δ). A low q value indicates that rare and dominant species were given about the same weight while a high q value indicates that more weight was given to dominant species. A low (respectively high) δ value indicates that small ( respectively large) species’ similarities were given more weight (Figure 1). The different lines represent the variance (Adjusted R2) of the β-distance matrix of communities explained by environment only (E/S), spatially autocorrelated environment (SxE) and a pure spatial effect (S/E). Black and white arrows points towards respectively local maxima and minima.
The environmental component interacting with space (SxE) explained a variable proportion of the inter-community pairwise functional diversities depending on the strength of the dominance and similarity effect (Figure 2B). A moderate amount of variation was explained for functional diversity for a weak dominance (q<2) or a moderate similarity effect (0.5<δ<4) with a maximal adjusted R2 of 0.20 (for q=1 and δ= 1.58). In comparison with the functional diversity pattern, the environmental component interacting with space (SxE) explained overall a low amount of variation of the phylogenetic diversity (Figure 2E) with a mean adjusted R2 of 0.04.
The purely spatial component (S/E) explained overall a moderate proportion of the variance in the inter-community pairwise functional diversities with a mean adjusted R2 of 0.12. . The purely spatial component (S/E) explained overall a moderate proportion of the variance in the intercommunity pairwise phylogenetic diversities matrices with a mean adjusted R2 of 0.19. It however reached high values of adjusted R2 for a weak dominance and similarity effect (δ<0.2, q<0.5) with a maximal adjusted R2 of 0.73 (for q=0 and δ=0.01). More in-depth analyses revealed that this combination tended to distinguish particular communities (mostly marshes) that contained species from the long branches of our phylogeny (spike moss and fern species) from the angiosperms. As these species were both infrequent and locally rare, their contribution was masked when the dominance and similarity effect were strong.
Discussion
The strong environmental gradients in alpine ecosystems are known to be important drivers of community structure (Mitchell et al. 2009, de Bello et al. 2012). In observational field studies they are often the only identified drivers whereas local experiments demonstrated the importance of positive and negative biotic interactions between plant neighbors (Choler et al. 2001, Callaway et al. 2002). This apparent discrepancy is rooted in the fact that most published studies have either focused on functional or phylogenetic diversity and have chosen, or implied, a single arbitrary dominance and similarity effect. Here we show that jointly investigating both functional and phylogenetic patterns together with a comprehensive inclusion of dominance and similarity effects can reveal multiple patterns likely due to either environmental filtering or negative biotic interactions.
Variable composition and stable phylogenetic dominance structure across communities
Patterns of functional and phylogenetic β-diversity were very different in the study region. This mismatch stemmed in the moderately low phylogenetic signal of the functional traits studied both taken individually and together. As a consequence, the two facets of diversity appeared quite decoupled in the study meta-community. Assuming that species’ niches can be abstracted as multi-dimensional hypervolumes (Hutchinson 1959), functional traits and phylogenetic identity can thus be interpreted as surrogates of distinct niche dimensions.
Interestingly, our analyses suggested that environmental filtering was the main driver of the patterns of functional and phylogenetic β-diversity when rare and dominant species were given the same weight (Figure 1 & 2). This suggested that the functional and phylogenetic compositions of the communities were filtered out by the strong abiotic gradients of the Guisane valley (albeit quite weakly for the phylogenetic identity of species, Figure 2D, 2E).
However for a stronger dominance effect, the imprint of environmental filtering was less pervasive on the functional β-diversity. More strikingly, the observed phylogenetic β-diversity was consistently low relative to random expectations, suggesting a high stability of the phylogenetic community structure across space. This was true only when the similarity effect was strong (δ >1), i.e. when small phylogenetic similarities were given more weight. We interpreted this pattern as the consequence of the dominance of angiosperms lineages over other lineages in alpine herbaceous communities. For a weak similarity effect (thus when angiosperms species are considered highly similar), the pattern of phylogenetic stability of dominant angiosperms lineages was blurred by the random turnover of non-angiosperms vs angiosperms between communities.
These last results showed that while communities differed strongly in terms of functional traits, their dominant species tended to come from the same lineages, suggesting a strong competitive advantage. In other words, the Leaf-Height-Seed strategy scheme (Westoby 1998) was mainly informative about environmental filters driven by climatic gradients. Thus communities strongly varied along the gradients in regard of the trait values of their constituent species (Figure 1A & Figure 2B). Conversely, species phylogenetic differences informed weakly about environmental filters (Figure 2D & 2E) supporting other local diversity patterns studies (Silvertown et al. 2006, Bernard-Verdier et al. 2013). However phylogenetic β-diversities seemed to capture niche information related to competitive hierarchy suggesting that competition was driven by unmeasured traits showing potentially strong phylogenetic signal.
Both the study of β-diversity SES and inter-community pairwise diversities yielded similar results about the action of environmental filtering on the functional and phylogenetiċ β-diversity patterns. The co-variation of the inter-community pairwise functional β-diversities with environmental variables suggested that the functional high β-diversity was driven by the steep local environmental gradients and was not solely due to spatial auto-correlation effects or to confounding assembly processes (Mayfield and Levine 2010). Our results further emphasized that the functional and phylogenetic β-diversity patterns were spatially auto-correlated; in particular within a specific window (low δ and q) for the phylogenetic diversity pattern. Our study area encompasses a single valley of limited area (260 km2) suggesting little influence of ecological drift. We thus associated the spatial auto-correlation to the influence of dispersal limitation or spatially structured environmental gradients that we did not directly account for. The dominance effect had an important impact on the detection of diversity patterns. When the dominance effect was weak, our results suggested a more pervasive print of environmental filtering (Figure 1A & Figure 2) while for a strong dominance effect, communities seemed to be more driven by stochastic processes (in regards of the functional traits we studied) and competition (in regards of their phylogenetic identity; Figure 1). We hypothesized that the environmental filters along the Guisane valley gradients primarily influenced which traits allowed species establishment within communities, but not which traits shaped species’ competitive hierarchies. In contrast, environmental gradients did not strongly influence the phylogenetic community structure. Regardless of the location along the gradients, the communities were structured by a few dominant species from the same lineages (e.g. Poaceae, Fabaceae, Asteraceae). In combination, these two patterns suggested that these lineages maintain their dominance across environmental gradients thanks to strong trait lability which has allowed (1) trait convergence and thus coexistence of distantly related species into communities despite strong environmental filters (Webb 2000) (2) the within-lineage emergence of niche-segregated species sorted out along gradients (Angert and Schemske 2005).
The similarity effect interacted with the dominance effect to reveal hidden features of the diversity patterns. The inter-community pairwise functional distances were more strongly linked to environment for a moderate similarity effect, i.e. when the functional tree branch lengths were almost unchanged (Figure 1). However, a weak or a strong similarity effect hinder the detection of an environmental effect showing that tree branch transformation was unsuitable to improve the understanding of community assembly along environmental gradients.
To summarize, the presence-absence structure of communities was mainly driven by the high turnover of species due to the environmental filtering of their traits and phylogenetic identity while the dominance structure was mainly driven by the high abundance of the same lineages over the gradient, likely because of unmeasured competitive advantages.
Emphasizing the different features of meta-communities
Our results emphasize the importance of studying together different types of diversity as the interpretation of diversity patterns changed according to the studied diversity. In that perspective, the family of Hill numbers and its extension to phylogenetic and functional distances provides a promising framework to analyze the spatial patterns of meta-communities (Arroyo-Rodriguez et al. 2013). It allows fine-tuning the effect of dominance and similarity, while retaining indices with similar mathematical properties (Chiu et al. 2014). Our results are particularly striking since the parameterization drastically changed the detection of functional and phylogenetic diversity patterns. In return, this allowed us to suggest that different ecological processes affected the occurrence of species (low q values) and the local dominance of species (higher q values), as also found in Boulangeat et al. (2012). The similarity effect tended to reveal hidden patterns, in particular for the phylogenetic β-diversity pattern by either putting the emphasis on ancient or recent species’ divergences. There are numerous discrepancies in the literature about the link between community assembly and functional or phylogenetic diversities. Among others, some functional traits can be associated to both environmental filtering and biotic interactions even in the same ecosystem (e.g. Gross et al. 2009, de Bello et al. 2012) and phylogenetic diversity has been associated to various patterns of diversity (Mouquet et al. 2012). While spatial or evolutionary scale have been proposed to explain these various outcomes (Cavender-Bares 2009), the impact of giving the same weight to all parts of the functional or phylogenetic tree is rarely tested (Thuiller et al. 2010, Cadotte et al. 2013). We argue here that the inclusion of the similarity effect in diversity patterns studies may help to clarify these discrepancies and provide more complete, if not clearer, diversity patterns. Other studies have done a similar job either through null model modifications (Hardy and Senterre 2007, Chalmandrier et al. 2013) or through other types of tree transformations (Rosauer et al. 2013). However, these frameworks ignored the parts of the functional and phylogenetic trees close to the root (which correspond to a moderate to a strong similarity effect) while ours can also do the reverse procedure and ignore the close-to-tips parts of the trees (weak similarity effect). Taken together, the exploration of the dominance and similarity effect can help to determine the window in which the diversity pattern is best predicted by variables such as environment and space (Figure 2), opening promising avenues to optimize the calibration of models of community turnover over space and environmental gradients (e.g. Dray et al. 2012, Rosauer et al. 2013).
Conclusion
The diversity patterns of meta-communities are the outcome of complex interactions between past evolution, current trait states and multiple assembly rules (Cavender-Bares et al. 2009, Lavergne et al. 2010). Using an integrative framework of diversity pattern analysis, we demonstrated how the consideration of the dominance structure of communities and species ecological similarity affects diversity patterns of alpine plant meta-community. We found that environment controlled the functional and (more modestly) the phylogenetic diversity of the meta-communities when focusing on presence-absence like patterns (i.e. low dominance effect), which is typical of a compressed environmental gradient. Additionally, considering phylogenetic diversity in our innovative framework allowed us to suggest that biotic interactions shaped the dominance pattern. Together these results let us to conclude that alpine plant species have both labile functional traits to adapt to environmental gradients and unknown evolutionary conserved traits that drive community assembly via inter-specific competition. Explicitly testing the effects of dominance and species ecological similarity can thus help disentangling the multiple assembly rules affecting the functional and phylogenetic structure of meta-communities along environmental gradients.
Supplementary Material
Appendix A: Detailed informations about the trait database and the phylogeny
Appendix B: Results using Leinster and Cobbold’s index and comparisons with Chao’s diversity index.
Supplement material: R scripts for performing the α, β, γ decomposition
Acknowledgments
We thank Anne Chao for her thorough comments on the properties of the diversity indices. The research leading to these results received funding from the European Research Council under the European Community’s Seven Framework Programme FP7/2007-2013 Grant Agreement no. 281422 (TEEMBIO). TM was funded by the ANR-BiodivERsA project CONNECT (ANR-11-EBID-002), as part of the ERA-Net BiodivERsA 2010 call. The LECA is part of Labex OSUG@2020 (ANR10 LABX56). The field research was conducted on the long-term research site Zone Atelier Alpes, a member of the ILTER-Europe network. ZAA publication n° xxx.
References
- Allen B, Kon M, Bar-Yam Y. A new phylogenetic diversity measure generalizing the Shannon index and its application to phyllostomid bats. The American Naturalist. 2009;174:236–243. doi: 10.1086/600101. [DOI] [PubMed] [Google Scholar]
- Angert A, Schemske DW. The evolution of species’ distributions: reciprocal transplants across the elevation ranges of Mimulus cardinalis and M. lewisii. Evolution. 2005;59:1671–1684. [PubMed] [Google Scholar]
- Arroyo-Rodriguez V, Rös M, Escobar F, Melo FPL, Satntos BA, Tabarelli M, Chazdon R. Plant beta-diversity in fragmented rain forests: testing floristic homogenization and differentiation hypotheses. Journal of Ecology. 101:1449–1458. [Google Scholar]
- De Bello, de F, Lavorel S, Lavergne S, Albert CH, Boulangeat I, Mazel F, Thuiller W. Hierarchical effects of environmental filters on the functional structure of plant communities: a case study in the French Alps. Ecography. 2012;36:393–402. [Google Scholar]
- Benichou P, Le Breton O. Prise en compte de la topographie pour la cartographie des champs pluviométriques statistiques. La Météorologie. 1987;7 [Google Scholar]
- Bernard-Verdier M, Flores O, Navas M-L, Garnier E. Partitioning phylogenetic and functional diversity into alpha and beta components along an environmental gradient in a Mediterranean rangeland. Journal of Vegetation Science. 2013;24:877–889. [Google Scholar]
- Blanchet FG, Legendre P, Borcard D. Forward selection of explanatory variables. Ecology. 2008;89:2623–2632. doi: 10.1890/07-0986.1. [DOI] [PubMed] [Google Scholar]
- Borcard D, Legendre P, Drapeau P. Partialling out the spatial component of ecological variation. Ecology. 1992;73:1045–1055. [Google Scholar]
- Boulangeat I, Gravel D, Thuiller W. Accounting for dispersal and biotic interactions to disentangle the drivers of species distributions and their abundances. Ecology letters. 2012;15:584–593. doi: 10.1111/j.1461-0248.2012.01772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun-Blanquet J. Über den Deckungswert der Arten in den Pflanzengesellschaften der Ordnung Vaccinio-Piceetalia. Jahresber. Naturforsch. Ges. Graubündens. 1946;130:115–119. [Google Scholar]
- Burns JH, Strauss SY. More closely related species are more ecologically similar in an experimental test. Proceedings of the National Academy of Sciences. 2011;108:5302. doi: 10.1073/pnas.1013003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadotte M, Albert CH, Walker SC. The ecology of differences: assessing community assembly with trait and evolutionary distances. Ecology letters. 2013 doi: 10.1111/ele.12161. [DOI] [PubMed] [Google Scholar]
- Cahill JF, Kembel SW, Lamb EG, Keddy PA. Does phylogenetic relatedness influence the strength of competition among vascular plants? Perspectives in Plant Ecology, Evolution and Systematics. 2008;10:41–50. [Google Scholar]
- Callaway RM, Brooker R, Choler P, Kikvidze Z, Lortie CJ, Michalet R, Paolini L, Pugnaire FI, Newingham B, Aschehoug ET. Positive interactions among alpine plants increase with stress. Nature. 2002;417:844–848. doi: 10.1038/nature00812. others. [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Keen A, Miles B. Phylogenetic structure of Floridian plant communities depends on taxonomic and spatial scale. Ecology. 2006;87:109–122. doi: 10.1890/0012-9658(2006)87[109:psofpc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. The merging of community ecology and phylogenetic biology. Ecology Letters. 2009;12:693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- Chalmandrier L, Münkemüller T, Gallien L, De Bello F, Mazel F, Lavergne S, Thuiller W. A family of null models to distinguish between environmental filtering and biotic interactions in functional diversity patterns. Journal of Vegetation Science. 2013;24:853–864. doi: 10.1111/jvs.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao A, Chiu C-H, Jost L. Phylogenetic diversity measures based on Hill numbers. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365:3599–3609. doi: 10.1098/rstb.2010.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C-H, Jost L, Chao A. Phylogenetic beta diversity, similarity, and differentiation measures based on Hill numbers. Ecological Monographs. 2014;84:21–44. [Google Scholar]
- Choler P, Michalet R, Callaway RM. Facilitation and competition on gradients in alpine plant communities. Ecology. 2001;82:3295–3308. [Google Scholar]
- Cody ML, Diamond JM. Ecology and evolution of communities. Belknap Press; 1975. [Google Scholar]
- Crisp MD, Arroyo MTK, Cook LG, Gandolfo MA, Jordan GJ, McGlone MS, Weston PH, Westoby M, Wilf P, Linder HP. Phylogenetic biome conservatism on a global scale. Nature. 2009;458:754–756. doi: 10.1038/nature07764. [DOI] [PubMed] [Google Scholar]
- Devictor V, Mouillot D, Meynard C, Jiguet F, Thuiller W, Mouquet N. Spatial mismatch and congruence between taxonomic, phylogenetic and functional diversity: the need for integrative conservation strategies in a changing world. Ecology letters. 2010;13:1030–1040. doi: 10.1111/j.1461-0248.2010.01493.x. [DOI] [PubMed] [Google Scholar]
- Dray S, Pélissier R, Couteron P, Fortin M-J, Legendre P, Peres-Neto PR, Bellier E, Bivand R, Blanchet FG, De Cáceres M. Community ecology in the age of multivariate multiscale spatial analysis. Ecological Monographs. 2012;82:257–275. [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biological Conservation. 1992;61:1–10. [Google Scholar]
- Graham CH, Fine PV. Phylogenetic beta diversity: linking ecological and evolutionary processes across space in time. Ecology Letters. 2008;11:1265–1277. doi: 10.1111/j.1461-0248.2008.01256.x. [DOI] [PubMed] [Google Scholar]
- Gross N, Kunstler G, Liancourt P, de Bello F, Suding KN, Lavorel S. Linking individual response to biotic interactions with community structure: a trait-based framework. Functional Ecology. 2009;23:1167–1178. [Google Scholar]
- Hardy OJ, Pavoine S. Assessing phylogenetic signal with measurement error: a comparison of Mantel tests, Blomberg et al.’s K, and phylogenetic distograms. Evolution. 2012;66:2614–2621. doi: 10.1111/j.1558-5646.2012.01623.x. [DOI] [PubMed] [Google Scholar]
- Hardy OJ, Senterre B. Characterizing the phylogenetic structure of communities by an additive partitioning of phylogenetic diversity. Journal of Ecology. 2007;95:493–506. [Google Scholar]
- Harmon LJ, Weir J, Brock C, Glor RE, Challenger W. GEIGER: investigating evolutionary radiations. Bioinformatics Applications Note. 2008;24:129131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
- Hill MO. Diversity and evenness: a unifying notation and its consequences. Ecology. 1973;54:427–432. [Google Scholar]
- Hutchinson GE. Homage to Santa Rosalia or why are there so many kinds of animals? The American Naturalist. 1959;93:145–159. [Google Scholar]
- Jost L. Partitioning diversity into independent alpha and beta components. Ecology. 2007;88:2427–2439. doi: 10.1890/06-1736.1. [DOI] [PubMed] [Google Scholar]
- Körner C. Alpine Plant Life. Springer; 2003. [Google Scholar]
- Kraft NJB, Comita LS, Chase JM, Sanders NJ, Swenson NG, Crist TO, Stegen JC, Vellend M, Boyle B, Anderson MJ, Cornell HV, Davies KF, Freestone AL, Inouye BD, Harrison SP, Myers JA. Disentangling the drivers of diversity along latitudinal and elevational gradients. Science. 2011;333:1755–1758. doi: 10.1126/science.1208584. [DOI] [PubMed] [Google Scholar]
- Lavergne S, Garnier E, Debussche M. Do rock endemic and widespread plant species differ under the Leaf–Height–Seed plant ecology strategy scheme? Ecology Letters. 2003;6:398–404. [Google Scholar]
- Lavergne S, Mouquet N, Thuiller W, Ronce O. Biodiversity and climate change: integrating evolutionary and ecological responses of species and communities. Annual Review of Ecology, Evolution, and Systematics. 2010;41:321–350. [Google Scholar]
- Leinster T, Cobbold CA. Measuring diversity: the importance of species similarity. Ecology. 2012;93:477–489. doi: 10.1890/10-2402.1. [DOI] [PubMed] [Google Scholar]
- Lortie CJ, Brooker RW, Choler P, Kikvidze Z, Michalet R, Pugnaire FI, Callaway RM. Rethinking plant community theory. Oikos. 2004;107:433–438. [Google Scholar]
- Losos JB. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecology Letters. 2008;11:995–1003. doi: 10.1111/j.1461-0248.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- Mason NWH, Mouillot D, Lee WG, Wilson JB. Functional richness, functional evenness and functional divergence: the primary components of functional diversity. Oikos. 2005;111:112–118. [Google Scholar]
- Mayfield MM, Levine JM. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecology letters. 2010;13:1085–1093. doi: 10.1111/j.1461-0248.2010.01509.x. [DOI] [PubMed] [Google Scholar]
- Mitchell MGE, Cahill JF, Jr, Hik DS. Plant interactions are unimportant in a subarctic-alpine plant community. Ecology. 2009;90:2360–2367. doi: 10.1890/08-0924.1. [DOI] [PubMed] [Google Scholar]
- Mouchet M, Guilhaumon F, Villéger S, Mason NWH, Tomasini J-A, Mouillot D. Towards a consensus for calculating dendrogram-based functional diversity indices. Oikos. 2008;117:794–800. [Google Scholar]
- Mouquet N, Devictor V, Meynard CN, Munoz F, Bersier L-F, Chave J, Couteron P, Dalecky A, Fontaine C, Gravel D, Hardy OJ, Jabot F, Lavergne S, Leibold M, Mouillot D, Münkemüller T, Pavoine S, Prinzing A, Rodrigues ASL, Rohr RP, Thébault E, Thuiller W. Phylogenetic ecology: advances and perspectives. Biological Reviews. 2012;87:769–785. doi: 10.1111/j.1469-185X.2012.00224.x. [DOI] [PubMed] [Google Scholar]
- Münkemüller T, Gallien L, Lavergne S, Renaud J, Roquet C, Dullinger S, Guisan A, Lenoir J, Svenning J-C, Vittoz P, Willner W, Wohlgemuth T, Zimmermann NE, Thuiller W. Scale decisions can reverse conclusions on community assembly processes. Global Ecology and Biogeography. 2014;23:620–632. doi: 10.1111/geb.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münkemüller T, Lavergne S, Bzeznik B, Dray S, Jombart T, Schiffers K, Thuiller W. How to measure and test phylogenetic signal. Methods in Ecology and Evolution. 2012;3:743–756. [Google Scholar]
- Myers JA, Chase JM, Jiménez I, Jørgensen PM, Araujo-Murakami A, Paniagua-Zambrana N, Seidel R. Beta-diversity in temperate and tropical forests reflects dissimilar mechanisms of community assembly. Ecology letters. 2013;16:151–157. doi: 10.1111/ele.12021. [DOI] [PubMed] [Google Scholar]
- Pagel M. Inferring evolutionary processes from phylogenies. Zoologica Scripta. 1997;26:331–348. [Google Scholar]
- Pakeman RJ, Garnier E, Lavorel S, Ansquer P, Castro H, Cruz P, Doležal J, Eriksson O, Freitas H, Golodets C, Kigel J, Kleyer M, Lepš J, Meier T, Papadimitriou M, Papanastasis VP, Quested H, Quétier F, Rusch G, Sternberg M, Theau J-P, Thébault A, Vile D. Impact of abundance weighting on the response of seed traits to climate and land use. Journal of Ecology. 2008;96:355–366. [Google Scholar]
- Pakeman RJ, Quested HM. Sampling plant functional traits: What proportion of the species need to be measured? Applied Vegetation Science. 2007;10:91–96. [Google Scholar]
- Pavoine S, Bonsall MB. Measuring biodiversity to explain community assembly: a unified approach. Biological Reviews. 2011;86:792–812. doi: 10.1111/j.1469-185X.2010.00171.x. [DOI] [PubMed] [Google Scholar]
- Pavoine S, Love MS, Bonsall MB. Hierarchical partitioning of evolutionary and ecological patterns in the organization of phylogenetically-structured species assemblages: application to rockfish (genus: Sebastes) in the Southern California Bight. Ecology letters. 2009;12:898–908. doi: 10.1111/j.1461-0248.2009.01348.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- Rao CR. Rao’s axiomatization of diversity measures. Encyclopedia of Statistical Sciences. 1986;7:614–617. [Google Scholar]
- Ricklefs RE. Disintegration of the Ecological Community. The American Naturalist. 2008;172:741–750. doi: 10.1086/593002. [DOI] [PubMed] [Google Scholar]
- Roquet C, Thuiller W, Lavergne S. Building megaphylogenies for macroecology: taking up the challenge. Ecography. 2013;36:13–26. doi: 10.1111/j.1600-0587.2012.07773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosauer DF, Ferrier S, Williams KJ, Manion G, Keogh JS, Laffan SW. Phylogenetic generalised dissimilarity modelling: a new approach to analysing and predicting spatial turnover in the phylogenetic composition of communities. Ecography. 2013;36:1–12. [Google Scholar]
- Safi K, Cianciaruso MV, Loyola RD, Brito D, Armour-Marshall K, Diniz-Filho JAF. Understanding global patterns of mammalian functional and phylogenetic diversity. Philosophical Transactions of the Royal Society B: Biological Sciences. 2011;366:2536–2544. doi: 10.1098/rstb.2011.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvertown J, McConway K, Gowing D, Dodd M, Fay MF, Joseph JA, Dolphin K. Absence of phylogenetic signal in the niche structure of meadow plant communities. Proceedings of the Royal Society B: Biological Sciences. 2006;273:39–44. doi: 10.1098/rspb.2005.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slingsby JA, Verboom GA. Phylogenetic Relatedness Limits Co-occurrence at Fine Spatial Scales: Evidence from the Schoenoid Sedges (Cyperaceae: Schoeneae) of the Cape Floristic Region, South Africa. The American Naturalist. 2006;168:14–27. doi: 10.1086/505158. [DOI] [PubMed] [Google Scholar]
- Spasojevic MJ, Copeland S, Suding KN. Using functional diversity patterns to explore metacommunity dynamics: a framework for understanding local and regional influences on community structure. Ecography. 2014;37:1–11. [Google Scholar]
- Thuiller W, Gallien L, Boulangeat I, de Bello F, Münkemüller T, Roquet C, Lavergne S. Resolving Darwin’s naturalization conundrum: a quest for evidence. Diversity and Distributions. 2010;16:461–475. [Google Scholar]
- Thuiller W, Guéguen M, Georges D, Bonet R, Chalmandrier L, Garraud L, Renaud J, Roquet C, Van Es J, Zimmermann NE, Lavergne S. Are different facets of plant diversity well protected against climate and land cover changes? A test study in the French Alps. Ecography. 2014a doi: 10.1111/ecog.00670. 10.1111/ecog.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuiller W, Lavorel S, Midgley G, Lavergne S, Rebelo T. Relating plant traits and species distributions along bioclimatic gradients for 88 Leucadendron taxa. Ecology. 2004;85:1688–1699. [Google Scholar]
- Thuiller W, Maiorano L, Mazel F, Guilhaumon F, Ficetola GF, Lavergne S, Renaud J, Roquet C, Mouillot D. Conserving the functional and phylogenetic trees of life of European tetrapods. Philosophical Transactions of the Royal Society B: Biological Sciences. 2014b doi: 10.1098/rstb.2014.0005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomisto H. A diversity of beta diversities: straightening up a concept gone awry. Part 1. Defining beta diversity as a function of alpha and gamma diversity. Ecography. 2010a;33:2–22. [Google Scholar]
- Tuomisto H. A diversity of beta diversities: straightening up a concept gone awry. Part 2. Quantifying beta diversity and related phenomena. Ecography. 2010b;33:23–45. [Google Scholar]
- Vamosi SM, Heard SB, Vamosi JC, Webb CO. Emerging patterns in the comparative analysis of phylogenetic community structure. Molecular Ecology. 2009;18:572–592. doi: 10.1111/j.1365-294X.2008.04001.x. [DOI] [PubMed] [Google Scholar]
- Violle C, Navas ML, Vile D, Kazakou E, Fortunel C, Hummel I, Garnier E. Let the concept of trait be functional! Oikos. 2007;116:882–892. [Google Scholar]
- Volkov I, Banavar JR, Hubbell SP, Maritan A. Neutral theory and relative species abundance in ecology. Nature. 2003;424:1035–1037. doi: 10.1038/nature01883. [DOI] [PubMed] [Google Scholar]
- Webb CO. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. American Naturalist. 2000;156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- Weiher E, van der Werf A, Thompson K, Roderick M, Garnier E, Eriksson O, O. Challenging Theophrastus: A common core list of plant traits for functional ecology. Journal of Vegetation Science. 1999;10:609–620. [Google Scholar]
- Westoby M. A leaf-height-seed (LHS) plant ecology strategy scheme. Plant and soil. 1998;199:213–227. [Google Scholar]
- Whittaker RH. Vegetation of the Siskiyou mountains, Oregon and California. Ecological Monographs. 1960;30:279–338. [Google Scholar]
- Zimmermann NE, Edwards TC, Moisen GG, Frescino TS, Blackard JA. Remote sensing-based predictors improve distribution models of rare, early successional and broadleaf tree species in Utah. Journal of Applied Ecology. 2007;44:1057–1067. doi: 10.1111/j.1365-2664.2007.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix A: Detailed informations about the trait database and the phylogeny
Appendix B: Results using Leinster and Cobbold’s index and comparisons with Chao’s diversity index.
Supplement material: R scripts for performing the α, β, γ decomposition