Abstract
Purpose
Non-invasive biomarkers for early detection of pancreatic ductal adenocarcinoma (PDAC) are currently not available. Here, we aimed to identify a set of urine proteins able to distinguish patients with early stage PDAC from healthy individuals (H).
Experimental design
Proteomes of 18 urine samples from healthy controls, chronic pancreatitis and PDAC patients (six/group) were assayed using GeLC/MS/MS analysis. The selected biomarkers were subsequently validated using ELISA assays using multiple logistic regression applied to a training dataset in a multicentre cohort comprising 488 urine samples.
Results
LYVE-1, REG1A and TFF1 were selected as candidate biomarkers. When comparing PDAC (n=192) to healthy (n=87) urines, the resulting areas under the receiver operating characteristic curves (AUCs) of the panel were 0.89 (95%CI 0.84-0.94) in the training (70% of the data), and 0.92 (95%CI 0.86-0.98) in the validation (30% of the data) datasets. When comparing PDAC stage I-II (n=71) to healthy urines, the panel achieved AUCs of 0.90 (95%CI 0.84-0.96) and 0.93 (95%CI 0.84-1.00) in the training and validation datasets, respectively. In PDAC stage I-II and healthy samples with matching plasma CA19.9 the panel achieved a higher AUC of 0.97 (95%CI 0.94-0.99) than CA19.9 (AUC=0.88, 95%CI 0.81-0.95, p=0.005). Adding plasma CA19.9 to the panel increased the AUC from 0.97 (95%CI 0.94-0.99) to 0.99 (95%CI 0.97-1.00, p=0.04) but did not improve the comparison of stage I-IIA PDAC (n=17) to healthy urine.
Conclusion
We have established a novel, three-protein biomarker panel that is able to detect patients with early stage pancreatic cancer in urine specimens.
Keywords: Pancreatic Ductal Adenocarcinoma, Diagnostic Biomarkers, Urine, LYVE1, REG1, TFF1
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the rare cancers for which no significant improvements in diagnosis and therapy have been made in the last 30 years. Despite considerable progress in our understanding of the disease at the molecular level, novel findings have not yet translated into clinical benefit, and the majority of patients are still faced with a grim median survival of 5 to 6 months. With over 38,000 PDAC-related deaths in the US and over 40,000 in Europe in 2013, this malignancy is currently the fourth leading cause of cancer-related death, but predicted to become the second by 2030 (1), (2), (3).
These worrying figures would change significantly with improved tool(s) for early detection, as 5-year survival approaching 70% has been reported after incidental diagnosis of stage I PDAC tumours, when they were still confined to the pancreas with a size <2 cm (4), (5), (6). Detection at an early stage is also crucial given the poor efficacy of current therapies for metastatic disease, when potentially curative surgery is no longer feasible.
Timely detection of PDAC is, however, hampered by several factors: lack of specific clinical symptoms in the early stage of the disease, insufficient sensitivity of current imaging modalities and, despite intensive efforts, lack of accurate body fluid-based biomarkers of early-stage disease (for review see (7)). Early stage PDAC is also difficult to differentiate from chronic pancreatitis (CP), a benign inflammatory disease of the pancreas and one of the risk factors for PDAC (8). Serum CA19.9, the only PDAC biomarker in widespread clinical use, suffers from false negative results in patients with Lewis-negative genotype, low sensitivity (79%-81%) in symptomatic patients, and its levels may be elevated in various other benign and malignant pancreatic and hepato-biliary diseases, as well as in unrelated cystic and inflammatory diseases (for review see (9)).
After a successful proof of concept study where we showed that the protein signatures of CP and PDAC can be identified in urine (10), we here describe the development of a three-biomarker panel that can detect early stage PDAC completely non-invasively, through analysis of urine samples.
Materials and Methods
Clinical specimens
Healthy, CP and PDAC (n=6/each group) urine specimens were obtained from the Royal London Hospital (RLH) and used for the discovery phase. For validation purposes, a total of 371 urine specimens (87 H, 92 CP and 192 PDAC urine samples from RLH and University College London (jointly referred to as ‘LON’), the Department of Surgery, Liverpool University (‘LIV’), and the CNIO Madrid, Spain (‘SPA’) were assayed. Demographics and clinical characteristics of patients and healthy participants included in the study are shown in Table 1. Additional 117 urine samples from patients with other benign and malignant hepatobiliary pathologies (33 from patients with intraductal papillary mucinous tumours (IPMNs) without associated adenocarcinoma, 18 from pancreatic neuroendocrine tumours (NETs), 16 from duodenal cancers (DuCa), 26 from ampullary cancers (AMP), and 24 from patients with cholangiocarcinomas (CHL)) were obtained from LIV (demographic details are provided in Supplementary Figure S5A). Matching plasma samples for measuring CA19.9 were available from RLH and LIV. Healthy individuals had no known pancreatic conditions or malignancies, and all samples were derived from individuals with no history of renal diseases. Dipstick test analysis (Bayer multistix SG 08935414) was also performed to exclude potential bilirubinemia, proteinuria, bacterial contamination and hematuria. The specimens in all participating centres were collected using the same standard operating procedures: clean-catch, midstream urine was collected, frozen within 2 hours of collection and stored at −80°C until utilized; samples were collected before any surgery or chemotherapeutic treatment. All samples were collected with full ethical approval from the involved centres, and with informed consent from all individuals who donated urine/blood samples.
Table 1. Demographics and clinical characteristics of the healthy and patient cohorts.
| Normal |
CP |
PDAC |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases (n) |
Gender | Age range (Median) |
Cases (n) |
Gender | Age range (Median) |
Cases (n) |
Gender | Age range (Median) |
Stage /n plasma$ |
|
| LON | 87 | M=46 F=41 |
28 - 87 (55) |
45 | M=32 F=13 |
29 – 82 (53) |
60 | M=38 F=22 |
29 – 82 (64) |
I=4/4 IIA=1/1 IIB=13/13 III=33/30 IV=6/5 U=3/3 |
| Plasma (CA19.9) |
28 | M=16 F=12 |
28-67 (46) |
19 | M=14 F=5 |
29-74 (54) |
56 | M=34 F=22 |
29-82 (64) |
|
|
| ||||||||||
| LIV | 0 | N/A | N/A | 41 | M=25 F=16 |
29 – 82 (51) |
91 | M=53 F=38 |
39 - 83 (68) |
I=3/3 IIA=8/8 IIB=42/42 III=38/38 IV=0/0 U=0/0 |
| Plasma (CA19.9) |
0 | N/A | N/A | 31 | M=17 F=14 |
37-73 (51) |
91 | M=53 F=38 |
39-83 (68) |
|
|
| ||||||||||
| SPA | 0 | N/A | N/A | 6 | M=4 F=2 |
54 – 68 (57) |
41 | M=23 F=18 |
43 – 94 (72) |
I=0/NA II=0/NA III=0/NA IV=0/NA U=41/NA |
| Plasma (CA19.9) |
0 | N/A | N/A | 0 | N/A | N/A | 0 | N/A | N/A | |
|
| ||||||||||
| Total | 87 | M=46 F=41 |
28 - 87 (55) |
92 | M=61 F=31 |
29 – 82 (54) |
192 | M=114 F=78 |
29 – 94 (68) |
I=7/7 IIA=9/9 IIB=55/55 III=71/68 IV=6/5 U=44/3 |
| Plasma (CA19.9) |
28 | M=16 F=12 |
28-67 (46) |
50 | M=31 F=19 |
29-74 (53) |
147 | M=87 F=60 |
29-83 (67) |
|
|
| ||||||||||
|
Total urine = 371
Total plasma = 147 | ||||||||||
number of cases with plasma CA19.9
GeLC-MS/MS (SDS-PAGE-Liquid Chromatography-Tandem Mass Spectrometry) analysis of urine proteomes
Six urine samples (three males and three females) for each group (H, CP and PDAC; in total 18 samples) were utilised: H males/females age 45,50,60/44,45,54 years; CP males/females age 46,48,51/47,69,74 years; PDAC males/females age 44,74,84/71,73,77 years; male PDAC stage all IIB/female two IB, one IIA. All urine samples were desalted and concentrated as described previously (10). 20μg of each pre-processed pool of three samples per group were separated in duplicate on 4-12% mini-gels (Invitrogen); female and male urines were analyzed separately. The gels were stained with Colloidal Coomassie, and each sample lane cut using a grid into 40 equally sized slices. Gel slices were digested robotically with trypsin and resultant peptides analyzed by nano LC/MS/MS using a nanoAcquity (Waters) interfaced to a LTQ Orbitrap XL tandem mass spectrometer (ThermoFisher). Product ion data were searched against the human IPI protein database using Mascot, and subsequently parsed into the Scaffold software (Proteome Software) for collation into non-redundant protein lists. Reversed database searching was used to assess false discovery rates, the target protein FDR being <0.5% per sample. A semi-quantitative assessment of relative protein abundance between PDAC, CP and Healthy samples was obtained using the spectral counting approach (11).
Urine biomarkers and CA19.9 measurements
Total protein concentration in urines was determined by Bradford assay (Coomassie Protein Assay Reagent, Pierce). The quantitative ELISA determination of human LYVE-1 (Cat# SEB049Hu, Uscn Life Science Inc.) and human TFF1 (Cat# ELH-LYVE1-001, RayBiotech Inc.) was performed according to the manufacturer’s instructions; human REG1A levels were initially assessed in our laboratory, and afterwards by BioVendor Analytical Testing Service (BioVendor - Laboratorní medicína a.s). Calibration curves were prepared using purified standards for each protein assessed. Curve fitting was accomplished by a four-parameter logistic regression following the manufacturer’s instructions. The limits of detection and the coefficient of variation (CV) for each of the ELISA assays were as follows: LYVE-1 - 8.19 pg/ml, intra-assay CV - 9%, inter-assay CV - 12%, TFF1 - 0.037 ng/ml, intra-assay CV -9%, inter-assay CV - 12%. REG1A - 0.094 ng/ml, intra-assay CV - 9%, inter-assay CV 20%; REG1B- 3.13 pg/ml, intra-assay CV - 3.9%, inter-assay CV - 2.7%. Urine creatinine was measured by the Jaffé method using the Roche Cobas 8000 system (Roche Diagnostics, Mannheim, Germany) and plasma CA19.9 using a Roche Modular E170 instrument according to the routine protocols at the Clinical Biochemistry Laboratory, RLH (London, UK).
Tissue microarrays and Immunohistochemistry (IHC)
The details of the tissue microarray and scoring procedure used in evaluating the expression of the biomarkers was described previously (12). IHC was performed with anti-REG1A (Abcam, Rabbit polyclonal, ab47099, 1:100 dilution), anti-TFF1 (Abcam, Rabbit polyclonal, ab50806, 1:100 dilution), and anti-LYVE1 (Acris, Rabbit polyclonal, DP3500PS, 1:100 dilution) antibodies using the Ventana Discovery system, according to standard protocols (sCC1, 1h incubation).
Statistical analysis
To identify potential urine biomarkers from the MS data, the statistical analysis was performed on the normalized data (based on the sum of spectral counts/sample) using Arraytrack software (http;edkb.fda.go/webstart/arraytrack) by t-test. The data were further filtered according to both p values and fold change between any two sample groups, separately in males and females.
The concentrations of the selected proteins (LYVE1, REG1A and TFF1) obtained by ELISA assays were compared using nonparametric Kruskal-Wallis tests. To adjust for inflated type I error due to multiple testing between PDAC patients and control groups (healthy and CP), i.e. 27 comparisons, the significance threshold for p-values was adjusted using the conservative Bonferroni correction, i.e. a threshold of α(0.05)/27 = 0.0018 was used to define a significant result at the 5% level after adjustment for multiplicity.
Correlation between the three biomarkers was assessed using Pearson’s correlation coefficient.
Each individual biomarker and the panel were investigated for their ability to discriminate between PDAC patients (all stages, or early stages I-II) and control samples (healthy and CP) using ROC analysis and a hold-out approach. For each comparison, 70% of the subjects in the patient and control datasets were randomly selected for inclusion in the training dataset. Logistic regression was then applied. All protein concentration data were natural log-transformed and mean-centered prior to regression analysis. In individual biomarker analyzes, creatinine-normalised data were used to correct for the urine dilution factor; for the panel analysis, the model included the three biomarkers (prior to creatinine normalisation) and was adjusted for creatinine and age (as the median age of PDAC patients was higher than that of healthy and CP individuals, Table 1), i.e. 5-parameter model. Separate models were applied to the training datasets for the comparison of PDAC all stages versus healthy, PDAC stages I-II versus healthy, PDAC all stages versus CP and PDAC stages I-II versus CP. ROC curves were generated for each of the above regression models; the area under the curve (AUC), and the sensitivity (SN) and specificity (SP) at the ‘optimal’ cut-point for discrimination between groups were obtained. The optimal cut-point corresponded to the point closest to the top-left part of the plot in the ROC plane (coordinates 0,1) with optimal SN and SP according to the following criterion:
as calculated by the ‘ci.threshold’ procedure of the R ‘pROC’ package (13). This approach has been shown to have good performance in the estimation of the optimal cut-point of a biomarker (14).
The rest of the subjects (30%) formed independent datasets which were used for model validation. For the primary analysis (all PDAC versus Healthy), 49 PDAC and 28 healthy samples give more than 90% power to detect a standardized difference of 1.0 (i.e. a difference between PDAC and healthy samples of at least one standard deviation) using a one-sided test.
Validation was performed by classifying each sample in the validation dataset according to the logistic regression model developed based on the training dataset, and comparing this classification with the actual diagnosis, hence deriving a new ROC curve. The optimal cut-points computed for the training sets were used to derive the SN and SP of the validation dataset. Confidence intervals (CI, 95%) for AUCs were derived based on DeLong’ asymptotically exact method to evaluate the uncertainty of an AUC (15); SN and SP, 95% CI were derived using non-parametric stratified resampling with the percentile method (2,000 bootstrap replicates) as described by (16). AUCs were compared using DeLong’s 1-sided test for correlated/paired AUCs (15).
For exploratory analyzes, ROC curves were derived for the comparison of PDAC stage I-IIA versus healthy or CP based on logistic regression modelling using all available samples.
ROC curve analyzes were performed in R version 2.13.0 (The R Foundation for Statistical Computing, http://www.r-project.org/foundation) using procedures from the Epi (14), pROC (13) and ROCR (17) packages.
Results
Urine proteomes
We undertook an in-depth proteomics analysis by GeLC/MS/MS of 18 urine specimens derived from PDAC, CP and healthy (H) individuals (6 per group, three males, three females) (Supplementary Figure S1A). This analysis resulted in the identification of around 1,500 (1,198 in male and 1,061 in female urine) non-redundant proteins (Supplementary Table 1). These proteins originated from all cellular compartments and were mapped using IPA (Ingenuity pathway analysis, http://www.ingenuity.com/) to a number of cellular functions and diseases (Supplementary Figure S1B and C, respectively), confirming that urine provides a rich source of diverse proteins with respect to their origin and functional roles.
Our MS analysis was performed separately on urine samples from male and female subjects. We noticed considerable gender-specific differences: of 997 proteins identified in healthy urine samples, 398 (40%) were unique to male urines, 118 (12%) were unique to female and 481 (48%) were common to both.
Three proteins commonly deregulated in both males and females: LYVE1, REG1A and TFF1, were selected for further evaluation based on the statistics (t-test, p-values <0.05; fold change >1.5), interrogation of Pancreatic Expression Database (http://www.pancreasexpression.org/) (18) and additional literature search for previous knowledge on the potential candidates, and also on the availability of commercial ELISA assays. While REG1B in our proteomics data appeared to be slightly better candidate, only REG1A ELISA assay was available commercially at the time. However, when REG1B ELISA became available, we tested a subset of urine samples and similar results were obtained (see later). The presence of our three selected biomarkers as full-size proteins: 35kDa for LYVE1, 19kDa for REG1A, and 9kDa for TFF1 in urine specimens was confirmed by Western blot (data not shown).
Biomarker panel in detecting PDAC
The selected biomarkers were subsequently assessed using ELISA assays on 371 urine samples collected from three centres: London and Liverpool, UK, and Madrid, Spain. Demographics and clinical characteristics of patients and healthy participants included in the study are shown in Table 1.
PDAC stage I-IV versus healthy
The ELISA analysis showed significantly higher urine concentrations for each of the candidate biomarkers in the urine of PDAC patients (n = 192) when compared to healthy samples (n = 87, all with p<0.0001, Figure 1). Of note, REG1B and REG1A ELISA assays produced similar results (Figure 1).
Figure 1. Urine concentration of the candidate protein biomarkers.
A, Scatter dot plots of LYVE1, REG1A, REG1B and TFF1 protein concentration (creatinine-normalised) analyzed by ELISA in healthy, chronic pancreatitis (CP) and pancreatic adenocarcinoma (PDAC) patients’ urine. Upper bars: Kruskal-Wallis test; ****: p<0.0001; ***: p<0.001. B, Statistical summary. Median and Interquartile range (IQR) of raw/creatinine-normalised data for the biomarkers, median and IQR of urine creatinine (mmol/L), as well as plasma CA19.9 (U/mL) by sample groups are shown.
In PDAC, LYVE1, REG1A and TFF1 were positively correlated with each other, while in healthy samples, only LYVE1 and REG1A were correlated (Supplementary Figure S2).
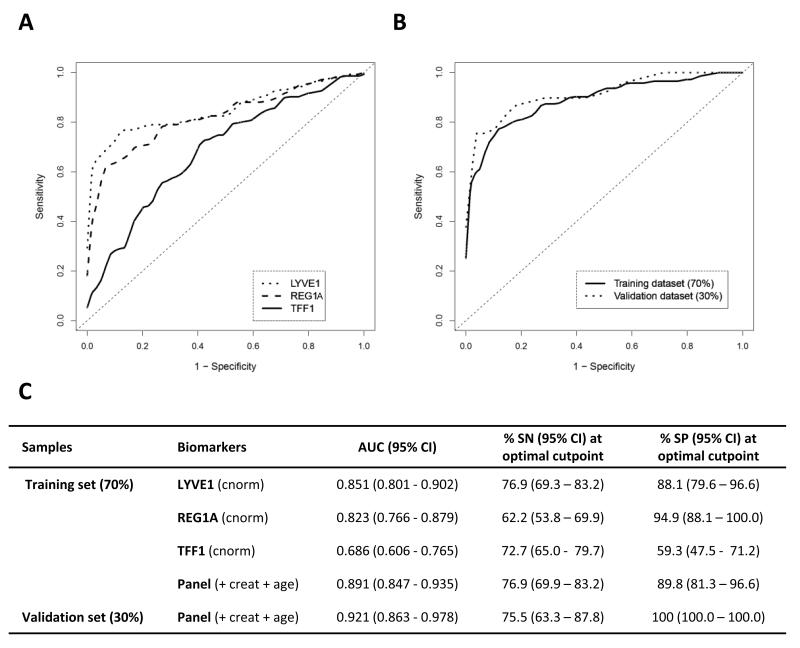
The diagnostic performance of LYVE1, REG1A and TFF1 was established using Receiver Operating Characteristic (ROC) curve analysis (Figure 2). We first assessed their individual performance in discriminating between PDAC stage I-IV and healthy urines in a training dataset (70% of the samples, n = 143 and n = 59, respectively). Individual (creatinine-normalised) urine biomarkers were able to discriminate between the two groups with AUC values of 0.851 (95%CI 0.801-0.902) for LYVE1, 0.823 (95%CI 0.766-0.879) for REG1A and 0.686 (95%CI 0.606-0.765) for TFF1, with respective SN of 76.9% (95%CI 69.3–83.2), 62.2% (95%CI 53.8–69.9) and 72.7% (95%CI 65.0- 79.7), and respective SP of 88.1% (95%CI 79.6–96.6), 94.9% (95%CI 88.1–100.0), and 59.3% (95%CI 47.5-71.2) (Figure 2A, C). The three biomarkers were then combined into a panel adjusted for creatinine and age (Figure 2B). The results of the logistic regression model underlying the ROC analysis in the training and validation (30% of the samples, PDAC n = 49, healthy n = 28) datasets are shown in Figure 2B and C. The panel achieved SN >75% and SP >85% for AUCs of 0.891 (95% CI 0.847-0.935) and 0.921 (95% CI 0.863-0.978) in the training and validation datasets, respectively, thus showing better performance than any of the individual biomarkers.
Figure 2. Diagnostic performance of urine biomarkers in discriminating pancreatic adenocarcinoma patients from healthy controls.
A, ROC curves of PDAC (n=143) versus healthy (n=59) subjects for individual creatinine-normalised urine biomarkers in the training set (70% of the data); B, ROC curves of PDAC versus healthy for the panel in the training set and in the independent validation set (30% of the data: PDAC n=49, healthy n=28); C, Summary table. AUC: area under the curve, SN: sensitivity, SP: specificity, with corresponding 95% Confidence Intervals (CI). SN and SP in the validation set are derived for optimal cutpoint determined in the training dataset. cnorm: creatinine-normalised, creat: creatinine.
PDAC early stages versus healthy
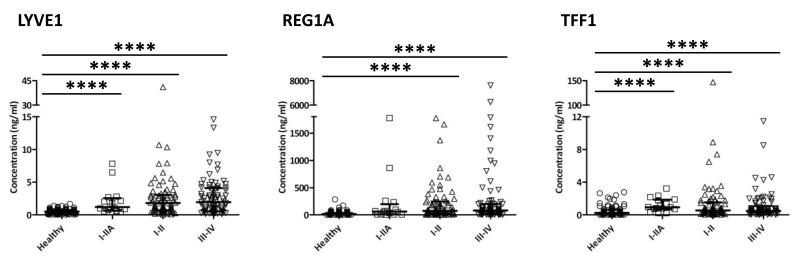
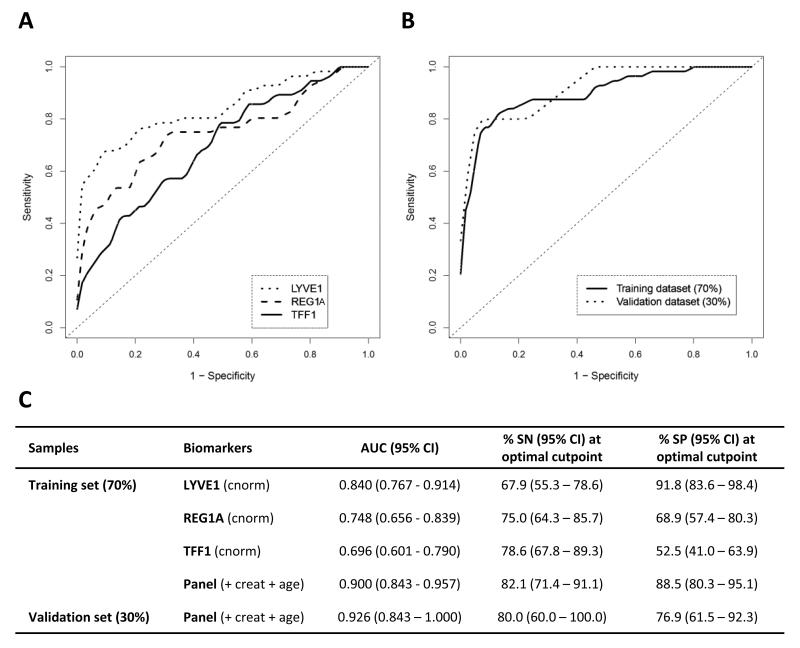
Next, we assessed the performance of our biomarkers in discriminating early stage cancers from healthy individuals. Tumour staging information was available for 148 (77%) of the PDAC patients. The concentrations of each of the biomarkers were significantly increased in later stages (stage III-IV, n = 77, all p<0.0001), in stages I-II (n = 71, all p<0.0001) and in stages I-IIA (locally invasive disease without lymph node metastases, n=16, p<0.0001, except for REG1A p = 0.007) compared to healthy people (n = 87) (Figure 3). The performance of the individual markers and the panel in discriminating between PDAC stage I-II from healthy urines was first assessed in a new training dataset (70% of the samples; PDAC stage I-II n = 56 and healthy n = 61, respectively). A new 5-parameter model was built using this training dataset and validated using the rest of the data (30% of the samples; PDAC stage I-II n = 15, Healthy n = 26) (Figure 4A, B). The panel achieved AUCs of 0.900 (95% CI 0.843-0.957) and 0.926 (95% CI 0.843-1.000) in the training and validation datasets, respectively (Figure 4C). Therefore, our urine biomarker panel can differentiate early PDAC from healthy samples with high accuracy.
Figure 3. Urine concentration of the three biomarkers in different stages of pancreatic adenocarcinoma.
Scatter dot plots of urine LYVE1, REG1A, TFF1 protein concentration (creatinine-normalised) in urines of healthy (n=87) and pancreatic adenocarcinoma patients at different stages of disease development (I-IIA n=16, I-II n=71, III-IV n=77). Bars indicate median and IQR values. Upper bars: Kruskal-Wallis test; ****: p<0.0001.
Figure 4. Diagnostic performance of urine biomarkers in discriminating early pancreatic adenocarcinoma patients form healthy individuals.
A, ROC curves of stages I-II PDAC (n=56) versus healthy (n=61) subjects for individual urine biomarkers in the training set (70% of the data); B, ROC curves of stage I-II PDAC versus healthy for the panel in the training set and in the independent validation set (30% of the data; PDAC n=15, healthy n=26); C, Summary table. AUC: area under the curve, SN: sensitivity, SP: specificity, with corresponding 95% Confidence Intervals (CI). SN and SP in the validation set are derived for optimal cutpoint determined in the training dataset. cnorm: creatinine-normalised, creat: creatinine.
As an exploratory analysis, we then selected the urine samples from individuals for which matched plasma samples were available so CA19.9 values could be obtained. The ROC curves were derived for plasma CA19.9 (as a categorical variable with a cut-off at clinically established threshold of 37 U/mL), the panel, and a combination of the panel and CA19.9. For the comparison of PDAC stage I-II (n=71) versus healthy (n=28) samples, we obtained AUCs of 0.880 (95%CI 0.947-0.999) for CA19.9, and 0.973 (95%CI 0.947-0.999) for the panel, which was significantly greater than plasma CA19.9 alone (p=0.005). The addition of plasma CA19.9 to the panel significantly increased the AUC to 0.991 (95%CI 0.979-1.000, p=0.04, Figure 5A/C). When PDAC stage I-IIA (n=16) were compared to healthy samples, AUCs were 0.839 (95%CI 0.719-0.959) for CA19.9, and 0.971 (95%CI 0.929-1.000) for the panel (p=0.006). The addition of plasma CA19.9 to the panel did not result in any improvement (AUC=0.969, 95%CI 0.924-1.000, p=0.7, Figure 5B/C).
Figure 5. Exploratory comparison of plasma CA19.9 and the urine biomarker panel in discriminating early pancreatic adenocarcinoma patients form healthy individuals.
A, ROC curves of the biomarker panel with corresponding plasma CA19.9 alone and in combination comparing healthy urine (n=28), and urines from PDAC stages I-II, n=71 and I-IIA, n=16 (B). C, Summary table. AUC: area under the curve, SN: sensitivity, SP: specificity with 95% Confidence Interval (CI). SN and SP in the validation set were derived for optimal cutpoint determined in the training dataset.
Biomarker panel in differentiating PDAC from CP
PDAC stage I-IV versus CP
Urine concentration for all three biomarkers was higher in PDAC (n = 192) compared to CP samples (n = 92, p<0.0001 for LYVE1 and REG1A and p = 0.0002 for TFF1, Figure 1) and as for PDAC, the biomarker concentrations were positively correlated with each other in the CP data (Supplementary Figure S2). In the training dataset (PDAC n = 143, CP n = 62) LYVE1 and REG1A were able to discriminate between the two groups with SN of 77-78% and SP of 66-69% (respective AUC values of 0.775 (95%CI 0.704 - 0.846) and 0.722 (95%CI 0.643 - 0.801, Supplementary Figure S3), while the SP of TFF1 only reached 50% for a similar SN. Combining the three biomarkers into a panel only improved marginally the performance of LYVE1 and REG1 alone as assessed in the training (AUC= 0.815, 95%CI 0.752-0.878), and validation (PDAC n = 49, CP n =30, AUC= 0.839, 95%CI 0.751-0.928) datasets.
PDAC early stages versus CP
Biomarker urine concentrations were significantly increased in stages I-II PDAC (n = 71) compared to CP (n = 87, with p<0.0001 for LYVE1 and REG1A and p = 0.0001 for TFF1, data not shown). The panel achieved high SN (>85%) in both the training (PDAC stage I-II n = 56, CP n = 66) and validation (PDAC stage I-II n = 15, CP n = 26) datasets, but relatively low SP (66.7% and 50%), similar to the SP observed for individual biomarkers, with respective AUCs of 0.831 (95%CI 0.762-0.901) and 0.846 (95%CI 0.730-0.963, Supplementary Figure S3D-F).
As before, we explored the panel in combination with plasma CA19.9. For the comparison of PDAC stage I-II (n=71) versus CP (n=50) samples, the ROC curves showed AUCs of 0.775 (95%CI 0.699-0.852) for CA19.9, 0.830 (95%CI 0.759-0.902) for the panel (p=0.1), and 0.885 (95%CI 0.825-0.945) for the panel in combination with CA19.9 (p=0.01 for superiority over the panel alone) (Supplementary Figure S4A/C). In the comparison of PDAC stage I-IIA (n = 16) versus CP, the ROC curves showed AUCs of 0.735 (95%CI 0.609-0.861) for CA19.9, 0.871 (95%CI 0.770-0.972) for the panel (p=0.004 for superiority over plasma CA19.9), and 0.866 (95%CI 0.749-0.984) for the combination (p=0.6) (Supplementary Figure S4B/C). Therefore, the panel performed better in differentiating stage I-IIA from CP than CA19.9.
Biomarker expression in urine of other hepatobiliary pathologies
We explored the expression of our biomarkers in urine specimens collected from patients with several other benign or malignant hepatobiliary pathologies and compared it to the expression in patients with early stage PDAC (Supplementary Figure S5). Levels of LYVE1 tended to be higher in PDAC stage I-II samples compared to IPMNs, AMP and pancreatic NETs specimens, while REG1A levels were only increased in comparison to IPMNs. Plasma CA19.9 levels were higher in PDACs stage I-II compared to pancreatic NETs and DuCA samples. This might suggest a potential utility for LYVE1 and REG1A in distinguishing other benign or malignant hepatobiliary pathologies from early stage PDACs.
Tissue origin of the three biomarkers
Having demonstrated a good performance of the panel in differentiating early cancer patients from healthy individuals, we next sought to establish the expression of our biomarkers in pancreatic tissue. We performed immunohistochemistry (IHC) using in-house constructed PDAC tissue microarrays. A strong expression of REG1A was seen in histologically normal adjacent acinar cells, but the staining was also seen in 44/60 tumours (73%) (Supplementary Figure S6A). TFF1 was absent in normal pancreas, but was expressed in 43/60 (72%) of PDACs (Supplementary Figure S6B). While no LYVE1 expression was seen in any of the cancer cells, it was seen in lymphatic vessels in eight PDAC tissues (Supplementary Figure S6C). The lack of LYVE1 immunoreactivity in the remaining cases, however, might be due to underrepresentation of relevant peripheral regions in our tissue microarray.
Next, we measured the levels of all three biomarkers in urines from seven PDAC patients for whom samples were collected prior to and after surgery (Supplementary Figure S6D). In all patients, levels of LYVE1 and REG1A decreased after surgery, and this was also seen in six out of seven patients for TFF1 (except for Patient 2, where the first post-surgical urine sample was collected four months after the procedure), likely due to substantial loss of tumour mass after surgery.
Discussion
PDAC is one of the most challenging cancers to detect; the majority of patients thus present at an advanced stage of the disease. Hence less than 20% of PDAC patients undergo potentially curative surgery, while the remainder can only be offered palliative treatment.
Here, we describe a three-biomarker urine panel that discriminates early stage PDAC patients from healthy subjects with high accuracy. We chose to develop a diagnostic test based on urine specimens, as this body fluid has several advantages over blood: it is far less complex, provides an ‘inert’ and stable matrix for analysis, and can be repeatedly and non-invasively sampled in sufficient volumes. So far, more than 2,300 proteins have been detected in urine (19), of which at least a third are of a systemic origin (20). As an ultrafiltrate of blood, it can be expected that at least some of the biomarkers will be found in higher concentration in urine than in blood.
Interestingly, when exploring the proteomes of healthy male and female urines, similarly as indicated previously (21), we observed large differences, indicating the need for the systematic exploration of gender-specific differences in biofluids.
Of the three biomarkers in our panel, REG1A and TFF1 have already been associated with PDAC. The REG1A gene product belongs to a family of REG (regenerating) glycoproteins, which are expressed in pancreatic acinar cells and acts as both autocrine and paracrine growth factor (22), (23). The REG protein(s) are expressed during islet cell regeneration (24), (25), but are also potentially associated with differentiation and maintenance of an exocrine phenotype (26), (27). REG1 (non-specified) expression has been reported previously in about 25% of PDACs (n=20) (28), while we observed strong expression of REG1A in 73% of the cases examined here. REG1A and REG1B proteins are almost 90% identical (29), and are difficult to distinguish. REG1A levels have previously been observed in the plasma of mice with PanIN (Pancreatic intraepithelial neoplasia) lesions compared to control mice (30), and REG1B has recently been reported as one of the four candidate serological biomarkers that can improve the performance of CA19.9 in PDAC (31). Both proteins were able to significantly differentiate healthy from PDAC urine samples, demonstrating thus for the first time their high discriminatory power in urine specimens. TFF1 belongs to a family of gastrointestinal secretory peptides, which interact with mucins and are expressed at increased levels during reconstitution and repair of mucosal injury. They protect epithelial cells from apoptotic death and increase their motility, but also play similar pivotal roles in cancer cells, and are thus involved in the development and progression of various cancer types (32), (33). In PDAC, TFF1 has been reported in both sporadic (34), (35) and familial PanINs (36), and it has been associated with early stages (I and II) of the disease and disease without LN involvement, i.e. stage IIA (37). This expression mimics the pattern seen in our urine samples, and confirms the biological importance of TFF1 as an early diagnostic biomarker. Transcripts of all three members of the family, and in particular TFF3, have been observed in the urinary tract of healthy subjects, but only TFF2 peptide was excreted in the urine (38). The increased TFF1 levels in urine samples from our cancer patients and its sharp decrease after surgical removal of the tumour further suggest that TFF1 protein originates from PDAC. LYVE1 (lymphatic vessel endothelial hyaluronan receptor) binds hyaluronan (HA), an extracellular matrix mucopolysaccharide, and transports it across the lymphatic vessel wall, particularly in the lymph nodes, a site of HA degradation (39). Most studies utilise LYVE1 as a marker of lymphatic endothelium in the context of lymphangiogenesis, however, markedly different findings regarding active lymphangiogenesis across different tumour types exist (40). No functional lymphangiogenesis is seen intratumourally in PDAC (41), (42), which may explain the infrequent presence of LYVE1 immunoreactive lymphatic vessels in our IHC analysis. It is also possible that LYVE1 expression was scarce due to the under-representation of extratumoral tissues within our case series, similarly to the findings of Carreira et al, who reported a complete absence of LYVE1-positive lymphatic vessels in 25 hepatocellular carcinomas and 17 metastatic adenocarcinomas, but detected LYVE1 in the regions further away from the tumour nodules (43). LYVE1 showed potential also for differentiating IPMN, ampullary cancer and NET from early stage PDAC samples; however, its role in PDAC still needs to be established.
When combined, REG1A, TFF1 and LYVE1 form a powerful urinary panel that can detect patients with stages I-II PDAC, with over 90% accuracy. To our knowledge, a panel with such performance has not been reported as yet. Our exploratory analyzes also suggest that when combined with CA19.9, accuracy may be increased, which may prove important in light of recent finding that serum CA19.9 is up-regulated up to two years prior to PDAC diagnosis (44). In addition, the panel may prove useful in discriminating patient in stages I-IIA from healthy ones, although this will need to be confirmed in a larger, independent study.
The strength of our study is manifested in multiple ways: the three biomarkers were discovered by MS analysis, validated using an independent technique (ELISA assays) and examined in urine specimens that originated from three different centres. This may facilitate the successful ‘portability’ of our biomarker panel in future follow-up studies.
The diagnostic performance of our biomarker panel now needs to be further validated: our healthy controls were younger on average than our cancer patients; an older control group would thus be more relevant. In addition, further comparison of the performance of urine markers with CA19.9 is needed. Finally, it is now essential to establish if/how early in the latency period our panel can detect PDAC, as soon as such a longitudinally collected urine cohort becomes available.
The high-risk groups for developing PDAC represent the priority cohort for screening strategies. These include pancreatic cancer families (FPC) with at least two affected first-degree relatives and individuals with hereditary syndromes with known underlying gene abnormalities, such as hereditary intestinal polyposis syndrome Peutz-Jeghers (STK11/LKB1), FAMMM, familial atypical multiple mole and melanoma (p16/CDKN2A), and hereditary pancreatitis (PRSS1/SPINK1) (for a recent review see (45)). While we have previously reported deregulation of TFF1 and several REG genes in PanINs from FPC tissues (36), it is now critical to test LYVE1, and the panel as a whole, directly on urines collected from such high-risk individuals. This would best be performed within the framework of already established surveillance programmes (46), in research setting. A detailed recommendation on management of individuals at high risk was recently reported by the Cancer of the Pancreas Screening (CAPS) consortium (47). We propose to include a urine test based on our three biomarkers into the pre-defined algorithms of the surveillance protocols to ultimately validate its performance. Ease of sampling and repeated testing using the urinary panel might also help shed some light on currently somewhat conflicting (but not mutually exclusive) data on the timescale of PDAC progression (48), (49).
It would also be worthwhile to explore the use of our urinary panel in clinical decision-making in individuals with various environmental exposures (smoking, obesity, new onset type II diabetes) which carry an increased risk of developing PDAC that could be predicted in primary care using models such as QCancer (Pancreas) (50) or derived from the UK THIN database (51), but for which a consensus on screening has yet to be reached. Similarly, as shown in our study, urinary panel testing may prove useful and possibly superior to plasma CA19.9, in identifying cancer patients among patients with CP, another risk factor for developing PDAC (8). This performance should be further strengthened with additional marker(s) that could be selected from our proteomics analysis.
Being completely non-invasive and inexpensive, this urine screening test could, upon further validation, and when coupled with timely surgical intervention, lead to a much improved outcome in patients with high-risk of developing pancreatic adenocarcinoma.
Supplementary Material
Statement of translational relevance.
Currently, 80% of patients diagnosed with pancreatic adenocarcinoma present with locally invasive and/or metastatic disease, resulting in a poor 5-year survival of <5%. The development of a diagnostic tool for early detection of patients with this malignancy may significantly impact their prognosis. We established a panel of three urine biomarkers that can distinguish patients with early stage disease from healthy people, which could enable completely non-invasive and inexpensive screening of patients at high risk of developing pancreatic adenocarcinoma.
Acknowledgements
We thank Chipo Chitsenga for help with sample collection. We are indebted to all the patients and healthy donors, without whom this study would not be possible. We also acknowledge the advice from Bendix Carstensen on the use of his Epi package.
Financial support: The study was funded by Pancreatic Cancer Research Fund (PCRF) and Queen Mary Innovation (TCJ), Cancer Research UK award 18975 (NL & TCJ), The NIHR Liverpool Pancreas Biomedical Research Unit (JN, EC, WG), The NIHR UCLH Biomedical Research Centre and NIH P01 CA084203 (SPP), and #PI09-02102, #PI12-00815, RD12/0036/0034 and RD12/0036/0050 from Instituto de Salud Carlos III, Madrid, Spain (FXR, NM). HMK, SPP, FXR, JN, EC, WG, TCJ and NM are members of an EU Pancreas - BM1204 COST Action (http://www.eupancreas.com).
Footnotes
Disclosure: No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Malvezzi M, Bertuccio P, Levi F, La Vecchia C, Negri E. European cancer mortality predictions for the year 2013. Ann Oncol. 2013;24:792–800. doi: 10.1093/annonc/mdt010. [DOI] [PubMed] [Google Scholar]
- 3.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 4.Tsuchiya R, Noda T, Harada N, Miyamoto T, Tomioka T, Yamamoto K, et al. Collective review of small carcinomas of the pancreas. Ann Surg. 1986;203:77–81. doi: 10.1097/00000658-198601000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furukawa H, Okada S, Saisho H, Ariyama J, Karasawa E, Nakaizumi A, et al. Clinicopathologic features of small pancreatic adenocarcinoma. A collective study. Cancer. 1996;78:986–90. doi: 10.1002/(SICI)1097-0142(19960901)78:5<986::AID-CNCR7>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 6.Shimizu Y, Yasui K, Matsueda K, Yanagisawa A, Yamao K. Small carcinoma of the pancreas is curable: new computed tomography finding, pathological study and postoperative results from a single institute. J Gastroenterol Hepatol. 2005;20:1591–4. doi: 10.1111/j.1440-1746.2005.03895.x. [DOI] [PubMed] [Google Scholar]
- 7.Kaur S, Baine MJ, Jain M, Sasson AR, Batra SK. Early diagnosis of pancreatic cancer: challenges and new developments. Biomark Med. 2012;6:597–612. doi: 10.2217/bmm.12.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malka D, Hammel P, Maire F, Rufat P, Madeira I, Pessione F, et al. Risk of pancreatic adenocarcinoma in chronic pancreatitis. Gut. 2002;51:849–52. doi: 10.1136/gut.51.6.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballehaninna UK, Chamberlain RS. Serum CA 19-9 as a Biomarker for Pancreatic Cancer-A Comprehensive Review. Indian J Surg Oncol. 2011;2:88–100. doi: 10.1007/s13193-011-0042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weeks ME, Hariharan D, Petronijevic L, Radon TP, Whiteman HJ, Kocher HM, et al. Analysis of the urine proteome in patients with pancreatic ductal adenocarcinoma. Proteom Clin Appl. 2008;2:1047–57. doi: 10.1002/prca.200780164. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 12.Ene-Obong A, Clear AJ, Watt J, Wang J, Fatah R, Riches JC, et al. Activated pancreatic stellate cells sequester CD8+ T cells to reduce their infiltration of the juxtatumoral compartment of pancreatic ductal adenocarcinoma. Gastroenterology. 2013;145:1121–32. doi: 10.1053/j.gastro.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rota M, Antolini L. Finding the optimal cut-point for Gaussian and Gamma distributed biomarkers. Comput Stat Data An. 2014;69:1–14. [Google Scholar]
- 15.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 16.Carpenter J, Bithell J. Bootstrap confidence intervals: when, which, what? A practical guide for medical statisticians. Stat Med. 2000;19:1141–64. doi: 10.1002/(sici)1097-0258(20000515)19:9<1141::aid-sim479>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 17.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–1. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 18.Dayem Ullah AZ, Cutts RJ, Ghetia M, Gadaleta E, Hahn SA, Crnogorac-Jurcevic T, et al. The pancreatic expression database: recent extensions and updates. Nucl. Acids Res. 2014;42:D944–9. doi: 10.1093/nar/gkt959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kentsis A, Monigatti F, Dorff K, Campagne F, Bachur R, Steen H. Urine proteomics for profiling of human disease using high accuracy mass spectrometry. Proteom Clin Appl. 2009;3:1052–61. doi: 10.1002/prca.200900008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jia L, Zhang L, Shao C, Song E, Sun W, Li M, et al. An attempt to understand kidney’s protein handling function by comparing plasma and urine proteomes. PloS One. 2009;4:e5146. doi: 10.1371/journal.pone.0005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Shao C, Wei L, Duan J, Wu S, Li X, et al. An individual urinary proteome analysis in normal human beings to define the minimal sample number to represent the normal urinary proteome. Proteome Science. 2012;10:70. doi: 10.1186/1477-5956-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, Zheng Q, Rengabhashyam P, Zenilman ME. A Brief History of Pancreatic Reg: Implications as to its Clinical Importance. Einstein Q J Biol Med. 2000;17:178. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YW, Ding LS, Lai MD. Reg gene family and human diseases. World J Gastroenterol. 2003;9:2635–41. doi: 10.3748/wjg.v9.i12.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe T, Yonemura Y, Yonekura H, Suzuki Y, Miyashita H, Sugiyama K, et al. Pancreatic beta-cell replication and amelioration of surgical diabetes by Reg protein. Proc Natl Acad Sci USA. 1994;91:3589–92. doi: 10.1073/pnas.91.9.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parikh A, Stephan AF, Tzanakakis ES. Regenerating proteins and their expression, regulation and signaling. Biomol Concepts. 2012;3:57–70. doi: 10.1515/bmc.2011.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez D, Mueller CM, Zenilman ME. Pancreatic regenerating gene I and acinar cell differentiation: influence on cellular lineage. Pancreas. 2009;38:572–7. doi: 10.1097/mpa.0b013e3181a1d9f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zenilman ME, Perfetti R, Swinson K, Magnuson T, Shuldiner AR. Pancreatic regeneration (reg) gene expression in a rat model of islet hyperplasia. Surgery. 1996;119:576–84. doi: 10.1016/s0039-6060(96)80270-4. [DOI] [PubMed] [Google Scholar]
- 28.Kimura N, Yonekura H, Okamoto H, Nagura H. Expression of human regenerating gene mRNA and its product in normal and neoplastic human pancreas. Cancer. 1992;70:1857–63. doi: 10.1002/1097-0142(19921001)70:7<1857::aid-cncr2820700708>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez D, Figarella C, Marchand-Pinatel S, Bruneau N, Guy-Crotte O. Preferential expression of reg I beta gene in human adult pancreas. Biochem Biophys Res Commun. 2001;284:729–37. doi: 10.1006/bbrc.2001.5033. [DOI] [PubMed] [Google Scholar]
- 30.Faca VM, Song KS, Wang H, Zhang Q, Krasnoselsky AL, Newcomb LF, et al. A mouse to human search for plasma proteome changes associated with pancreatic tumor development. PLoS Med. 2008;5:e123. doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makawita S, Dimitromanolakis A, Soosaipillai A, Soleas I, Chan A, Gallinger S, et al. Validation of four candidate pancreatic cancer serological biomarkers that improve the performance of CA19.9. BMC Cancer. 2013;13:404. doi: 10.1186/1471-2407-13-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emami S, Rodrigues S, Rodrigue CM, Le Floch N, Rivat C, Attoub S, et al. Trefoil factor family (TFF) peptides and cancer progression. Peptides. 2004;25:885–98. doi: 10.1016/j.peptides.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Perry JK, Kannan N, Grandison PM, Mitchell MD, Lobie PE. Are trefoil factors oncogenic? Trends Endocrinol Metab. 2008;19:74–81. doi: 10.1016/j.tem.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Arumugam T, Brandt W, Ramachandran V, Moore TT, Wang H, May FE, et al. Trefoil factor 1 stimulates both pancreatic cancer and stellate cells and increases metastasis. Pancreas. 2011;40:815–22. doi: 10.1097/MPA.0b013e31821f6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prasad NB, Biankin AV, Fukushima N, Maitra A, Dhara S, Elkahloun AG, et al. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res. 2005;65:1619–26. doi: 10.1158/0008-5472.CAN-04-1413. [DOI] [PubMed] [Google Scholar]
- 36.Crnogorac-Jurcevic T, Chelala C, Barry S, Harada T, Bhakta V, Lattimore S, et al. Molecular analysis of precursor lesions in familial pancreatic cancer. PloS One. 2013;8:e54830. doi: 10.1371/journal.pone.0054830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sagol O, Tuna B, Coker A, Karademir S, Obuz F, Astarcioglu H, et al. Immunohistochemical detection of pS2 protein and heat shock protein-70 in pancreatic adenocarcinomas. Relationship with disease extent and patient survival. Pathol Res Pract. 2002;198:77–84. doi: 10.1078/0344-0338-00190. [DOI] [PubMed] [Google Scholar]
- 38.Jackson DG. Immunological functions of hyaluronan and its receptors in the lymphatics. Immunol Rev. 2009;230:216–31. doi: 10.1111/j.1600-065X.2009.00803.x. [DOI] [PubMed] [Google Scholar]
- 39.Rinnert M, Hinz M, Buhtz P, Reiher F, Lessel W, Hoffmann W. Synthesis and localization of trefoil factor family (TFF) peptides in the human urinary tract and TFF2 excretion into the urine. Cell Tissue Res. 2010;339:639–47. doi: 10.1007/s00441-009-0913-8. [DOI] [PubMed] [Google Scholar]
- 40.Jackson DG, Prevo R, Clasper S, Banerji S. LYVE-1, the lymphatic system and tumor lymphangiogenesis. Trends Immunol. 2001;22:317–21. doi: 10.1016/s1471-4906(01)01936-6. [DOI] [PubMed] [Google Scholar]
- 41.Sipos B, Kojima M, Tiemann K, Klapper W, Kruse ML, Kalthoff H, et al. Lymphatic spread of ductal pancreatic adenocarcinoma is independent of lymphangiogenesis. J Pathol. 2005;207:301–12. doi: 10.1002/path.1840. [DOI] [PubMed] [Google Scholar]
- 42.Schneider M, Buchler P, Giese N, Giese T, Wilting J, Buchler MW, et al. Role of lymphangiogenesis and lymphangiogenic factors during pancreatic cancer progression and lymphatic spread. Int J Oncol. 2006;28:883–90. [PubMed] [Google Scholar]
- 43.Mouta Carreira C, Nasser SM, di Tomaso E, Padera TP, Boucher Y, Tomarev SI, et al. LYVE-1 is not restricted to the lymph vessels: expression in normal liver blood sinusoids and down-regulation in human liver cancer and cirrhosis. Cancer Res. 2001;61:8079–84. [PubMed] [Google Scholar]
- 44.O’Brien DP, Sandanayake NS, Jenkinson C, Gentry-Maharaj A, Apostolidou S, Fourkala EO, et al. Serum CA19-9 is significantly upregulated up to 2 years before diagnosis with pancreatic cancer: implications for early disease detection. Clin Cancer Res. 2014;21:622–31. doi: 10.1158/1078-0432.CCR-14-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Templeton AW, Brentnall TA. Screening and surgical outcomes of familial pancreatic cancer. Surg Clin North Am. 2013;93:629–45. doi: 10.1016/j.suc.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Wada K, Takaori K, Traverso LW, Hruban RH, Furukawa T, Brentnall TA, et al. Clinical importance of Familial Pancreatic Cancer Registry in Japan: a report from kick-off meeting at International Symposium on Pancreas Cancer 2012. J Hepatobiliary Pancreat Sci. 2013;20:557–66. doi: 10.1007/s00534-013-0611-5. [DOI] [PubMed] [Google Scholar]
- 47.Canto MI, Harinck F, Hruban RH, Offerhaus GJ, Poley JW, Kamel I, et al. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut. 2013;62:339–47. doi: 10.1136/gutjnl-2012-303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu J, Blackford AL, Dal Molin M, Wolfgang CL, Goggins M. Time to progression of pancreatic ductal adenocarcinoma from low-to-high tumour stages. Gut. 2015;0:1–7. doi: 10.1136/gutjnl-2014-308653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hippisley-Cox J, Coupland C. Identifying patients with suspected pancreatic cancer in primary care: derivation and validation of an algorithm. British Journal of General Practice. 2012;62:e38–45. doi: 10.3399/bjgp12X616355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keane MG, Horsfall L, Rait G, Pereira SP. A case-control study comparing the incidence of early symptoms in pancreatic and biliary tract cancer. BMJ Open. 2014;4:e005720. doi: 10.1136/bmjopen-2014-005720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.