Abstract
Background
The filamentous fungus Neurospora crassa efficiently utilizes plant biomass and is a model organism for genetic, molecular and cellular biology studies. Here, a set of 567 single-gene deletion strains was assessed for cellulolytic activity as compared to the wild-type parental strain. Mutant strains included were those carrying a deletion in: (1) genes encoding proteins homologous to those implicated in the Saccharomyces cerevisiae secretion apparatus; (2) genes that are homologous to those known to differ between the Trichoderma reesei hyper-secreting strain RUT-C30 and its ancestral wild-type strain; (3) genes encoding proteins identified in the secretome of N. crassa when cultured on plant biomass and (4) genes encoding proteins predicted to traverse the secretory pathway.
Results
The 567 single-gene deletion collection was cultured on crystalline cellulose and a comparison of levels of secreted protein and cellulase activity relative to the wild-type strain resulted in the identification of seven hyper-production and 18 hypo-production strains. Some of these deleted genes encoded proteins that are likely to act in transcription, protein synthesis and intracellular trafficking, but many encoded fungal-specific proteins of undetermined function. Characterization of several mutants peripherally linked to protein processing or secretion showed that the hyper- or hypo-production phenotypes were primarily a response to cellulose. The altered secretome of these strains was not limited to the production of cellulolytic enzymes, yet was part of the cellulosic response driven by the cellulase transcription factor CLR-2. Mutants implicated the loss of the SREBP pathway, which has been found to regulate ergosterol biosynthesis genes in response to hypoxic conditions, resulted in a hyper-production phenotype. Deletion of two SREBP pathway components in T. reesei also conferred a hyper-production phenotype under cellulolytic conditions.
Conclusions
These studies demonstrate the utility of screening the publicly available N. crassa single-gene deletion strain collection for a particular phenotype. Mutants in a predicted E3 ligase and its target SREBP transcription factor played an unanticipated role in protein production under cellulolytic conditions. Furthermore, phenotypes similar to those observed in N. crassa were seen following the targeted deletion of orthologous SREBP pathway loci in T. reesei, a fungal species commonly used in industrial enzyme production.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-015-0297-9) contains supplementary material, which is available to authorized users.
Keywords: Neurospora crassa, Secretion, Cellulase, Cellulose, Trichoderma reesei, Lignocellulose, SREBP, Dsc E3 ligase
Background
Neurospora crassa was first described following the investigation of a mold infestation in French bakeries during the 1840s [1]. The organism can be readily isolated from the environment, often in association with burned grasses, sugar cane or trees [2, 3]. N. crassa has been a well-established model system in the laboratory for over a century [4] and as a result there are numerous tools available for its study, including a nearly complete single-gene deletion strain collection [5, 6].
Observations of enzyme activities related to the ability of Neurospora to utilize plant biomass were first reported in the 1950s [7]. A systems biology analysis of the N. crassa transcriptome and secretome associated with utilization of plant biomass and cellulose revealed a robust enzymatic response [8]. A regulon of 212 genes that are transcriptionally induced in N. crassa in the presence of a high-purity crystalline cellulose (Avicel) as compared to starvation or rich media culture conditions has been defined [9]. The secretion of 38 proteins, as identified by mass spectrometry, was also associated with these cellulosic culture conditions [8]. Almost 40% of the total secretome by weight is attributable to a single cellulolytic enzyme, cellobiohydrolase-1 (CBH-1 or NCU07340) [10]. Additional studies exploring Neurospora’s response to xylan [11] and pectin [12] have furthered our understanding of a filamentous fungus’ systematic response to plant biomass.
In the current model, under conditions of starvation, N. crassa produces low levels of hydrolytic enzymes capable of digesting a broad range of nutrient sources. When these ‘scout’ enzymes encounter plant cell wall material, specific breakdown products are generated that signal to N. crassa the presence of an utilizable carbon source [13, 14]. In the case of cellulose, the main byproducts are cellodextrins, which can induce the production of lignocellulose-degrading enzymes [15]. The N. crassa transcriptional response to cellulose is primarily controlled by the transcription factors CLR-1 (NCU07705) and CLR-2 (NCU08042) [9], which regulate the production of cellulases and accessory proteins. Studies have shown that the mis-expression of CLR-2 is sufficient, even under non-inducing conditions, for the production of cellulases and some hemicellulases [16].
The N. crassa enzymes involved in extracellular plant cell wall deconstruction must traverse the secretory pathway in order to be transported across the fungal plasma membrane. Transcription of genes encoding translocation machinery that traffic mRNA/proteins destined for the endoplasmic reticulum is significantly increased during growth of N. crassa on Avicel [9, 12], suggesting a specific role for components of the secretory pathway in the trafficking of proteins needed for plant cell wall deconstruction and utilization. In the yeast Saccharomyces cerevisiae, the secretory apparatus has been extensively characterized [17] and is considered a model for eukaryotic protein secretion. However, recent studies in the filamentous fungal species Trichoderma reesei, Aspergillus sp. and Neurospora crassa have begun to highlight not only the similarities, but also the differences in function of the secretory pathway in filamentous fungi as compared to S. cerevisiae [18–21]. As a result, investigations into protein trafficking in filamentous fungal species are needed to understand the secretion of plant biomass degrading enzymes by these multicellular organisms.
In this study, strains of N. crassa with single-gene deletions in loci thought likely to play a role in cellulase activity or secretion were assayed for protein production and enzyme activity when grown on a cellulosic substrate (Avicel). Initial work with several hyper-production deletion strains has implicated genes encoding components of the Sterol Regulatory Element Binding Protein (SREBP) pathway, which has not been previously linked to protein secretion. Deeper characterization of these mutants will enhance our knowledge of the underpinnings and mechanisms associated with the secretion of proteins involved in plant biomass deconstruction and utilization of complex carbon sources by filamentous fungi, which can be further manipulated in industrial settings to enable increased protein production.
Results
Screening of deletion strains for protein secretion and cellulase activity
Single-gene deletion strains were selected from the N. crassa full genome deletion collection available through the Fungal Genetics Stock Center [5, 6] based on four criteria. First, genes encoding proteins that potentially function in the secretory apparatus (such as SNARE proteins involved in intracellular vesicle docking and fusion, GTPases that play a role in vesicle formation, and GPI-anchored or myristoylated proteins that link to membranes) were selected based on homology to known proteins in S.cerevisiae (http://www.yeastgenome.org). This process led to the inclusion of 103 deletion strains (Additional file 1: Table S1; Category A). Second, N. crassa homologs to genes in T. reesei that varied in DNA sequence between the hyper-cellulolytic RUT-C30 strain and its parental wild isolate QM6a [22, 23] were identified. From this analysis, another 47 deletion strains were included (Additional file 1: Table S1; Category B). Third, deletion strains for known and predicted cellulases and hemicellulases of N. crassa as well as those proteins identified in the Neurospora secretome when grown on either crystalline cellulose (Avicel) or ground Miscanthus x gigantaeus [8] were included (34 deletion strains; Category C; Additional file 1: Table S1). Fourth, additional genes encoding secreted proteins were identified by evaluating the predicted Neurospora proteome via SignalP (http://www.cbs.dtu.dk/services/SignalP) and TargetP (http://www.cbs.dtu.dk/services/TargetP) algorithms. The 596 loci with high scores from both programs (SignalP score >0.5 and TargetP score = 1) were considered likely to encode proteins that traversed the secretory pathway. Deletion strains were available for 384 of these identified genes (Additional file 1: Table S1; Category D); the remaining 212 loci either had no deletions or were deposited as a heterokaryon. The combination of all four categories led to a total of 567 strains (Additional file 1: Table S1), with almost half carrying deletions in genes that encode ‘hypothetical proteins’ (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html).
Conidial suspensions from each of the 567 deletion strains listed in Additional file 1: Table S1 were randomly arrayed in deep-well, 96-well plates. For each plate, duplicate wells of the parental strain for the deletion collection (FGSC 2489 [5]), a positive control strain (ΔNCU06650—a strain previously identified as a hyper-producer of cellulases [8, 24]) and a negative control strain (ΔNCU07340 or Δcbh-1—a strain with limited cellulolytic activity due to the loss of cellbiohydrolase-1 [8]) were included. Strains were propagated on plugs of sucrose-based Vogel’s minimal medium (VMM [25]) agar in deep-well, 96-well plates and conidia harvested from these plates were used to inoculate cellulosic medium (Avicel in combination with Vogel’s salts) in deep-well, 24-well plates. After 4 days of growth, the supernatant from each culture was harvested and assayed for total protein level and enzyme activity (endoglucanase and Avicelase assays [8]).
Each 96-well plate of deletion and control strains was processed independently and in triplicate and the results of the protein and enzyme activity assays were evaluated relative to the internal control strains on each plate (FGSC 2489, ΔNCU06650, Δcbh-1). Strains that consistently ranked in the top or bottom 20% of each plate for secreted protein and cellulolytic activity were subsequently arrayed in glycerol stocks in new deep-well, 96-well plates along with the same three control strains. If available, genetically identical deletion strains in both the mat A and mat a mating type background were included. Strains were re-evaluated for secreted protein and enzyme activity levels as compared to the included control strains, again in triplicate.
In addition to culturing on Avicel, each strain that came through the primary screen was also inoculated into sucrose-based VMM to assess overall growth phenotypes. Strains that conidiated poorly, showed limited growth in VMM, did not have matching phenotypes between matA and mata strains or did not demonstrate consistent results between experimental replicates were discarded. From these secondary screens, 25 strains of interest were identified (Table 1): seven deletion strains that showed approximately double the wild-type level of secreted protein and associated cellulolytic activity (hyper-production strains) and 18 deletion strains that showed roughly less than half the amount of protein secretion and cellulolytic activity of wild type (hypo-production strains).
Table 1.
Single-gene deletion strains of N. crassa with altered cellulase production compared to the parental wild-type strain
| Phenotype | Deleted locus | Categorya | Fold changeb | Gene annotation | Conservation | S. cerevisiae homologc |
|---|---|---|---|---|---|---|
| Increased secreted cellulases | NCU00541 | B | 1.9 | Hypothetical protein | Asco | None |
| NCU01291 | D | 2.5 | Hypothetical protein | Asco | None | |
| NCU02336 | D | 1.9 | Capsule associated protein | Asco | None | |
| NCU03459 | D | 2.9 | UBA domain-containing protein Ucp14 | Asco, Basidio | DSC2 (4.7e−17)d | |
| NCU03740 | D | 4.2 | Hypothetical protein | Asco | TUL1 (1.6e−28) | |
| NCU06650 | D | 2.3 | Secretory phospholipase A2 | Asco, Gram+ | None | |
| NCU07788 | B | 1.9 | Colonial-26 | Asco | YFL052W (9.8e−16) | |
| Decreased secreted cellulases | NCU00566 | A | 0.4 | Synaptobrevin | Asco, Basidio | SNC2 (1.2e−20) |
| NCU00600 | A | 0.2 | Rho-type GTPase-3 | Asco | RHO3 (6.7e−66) | |
| NCU00761 | D | 0.6 | Triacylglycerol lipase | Asco, Basidio | None | |
| NCU01739 | D | 0.3 | Hypothetical protein | Asco, Basidio | None | |
| NCU02017 | B | 0.0 | All development altered-2 | Asco, Basidio | NCB2 (3.7e−26) | |
| NCU02152 | B | 0.4 | RRM domain-containing protein | Asco, Basidio | None | |
| NCU02182 | B | 0.7 | Hypothetical protein | Asco, Basidio | None | |
| NCU04593 | D | 0.6 | Lcc9 | Asco | FET5 (2.9e−100) | |
| NCU04952 | C | 0.2 | Glycosylhydrolase family 3–4 | Asco, Basidio | None | |
| NCU05213 | A | 0.2 | Hypothetical protein | Asco | TRS85 (4.8e−18) | |
| NCU05547 | D | 0.5 | Hypothetical protein | Asco | None | |
| NCU05720 | A | 0.6 | Chitin synthase export chaperone | Asco, Basidio | CHS7 (1.6e−62) | |
| NCU05890 | D | 0.3 | Hypothetical protein | Euk | None | |
| NCU06086 | D | 0.1 | Regulatory protein suaprga1 | Asco, Basidio | MAM33 (2.3e−31) | |
| NCU06703 | D | 0.4 | Mating pheromone-induced death-1 | Asco | MID1 (3.4e−28) | |
| NCU07062 | A | 0.3 | Serine/threonine protein kinase-49 | Asco, Basidio | SCH9 (4.9e−48) | |
| NCU08339 | D | 0.4 | Endosomal P24B protein | Asco | EMP24 (9.8e−49) | |
| NCU08909 | C | 0.6 | Beta-1,3-glucanosyltransferase | Asco | GAS1 (3e−111) |
Neuro Neurospora genus, Asco ascomycete fungi, Basidio basidiomycete fungi, Gram + Gram-positive bacteria, Euk higher eukaryotes.
aCategory A: Proteins potentially involved in cellular secretion apparatus (e.g., tether and SNARE proteins, GTPases, GPI-anchored and myristoylated proteins). Category B: Homologs of T. reesei genes that vary between wild type and the hyper-producing RUT-C30 strain as identified in [22, 23]. Category C: Known and predicted cellulases, hemicellulases and other proteins secreted in response to cellulose as identified in [8]. Category D: Proteins that traverse the secretory pathway as predicted using the SignalP (score >0.5) and TargetP (score = 1) algorithms.
bFold-change relative to wild type for secreted protein as measured by Bradford assay (n = 6). When deletions were available in both the mat A and mat a mating type backgrounds, the two values were averaged.
cDefined as nearest homolog with an E value <1e−10 using BlastP.
dOnly protein for which reciprocal BlastP using S. cerevisiae sequence against N. crassa genome does not return original locus; homology with Ucp14p of Schizosaccharomyces pombe.
Identification of hyper- and hypo-production strains
The 25 hyper- and hypo-production strains were distributed across the four categories (A–D) originally used to identify the strains included in the screen (Table 1). The genes deleted in each of these strains were conserved across the Ascomycota and many were also maintained throughout the Basidiomycota (Table 1). However, 11 of the deleted loci did not have identifiable homologs (<1e−10) in the S. cerevisiae genome, suggesting that their function may be specific to the biology of filamentous fungi.
The Category A deletion strains in Table 1, which are predicted to encode elements of the secretory apparatus, showed reduced levels of secreted protein and cellulase activity as compared to the wild-type strain. These encoded proteins were homologous to known secretion components in S. cerevisiae, such as NCU00566 (SNC2 homolog), NCU00600 (RHO3 homolog), NCU05213 (TRS85 homolog), NCU05720 (CHS7 homolog) and NCU08339 (EMP24 homolog).
The genes identified in Category B—N. crassa homologs of genes carrying mutations in the T. reesei RUT-C30 hyper-production strain [22, 23]—were from a variety of functional categories. Mutations in two genes, one encoding a hypothetical protein (NCU00541) and one encoding the transcription factor COL-26 (NCU07788) [5], showed increased secretion and cellulase activity. col-26 is a homolog to bglR in T. reesei [26], which has been implicated in the regulation of β-glucosidase genes. In N. crassa, col-26 is important for nutrient sensing and genetically interacts with vib-1 and cre-1, which are important for utilization of cellulose and carbon catabolite repression, respectively [27]. The remaining three Category B mutants showed decreased cellulase activity and secreted protein levels, including the deletion of the transcription factor ADA-2 (NCU02017) [5], a locus predicted to encode an RNA-binding protein (NCU02152) and one gene encoding a hypothetical protein (NCU02182).
Two of the identified hypo-production strains carried deletions in genes encoding proteins present in the N. crassa cellulolytic secretome (Table 1; Category C). One, NCU08909 (gel-3), encodes a GPI-anchored β-1,3-glucanosyltransferase implicated in cell wall biosynthesis [28]. The other, NCU04952 (gh3-4), encodes a glycosylhydrolase family 3 member that functions as an extracellular β-glucosidase [15].
Thirteen strains that displayed a secretion phenotype contained a deletion of a gene encoding a protein predicted to traverse the secretory pathway based on its scores using the SignalP and TargetP algorithms (Table 1; Category D). Five of the Category D mutants encode hypothetical proteins, all of which lack significant homology to genes in S. cerevisiae. The remaining eight genes showed sequence homology to proteins with identified function in S. cerevisiae or other fungal species (Table 1). Of the 13 Category D mutants in Table 1, only three have been previously characterized in N. crassa: NCU06650, which encodes a phospholipase A [8, 24], NCU06703 (mid-1), which encodes a mechanosensitive calcium channel [29] and NCU06086, which is predicted to be part of the N. crassa mitochondrial proteome [30].
Phenotypic characterization of select hyper- and hypo-production strains
For proof of principle, we focused our studies on four mutants: two showing consistently increased protein secretion and cellulase activity from the hyper-production mutants (ΔNCU03459 and ΔNCU3740) and two that showed consistently decreased protein secretion and cellulase activity relative to the other hypo-production mutants (ΔNCU00600 and ΔNCU05213) (Table 1; italics). The targeted proteins in each of these four deletion mutants are predicted to play various roles in protein processing and secretion. NCU03459 is annotated as encoding an ubiquitin binding associated (UBA) protein and has sequence similarity to Dsc2p (formerly YOL073C) in S. cerevisiae and Dsc2 (formerly Ubc14) in S. pombe, both of which function in association with a Golgi E3 ligase complex [31, 32]. The second mutant associated with hyper-production, ΔNCU03740, has significant similarity to ubiquitin ligases, most closely related to that of Tul1p in S. cerevisiae and Dsc1 in S. pombe. One of the mutants displaying a hypo-production phenotype, ΔNCU00600, encodes a protein that is similar to a GTPase, Rho3p, which is involved in polarized secretion in S. cerevisiae [33, 34]. The remaining mutant with a hypo-production phenotype, ΔNCU05213, shows similarity to Trs85p, which is part of the TRAPP-III complex involved in ER-Golgi trafficking and autophagy in S. cerevisiae [35].
On sucrose-based VMM agar, the ΔNCU00600 (Δrho-3) and ΔNCU05213 (Δtrs85) hypo-production mutants grossly resembled FGSC 2489 (wild type) in morphology (Additional file 2: Figure S1A), although under higher magnification these two mutants showed a hyper-branching hyphal structure (Additional file 2: Figure S1B). For the hyper-production strains, both ΔNCU03740 (Δtul-1) and ΔNCU03459 (Δdsc-2) showed reduced aerial hyphae compared to wild type (Additional file 2: Figure S1A), but when examined at a higher magnification, these two hyper-production strains displayed growth and hyphal branching patterns that were similar to the wild-type parent (Additional file 2: Figure S1B). On Avicel agar, all strains, including the wild-type strain, showed a reduced hyphal diameter as compared to their growth on VMM agar (Additional file 2: Figure S1C). However, obvious differences in hyphal tip morphology or internal organelle structure in the hyphae of the four mutant strains were not observed using the plasma membrane endocytic dye FM4-64 or an ER dye (ER-Tracker Red) under either VMM or Avicel conditions (Additional file 2: Figure S1C and D). All four strains appeared to grow equally well when cultured in VMM, xylan and Avicel media, with the possible exception of Δrho-3 mutant on Avicel (Additional file 2: Figure S1E).
Protein secretion and enzyme activity in the hyper- and hypo-production strains
None of the four selected deletion strains demonstrated a statistically significant difference relative to the wild-type strain in the amount of protein present in culture supernatants when grown in either sucrose- or xylan-based media (Fig. 1a). However, the Δdsc-2 and Δtul-1 strains showed significant increases (p ≤ 0.0001) in secreted protein levels as compared to wild type when cultured in Avicel, while both the Δrho-3 and Δtrs85 strains showed consistently lower secreted protein levels than the parental wild-type strain. When equal volumes of culture supernatants were analyzed by SDS-PAGE, an increased level of all secreted proteins was observed for the Δdsc-2 and Δtul-1 mutants compared to wild type under only Avicel conditions (Fig. 1b). In addition, the Δrho-3 and Δtrs85 mutants showed reduced levels of secreted proteins under Avicel conditions, though the protein banding pattern remained similar to the wild-type strain. Under xylan conditions, the pattern and levels of proteins observed were similar for all strains (Fig. 1b). When cultured in the presence of sucrose, the composition of the secretome from the Δdsc-2 and Δtul-1 mutants differed from that of the wild-type strain, while the secretome of the Δrho-3 and Δtrs85 mutants were similar to each other and to the wild-type strain.
Fig. 1.
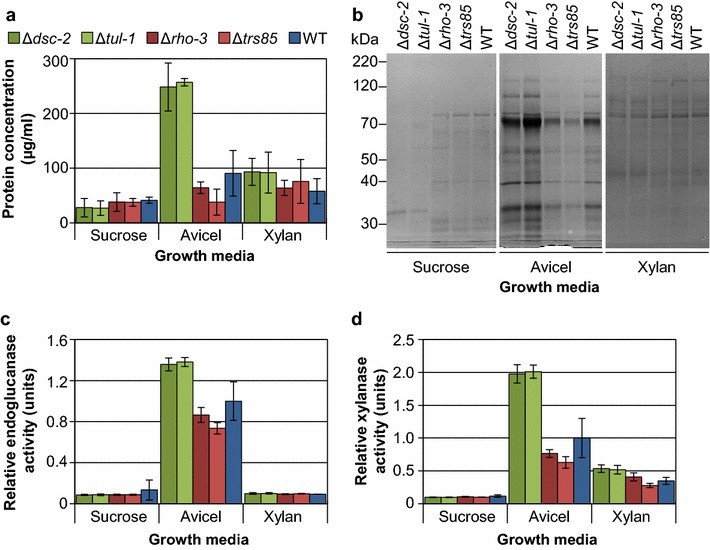
Secretion phenotypes are cellulose-specific. a Total secreted protein as measured by Bradford assay. b SDS-PAGE analysis of total secreted protein, equal volumes loaded. c, d Endoglucanase and xylanase activities, respectively, as measured using dye-release assays. Conidia from wild type and the deletions strains were inoculated directly into growth media containing sucrose (24 h), Avicel (96 h) or xylan (48 h). Protein and enzyme activity levels in the culture supernatants relative to the parental wild-type strain under each condition were analyzed by one-way ANOVA using the Dunnett’s multiple comparisons test. Data drawn from biological replicates (n = 4); error bars indicate standard deviation.
Supernatants from the Avicel-grown Δdsc-2 and Δtul-1 cultures showed significantly elevated endoglucanase activity levels relative to the wild-type strain (p ≤ 0.0001), while the Δrho-3 and Δtrs85 mutants were lower (Fig. 1c). Endoglucanase activity was virtually undetectable for all strains cultured in either VMM or xylan medium, indicating that a cellulosic inducer was still required by each of these strains to produce cellulases. In wild-type supernatants, hemicellulolytic (xylanase) activity is detectable from cultures grown on xylan or Avicel. The latter is primarily due to the induction of several xylanases by the cellulose-specific transcription factor CLR-2 [9, 16], but also the minor contamination of commercially available Avicel with hemicellulose [27]. When cultured on Avicel, xylanase activity in the Δdsc-2 and Δtul-1 strains was also significantly higher (p ≤ 0.0001) than that for the wild-type strain, whereas the Δrho-3 and Δtrs85 strains showed reduced xylanase activity (Fig. 1d). Although no major difference in protein secretion levels was apparent when the mutant strains were cultured on xylan (Fig. 1a), the level of xylanase activity in the hyper-production strains Δdsc-2 and Δtul-1 was slightly higher relative to the wild-type strain (p ≤ 0.01).
The enzyme activity and protein secretion data (Fig. 1) found that the effects associated with the hyper- and hypo-production phenotype of the deletion mutants were more dramatic when grown on a cellulosic substrate. The production of cellulases in N. crassa is directly regulated by the cellulose-responsive transcription factor, CLR-2 [9, 16]. We therefore evaluated protein levels in the mutants using antibodies targeted to enzymes whose production is regulated by CLR-2 under cellulosic conditions, including: cellobiose dehydrogenase (CDH-1; NCU00206), an endoglucanase (GH5-1; NCU00762), an endoxylanase (GH10-1; NCU05924) and a polysaccharide monooxygenase (GH61-5; NCU08760) (Fig. 2). In addition, we used antibodies to enzymes known to be independent of CLR-2 regulation, such as an alpha-l-arabinofuranosidase (GH51-1; NCU02343) and an endoxylanase (GH10-2; NCU08189). We also included an antibody to glucoamylase (GLA-1; NCU01517), an enzyme that is constitutively produced by wild-type N. crassa under sucrose, Avicel and xylan growth conditions [36].
Fig. 2.

Secretion phenotypes are not limited to cellulases. a Immunoblot analysis of total secreted protein from experiment outlined in Fig. 1. Equal volumes of culture supernatant were loaded and separated by electrophoresis on 10% polyacrylamide gels, then transferred to nitrocellulose membranes for antibody detection. Primary antibodies were directed against the following: CDH-1 (cellobiose dehydrogenase), GH5-1 (an endoglucanase), GH10-1 (an endoxylanase), GH61-5 (a polysaccharide monooxygenase), GH51-1 (an arabinofuranosidase), GH10-2 (an endoxylanase) and GLA-1 (glucoamylase). b Quantitation of detected proteins. For CDH-1, GH5-1, GH10-1 and GH61-5 values are given relative to WT in Avicel; for GH51-1 and GH10-2 values are given relative to WT in xylan; for GLA-1 values are given as relative to WT in sucrose.
Glucoamylase-1 was detected in the wild type, Δrho-3 and Δtrs85 supernatants for all of the growth media used (Fig. 2). However, in the hyper-production strains Δdsc-2 and Δtul-1, GLA-1 was undetectable when the mutants were grown under sucrose conditions despite visibly apparent fungal cell biomass (Additional file 2: Figure S1E). When grown on Avicel, the protein levels of the CLR-2-dependent proteins CDH-1, GH5-1, GH10-1 and GH61-5 recapitulated the hyper-production phenotype of the Δdsc-2 and Δtul-1 mutants, as well as the hypo-production phenotype of the Δrho-3 and Δtrs85 mutants (Fig. 2). This finding was also true for the GLA-1 enzyme under cellulolytic growth conditions. The two CLR-2-independent proteins, GH51-1 and GH10-2, were detected only under xylan conditions. Importantly, neither the hyper-production phenotype of the Δdsc-2 and Δtul-1 mutants, nor the hypo-production phenotype of the Δrho-3 or Δtrs85 mutants, was recapitulated under xylan growth conditions: GH51-1 was present at roughly equivalent levels for all of the strains tested, while GH10-2 levels were only decreased in the Δrho-3 mutant. Collectively, these data indicate that the hyper-production phenotype of Δdsc-2 and Δtul-1 mutants was only apparent under cellulolytic conditions, but that this phenomenon was not limited to cellulosic enzymes.
The hyper-production phenotype of the Δdsc-2 and Δtul-1 mutants is retained on plant biomass
Our data showed that the hyper-production phenotypes of the Δdsc-2 and Δtul-1 mutants and the hypo-production phenotype of the Δrho-3 and Δtrs85 strains were strongest on crystalline cellulose (Figs. 1a–c, 2). We next evaluated whether these phenotypes were restricted to Avicel or could be observed when N. crassa was exposed to other cellulosic substrates, including the soluble cellulose substrate carboxymethyl cellulose (CMC) and plant biomass (Miscanthus x giganteus). For these studies, conidia from the four mutant strains were first inoculated into VMM for 48 h to accumulate biomass before being transferred to either Avicel, CMC or ground Miscanthus for 48 h. This approach was used to eliminate growth rate differences between the Avicel, CMC and Miscanthus-grown cultures, as well as any conidial germination differences, such as those previously reported for N. crassa grown in Avicel versus Miscanthus media [8].
The cellulase and protein hyper-production phenotype of the Δdsc-2 and Δtul-1 mutants was recapitulated when these mutants were switched into Avicel, CMC or Miscanthus media (p ≤ 0.01; Fig. 3). However, the reduced protein secretion observed in the Δtrs85 and Δrho-3 mutants when conidia were directly inoculated into Avicel (Fig. 1) was not recapitulated when mycelia of these mutants grown on VMM was switched to Avicel, CMC or Miscanthus, although the Δtrs85 mutant consistently trended below that of wild type with regard to protein secretion and enzyme activity levels. These observations suggested that the Δrho-3 and Δtrs85 mutants might be defective for germination and/or colony establishment when conidia were directly inoculated into Avicel, but that this phenotype disappeared if fungal biomass was first established in sucrose-based VMM. We therefore evaluated germination frequency of conidia from the Δdsc-2, Δtul-1, Δrho-3 and Δtrs85 mutants as compared to the wild-type parental strain in both VMM and Avicel broth. No correlation was found between the hyper- or hypo-production phenotypes and the conidial germination rates of the mutants versus the parental wild-type strain (Fig. 3d). In fact, while the Δrho-3 and Δtrs85 hypo-production mutants showed a similar germination rate to the wild-type strain under all conditions, the Δdsc-2 and Δtul-1 hyper-production strains germinated much more slowly than the wild-type strain in VMM. From these data, we hypothesize that the hypo-production phenotype identified in the Δtrs85 and Δrho-3 mutants is attributable to a reduced establishment of hyphal growth after conidial germination of these mutants under cellulolytic conditions.
Fig. 3.
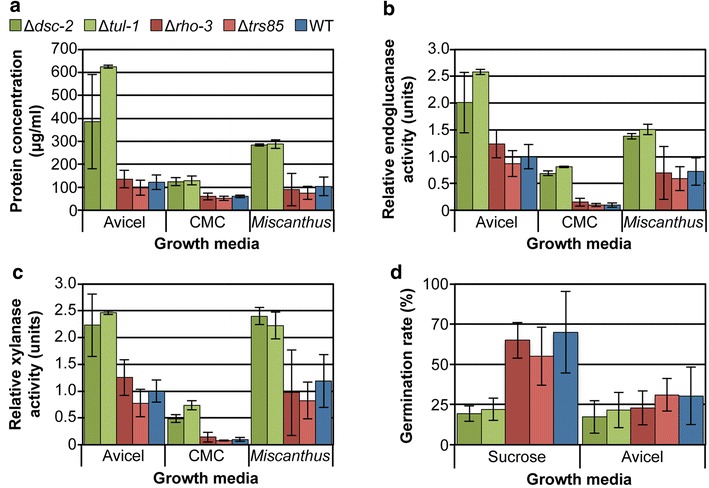
Secretion phenotypes are maintained on cellulosic substrates. a Total secreted protein. b, c Endoglucanase and xylanase activities, respectively. Conidia from wild type and the mutant strains were first inoculated into VMM (48 h) and the resulting biomass subsequently switched to Avicel, CMC, or Miscanthus (48 h). d Germination frequency. Conidia were cultured in sucrose (4 h) or Avicel broth (8 h) and the numbers of germinated conidia were evaluated by light microscopy. Statistical analyses for protein and enzyme activity levels were relative to WT within each condition (n = 3). Error bars indicate standard deviation.
In our screen we identified 18 mutants that showed a cellulolytic hypo-production phenotype when conidia from these strains were directly inoculated into Avicel medium (Table 1). We hypothesized that some of these mutants, like Δrho-3 and Δtrs85, might also lose their hypo-production phenotype when the mutants were first allowed to establish fungal biomass under sucrose-based VMM conditions. To test this hypothesis, the hypo-production strains were allowed to generate biomass in VMM and subsequently switched to Avicel medium. The culture supernatants were then evaluated for secreted protein and enzyme activity levels (data not shown). All but a few of the hypo-production mutants (ΔNCU02017, ΔNCU04952 and ΔNCU07062) lost their hypo-production phenotypes when pre-incubated in VMM prior to exposure to Avicel (Table 1). These data suggest that biomass establishment under recalcitrant carbon conditions is a major factor in the reduced secretion phenotype observed in many of the N. crassa hypo-production mutants.
The Δdsc-2 and Δtul-1 strains maintain their hyper-production phenotype under constitutive CLR-2-driven cellulase production
The hyper-production phenotype of the Δdsc-2 and Δtul-1 strains was most prominent when these strains were grown in Avicel medium, but was also maintained in the presence of the cellulosic substrates CMC and Miscanthus (Figs. 1, 3). These data suggested that either the phenotype of these mutants was the result of conditions generated by growth on cellulose or was specifically related to the induction and secretion of cellulases that occurs in response to cellulose exposure. To differentiate between these two possibilities, we utilized a N. crassa strain in which the transcriptional activator, clr-2, is expressed in the absence of a cellulose inducer. Mis-expression of clr-2 under non-cellulolytic conditions recapitulates the wild-type Neurospora response to cellulose, resulting in the production and secretion of cellulases when the strain is grown in VMM [16]. We therefore crossed the Δdsc-2, Δtul-1, Δrho-3 and Δtrs85 mutants to the clr-2 mis-expression strain Δclr-2 (Pclr-2) [16] and recovered Δdsc-2; Δclr-2 (Pclr-2), Δtul-1; Δclr-2 (Pclr-2), Δrho-3; Δclr-2 (Pclr-2) and Δtrs85; Δclr-2 (Pclr-2) mutants. These strains were then evaluated for their hyper- or hypo-production phenotypes under VMM (sucrose) conditions.
The Δdsc-2; Δclr-2 (Pclr-2) and Δtul-1; Δclr-2 (Pclr-2) strains showed a significant increase in protein secretion as compared to the Δclr-2 (Pclr-2) parental background under sucrose-based VMM conditions and with a corresponding increase in endoglucanase activity (p ≤ 0.001 and p ≤ 0.01, respectively; Fig. 4), similar to what was observed when the Δdsc-2 and Δtul-1 mutants were cultured on Avicel. These data were consistent with the hypothesis that increased protein secretion and enzyme activity was specifically related to the induction and secretion of cellulases as regulated by CLR-2. Unlike the hyper-production mutations, the Δtrs85; Δclr-2 (Pclr-2) and Δrho-3; Δclr-2 (Pclr-2) strains resembled the Δclr-2 (Pclr-2) strain (WT), supporting the hypothesis that the hypo-production phenotype of these mutants is due to a delay in fungal biomass formation in the presence of Avicel, which was alleviated when grown in VMM (Fig. 3). Thus, the hyper-production phenotypes of the Δdsc-2 and Δtul-1 mutants were recapitulated by inducer-independent expression of the major cellulase transcriptional regulator, CLR-2.
Fig. 4.
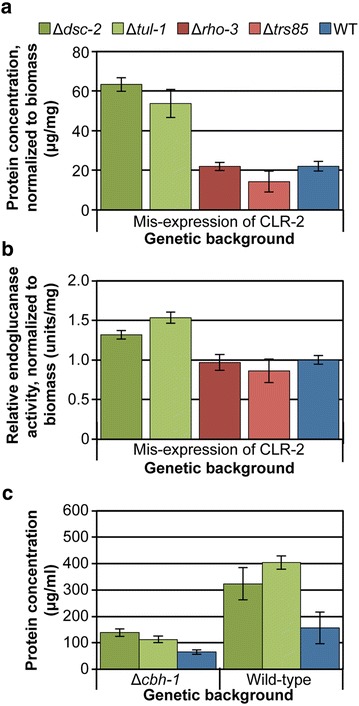
CLR-2-dependence and cellulose and secretory load independence of mutant phenotypes. a Total secreted protein and b endoglucanase activity normalized for total fungal cell biomass in each culture. Conidia from the indicated strains were cultured in growth media containing sucrose (48 h). c Total secreted protein; conidia from the indicated strains were inoculated into VMM (48 h) the resulting biomass then switched to Avicel (48 h). Statistical analyses for protein and enzyme activity levels were relative to WT within each condition (n = 3 for CLR-2 mis-expression study; n = 4 for Δcbh-1 study). Error bars indicate standard deviation.
The hyper-production phenotype of Δdsc-2 and Δtul-1 is retained under reduced secretion load
The cellular response of N. crassa to the presence of cellulose involves an impressive upregulation in transcription, translation, synthesis and secretion of proteins associated with plant biomass deconstruction [8, 9, 16]. While use of the Pclr-2 mis-expression construct allowed for the examination of the Δdsc-2, Δtul-1, Δrho-3 and Δtrs85 secreted protein phenotypes separate from the physical presence of cellulose, these strains still undergo a CLR-2-dependent cellulolytic response, including a large increase in the amount of protein transiting through the secretory pathway [9, 16]. We considered that the hyper-production phenotypes observed in the Δdsc-2 and Δtul-1 mutants could be a nonspecific consequence of the increased secretory load that results from CLR-2 induction without being directly dependent on the CLR-2 regulon. If this were true then a high secretory load of CLR-2-independent proteins should also result in the hyper-production of proteins in these mutants. Unfortunately, no other carbon source (including xylan, pectin and BSA) was identified that mimicked the high level of secreted protein observed under cellulosic conditions or when clr-2 is mis-expressed in VMM (data not shown). As an alternative approach, we crossed the Δdsc-2 and Δtul-1 mutants to a mutant containing a deletion of the major secreted protein under cellulolytic conditions, cellobiohydrolase-1 (CBH-1) and recovered Δdsc-2; Δcbh-1 and Δtul-1; Δcbh-1 mutants. CBH-1 comprises ~40% of the total Avicel secretome [10] and its absence from the genome significantly reduces the protein load of the secretory apparatus during the cellulolytic response.
The Δdsc-2; Δcbh-1 and Δtul-1; Δcbh-1 mutants were first cultured in VMM in order to generate biomass and then transferred to Avicel medium to induce a cellulolytic response. As previously shown [8], the level of protein secreted in the Δcbh-1 mutant was approximately half that of the wild-type strain (Fig. 4c). However, the hyper-production phenotype of the Δdsc-2; Δcbh-1 and Δtul-1; Δcbh-1 strains was maintained relative to the Δcbh-1 parental strain (p ≤ 0.001). These data indicated that the hyper-production phenotype of the Δdsc-2 and Δtul-1 mutants was not solely due to the secretion load seen in fungal biomass exposed to cellulosic substrates, but was a consequence of induction by cellulosic substrates.
Identification of the SREBP pathway components that affect secreted protein production under cellulosic conditions
The N. crassa dsc-2 and tul-1 are possible orthologs of dsc2 and dsc1 in S. pombe, although only moderate sequence conservation was observed (Additional file 3: Figure S2A, B). The Dsc complex in S. pombe is composed of five proteins: Dsc1, Dsc2, Dsc3, Dsc4 and Dsc5 [37]. The Dsc proteins form a complex in the Golgi membrane that resembles E3 ligases involved in ER-associated protein degradation [32]. In Aspergillus fumigatus, homologs of dsc1, dsc2, dsc3 and dsc4 are required for adaption to hypoxic conditions and virulence in a mouse model [38]. The Dsc complex in S. pombe and A. fumigatus is required for proteolytic cleavage of an ER-membrane bound transcription factor (Sre1 and SreA, respectively) involved in adaptation to hypoxic conditions. The N. crassa genome contains predicted proteins with low, but significant similarity to Dsc3 (NCU11245) and Dsc4 (NCU02676) (Additional file 3: Figure S2C, D) and a transcription factor similar to Sre1/SreA (NCU04731) (Additional file 3: Figure S2E). A strain carrying a deletion of NCU04731 was previously characterized in a transcription factor deletion strain set collection and was named for having short aerial hyphae (sah-2) [5]. We hypothesized that strains carrying mutations in additional components of the predicted Dsc complex and the transcription factor that is the predicted target of this E3 ligase would also show a cellulolytic hyper-production phenotype. To test this hypothesis, single-gene deletion mutant strains for NCU02676 (dsc-4) and sah-2 (NCU04731) were assessed for their protein production under cellulolytic conditions.
As observed for the Δdsc-2 and Δtul-1 mutants, strains carrying a deletion of Δdsc-4 or Δsah-2 showed significantly increased levels of secreted protein and cellulolytic enzyme activity (endoglucanase and xylanase) when cultured on Avicel as compared to the wild-type parental strain (p ≤ 0.001 and p ≤ 0.01, respectively; Fig. 5). When cultured on sucrose-based VMM, the protein secretion levels of the Δdsc-4 and Δsah-2 mutants were identical to the wild-type strain (and the Δdsc-2 and Δtul-1 mutants), although under xylan conditions both the Δdsc-4 and Δsah-2 mutants secreted slightly less protein than the wild-type strain (Fig. 5a). We subsequently evaluated whether the hyper-production phenotype observed in the Δtul-1, Δdsc-2 and Δsah-2 mutants was abolished by the introduction of wild-type copies of the tul-1, dsc-2 and sah-2 genes driven by the tef-1 promoter. As shown in Additional file 4: Figure S3, the morphological phenotype (short aerial hyphae) of the Δtul-1, Δdsc-2 and Δsah-2 mutants was fully complemented by the introduction of the wild-type allele of tul-1, dsc-2 and sah-2, respectively. In addition, the protein hyper-production phenotype of the Δtul-1, Δdsc-2 and Δsah-2 mutants was also abolished by the introduction of the wild-type alleles, as was the corresponding endoglucanase activity (Additional file 4: Figure S3). These data indicate that that SREPB pathway plays an important role in regulating the amount of protein secreted by N. crassa under cellulolytic conditions.
Fig. 5.
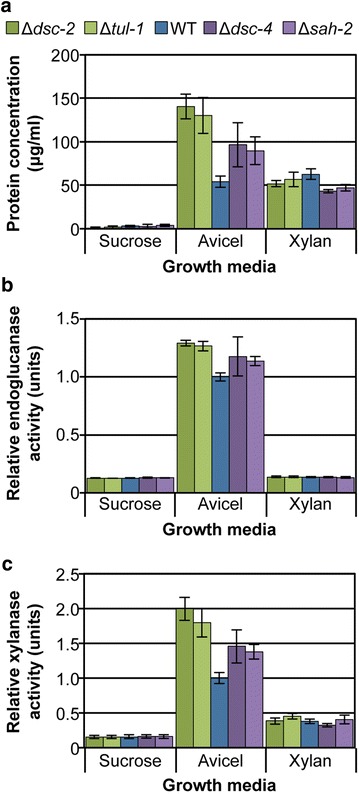
Strains containing mutations in an SREBP homolog, sah-2, and additional Dsc E3 ligase components show a cellulase hyper-production phenotype. a Total secreted protein. b, c Endoglucanase and xylanase activities, respectively. Conidia from wild type and the deletion strains were inoculated directly into growth media containing sucrose (24 h), Avicel (96 h), or xylan (48 h). Statistical analyses for protein and enzyme activity levels were relative to WT within each condition (n = 6). Error bars indicate standard deviation.
Our initial microscopic analyses using the membrane selective dyes FM4-64 and ER-Tracker Red did not show obvious differences in structure between the Δdsc-2 and Δtul-1 hyper-production mutants and the wild-type strain (Additional file 2: Figure S1C, D). To further explore this issue, we introduced an ER cellular marker, NCA-1-sGFP or a Golgi marker, VPS52-sGFP [39] into the Δtul-1 and Δsah-2 mutants to generate Δtul-1; nca-1-sGFP, Δtul-1; vps52-sGFP, Δsah-2; nca-1-sGFP and Δsah-2; vps52-sGFP strains. As with the ER-Tracker Red and FM4-64, no obvious differences in ER or Golgi morphology was observed in the Δtul-1; nca-1-sGFP, Δtul-1; vps52-sGFP, Δsah-2; nca-1-sGFP and Δsah-2; vps52-sGFP mutants under hyper-production conditions (Avicel) relative to the parental strains (Additional file 2: Figure S1E).
The cellulase hyper-production phenotype of the SREBP pathway mutants is not simply due to an increased expression level of cellulases
Our data with the mis-expressed clr-2 strain (Fig. 4) showed that the hyper-production phenotype of the Δdsc-2 and Δtul-1 strains is related to the CLR-2-dependent regulon. We therefore assessed expression levels of dsc-2 and tul-1 in existing datasets generated for the Δclr-2 strain when exposed to Avicel relative to the wild-type strain or in strains grown in VMM, but where clr-2 was mis-expressed; neither dsc-2 nor tul-1 showed evidence of transcriptional regulation by CLR-2 [9, 16]. These observations suggested that factors in addition to CLR-2 are important for modulating expression levels of the SREBP pathway components during plant biomass deconstruction. We hypothesized that the hyper-production of cellulases in the SREBP pathway mutants could be due to increased expression of cellulolytic genes in the mutants. We therefore assessed expression levels of the major cellulases (cbh-1, cbh-2 and gh5-1) [10] in N. crassa over time in the wild-type strain versus the Δtul-1 and Δsah-2 mutants by qRT-PCR. As shown in Fig. 6, the expression of all three of these cellulases was very similar to each other at the 48 and 120 h time point. At the 96 h time point, however, the Δtul-1 mutant actually showed lower expression levels for all three cellulase genes, especially for cbh-1, which encodes the major cellulase in N. crassa [10] (Fig. 6). Thus, these data do not support the hypothesis that a simple increase in the expression levels of the major cellulase genes in the SREBP pathway mutants is the cause of the hyper-production phenotype.
Fig. 6.
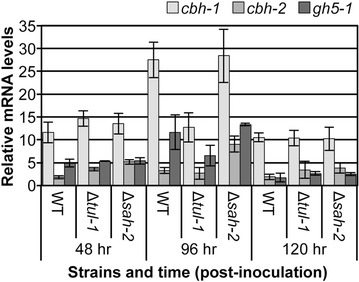
Time course of expression levels of major cellulase genes in the Δtul-1 and Δsah-2 mutants relative to the parental wild-type strain under cellulolytic conditions. qRT-PCR experiments measuring gene expression levels from Avicel broth cultures inoculated with conidia from wild type or the Δtul-1 or Δsah-2 mutants and grown over a 120 h time course. Relative transcript levels of major cellulase genes cbh-1 (NCU07340), cbh-2 (NCU09680)and gh5-1 (NCU00762) were normalized to expression levels of actin (NCU04173). Primer sequences used are given in Additional file 5: Table S2. Data from n = 3 biological replicates; error bars indicate standard deviation.
Role of Dsc/Sre1 pathway in Trichoderma reesei
Single sah-2 homologs were identified across many species of filamentous ascomycete fungi (Fig. 7), suggesting conservation of function. We therefore hypothesized the hyper-production phenotype of the SREBP pathway mutants would not be specific for N. crassa and that mutations in these genes in other filamentous fungi that degrade plant biomass would show a similar cellulase hyper-production phenotype. We chose to test this hypothesis in T. reesei as hyper-production mutants in this species are used in industrial settings for cellulase enzyme production [40]. A search of the T. reesei QM6a genome and its predicted encoded proteins using the N. crassatul-1 and sah-2 protein sequences as queries identified a single ortholog for each. The TUL1 protein in T. reesei (Trire2_21342) showed 40% amino acid sequence identity to N. crassa TUL-1 (NCU03740) and was predicted as a transmembrane Golgi-localized Ring finger ubiquitin ligase, while SAH2 (Trire2_22774) showed 51% amino acid sequence identity to N. crassa SAH-2 (NCU04731) with no annotated functions (Additional file 6: Figure S4). Thus, the tul1 and sah2 loci were targeted for deletion in a QM6a strain (Additional file 7: Figure S5).
Fig. 7.
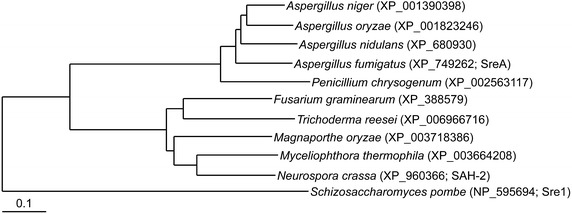
Conservation of Sre1 among industrially relevant fungi and closely related fungal species. Phylogenetic tree of putative Sre1 homologs was generated using MEGA6 [56]. Scale bar 0.1 substitutions per amino acid site. Accession numbers are noted in the tree.
Three independent homokaryotic T. reesei Δtul1 and Δsah2 mutants were selected for morphological and cellulase production analyses. All of the Δtul1 and Δsah2 mutants showed a similar morphological phenotype to the parental QM6a strain when the only available carbon source was glucose. However, increased hyphal biomass was apparent in the QM6aΔtul1 and QM6aΔsah2 cultures as compared to the QM6a parental strain when Avicel was used as the single carbon source (data not shown). All three independently derived Δtul1 mutants and all three independently derived Δsah2 mutants showed increased levels of secreted proteins and endoglucanase activity under Avicel conditions (Additional file 7: Figure S5). The Δtul1 and Δsah2 mutants displayed higher secreted protein levels and higher enzyme activity during a 7-day time course as compared to the QM6a parental strain (Fig. 8). Similar to what was observed in N. crassa, the hyper-production phenotype of the T. reesei QM6aΔtul1 mutant showed a stronger hyper-production phenotype than that of the QM6aΔsah2 mutant (compare Figs. 5, 8 and Additional file 7: Figure S5). Collectively, these data demonstrate that deletion of predicted components of the SREBP pathway results in hyper-production of cellulolytic proteins in two distantly related filamentous fungal species, suggesting that this function is highly conserved among fungi capable of degrading plant biomass.
Fig. 8.

The T. reesei tul1 and sah2 deletion mutants show a hyper-production phenotype under cellulolytic conditions. Avicel broth cultures were inoculated with conidia and grown for 7 days in shake cultures. Samples taken during the time course were evaluated for a total secreted protein and b endoglucanase activity. Statistical analysis for protein and enzyme activity levels were relative to the parental QM6aΔpyr4 (n = 3). Error bars indicate standard deviation.
Discussion
To identify elements involved in the ability of N. crassa to utilize cellulose, we took advantage of a publically available collection of single-gene deletion strains representing nearly every gene in the genome and generated under the auspices of the Neurospora Genome Project [5, 41], a resource currently unique among filamentous fungi. We selected deletion strains based on those genes encoding proteins likely to play a role in cellulase activity or production, including homologs to genes encoding components of the S. cerevisiae secretion apparatus, N. crassa homologs of genes that showed mutations in the T. reesei hyper-production strain RUT-C30, genes encoding the proteins identified in the secretome of N. crassa when cultured on plant biomass and genes encoding proteins predicted to traverse the secretory pathway. From this initial set of 567 strains (Additional file 1: Table S1), 25 strains—including seven hyper-production mutants and 18 hypo-production mutants (Table 1)—were identified. Sequence analysis suggested that while some of the deleted genes likely act in transcription, protein synthesis and intracellular trafficking, many of the loci encode fungal-specific proteins of undetermined functions. While the effect of some of these genes/proteins on cellulase secretion is not immediately apparent, for others it is more obvious given their homology to proteins known to impact biomass degradation in other filamentous fungi. For example, the hyper-production ΔNCU07788 (Δcol-26) mutant was included here because the T. reesei RUT-C30 hyper-production strain bears a mutation in bglR [22], the ortholog of col-26. In T. reesei, the bglR locus is known to play a role in nutrient signaling and cellulase production [26]. It was recently shown that col-26 in N. crassa also functions in nutrient signaling and that a Δcol-26 mutant, in conjunction with a deletion of the carbon catabolite repressor, cre-1, suppresses the inability of a Δvib-1 (a NDT80-like transcription factor) mutant to utilize cellulose [27].
While each of the identified mutants in Table 1 warrant further characterization, in this study we focused on four deletion strains: Δdsc-2 (ΔNCU03459) and Δtul-1 (ΔNCU03740), which were included based on their consistent hyper-production phenotype and their predicted role in the secretory pathway, particularly ubiquitination, and the hypo-producers Δrho-3 (ΔNCU00600) and Δtrs85 (ΔNCU05213), which are homologous to known elements in S. cerevisiae involved in protein trafficking. In the course of our studies, we observed that the hypo-production phenotype of the Δrho-3 and Δtrs85 mutants was abolished if first allowed to accumulate biomass in a preferable carbon source (Fig. 3). These results led us to re-evaluate the phenotype of all 18 hypo-production strains identified in Table 1. When first cultivated on sucrose-based VMM before switching to Avicel medium, the majority of these hypo-production strains were found to have similar levels of protein present in the culture supernatant as wild type or a much reduced hypo-production phenotype; this has implications for assessing deletion strain phenotypes in high-throughput screens using conidial suspensions in N. crassa and perhaps other filamentous fungi as well. However, three of the identified hypo-production mutants—Δada-2 (NCU02017; a transcription factor) [5], Δgh3-4 (NCU04952; a secreted β-glucosidase) [15] and Δstk-49 (NCU07062; an uncharacterized protein kinase) [42]—continued to demonstrate significantly reduced protein secretion and cellulolytic activity even when allowed to establish biomass before exposure to cellulose. It is possible that a deletion of Δgh3-4 may interfere with signaling, since cellobiose (and other cellodextrins) function as inducers for cellulose utilization in N. crassa [15]. How the ada-2 deletion strain (which was included in this screen because the T. reesei RUT-C30 hyper-production strain also carries a mutation in the ada-2 ortholog [22]) and the stk-49 deletion strain (which was included for its potential role in secretion) impact cellulase secretion remains to be determined.
The hyper-production strains Δdsc-2 and Δtul-1 showed a consistent increase in protein production and cellulolytic activity in the presence of cellulose and plant biomass as well as when CLR-2 was mis-expressed in sucrose medium (Figs. 1, 3, 4a, b). Although the hyper-production phenotype is linked to the cellulosic response, it is not limited to the production of cellulases (Fig. 2). This effect was not specifically related to an increase in the protein load traversing the secretory pathway under these conditions as an associated increase in secretion is observed even when the overall protein load is reduced, as in a Δcbh-1 mutant carrying the Δdsc-2 and Δtul-1 mutations (Fig. 4c). These observations suggest that DSC-2 and TUL-1 play an important role in regulating protein production/secretion under cellulosic conditions, when the organism secretes large amounts of enzymes needed to deconstruct recalcitrant plant biomass substrates.
Subsequent to our initial selection and characterization of the Δdsc-2 (ΔNCU03459) and Δtul-1 (ΔNCU03740) hyper-production mutants, it was shown in S. pombe that homologs to NCU03459 (Dsc2) and NCU03740 (Dsc1) function in a Golgi-localized E3 ligase complex [22]. These two proteins, in combination with three others (Dsc3-5), act upon the transcription factor Sre1 and regulate responses to hypoxia [32, 43]. Sre1 is a homolog of the mammalian SREBP transcription factors [44], which regulate genes encoding enzymes involved in sterol biosynthesis. A number of additional proteins important for SREBP pathway function have been identified and characterized in S. pombe [44, 45]. Later studies with the human fungal pathogen A. fumigatus showed that homologs to the Dsc complex (DscA-D) and Sre1 (SrbA) also functioned in adaptation to hypoxia, resistance to antifungal drugs and virulence [38, 46]. More recently, it was shown that a direct target of A. fumigatus SrbA was another transcription factor, SrbB, which has been implicated in the regulation of carbohydrate metabolism [47]. In mammalian cells, loss of SREBP caused changes in lipid composition and induced accumulation of reactive oxygen species, cell death (apoptosis) and a reduction in protein synthesis as a result of ER stress [48]. The linkage we identified between the loss of the SREBP pathway, which was not previously known to be involved in protein secretion, and the hyper-production of cellulases demonstrates the strength of screening deletion strain collections in an unbiased manner. Future studies in N. crassa will dissect the relationship between regulation of secretion by SAH-2, the Dsc E3 ligase complex and accessory proteins under cellulolytic and plant biomass conditions versus the role of this pathway under hypoxic conditions.
Although homologs of SAH-2 are well conserved among filamentous fungi (Fig. 7), it was possible that the hyper-production phenotype associated with mutations in the SREBP pathway components would be restricted to N. crassa. To test this hypothesis, we constructed strains carrying deletions of dsc1 and sah2 in the T. reesei strain QM6a. Neither of these mutations occur in the hyper-production RUT-C30 strain [22, 23]. The QM6aΔdsc1 and QM6aΔsah2 strains secreted significantly higher quantities of enzymatically active cellulases into the culture medium as compared to their parental strain (Fig. 8, Additional file 7: Figure S5). These data suggest that the role of the Dsc/SREBP pathway in regulating secretion of enzymes needed for plant biomass degradation is conserved between two distantly related filamentous fungi, N. crassa and T. reesei. Better understanding and modification of the components of this pathway could allow for further engineering of increased protein production in both these species as well as other industrially relevant filamentous fungi.
Conclusions
In screening a panel of N. crassa single-gene deletion mutants, we identified an association between the hyper-production of secreted cellulases and components of the SREBP pathway that is apparent only under cellulosic conditions. Previous studies on mutants in this pathway had not linked it to a role in protein secretion. Importantly, the hyper-production phenotype that accompanies the deletion of DSC and SAH-2/SRE-1 is conserved between distantly related species of filamentous fungi. The continuing characterization of sah-2, the dsc E3 ligase and accessory protein mutants will enhance our understanding of the ability of N. crassa to utilize complex carbon sources present in its natural environment, while broadening our knowledge of protein secretion in filamentous fungi, including industrially relevant species.
Methods
Neurospora strains and growth conditions
Single-gene deletion strains were obtained from the Fungal Genetics Stock Center [5, 6]. The Δclr-2 (Pclr-2) mis-expression strain [16] was used for crosses to construct double mutants carrying the Δclr-2 (Pclr-2) and Δdsc-2, Δtul-1, Δrho-3 or Δtrs85 mutation, which were verified by phenotypic screening and PCR. The Δtul-1; nca-1-sGFP and Δtul-1; vps52-sGFP strains were constructed by transforming a his-3; Δtul-1 strain with the plasmids nca-1-sGFP and vps52-sGFP [39], respectively. The Δsah-2; nca-1-sGFP and Δsah-2; vps52-sGFP strains were created by crossing strains bearing NCA-1-sGFP or VPS52-sGFP into the Δsah-2 mutant. Genotypes were verified by phenotypic screening and PCR. Complementation was performed by inserting the tul-1, dsc-2 or sah-2 gene into the Ptef1-Tcyc1 plasmid and transforming it into the csr-1 locus by selection for cyclosporin resistance [49]. Genotypes were verified by phenotypic screening and PCR.
Liquid growth media comprised 1X Vogel’s salts in ddH2O plus 2% weight-per-volume carbon source, while all solid media contained an additional 1.5% agar. Carbon sources included: sucrose, Avicel PH-101 (Sigma 11365), xylan (from birchwood, Sigma X0502 or beechwood, Sigma X4252), sodium carboxymethyl cellulose (CMC; Sigma 419273) or ground Miscanthus x giganteus (0.25 mm diameter). N. crassa strains were inoculated from freezer stocks into slant tubes of sucrose-based Vogel’s minimal medium (VMM) agar [25] and incubated at 30°C in the dark for 2 days followed by at least 4 days incubation at 25°C in the light. Conidia were harvested in sterile water and then inoculated at 106 per milliliter into 3 mL of growth medium aliquoted into 10-mL deep-well, square-side, round-bottom 24-well plates (GE Healthcare 7701–5102) or 100 mL growth medium in 250-mL Erlenmeyer flasks. Plates were covered with a breathable, sterile sealing tape while flasks were capped with a sterile foam stopper; both were then incubated at 25°C in constant light with rotation (200 rpm) for 24–96 h.
For switch experiments, conidia were inoculated into 3 mL VMM and cultured for 48 h as above. The fungal biomass was then washed with a solution of 1X Vogel’s salts in ddH2O before transferring to 3 mL of freshly aliquoted growth medium. The cultures were then incubated at 25°C in constant light with rotation (200 rpm) for 48 h.
For all experiments, N. crassa strains were cultured in replicates of at least three for statistical analysis by one-way ANOVA using the Dunnett’s multiple comparisons test.
Construction of Trichoderma mutant strains and growth conditions
A wild-type strain of T. reesei, QM6a (ATCC 13631), was obtained from the American Type Culture Collection. An uridine auxotroph strain, QM6aΔpyr4, was generated via the targeted deletion of pyr4 and used as the recipient for generating the tul1 and sah2 deletion mutants, QM6aΔtul1 and QM6aΔsah2. The pyr4 deletion vector, pSCpyr4, was constructed by ligation of 5′ flanking DNA, the hph marker gene for hygromycin resistance, and 3′ flanking DNA into a vector backbone. The sequences of primers used in the construction of the T. reesei deletion vectors are provided in Additional file 5: Table S2. The following fragments were amplified from QM6a genomic DNA: the 5′ flank of pyr4 (2.1 kb) was amplified using primers pSC-pyr4A-F and pSC-pyr4A-R, whereas the 3′ flank of pyr4 (1.9 kb) was amplified using primers pSC-pyr4C-F and pSC-pyr4C-R. A hygromycin-resistance cassette (2.3 kb) including hph was amplified from the genomic DNA of the N. crassa Δdsc-1 strain using primers pSC-pyr4B-F and pSC-pyr4B-R. The backbone of the vector (3 kb) was amplified from the plasmid pMF272 [50] using primers pSC-pyr4D-F and pSC-pyr4D-R.
For deletion of pyr4, the split marker technique was used to improve homologous recombination [51]. Split-marker fragments of the pyr4 deletion cassette were amplified from pSCpyr4 by PCR using the primer pairs pSC-pyrA-F with pSC-SM-R and pSC-SM-F with pSC-pyr4C-R. Strain QM6a was transformed as described [52]. Transformants were screened on glucose-based minimal medium (MM) agar [53] supplemented with hygromycin and deletion of pyr4 was verified by PCR (Additional file 5: Table S2).
To generate the QM6aΔtul1 and QM6aΔsah2 deletion strains, a tul1 deletion vector, pSCtul1, was constructed by ligation of 5′ flanking DNA, the pyr4 marker gene, 3′ flanking DNA and a vector backbone. The following fragments were amplified from QM6a genomic DNA: sequence 5′ to tul1 (2.4 kb) was amplified using primers pSC-tul1A-F and pSC-tul1A-R; the pyr4 cassette (1.8 kb) was amplified using primers pSC-tul1B-F and pSC-tul1B-R; and sequence 3′ to tul1 (2.5 kb) was amplified using primers pSC-tul1C-F and pSC-tul1C-R. The backbone of the vector (3 kb) was amplified from the plasmid pMF272 using primers pSC-tul1D-F and pSC-tul1D-R. Split-marker fragments of the tul1 deletion cassette were amplified from pSCtul1 by PCR using the primer pairs pSC-tul1A-F with pSC-SM-R and pSC-SM-F with pSC-tul1C-R. The sah2 deletion vector, pSCsah2, was constructed in a manner similar to pSCtul1. Three fragments were amplified from QM6a genomic DNA: sequence 5′ to sah2 (2.0 kb) was amplified using primers pSC-sah2A-F and pSC-sah2A-R; the pyr4 cassette (1.8 kb) was amplified using primers pSC-sah2B-F and pSC-sah2B-R; and sequence 3′ to sah2 (2.5 kb) was amplified using primers pSC-sah2C-F and pSC-sah2C-R. The backbone of the vector (3 kb) was amplified from the plasmid pMF272 using primers pSC-sah2D-F and pSC-sah2D-R. Split-marker fragments of the sah2 deletion cassette were amplified from pSCsah2 by PCR using the primer pairs pSC-sah2A-F with pSC-SM-R and pSC-SM-F with pSC-sah2C-R. The split-marker fragments of the tul1 and sah2 deletion cassettes were transformed into QM6aΔpyr4 as described [52]. Transformants were screened on MM agar and the deletion tul1 and sah2 was verified by PCR (Additional file 5: Table S2, Additional file 7: Figure S5).
Trichoderma reesei strains were inoculated onto potato dextrose agar (PDA) plates, supplemented with 5 mM uridine or 300 μM hygromycin when necessary, and incubated at 30°C for 5–7 days. Conidia were harvested using a solution of 0.9% sodium chloride and 0.1% Tween 80 and then inoculated at 4 × 106 per milliliter into 50 mL MM supplemented with 1% Avicel prepared in 250-mL Erlenmeyer flasks. The flasks were incubated at 28°C with rotation (200 rpm). Samples of the culture supernatant (1.5 mL) were taken for protein level and enzyme activity assays every 24 h. T. reesei strains were cultured in biological replicates of at least three for statistical analysis by one-way ANOVA using the Dunnett’s multiple comparisons test.
Protein and enzyme assays
Culture supernatants were harvested from the N. crassa and T. reesei cultures; when necessary, particulate matter was removed by flushing the supernatants through a 0.2-μm filter. Total secreted protein levels were determined by Bradford assay (Bio-Rad Protein Assay). The Azo-CM-Cellulose reagent (from Megazyme) was used to determine endo-1,4-β-glucanase activity levels; endo-1,4-β-xylanase activity levels were determined using Azo-Xylan (Birchwood) (also from Megazyme). Avicelase assays measuring the release of glucose and cellobiose from Avicel substrate were performed as in [8]. All enzyme activity assays were scaled down in volume to be run in 96-well or deep-well 96-well plates.
Microscopy
For microscopy studies, conidia were inoculated onto agar plates or liquid Vogel’s with 2% Avicel and then incubated overnight at room temperature. Microscopy studies were performed using a Leica MZ 16F stereomicroscope or a Leica SD6000 spinning disk confocal microscope with a 488 or 561 nm laser. Fluorescent dyes included FM® 4–64 for staining plasma and vesicular membranes and ER-Tracker™ Red for staining the ER (both from Molecular Probes).
SDS-PAGE and immunoblots
Protein samples were combined with Laemmli sample buffer and then loaded on 10% Criterion™ Tris–HCl Precast Gels (from Bio-Rad) and run at a constant voltage. Coomassie staining was achieved using GelCode® Blue Stain Reagent (from Thermo Scientific). For immunoblot, proteins were transferred to nitrocellulose membrane using a Trans-Blot® SD Semi-Dry Transfer Cell system (from Bio-Rad). Specific proteins were detected using peptide antibodies generated by Thermo Scientific’s Pierce Custom Antibody Services: CDH-1 (NCU00206), GH5-1 (NCU00762), GH10-1 (NCU05924), GH61-5 (NCU08760), GH51-1 (NCU02343) and GH10-2 (NCU08189). In addition, an antibody directed against GLA-1 (NCU01517) was kindly provided by Stephen Free. These primary antibodies were detected using a goat anti-rabbit IgG (whole molecule)-peroxidase antibody (from Sigma) secondary antibody. The specific protein bands were visualized using the LumiSensor™ Chemiluminescent HRP Substrate Kit (from GenScript) and ChemiDoc™ XRS+ molecular imager with Image Lab™ software (from Bio-Rad).
Sequence analysis
When possible, protein sequences were obtained from either the N. crassa genome database maintained by the Broad Institute (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html); the T. reesei genome database (http://genome.jgi-psf.org/Trire2/Trire2.home.html); the S. cerevisiae genome database (http://www.yeastgenome.org); the S. pombe genome database (http://www.pombase.org); or the A. fumigatus genome database (http://www.aspergillusgenome.org). Sequences from other select fungal species were identified using the BlastP algorithm [54], which was also used to evaluate similarity between two sequences. Alignments of multiple protein sequences were performed using MUSCLE [55]. Evolutionary relationships between multiple protein sequences were analyzed with MEGA6 [56]. The phylogenetic tree for putative SAH-2 homologs was built using the neighbor-joining method: evolutionary distances were computed using Poisson correction and all gaps were eliminated from the data set.
Quantitative RT-PCR analysis
An aliquot of 2 × 106/mL of N. crassa conidial suspension was directly inoculated into 100 mL liquid Vogel’s with 2% Avicel as carbon source in 250-mL Erlenmeyer flasks and cultured at 25°C in constant light with rotation (200 rpm) for 120 h. Biological triplicates were prepared for each strain. Total RNA was isolated from samples taken at 48, 96 and 120 h post-inoculation using zirconia/silica beads and a Mini-Beadbeater with 1 mL TRIzol reagent (from Invitrogen). RNA purification and DNase I treatment were performed using RNeasy Mini Kit (from QIAGEN). qRT-PCR was performed using EXPRESS One-Step SYBR® GreenER™ Kit (from Invitrogen) on the CFX Connect™ Real-Time PCR Detection System (from Bio-Rad). Primer sequences used for amplification are described in Additional file 5: Table S2. The relative transcript level of major cellulase genes cbh-1 (NCU07340), cbh-2 (NCU09680), and gh5-1 (NCU00762) were normalized to expression from the actin gene (NCU04173) and calculated by as a relative expression level.
Authors’ contributions
MCR carried out the majority of the N. crassa experiments and prepared the manuscript. LQ performed the T. reesei work and some of the N. crassa work described in this study. JPC identified loci targeted for deletion in Table S1, Categories B and D; TLS identified loci targeted for deletion in Table S1, Category A. NLG participated in conception of the study and edited/revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was funded by the Energy Biosciences Institute. The authors wish to thank the following for their contributions: Kevin McCluskey and the Fungal Genetics Stock Center for the arrayed mutants; Samuel Coradetti and Elizabeth Znameroski for useful N. crassa strain backgrounds; Stephen Free for the GLA-1 antibody; and Philipp Benz for helpful discussion.
Compliance with ethical guidelines
Competing interests A patent application has been filed by the University of California-Berkeley on the identification of single-gene deletion mutants in Neurospora crassa associated with increased production of cellulases.
Abbreviations
- CBH
Cellobiohydrolase
- CLR
Cellulose degradation regulator
- DSC
Defective for SREBP cleavage
- GH
Glycosyl hydrolase
- SREBP
Sterol regulatory element binding protein
- VMM
Vogel’s minimal medium
- MM
Minimal medium
Additional files
Additional file 1: Table S1. Single-gene deletion strains available from the Fungal Genetics Stock Center that were included in screen.
Additional file 2: Figure S1. Phenotypic characterization of hypo- and hyper-production strains.
Additional file 3: Figure S2. Dsc protein complex sequence alignments.
Additional file 4: Figure S3. Deletion of Dsc E3 ligase complex and SREBP1 component homologs showed a short aerial hyphae and resulted in higher cellulase production.
Additional file 5: Table S2. Primers used in this study.
Additional file 6: Figure S4. DSC-1 and SAH-2 alignments with T. reesei protein sequences.
Additional file 7: Figure S5. Generation of tul1 and sah2 deletion mutants in T. reesei and their corresponding hyper-production phenotypes.
Contributor Information
Morgann C Reilly, Email: morgann.reilly@gmail.com.
Lina Qin, Email: bioqinln@berkeley.edu.
James P Craig, Email: jimcraig134@gmail.com.
Trevor L Starr, Email: t_starr@berkeley.edu.
N Louise Glass, Email: lglass@berkeley.edu.
References
- 1.Payen (1843) Extrait d’un adressé à m. le maréchal duc de dalmatie, ministre de lay guerre, président du conseil, sur une altération extraordinaire du pain de munition. Ann de Chìmie et de Phys 9:5–22
- 2.Ellison CE, Hall C, Kowbel D, Welch J, Brem RB, Glass NL, et al. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Natl Acad Sci USA. 2011;108:2831–2836. doi: 10.1073/pnas.1014971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perkins DD, Turner BC, Barry EG. Strains of Neurospora collected from nature. Evolution. 1976;30:281–313. doi: 10.2307/2407702. [DOI] [PubMed] [Google Scholar]
- 4.Beadle GW, Tatum EL. Genetic control of biochemical reactions in Neurospora. Proc Nat Acad Sci USA. 1941;27:499–506. doi: 10.1073/pnas.27.11.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, et al. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA. 2006;103:10352–10357. doi: 10.1073/pnas.0601456103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCluskey K. The Fungal Genetics Stock Center: from molds to molecules. Adv Appl Microbiol. 2003;52:245–262. doi: 10.1016/S0065-2164(03)01010-4. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch HM. Temperature-dependent cellulase production by Neurospora crassa and its ecological implications. Experientia. 1954;10:180–182. doi: 10.1007/BF02157201. [DOI] [PubMed] [Google Scholar]
- 8.Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, Cate JH, et al. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc Natl Acad Sci USA. 2009;106:22157–22162. doi: 10.1073/pnas.0906810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coradetti ST, Craig JP, Xiong Y, Shock T, Tian C, Glass NL. Conserved and essential transcription factors for cellulase gene expression in ascomycete fungi. Proc Natl Acad Sci USA. 2012;109:7397–7402. doi: 10.1073/pnas.1200785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips CM, Iavarone AT, Marletta MA. Quantitative proteomic approach for cellulose degradation by Neurospora crassa. J Proteome Res. 2011;10:4177–4185. doi: 10.1021/pr200329b. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, Tian C, Diamond S, Glass NL. Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryot Cell. 2012;11:482–493. doi: 10.1128/EC.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benz JP, Chau BH, Zheng D, Bauer S, Glass NL, Somerville CR. A comparative systems analysis of polysaccharide-elicited responses in Neurospora crassa reveals carbon source-specific cellular adaptations. Mol Microbiol. 2013;91:275–299. doi: 10.1111/mmi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glass NL, Schmoll M, Cate JH, Coradetti S. Plant cell wall deconstruction by ascomycete fungi. Annu Rev Microbiol. 2013;67:477–498. doi: 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- 14.Znameroski EA, Glass NL. Using a model filamentous fungus to unravel mechanisms of lignocellulose deconstruction. Biotechnol Biofuels. 2013;6:6. doi: 10.1186/1754-6834-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Znameroski EA, Coradetti ST, Roche CM, Tsai JC, Iavarone AT, Cate JH, et al. Induction of lignocellulose-degrading enzymes in Neurospora crassa by cellodextrins. Proc Nat Acad Sci USA. 2012;109:6012–6017. doi: 10.1073/pnas.1118440109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coradetti ST, Xiong Y, Glass NL. Analysis of a conserved cellulase transcriptional regulator reveals inducer-independent production of cellulolytic enzymes in Neurospora crassa. Microbiol Open. 2013;2:595–609. doi: 10.1002/mbo3.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schekman R. Charting the secretory pathway in a simple eukaryote. Mol Biol Cell. 2010;21:3781–3784. doi: 10.1091/mbc.E10-05-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penalva MA, Galindo A, Abenza JF, Pinar M, Calcagno-Pizarelli AM, Arst HN, et al. Searching for gold beyond mitosis: mining intracellular membrane traffic in Aspergillus nidulans. Cell Logist. 2012;2:2–14. doi: 10.4161/cl.19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Read ND. Exocytosis and growth do not occur only at hyphal tips. Mol Microbiol. 2011;81:4–7. doi: 10.1111/j.1365-2958.2011.07702.x. [DOI] [PubMed] [Google Scholar]
- 20.Kubicek CP, Starr TL, Glass NL. Plant cell wall degrading enzymes and their secretion in plant pathogenic fungi. Annu Rev Phytopathol. 2014;52:427–451. doi: 10.1146/annurev-phyto-102313-045831. [DOI] [PubMed] [Google Scholar]
- 21.Saloheimo M, Pakula TM. The cargo and the transport system: secreted proteins and protein secretion in Trichoderma reesei (Hypocrea jecorina) Microbiology. 2012;158:46–57. doi: 10.1099/mic.0.053132-0. [DOI] [PubMed] [Google Scholar]
- 22.Le Crom S, Schackwitz W, Pennacchio L, Magnuson JK, Culley DE, Collett JR, et al. Tracking the roots of cellulase hyperproduction by the fungus Trichoderma reesei using massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106:16151–16156. doi: 10.1073/pnas.0905848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vitikainen M, Arvas M, Pakula T, Oja M, Penttila M, Saloheimo M. Array comparative genomic hybridization analysis of Trichoderma reesei strains with enhanced cellulase production properties. BMC Genom. 2010;11:441. doi: 10.1186/1471-2164-11-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J, Glass NL. Identification of the CRE-1 cellulolytic regulon in Neurospora crassa. PLoS One. 2011;6:e25654. doi: 10.1371/journal.pone.0025654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogel HJ. A convenient growth medium for Neurospora. Microbiol Genet Bull. 1956;13:42–46. [Google Scholar]
- 26.Nitta M, Furukawa T, Shida Y, Mori K, Kuhara S, Morikawa Y, et al. A new Zn(II)(2)Cys(6)-type transcription factor BglR regulates beta-glucosidase expression in Trichoderma reesei. Fungal Genet Biol. 2012;49:388–397. doi: 10.1016/j.fgb.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Xiong Y, Sun J, Glass NL. VIB1, a link between glucose signaling and carbon catabolite repression, is essential for plant cell wall degradation in Neurospora crassa. PLoS Genet. 2014;10:e1004500. doi: 10.1371/journal.pgen.1004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamei M, Yamashita K, Takahashi M, Fukumori F, Ichiishi A, Fujimura M. Deletion and expression analysis of beta-(1,3)-glucanosyltransferase genes in Neurospora crassa. Fungal Genet Biol. 2013;52:65–72. doi: 10.1016/j.fgb.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Lew RR, Abbas Z, Anderca MI, Free SJ. Phenotype of a mechanosensitive channel mutant, mid-1, in a filamentous fungus, Neurospora crassa. Eukaryot Cell. 2008;7:647–655. doi: 10.1128/EC.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keeping A, Deabreu D, Dibernardo M, Collins RA. Gel-based mass spectrometric and computational approaches to the mitochondrial proteome of Neurospora. Fungal Genet Biol. 2011;48:526–536. doi: 10.1016/j.fgb.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 31.Hartmann-Petersen R, Semple CA, Ponting CP, Hendil KB, Gordon C. UBA domain containing proteins in fission yeast. Int J Biochem Cell Biol. 2003;35:629–636. doi: 10.1016/S1357-2725(02)00393-X. [DOI] [PubMed] [Google Scholar]
- 32.Stewart EV, Nwosu CC, Tong Z, Roguev A, Cummins TD, Kim DU, et al. Yeast SREBP cleavage activation requires the Golgi Dsc E3 ligase complex. Mol Cell. 2011;42:160–171. doi: 10.1016/j.molcel.2011.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- 34.Roumanie O, Wu H, Molk JN, Rossi G, Bloom K, Brennwald P. Rho GTPase regulation of exocytosis in yeast is independent of GTP hydrolysis and polarization of the exocyst complex. J Cell Biol. 2005;170:583–594. doi: 10.1083/jcb.200504108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meiling-Wesse K, Epple UD, Krick R, Barth H, Appelles A, Voss C, et al. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J Biol Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- 36.Stone PJ, Makoff AJ, Parish JH, Radford A. Cloning and sequence analysis of the glucoamylase gene of Neurospora crassa. Curr Genet. 1993;24:205–211. doi: 10.1007/BF00351793. [DOI] [PubMed] [Google Scholar]
- 37.Lloyd SJ, Raychaudhuri S, Espenshade PJ. Subunit architecture of the Golgi Dsc E3 ligase required for sterol regulatory element-binding protein (SREBP) cleavage in fission yeast. J Biol Chem. 2013;288:21043–21054. doi: 10.1074/jbc.M113.468215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willger SD, Cornish EJ, Chung D, Fleming BA, Lehmann MM, Puttikamonkul S, et al. Dsc orthologs are required for hypoxia adaptation, triazole drug responses, and fungal virulence in Aspergillus fumigatus. Eukaryot Cell. 2012;11:1557–1567. doi: 10.1128/EC.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowman BJ, Draskovic M, Freitag M, Bowman EJ. Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa. Eukaryot Cell. 2009;8:1845–1855. doi: 10.1128/EC.00174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kubicek CP. Systems biological approaches towards understanding cellulase production by Trichoderma reesei. J Biotechnol. 2012;163:133–142. doi: 10.1016/j.jbiotec.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collopy PD, Colot HV, Park G, Ringelberg C, Crew CM, Borkovich KA, et al. High-throughput construction of gene deletion cassettes for generation of Neurospora crassa knockout strains. Methods Mol Biol. 2010;638:33–40. doi: 10.1007/978-1-60761-611-5_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park G, Servin JA, Turner GE, Altamirano L, Colot HV, Collopy P, et al. Global analysis of serine-threonine protein kinase genes in Neurospora crassa. Eukaryot Cell. 2011;10:1553–1564. doi: 10.1128/EC.05140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes AL, Todd BL, Espenshade PJ. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 2005;120:831–842. doi: 10.1016/j.cell.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 44.Bien CM, Espenshade PJ. Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot Cell. 2010;9:352–359. doi: 10.1128/EC.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart EV, Lloyd SJ, Burg JS, Nwosu CC, Lintner RE, Daza R, et al. Yeast sterol regulatory element-binding protein (SREBP) cleavage requires Cdc48 and Dsc5, a ubiquitin regulatory X domain-containing subunit of the Golgi Dsc E3 ligase. J Biol Chem. 2012;287:672–681. doi: 10.1074/jbc.M111.317370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blatzer M, Barker BM, Willger SD, Beckmann N, Blosser SJ, Cornish EJ, et al. SREBP coordinates iron and ergosterol homeostasis to mediate triazole drug and hypoxia responses in the human fungal pathogen Aspergillus fumigatus. PLoS Genet. 2011;7:e1002374. doi: 10.1371/journal.pgen.1002374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chung D, Barker BM, Carey CC, Merriman B, Werner ER, Lechner BE, et al. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog. 2014;10:e1004487. doi: 10.1371/journal.ppat.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Griffiths B, Lewis CA, Bensaad K, Ros S, Zhang Q, Ferber EC, et al. Sterol regulatory element binding protein-dependent regulation of lipid synthesis supports cell survival and tumor growth. Cancer Metab. 2013;1:3. doi: 10.1186/2049-3002-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bardiya N, Shiu PK. Cyclosporin A-resistance based gene placement system for Neurospora crassa. Fungal Genet Biol. 2007;44:307–314. doi: 10.1016/j.fgb.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 50.Freitag M, Hickey PC, Raju NB, Selker EU, Read ND. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol. 2004;41:897–910. doi: 10.1016/j.fgb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 51.Catlett NL, Lee B-N, Yoder O, Turgeon BG. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet Newslett. 2003;50:9–11. [Google Scholar]
- 52.Qin LN, Cai FR, Dong XR, Huang ZB, Tao Y, Huang JZ, et al. Improved production of heterologous lipase in Trichoderma reesei by RNAi mediated gene silencing of an endogenic highly expressed gene. Bioresour Technol. 2012;109:116–122. doi: 10.1016/j.biortech.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 53.Penttila M, Nevalainen H, Ratto M, Salminen E, Knowles J. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene. 1987;61:155–164. doi: 10.1016/0378-1119(87)90110-7. [DOI] [PubMed] [Google Scholar]
- 54.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 55.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]


