Abstract
Background
Escherichia coli and Staphylococcus aureus are the main pathogens infectious to poultry, and their resistances against antibiotics have become troublesome currently. Biofilm formation is an important reason for drug resistance. Our previous research has found that the extract of Camellia oleifera seeds has lots of pharmacological effects. In order to find the substitute for antibiotics, the saponin was isolated from the defatted C. oleifera seeds with structural identification. Its efficacy was evaluated by the inhibition on amoxicillin-resistant E. coli and erythromycin-resistant S. aureus and therapeutic effect on chicks infected by the two bacteria.
Results
The bacterial growth inhibition rate increased and the bacterial count in vivo decreased significantly in dose dependence after administration of the saponin and its combination with amoxicillin or erythromycin, suggesting its antibacterial effect. The saponin identified as camelliagenin shows significant inhibition on the biofilm of E. coli and S. aureus, and it is related to the decrease of mannitol dehydrogenase (MDH) activity and extracellular DNA (eDNA) content. Molecular simulation reveals the strong interaction existing between the saponin and MDH or eDNA.
Conclusions
The mechanism of camelliagenin’s improvement on antibiotic effects is its interaction with MDH and eDNA in biofilm. The saponin is a prospective substitute of antibiotics, and molecular simulation is a convenient alternative method to find out hopeful candidates of antibiotics substitute.
Keywords: Camellia oleifera, Camelliagenin, Antibacterial effect, Bacterial biofilm, Antibiotic substitute
Background
Escherichia coli and Staphylococcus aureus are major pathogenic bacteria infectious to poultry. E. coli causes the colibacillosis such as acute septicaemia, vitelline peritonitis, enteritis, genital diseases, and so forth, its morbidity and lethality are the highest in bacterial diseases of chicks in China [1, 2]. The pathogen is currently treated by antibiotics, but its resistance against antibiotics can be easily acquired, and antibiotics lead to the deficiency of therapeutic effects [3]. S. aureus can cause acute septicaemia, arthritis, chick omphalitis, cutaneous necrosis and periostitis, and it is another pathogen leading great economic losses to the animal husbandry in China [4]. It is also easy to acquire drug resistance; especially methicillin resistant S. aureus (MRSA) is resistant to most of antibiotics. The resistance against antibiotics becomes great threat to animals and human being, and it is meaningful to find out the antibiotic substitute to reduce the usage of antibiotics. Herbs and plants offer plenty of compounds, which may replace antibiotics with strong antibacterial activities.
Camellia oleifera, an evergreen plant, mainly grows in the middle region of China. Its seeds are used for the extraction of edible oil. The yield of the seeds dramatically increases in recent years because of plant edible oil requirement and cultivation, the production of defatted seeds is up to 800,000 tons per year [5]. Although the defatted seeds are rich in active compounds [6, 7], they have been discarded without isolation and further exploitation. The total flavonoids, saponins and polysaccharides were detected in seeds of Camellia oleifera with the contents of 1–3, 10–14 and 15–20 % respectively [8–10]. The effective utilization of them will provide large amount of bioactive products. It is valuable to extract them for industrialization.
Our previous research has found that the extract of Camellia oleifera seeds have lots of pharmacological effects such as antioxidation, anti-inflammation and analgesia, which are mainly due to the saponin [11]. It is possible to develop the saponin extract as antibiotic substitute for animal husbandry because of its abundant and cheap resource.
In order to evaluate its effect and discuss the mechanism, the saponin was isolated from the defatted seeds of C. oleifera with structural identification. Its efficacy was evaluated by amoxicillin-resistant E. coli and erythromycin-resistant S. aureus induced infection in chicks. Bacterial biofilm formation is an important reason for the resistance against antibiotics, recurrence and difficulty to control by chemicals [12]. The action mechanism was revealed by inhibitory effect of the saponin on biofilm formation through interaction with mannitol dehydrogenase (MDH) activity and extracellular DNA (eDNA).
Results and discussion
Purity of the extract and structure of the purified compound
Natural saponins generally exist in forms of glycosides, and different kinds of glycosides of saponin have been found in seeds of C. oleifera [13]. Saponin glycosides are soluble in water and aqueous solvents. The extracts are the mixture of different glycosides, which can be achieved by current technique [14]. HCl aqueous solution with ultrasonic extraction applied in our research can hydrolyze saponin glycosides into sapogenin, which are insoluble in water and easily isolated by precipitation to get the purified products.
Average yield of the saponin extract was (12.5 ± 0.7) % by the separation technique in three repetitions. Purity of the extract was calculated by peak areas in HPLC (Fig. 1). Relative percentage of saponin is (82.3 ± 4.2) %. The yield and purity suggests that the technique is practical and cost effective because the ultrasonic assisted acid-base alternative extraction does not need organic solvent and expensive equipment. Ultrasonic can expedite dissolution in lower temperature, which protects activity of thermosensitive compounds in the extraction [15].
Fig. 1.
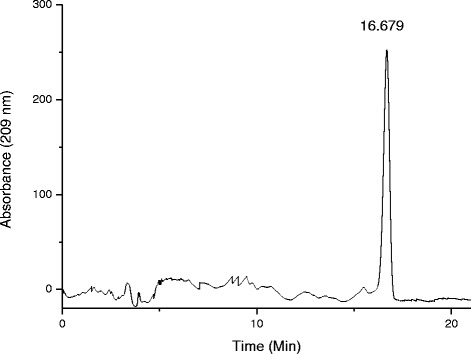
HPLC chromatograph of the saponin extract
The purified saponin was amorphous powder, insoluble in water, and soluble in ethanol, acetone and DMSO. mp 251.2–251.7 °C, m/z 488.1 (M+). There was an absorption peak of UV at 207 nm. IR spectra: hydroxyl (3435 cm−1), no characteristic absorption of α, β-unsaturated ester and ether. There were one carbonyl (d 9.49, J = 12.48), two olefinic protons (d 8.01, 7.49) but no Tig moiety signal in 1H NMR spectra. The signals of 13C NMR spectra were shown in Fig. 2 and attributable to the following: δ206.5 (C-25), 143.5 (C-12), 121.6 (C-13), 76.7 (C-19), 74.1 (C-2), 69.7 (C-31), 66.8 (C-16), 57.4 (C-3), 47.5 (C-4, 10, 22), 44.7 (C-17), 42.6 (C-11, 20), 40.3 (C-9, 18), 37.9 (C-5, 6, 15), 31.7 (C-8, 20), 28.4 (C-28, 29), 24.2 (C-14, 34), 15.3 (C-24, 26, 27). It is a sapogenin structure named camelliagenin (C30H48O5), which is consistent with literature [16]. Its structure is shown in Fig. 3.
Fig. 2.
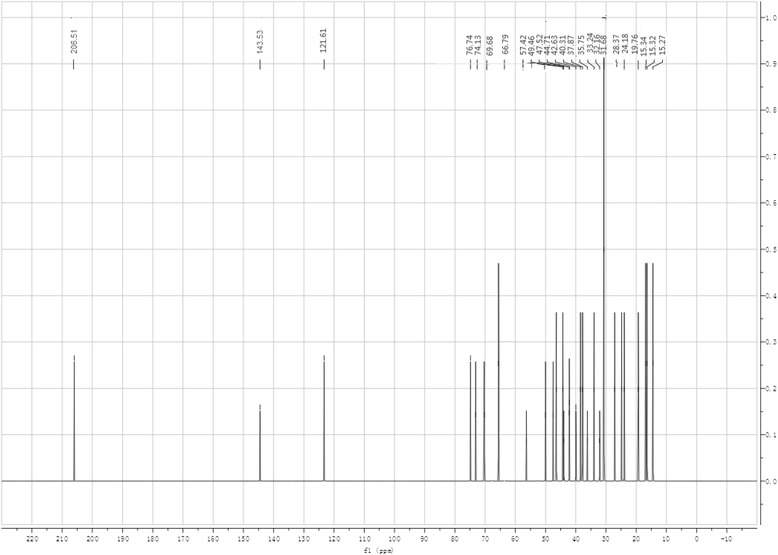
NMR spectra of the camelliagenin
Fig. 3.
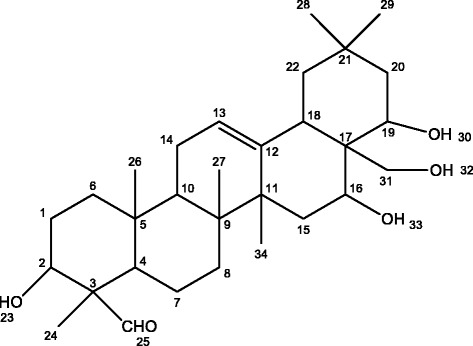
Structures of the saponin isolated from the defatted seeds of Camellia oleifera
Inhibitory effect of the saponin on bacteria in vitro and in vivo
Inhibitory effect of the saponin on bacteria in vitro was measured by MIC90 and IC50 of the camelliagenin on bacterial growth in culture medium. MIC90 of the saponin on 35 strains of E. coli and 30 strains of S. aureus was separately 71.4 ± 6.3 μg/ml and 94.5 ± 9.7 μg/ml. The camelliagenin had significant inhibition on growth of both E. coli and S. aureus, but its effect on E. coli was stronger than S. aureus suggestive of its selectivity. Amoxicillin and erythromycin had no obvious inhibitory effects on the two bacteria, indicating that the bacteria were resistant against the antibiotics. The camelliagenin plus antibiotics had better inhibition on bacteria than the camelliagenin or antibiotics alone, showing that the camelliagenin can strengthen bacterial sensitivity to antibiotics (Table 1).
Table 1.
Inhibition concentration (μg/ml) of the saponin extract on 50 % bacterial growth and biofilm formation of Escherichia coli and Staphylococcus aureus
| Groups | E. coli | S. aureus | ||
|---|---|---|---|---|
| Growth | Biofilm | Growth | Biofilm | |
| Amo | 72.6 ± 7.9 | 72.8 ± 9.6 | 138.6 ± 11.0 | 129.8 ± 10.5 |
| Sap | 50.2 ± 5.7a | 43.6 ± 4.2a | 79.6 ± 9.1a | 70.2 ± 10.2a |
| Sap : Amo (1:1) | 30.4 ± 6.5a,b | 22.8 ± 5.2a,b | 59.0 ± 9.6a,b | 52.4 ± 8.0a,b |
| Sap : Amo (5:1) | 23.4 ± 5.6a,b | 18.6 ± 5.7a,b | 43.2 ± 5.9a,b | 39.2 ± 3.5a,b |
| Sap : Amo (10:1) | 21.6 ± 5.1a,b | 15.6 ± 3.5a,b | 35.2 ± 4.5a,b | 30.2 ± 7.0a,b |
| Ery | 69.2 ± 6.3 | 65.6 ± 10.5 | 126.2 ± 6.7 | 122.8 ± 8.3 |
| Sap : Ery (1:1) | 28.2 ± 5.2b,c | 26.8 ± 6.8b,c | 51.6 ± 8.0b,c | 44.4 ± 6.1b,c |
| Sap : Ery (5:1) | 24.4 ± 4.7b,c | 19.2 ± 4.8b,c | 35.4 ± 5.3b,c | 28.6 ± 6.3b,c |
| Sap : Ery (10:1) | 13.4 ± 3.9b,c | 12.8 ± 5.1b,c | 27.6 ± 5.7b,c | 22.8 ± 6.2b,c |
Data were presented as mean ± standard deviation (n = 5)
Amo amoxicillin, Sap saponin, Ery erythromycin
a p < 0.01, compared with amoxicillin
b p < 0.01, compared with the saponin
c p < 0.01, compared with erythromycin
Inhibition of the camelliagenin on bacteria in vivo was evaluated by its therapeutic effect on chicks induced by E. coli and S. aureus. Chicks in negative groups showed serious symptoms of depression, anorexia, dullness, and diarrhea. Amoxicillin and erythromycin did not relieve these symptoms obviously, suggesting bacterial resistance against the antibiotics. The camelliagenin and combined administration of the camelliagenin and antibiotics significantly (p < 0.01) increased the body weight, immune organ index and reduced bacterial counts of liver in dose dependence, indicating that the camelliagenin enhances bacterial sensitivity to antibiotics and improves chicks immunity. The results are shown in Fig. 4. It further proves that the camelliagenin can not only substitute antibiotics, but also enhance antibiotic effects.
Fig. 4.
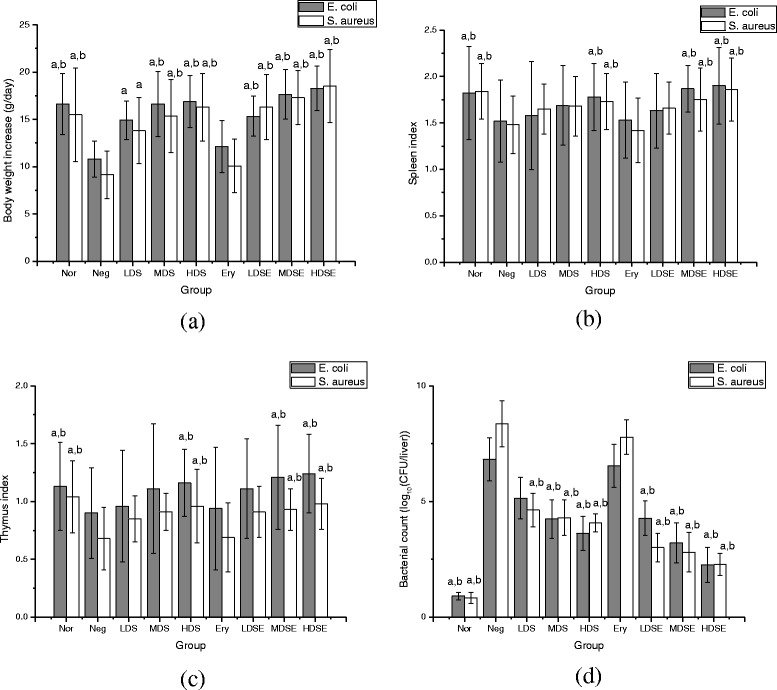
The effects of the saponin on body weight increase (a), spleen index (b), thymus index (c) and bacterial count (d) in liver of chicks infected by Escherichia coli and Staphylococcus aureus. Chicks in each group were injected intraperitoneally by 0.2 ml (1 × 108 CFU/ml) of E. coli or S. aureus suspension except normal group, 10 h later administered with feed containing erythromycin (50 mg/kg) or the saponin (50 mg/kg in low dose, 250 mg/kg in middle dose, 500 mg/kg in high dose) respectively for consecutive 3 d. The chicks were weighed, monitored for 14 d. a, p < 0.01, compared with negative group; b, p < 0.01, compared with erythromycin group. Nor: normal group; Neg: negative group; LDS: low dose of the saponin; MDS: middle dose of the saponin; HDS: high dose of the saponin; Ery: erythromycin; LDSE: low dose of the saponin + erythromycin; MDSE: middle dose of the saponin + erythromycin; HDSE: high dose of the saponin + erythromycin
In order to reduce the number of experimental animals and limitation of models, inhibitory tests were both carried out on bacteria in vitro and in vivo. The results show positive effects on inhibition of bacterial growth, indicating that the camelliagenin is promising as an effective antibacterial agent for E. coli and S. aureus. It may take effects on other kinds of bacteria, but needs further evaluation.
Effects of the camelliagenin on biofilm formation and exudates
Bacterial biofilm is a key factor to induce resistance against antibiotics and a target of new antimicrobials [17]. Thus the biofilms of E. coli and S. aureus were analyzed in our research. The results showed that amoxicillin or erythromycin had no different effect on biofilms from negative group (Table 1), and IC50 of the camelliagenin on biofilm of the two bacteria were lower than that of amoxicillin or erythromycin at significant level (p < 0.01). It indicates that antibacterial effects of the camelliagenin are related to its inhibition on bacterial biofilm formation.
The expression of specific genes is involved in biofilm formation and responsible for bacterial drug resistance [18]. The main difference between biofilm bacteria and planktonic bacteria is that biofilm bacteria are tightly packed and wrapped in their own secreted extracellular polysaccharide matrix called extracellular polymeric substances (EPS). The main component of EPS is alginate [19]. Mannitol dehydrogenase is a key enzyme in alginate synthesis process of biofilm [20]. Some antimicrobials can destroy biofilm formation since it inhibits activity of mannitol dehydrogenase in biosynthetic pathway of alginate [21]. In addition, large amount of extracellular DNA (eDNA) is found in the biofilm [22], it not only affects the formation of biofilms, but also increases the resistance of biofilms by chelating cation [23]. It is proved that eDNA enzymes can clear immature biofilm in vitro [24], and biofilm formation can be regulated by mannitol dehydrogenase and eDNA [25].
MDH and eDNA in biofilm were measured respectively by a decrease in the absorbance of reactive mixture at 340 nm and biofilm lysate at 260 nm compared to untreated controls. ∆A340 and ∆A260 increased significantly (p < 0.01) after the treatments of the camelliagenin at concentration dependence (Fig. 5). It suggests that they play a role in the inhibition of MDH and eDNA in the biofilms.
Fig. 5.
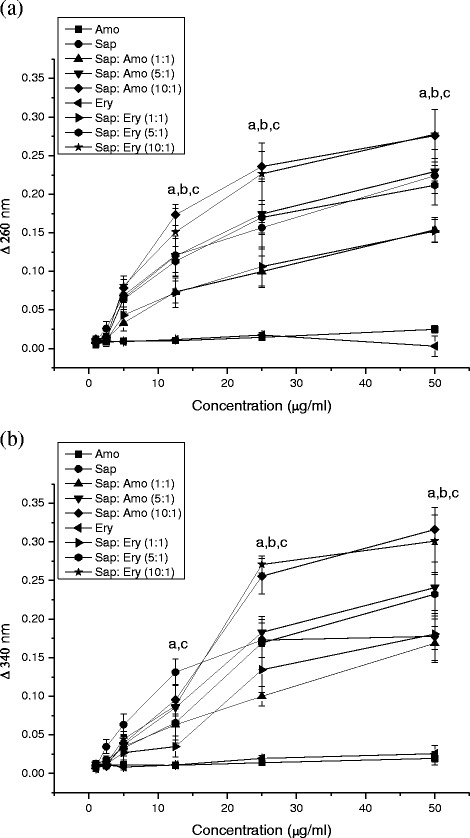
Activity of mannitol dehydrogenase (a) and content of extracellular DNA (b) in bacterial biofilm affected by the saponin and antibiotics (, n = 5). Data were measured respectively by a decrease in the absorbance of reactive mixture at 340 nm and biofilm lysate at 260 nm compared to untreated controls. ∆ 340 nm reflects decrease of mannitol dehydrogenase activity, and ∆ 260 nm reflects decrease of eDNA in biofilm. a, p < 0.01, compared with amoxicillin; b, p < 0.01, compared with the saponin; c, p < 0.01, compared with erythromycin. Amo: amoxicillin; Sap: saponin; Ery: erythromycin
Interaction of the camelliagenin with mannitol dehydrogenase and eDNA
Molecular docking as a better method of molecular simulation is applied in this research. Docking simulation shows that the saponin can interact with mannitol dehydrogenase (MDH) and eDNA. Each molecule successfully docks in 10 poses. The average binding energy is separately −86.94 ± 1.99 kcal/mol and −105.01 ± 1.19 kcal/mol, the average interactive energy is respectively 39.70 ± 2.28 kcal/mol and 19.77 ± 1.64 kcal/mol. It suggests that the camelliagenin can spontaneously bind to MDH and eDNA and exerts stronger interaction. Mimic diagrams of the camelliagenin binding to mannitol dehydrogenase and DNA are shown in Fig. 6, illustrating that they can well bind and interact with each other.
Fig. 6.
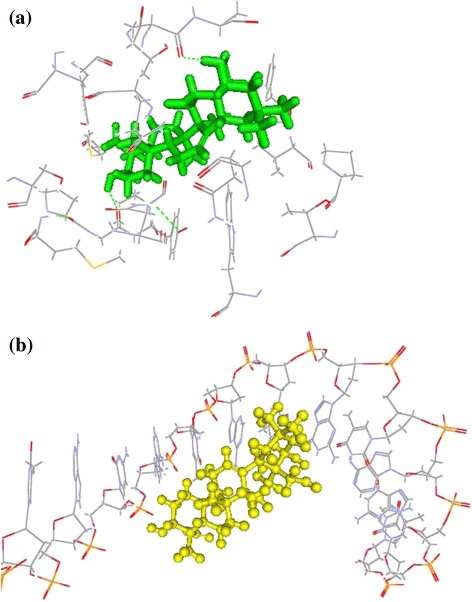
Interaction of the saponin with mannitol dehydrogenase (a) and extracellular DNA (b)
Based on the lock-key principle and complementary structural hypothesis, molecular docking simulates mutual interaction between ligand and receptor [26]. If receptor and ligand can interact, they must approach each other, and then combine in a particular conformation of the binding site, finally reach stable complex by adjusting conformation. Correct affinity prediction is conducive to drug design and screening. The main factors affecting the binding stability of ligand and receptor are hydrophobic force and bonding force. Free energy value is an important parameter for evaluation of docking affinity, binding activity and stability of receptor and ligand, and it can be used to judge the interaction of ligand and receptor. We simulated the interaction of the camelliagenin with mannitol dehydrogenase and eDNA, the result shows a good correlation between the interaction and its anti-biofilm effect. It suggests that molecular docking can be used to predict the saponin effect on biofilm inhibition.
Saponins are detergent-like substances showing antibacterial potential, and the mechanism deserves discussion. The critical micelle concentration of the camelliagenin is 0.5 % [27], but its MIC90 and IC50 are far away from the concentration whereas it shows detergent activity, demonstrating that its antibacterial activity is not due to the surface property. It is reported that the effect of saponin is associated with the changing of membrane permeability of Gram-negative cells in contrast to Gram-positive cells. Saponin might interact with Gram-negative cells components, like lipid A and thereby increase the permeability of bacterial cell wall [28]. Because the camelliagenin can reduce mannitol dehydrogenase and eDNA in biofilm, we provide a new hypothesis that the camelliagenin interacts with key components in biofilm, and blocks their activities in biofilm formation. The interaction of the camelliagenin with mannitol dehydrogenase and eDNA contributes to its anti-biofilm activity.
Conclusions
The camelliagenin is isolated from the defatted seeds of C. oleifera by ultrasonic assisted acid–base alternative technique. This is cost effective for industrialization because no organic solvent and expensive equipment are applied in the process. It is identified as camelliagenin and has significant antibacterial activities on E. coli and S. aureus in vitro and in vivo. It suggests that the camelliagenin is hopeful candidate for prevention of antibiotic resistance. The effects of the camelliagenin include its inhibition on bacterial biofilm formation, which is related to MDH and eDNA in the biofilm. The camelliagenin can spontaneously bind to and interact with MDH and eDNA. That could be the mechanism of the camelliagenin action on the two bacteria.
Methods
Drugs and bacteria
The defatted seeds of Camellia oleifera Abel were collected from oil manufacturing company (Meizhou, China). Escherichia coli was collected from fecal samples of chicks with generalized colibacillosis in Guangdong Chicken Farm, and identified as amoxicillin-resistant E. coli using the reported method [29]. Staphylococcus aureus was collected from nasal swab species of chicks with staphylocosis in Guangxi Chicken Farm, and identified as erythromycin-resistant S. aureus by the method in the reference [30]. The fecal and nasal samples from the chickens were taken after we obtained consent from the farm owners. E. coli control strain ATCC25922 and S. aureus control strain ATCC25923 were purchased from Guangdong Microbial Institute (Guangzhou, China). Reagents for biofilm assay were NH broth, crystal violet, amoxicillin and erythromycin purchased from Shanghai Hualan Biochemical Company (Shanghai, China). Chemical reference substance camelliagenin was bought from Shanghai Chemicals Company (Shanghai, China). Other reagents were purchased from Guangzhou Reagent Company (Guangzhou, China).
Animals
The experiments were carried out on ten-day-old male Roman chicks. The chicks were housed under conditions of 24 ± 2 °C, 50 ± 10 % humidity with a 12 h light/ dark cycle for 1 week adaptation. Food and water were accessible ad libitum. The experiments have been performed in accordance with the Chinese guidelines for the use of laboratory animals, and received approval from the animal experimentation ethic committee of South China University of Technology. All efforts were made to minimize animal suffering and to reduce the number of animals used. All sections of this report adheres to the ARRIVE guidelines for reporting animal research. A completed ARRIVE guidelines checklist is included in Checklist.
Procedure of the isolation
1 kg of the defatted seeds of C. oleifera were crushed to pass through 20 mesh sieve, submerged in 2 % HCl aqueous solution (water / seeds = 20 / 1 in ml / g) with ultrasonic at 300 W for 1 h. In order to protect the compounds from heat, temperature was controlled below 60 °C with cooling water. The extract solution was stood for 5 h. The precipitate was washed with 1000 ml of 2 % NaOH and 500 ml of water, and dried in vacuum to get the saponin extract.
Determination of purity by HPLC
The analysis of saponin was run on HP 1100 HPLC (Agilent Company, USA) in the following operating conditions: column: Hypersil ODS (250 × 4.6 mm, 5 μm); flow phase: methanol/water (80/20); injection volume: 10 μl; flow rate: 1 ml/min; temperature: 25 °C; wavelength: 209 nm [31].
Structure determination
1 g of saponin extract was dissolved in 5 ml of 80 % methanol, and purified by silica gel chromatograph with elution by chloroform/methanol (8/2). The peaks were collected and dried in vacuum, and 0.5 g saponin was obtained for structural analysis.
UV spectra analysis was carried out on UV-3010 Ultra violet spectrometer (Hitachi Company, Japan) scanning from 200 to 600 nm. IR spectra were measured on Nicolet 380 FI-IR spectrograph (Nicolet Apparatus Company, USA) with KBr tablets from 4000 to 400 cm−1 with resolution 2 cm−1. Mass spectra were recorded on Bruker Esquire Hct Plus Mass spectrometer with ESI (Bruker Company, Germany) in m/z of cation model scanning from 150 to 1200 for 60 min. NMR spectra were determined on 400 MHz AM NMR (Bruker Company, Switzerland) in DMSO-d6 operating at 101 MHz for 13C NMR and 400 MHz for 1H NMR.
Antibacterial activity
The antibacterial activity of the saponin against E. coli and S. aureus was examined using the microbroth dilution method according to the CLSI standard [32]. Thirty-five strains of E. coli and 30 strains of S. aureus were isolated using the method reported [33, 34], they were respectively inoculated in broth medium, which was cultured for 24 h at 37 °C, and diluted to 105 CFU/ml. Serial dilutions were prepared from 500 μg/ml of the saponin using DMSO to make 500, 250, 125, 62.5, 31.25, and 15.625 μg/ml. The wells were inoculated with 0.1 ml aliquot of test bacteria (105 CFU/ml) having serial dilutions of the saponin (50 μl, each). The micro plate was incubated at 37 °C ± 1 °C for 24 h. Dilution of the saponin to respective test organism showing no visible growth was considered as MIC. The minimum inhibitory concentration on 90 % bacterial strains was calculated as MIC90.
Bacterial biofilm experiment
Bacterial biofilm experiments were carried out according to the reference [35]. The diluted bacteria (105 CFU/ml) of E. coli and S. aureus were divided into the saponin groups, negative control (no drug), antibiotic groups (amoxicillin and erythromycin) and groups of the saponin plus antibiotics (1:1, 5:1, 10:1) with 5 repetition wells for each. The extracts were dissolved with water to 500 μg/ml. 50 μl of drug solution and 450 μl of diluted bacteria (final drug concentration 50 μg/ml) were mixed in one well of 24 well plates, cultured for 24 h, and then absorbance in 490 nm (OD490) was measured with water as blank to calculate the bacterial growth inhibition rate. The medium in the cell was carefully removed and washed 3 times with 1 ml of distilled water, and 1 % crystal violet solution (500 μl) was added to dye the biofilm. 30 min later, the solution was aspirated, and 500 μl of ethanol was added to extract crystal violet from the biofilm. The absorbance in 560 nm (OD560) was determined with ethanol as blank to calculate the biofilm inhibition rate.
Inhibitory concentrations (IC50) of the drugs on 50 % bacterial growth and biofilm formation were determined by dilution until the inhibition rate was 50 %. It is used as the index to evaluate the inhibitory effects on bacteria.
Assay of mannitol dehydrogenase and extracellular DNA in biofilm
The saponin extract and the antibiotics were diluted to final drug concentration in culture medium at 50, 25, 12.5, 5, 2.5, 1 μg/ml for 48 h bacteria culture. After removing the medium, the biofilm was homogenized in 500 μl of 50 mmol/l phosphate buffer (pH 5.5), 10 μl of the supernatant was taken for enzyme activity assay, and the rest was used for extracellular DNA determination.
Mannitol dehydrogenase (MDH) activity was determined according to a modified method [36]. Briefly, MDH was measured by the decrement of NADH, which was monitored by the absorbance at 340 nm. The reaction mixture contained 50 μl of 200 mM sodium phosphate buffer (pH 5.5), 50 μl of 2 mM NADH, 50 μl of water and 10 μl of the biofilm extract. The mixture was maintained at 32 °C for 2 min, and the reaction was started by adding 40 μl of 1 M fructose and lasted for 5 min. The absorbance at 340 nm was detected by UV-3010 spectrometer (Hitachi Company, Japan).
Extracellular DNA (eDNA) was measured in the following protocol [37]. The biofilm extract was mixed with 10 U/ml cellulase at 37 °C for 1 h, followed by treatment with 10 U/ml proteinase K for another 1 h. Treated samples were centrifuged at 10, 000 g for 10 min. The supernatant was collected and the absorbance was detected at 260 nm by UV spectrometer to deduce eDNA content in biofilm.
Antibacterial test in vivo
The antibacterial tests were performed as the reported with some modification [38]. Two tests including E. coli and S. aureus were carried out separately. For one test, the chicks were randomly divided into 9 groups with 30 chicks in each group, including normal group, negative group, antibiotic group, the saponin extract and the saponin extract plus antibiotic groups in high, middle and low dose. The groups of chicks except normal group were injected intraperitoneally with 0.2 ml of 1 × 108 CFU/ml bacterial suspension, 10 h later they were administered with 1 kg of feed containing 50 mg amoxicillin or erythromycin in antibiotic group, 50 mg, 250 mg and 500 mg in low, middle and high dose of the saponin extract respectively for consecutive 3 d. The doses were based on regular use of antibiotics and safety of the saponin [39]. The chicks were weighed, monitored for 14 d, and evaluated by relief of symptoms of anorexia, lassitude, diarrhea, etc. [40]. The chicks were euthanized on the fifteenth day, thymus and spleen were removed and weighed to calculate immune organ index (organ weight / body weight). Chick liver was plated onto Tryptose Soya Agar (TSA) plates for bacterial colony count, and the bacterial density was expressed as the number of CFU per liver.
Simulation of molecular interaction
Molecular docking was simulated by Discovery Studio V2.5 software (Accelry Inc., CA, USA) in the following procedure [41]. Structural data of mannitol dehydrogenase and eDNA were downloaded from Brookhaven Library. The missing amino acids and hydrogens were supplemented, and excessive protein conformation was removed. Mannitol dehydrogenase and eDNA were separately defined as the receptor, and then binding sites and coordinates were defined. A new plot window was open to draw structure of the extracts, optimize 3 D geometric structures, and apply CHARMm force field to ensure correct bond length, bond angle in a state of energy stability. CDOCKER Protocol was run to obtain binding parameters, which are used to evaluate the interaction. CDOCKER energy and CDOCKER interaction energy are two important parameters, and their absolute values are in according with the affinity and action force between the receptor and ligand.
Statistical analysis
Data was presented as mean ± standard deviation (). SPSS 11.0 software (SPSS Inc., USA) was used to analyze the data of animal tests in groups by one-way ANOVA and Dunnett’s test.
Acknowledgements
The authors thank the staff in the laboratory centre of South China University of Technology for analyzing spectroscopic data. The financial support for this work from the NSFC (No. 81173646) and Guangzhou Scientific Plan Project (No. 201510010065) are also gratefully acknowledged.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YY designed the study and wrote and critically revised the manuscript. QY, FF, and YL performed the experiments, analyzed the data, and drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Yong Ye, Email: yeyong@scut.edu.cn.
Qian Yang, Email: 2223841517@qq.com.
Fei Fang, Email: fang100fei@126.com.
Yue Li, Email: liyue248@126.com.
References
- 1.Jing MJ, Jing XJ, Wang YJ. Investigation on epidemiology of chicken colibacillosis. Chin Livest Poult Breed. 2011;7:151–152. [Google Scholar]
- 2.Tian W, Zhao JJ, Deng YT, Yao QF, Chen XJ, Liu JH. Prevalence of oqxAB multidrug efflux pump in Escherichia coli isolated from animals. Chin Anim Husb Vet Med. 2011;38:156–159. [Google Scholar]
- 3.Zhang T, Wang CG, Lv JC, Wang RS, Zhong XH. Survey on tetracycline resistance and antimicrobial-resistant genotype of avian Escherichia coli in North China. Poult Sci. 2012;91:2774–2777. doi: 10.3382/ps.2012-02453. [DOI] [PubMed] [Google Scholar]
- 4.Dai FW, Ke XF, Zhou SS, Song XM, Sa XY. The epidemiology of methicillin-resistant staphylococcus aureus (MRSA) in animals. Chin J Compar Med. 2010;20:81–85. [Google Scholar]
- 5.Li WL, Huang FH, Wang LB. Review on processing and comprehensive utilization of oil tea camellia seed. China Oils Fats. 2011;36:55–57. [Google Scholar]
- 6.Li XZ, Zhang S, Li WS. α-Glucosidase inhibitory activity of Camellia cake polysaccharides in vitro. J Hunan Univ (Nat Sci) 2012;39:71–73. [Google Scholar]
- 7.Luo YM, Li B, Xie YH. Study on the chemical constituents of Camellia oleifera Abel. Chin Trad Herbal Drugs. 2003;34:117–118. [Google Scholar]
- 8.Chen YF, Yang CH, Chang MS, Ciou YP, Huang YC. Foam properties and detergent abilities of the saponins from Camellia oleifera. Int J Mol Sci. 2010;11:4417–4425. doi: 10.3390/ijms11114417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du LC, Wu BL, Chen JM. Flavonoid triglycosides from the seeds of Camellia oleifera Abel. Chin Chem Lett. 2008;19:1315–1318. doi: 10.1016/j.cclet.2008.09.011. [DOI] [Google Scholar]
- 10.Wei XL, Mao FF, Cai X, Wang Y. Composition and bioactivity of polysaccharides from tea seeds obtained by water extraction. Int J Biol Macromol. 2011;49:587–590. doi: 10.1016/j.ijbiomac.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 11.Ye Y, Xing HT, Chen XL. Anti-inflammatory and analgesic activities of the hydrolyzed sasanquasaponins from the defatted seeds of Camellia oleifera. Arch Pharm Res. 2013;36:941–951. doi: 10.1007/s12272-013-0138-y. [DOI] [PubMed] [Google Scholar]
- 12.Blackledge MS, Worthington RJ, Melander C. Biologically inspired strategies for combating bacterial biofilms. Curr Opin Pharmacol. 2013;13:699–706. doi: 10.1016/j.coph.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao DF, Xu M, Zhao P, Zhang XY, Wang YF, Yang CR, Zhang YJ. Kaempferol acetylated glycosides from the seed cake of Camellia oleifera. Food Chem. 2011;124:432–436. doi: 10.1016/j.foodchem.2010.06.048. [DOI] [Google Scholar]
- 14.Chen JH, Liau BC, Jong TT, Chang CM. Extraction and purification of flavanone glycosides and kaempferol glycosides from defatted Camellia oleifera seeds by salting-out using hydrophilic isopropanol. Sep Purif Technol. 2009;67:31–37. doi: 10.1016/j.seppur.2009.02.020. [DOI] [Google Scholar]
- 15.Ghafoor K, Choi YH, Jeon JY, Jo IH. Optimization of ultrasound-assisted extraction of phenolic compounds, antioxidants, and anthocyanins from grape (Vitis vinifera) seeds. J Agri Food Chem. 2009;57:4988–4994. doi: 10.1021/jf9001439. [DOI] [PubMed] [Google Scholar]
- 16.Toshiyuki M, Junko N, Hisashi M, Masayuki Y. Bioactive saponins and glycosides. XV.1) saponin constituents with gastroprotective effect from the seeds of tea plant, Camellia sinensis L. var. assamica, cultivated in Sri Lanka: structures of assamsaponins A, B, C, D, and E. Chem Pharm Bull. 1999;47:1759–1764. doi: 10.1248/cpb.47.1759. [DOI] [PubMed] [Google Scholar]
- 17.Haussler S, Parsek MR. Biofilms 2009: new perspectives at the heart of surface-associated microbial communities. J Bacteriol. 2010;192:2941–2949. doi: 10.1128/JB.00332-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen PØ, Bjarnsholt T, Phipps R, Rasmussen TB, Calum H, Christoffersen L, Moser C, Williams P, Pressler T, Givskov M, Høiby N. Rapid necrotic killing of poly-morphonuelear leukocytes is caused by quorum –sensing-controlled production of rhamnolipid by Escherichia Coli. J Microbiol. 2007;53:1329–1338. doi: 10.1099/mic.0.2006/003863-0. [DOI] [PubMed] [Google Scholar]
- 19.Orgad O, Oren Y, Walker SL, Herzberg M. The role of alginate in Escherichia Coli EPS adherence, viscoelastic properties and cell attachment. Biofouling. 2011;27:787–798. doi: 10.1080/08927014.2011.603145. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee PC, Vanags RI, Chakrabarty AM. Alginic acid synthesis in Escherichia Coli mutants defective in carbohydrate metabolism. J Bacteriol. 1983;155:238–245. doi: 10.1128/jb.155.1.238-245.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin CA, Hoven AD, Cook AM. Therapeutic frontiers: preventing and treating infectious diseases by inhibiting bacterial quorum sensing. Eur J Clin Microbiol Infect Dis. 2008;27:635–642. doi: 10.1007/s10096-008-0489-3. [DOI] [PubMed] [Google Scholar]
- 22.Nur A, Hirota K, Yumoto H. Effects of extracellular DNA and DNA-binding protein on the development of a Streptococcus intermedius biofilm. J Applied Microbiol. 2013;115:260–270. doi: 10.1111/jam.12202. [DOI] [PubMed] [Google Scholar]
- 23.Mulcahy H, Charron ML, Lewenza S. Extracellular DNA chelates cations and induces antimicrobial resistance in Escherichia Coli biofilms. PLoS Pathog. 2008;4:1213–1218. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ou YZ, Zhu YL, Chen JM. The mechanism of JSfnp Zococc epidemics AtlE protein mediating primary attachment of biofilm formation. Fudan Univ J Med Sci. 2006;33:569–573. [Google Scholar]
- 25.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan AX, Chen J, Liu HD. Applications and developments of molecular docking method. J Anal Sci. 2009;25:473–477. [Google Scholar]
- 27.Chen YF, Yang CH, Chang MS, Ciou YP, Huang YC. Foam properties and detergent abilities of the saponins from Camellia oleifera. Int J Mol Sci. 2010;11:4417–4425. doi: 10.3390/ijms11114417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arabski M, Wasik S, Dworecki K, Kaca W. Laser interferometric and cultivation methods for measurement of colistin/ampicilin and saponin interactions with smooth and rough of Proteus mirabilis lipopolysaccharides and cells. J Microbiol Methods. 2009;77:178–183. doi: 10.1016/j.mimet.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 29.El-Rami FE, Sleiman FT, Abdelnoor AM. Identification and antibacterial resistance of bacteria isolated from poultry. Pol J Microbiol. 2012;61:323–6. doi: 10.1099/jmm.0.035550-0. [DOI] [PubMed] [Google Scholar]
- 30.Wang GQ, Xing ZH, Wei YZ. Antimicrobial resistance of Staphylococcus aureus isolated from chicken. J Agri Sci. 2011;32:43–49. [Google Scholar]
- 31.Zhao JJ, Du XF. Sasanquasaponin reference substance preparation and quantitative analysis by HPLC. China Oil Fats. 2009;34:68–7. [Google Scholar]
- 32.Umer S, Tekewe A, Kebede N. Antidiarrhoeal and antimicrobial activity of Calpurnia aurea leaf extract. BMC Complem Altern Med. 2013;13:21–25. doi: 10.1186/1472-6882-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang JL. Isolation and identification of drug-resistant Escherichia coli strains from chickens. Agri Sci Tech. 2012;13:1699–1701. [Google Scholar]
- 34.Qiu M, Hao Z, Zhang WJ, Shen W, Wu B, Li YH, Zhang GX. Isolation and identification of Staphylococcus aureus from animals and drug resistance analysis. Chin Agri Sci Bull. 2011;27:356–359. [Google Scholar]
- 35.Robert WH, Justin JR. Inhibition of Escherichia Coli biofilm formation with bromoageliferin analogues. J Am Chem Soc. 2007;129:6966–6967. doi: 10.1021/ja069017t. [DOI] [PubMed] [Google Scholar]
- 36.Ortiz ME, Fornaguera MJ, Raya RR, Mozzi F. Lactobacillus reuteri CRL 1101 highly produces mannitol from sugarcane molasses as carbon source. Appl Microbiol Biotechnol. 2012;95:991–999. doi: 10.1007/s00253-012-3945-z. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, Xi C. Evaluation of different methods for extracting extracellular DNA from the biofilm matrix. Appl Environ Microb. 2009;75:5390–5395. doi: 10.1128/AEM.00400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haritova A, Urumova V, Lutckanov M, Petrov V, Lashev L. Pharmacokinetic-pharmacodynamic indices of enrofloxacin in Escherichia coli O78/H12 infected chicks. Food Chem Toxicol. 2011;49:1530–1536. doi: 10.1016/j.fct.2011.03.044. [DOI] [PubMed] [Google Scholar]
- 39.Hu SH, Wei J, Yan XW, Li QM. The poisonous study of Sasanqua saponin-the acute poisonous and accumulated poisonous of Sasanqua saponin to SD mouse through its mouth and skin. China Oils Fats. 1998;23:47–48. [Google Scholar]
- 40.Liang Y. Prevention and treatment on colibacillosis & pullorosis with extracts from Scutellaria baicalensis Georgi in chickens. J Trad Chin Vet Med. 2007;2:20–22. [Google Scholar]
- 41.Uthaman G, Mannu J, Durai S. Molecular docking studies of dithionitrobenzoic acid and its related compounds to protein disulfide isomerase: computational screening of inhibitors to HIV-1 entry. BMC Bioinf. 2008;9:S14–16. doi: 10.1186/1471-2105-9-S12-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]


