Abstract
Background &Objective:
Breast cancer is the most common female malignancy. Detection of DNA of human papillomaviruses (HPVs) in breast carcinomas suggests that the virus may play a role in the pathogenesis of this disease. The aim of this study was to evaluate the frequency of HPVs genotypes 6, 11, 16, 18 and 31 in paraffin-embedded tissue samples of invasive breast carcinomas.
Methods:
Three hundred and twenty six paraffin-embedded tissue samples of breast cancer were studied. PCR was performed using specific primers for HPV genotypes.
Results:
Of total 206 (63.2%) samples positive for Beta-globin gene, 54 (26.2%) were HPV-positive and 152 (73.8%) were negative for HPV. Distribution of HPV genotypes were as follows: 19 (25.7%) were positive for genotype 11, 5 (6.8%) were positive for genotype 6; and 2 cases (2.7%) were positive for both genotypes 6 and 11. Samples were also screened for HPV genotypes 16, 18 and 31 but none was positive.
Conclusion:
The current study confirmed the association of HPV and breast cancer. However, all samples were negative for high-risk HPV types 16, 18 and 31.
Key Words: Human Papillomavirus (HPV), Breast Cancer, Polymerase Chain Reaction (PCR)
Introduction
Human Papillomavirus (HPVs) can be transmitted between individuals through genitals, anal, mouth or breast (1). Breast cancer is the most common malignant tumor among women and the second highest cause of cancer-related death (2,3). Among Iranian women, mortality due to breast cancer is higher than deaths due to other kinds of cancer (4). More than 1,000,000 new cases are diagnosed each year in the United States of America and it caused about 458,000 deaths worldwide alone in 2008(5). Although the incidence of breast cancer has almost doubled in the past four years in the United States, the death rate remained noticeably constant. The loss of cellular regulation that gives rise to most of cancers is due to genetic damages. Numerous factors, especially mutations, are considered the risk factors in the onset of cancer (6,7). These are often unknown mutations, which are inherited or acquired. Breast cancer is rare in people under 25 years unless a family history of breast cancer exist (8). The risk of breast cancer increases with age. Although there are many patients without a family history of breast cancer, 13% of women with a family history are found to develop breast cancer. This finding reinforces the probability of the involvement of other non-genetical factors (9,10). Exercising, using medication, breastfeeding, life style, stress, genetic aptitude for the disease and other factors may have role in the genesis of breast cancer (9, 11,12). Virus infection may be responsible for breast cancer. However, more studies are required to confirm their associations.
HPVs are small and are capable of causing warts in some vertebrates, including humans. Based on phylogenetic relationships and their role in benign and malignant cervical cancers, HPVs are classified into three categories: high-risk, medium-risk and low-risk. HPV types 16, 18 and 45 fall into high-risk category while intermediate-risk group includes HPV types 31, 33, 35, 39, 51, 52, 53, 56, 58, 59, 66, 68, 73 and 82. Low-risk types include types 6, 11, 40, 42, 43, 44, 54, 61, 70, 72 and 81. These viruses have the ability to cause cancer in humans and animals (13). Some HPVs cause cervical cancer and epithelial tumors (14-17).
In the present study, the authors evaluated 326 paraffin-embedded samples in order to identify possible relationships between HPV infection and manifestations and progressions of breast cancer.
Materials and Methods
Sample collection
After obtaining approval from ethics committee of Mashhad University of Medical Sciences, samples collected between August 2002 and September 2012 from pathology departments of Imam Reza and Qaem hospitals in the city of Mashhad, northeast of Iran were studied in a cross-sectional study. Three hundred and twenty six paraffin-embedded samples of breast carcinoma were chosen and confirmed pathologically. Five sections of 10-20 micron were extracted from paraffin blocks in sterile conditions.
DNA extraction and PCR
Zylol-Ethanol method was used for deparaffinization of samples. For this purpose, paraffin-embedded samples were sliced in diameter of 20 micrometers using microtome and were kept in 1.5 ml micro-tubes. In the next step, 1 ml of zylol was added and the mixture was shaken for 30 min at 25 °C. Following that, it was centrifuged at 13000 rpm for 10 min. The supernatant was removed. This step was repeated for several times. Five hundred microliters of absolute ethanol was added to the pellet and it was shaken and centrifuged at 13000 rpm for 10 min. In the final step, the supernatant was removed again and this step was repeated for the second time. The pellet was kept at 25 °C so that it could lose ethanol but it was not allowed to dry completely. PCR primers were designed by Gene Runner (Hastings Software Inc.) software according to the pattern sequences in GenBank.
β-Globin gene PCR to assess the quality of extracted DNA
The quality of the extracted DNA from paraffin-embedded tissue samples was assessed by primers of β-globulin gene. PC04 and GH20 primers were used, which gave rise to PCR products of 260 bp in length (Table 1).
Table 1.
Primer sequences and the corresponding PCR product of Beta globin and L1
| Gene | Product size(bp) | Reference | ||
|---|---|---|---|---|
| Beta globin | GH20 | GAAGAGCCAAGGACAGGTAC | 260 | 16 |
| PC04 | CAACTTCATCCACGTTCACC | |||
| L1 (HPV) | + GP6 | GAAAAATAAACTGTAAATCATATTC | 142 | 18 |
| + GP5 | TTTGTTACTGTGGTAGATACTAC | |||
In order to amplify beta globin gene, the reaction mixture contained 0.8 μl of DNA, 0.4 μl of dNTPs (10mM), 0.8 μl of Taq DNA polymerase (5 U/μl), 1.6 μl of MgCl2 (25mM) , 10 pmol/μl of each primer, and 12.2 μl of H2O (DNase-RNase free). PCR program was 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 55 °C for 45 s, 72 °C for 45 s and a final extension of 72 C for 5 min.
PCR-based method for HPV genotyping
Genotyping was performed by PCR using virus-specific primers for different subtypes (Table 2). The reaction mixture for PCR reaction of HPV types 6, 16, 18 and 31 contained 2 µl of DNA, 1x reaction buffer, 10 µmol dNTPs, 50 mmol MgCl2, 10 pmol of each primers and 5 u/µl Taq DNA polymerase (Cinnagen, Iran). However, for type 11, 25mmol MgCl2 and 3 µl of DNA was used. PCR program was 94 °C for 5 min; 35 cycles including 94 °C for 45 s; Melting temperatures of 55 °C for type 11, 57 °C for type 6, 60.8 °C for types 18 and 31, and 63 °C for type 16; 72 °C for 45 s and was followed by final extension of 72 °C for 10 min. It produced the PCR products of 360 bp for type 11, 280 bp for type 6, 420 bp for types 18 and 31, and 400 bp for type 16. Finally, PCR products were visualized on a 2% agarose gel. Using specific primers, samples, which produced the corresponding fragments, were considered positive for that type of HPV.
Table 2.
Sequence of genotype-specific primers used in the PCR
| Primer | Sequence | Genotype | Reference |
|---|---|---|---|
| HPV 6 F | TAG TGG GCC TAT GGC TCGTC | 6 | (17) |
| HPV 6 R | TCC ATT AGC CTC CAC GGG TG | ||
| HPV 11 F | GGA ATA CAT GCG CCA TGT GG | 11 | (17) |
| HPV 11 R | CGA GCA GAC GTC CGT CCT CG | ||
| HPV 16- E6 F | CAG GAC CCA CAG GAG CGA CC | 16 | (18) |
| HPV 16 –E6 R | ATC GAC CGG TCC ACC GAC CC | ||
| HPV 18 –E6 F | GCT TTG AGG ATC CAA CAC GG | 18 | (18) |
| HPV18 – E6 R | TGC AGC ACG AAT GGC ACT GG | ||
| HPV 31-E6/E7 F | GAA ATT GCA TGA ACT AAG CTC G | 31 | (18) |
| HPV 31-E6/E7 R | CAC ATA TAC CTT TGT TTG TCA A |
Data Analysis
Frequency charts and diagrams together with SPSS v.20 (Chicago, IL, USA) was used for data explanation purposes.
Results
Results of beta-globin
Overall, 326 samples were used. Age ranged between 24 and 74 years. The maximum age of positive samples was between 35 and 40 years. Samples from 326 patients with invasive breast carcinoma showed that 206 cases (63.2%) were positive for PCR of Beta-globin gene and 120 cases (36.8%) were negative. Among positive samples, 54 cases (26.2%) were positive for HPV and 152 cases (73.8%) were negative. While among Beta globin-negative samples, 20 cases (16.7%) were HPV-positive and 100 (83.3%) were negative. Totally, 74 out of 326 samples (22.7%) were positive for HPV and 252 cases (77.3%) were HPV-negative.
Distribution of HPV genotypes
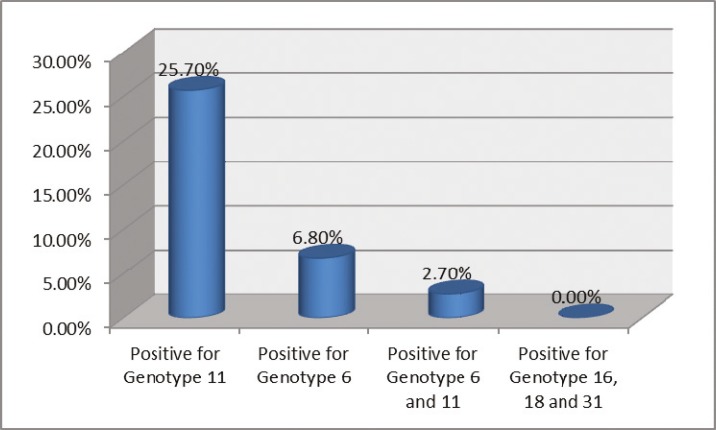
Nineteen cases (25.7%) were positive for genotype 11, and 5 cases (6.8%) were positive for genotype 6. Two cases (2.7%) were found to be positive for both genotypes 6 and 11 (Fig.1). Samples were also screened for genotypes 16, 18 and 31 and showed no positive result. Genotype of the virus was unknown for 48 cases (64.9%). In addition, there were 5 beta-globin negative cases among the 19 patients with genotype 11, and one out of five patients who was positive for genotype 6, was negative for beta-globin.
Fig. 1.
Number of patients according to HPV genotypes
Distribution based on tumor grade in HPV positive specimens
Among all HPV-positive cases, 1.4% had Grade A, 8.1% had grade B, 10.8% had grade C and 79.7% had unknown grade.
Distribution according to involvement of axillary lymph node
Of total HPV-positive samples, 16.2% had axillary lymph node involvement, 1.4% had no involvement and for the remaining 82.4%, the involvement was unclear.
Discussion
Breast cancer is the most common malignant tumor in women. HPV is a small, non-enveloped virus containing DNA, which is highly specific for both species and tissue. Several studies supported the role of HPV in breast cancer. Seyed Alavi and colleagues studied the role of HPV infection in breast cancer and its correlation with clinical parameters. The role of HPV in 24 cases (48%) of the total cases of breast cancer was identified. They also showed that 13 of them (26%) were infected with high-risk types of whereas in 8 cases (16%), they were infected with low-risk types of HPVs. In three breast cancer tissues (6%), both types of HPVs (High-risk and low-risk) were detected (3). Widschwendter et al. studied 11 patients with simultaneous cervical and breast cancers and observed that seven cases were positive for HPV (18). Kan et al. performed a study on DNA sequences of HPVs involved in breast cancers to assess possible correlations. They demonstrated that 24 samples (48%) of total 50 samples were positive for HPV (19). Khan and colleagues studied the association of HPV in breast carcinoma in Japan. In 26 patients (21%), breast carcinomas were observed. Among them, HPV type 16 was the most prevalent genotype, which has been considered as the main culprit (92%) for manifestation of disease (20). HPV DNA is highly associated with breast cancer (21, 22). Yasmeen and colleagues conducted a study to evaluate the role of onco-proteins E6 and E7 of HPV type 16 in cell invasion and metastasis of breast cancer. They reported that E6/E7 of genotype 16 is capable of making two invasive and metastatic cell lines: BT20 and MCF7 (23). HPV infection may have a significant role in the development of breast cancer (20, 24-27). Lee and colleagues used PCR method to study the role of HPV DNA in breast carcinoma. 24.49% of the cases of breast carcinomas were associated with HPV virus. In addition they also detected 40 types of HPVs with HPV type 33, 18, 16 and 35 being the most prevalent ones (28).
Some studies, on the other hand, did not provide evidence in favor of the role of HPV in breast cancer. Wrede et al. and Gopalkrishna et al. conducted studies to evaluate the presence of HPV types 16 and 18 in breast carcinomas. However, none of the samples showed any correlations (29, 30). A study was conducted to investigate the presence of HPV DNA in breast carcinoma in Korean women and the relationship between HPV and breast cancer progression. However, no noticeable correlations were observed (31). Throughout the last decade, several independent studies in different regions reported no significant correlation between HPV and the incidence and progression of breast cancer (32-35). The role of HPV in cancers such as uterine cervix has been widely accepted.
Conclusion
The current study confirmed the role of HPVs in breast carcinoma among embedded tissue samples of invasive breast carcinoma in northeast of Iran. However, further investigations with a much larger sample size are required to validate the exact role of HPVs in the prevalence and progression of this cancer.
Acknowledgment
This study was from a thesis presented for obtaining the medical doctor degree from Mashhad University of Medical Sciences, Mashhad, Iran (Thesis No. 6634). This study was financially supported by Mashhad University of Medical Sciences.
Conflict of interest statement
The authors declare that there is no conflict of interests.
References
- 1.Bosch FX, Manos MM, Muñoz N, Sherman M, Jansen AM, Peto J, et al. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J Natl Cancer Inst. 1995;87(11):796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 2.Acharya UR, Ng E, Tan J-H, Sree SV. Thermography based breast cancer detection using texture features and support vector machine. J Med Syst. 2012;36(3):1503–10. doi: 10.1007/s10916-010-9611-z. [DOI] [PubMed] [Google Scholar]
- 3.Alavi G, Sharifi N, Sadeghian A, Jabari H, Bahreyni M, Bagheri H. Presence of human papilloma virus sequences in breast cancer tissues and association with histopathological features. Iranian Journal of Obstetrics, Gynecology and Infertility. 2009;12(2):1–4. [Google Scholar]
- 4.Parkin DM, Bray F. The burden of HPV-related cancers. Vaccine. 2006;(3):24 –25. doi: 10.1016/j.vaccine.2006.05.111. [DOI] [PubMed] [Google Scholar]
- 5.Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–9. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batschauer AP, Cruz NG, Oliveira VC, Coelho FF, Santos IR, Alves MT, et al. HFE, MTHFR, and FGFR4 genes polymorphisms and breast cancer in Brazilian women. Mol Cell Biochem. 2011;357(1-2):247–53. doi: 10.1007/s11010-011-0895-1. [DOI] [PubMed] [Google Scholar]
- 7.Romanowicz-Makowska H, Smolarz B, Zadrozny M, Westfa B, Baszczyński J, Kokołaszwili G, et al. The association between polymorphisms of the RAD51-G135C, XRCC2-Arg188His and XRCC3-Thr241Met genes and clinico-pathologic features in breast cancer in Poland. Eur J Gynaecol Oncol. 2012;33(2) [PubMed] [Google Scholar]
- 8.Unic I, Stalmeier PF, Peer PG, van Daal WA. A Review on Family History of Breast Cancer: Screening and Counseling Proposals for Women with Familial (Non-Hereditary) Breast Cancer. Patient Educ Couns. 1997;32(1-2):117–27. doi: 10.1016/s0738-3991(97)00062-1. [DOI] [PubMed] [Google Scholar]
- 9.Hilakivi-Clarke L, Rowland J, Clarke R, Lippman ME. Psychosocial factors in the development and progression of breast cancer. Breast Cancer Res Treat. 1994;29(2):141–60. doi: 10.1007/BF00665676. [DOI] [PubMed] [Google Scholar]
- 10.Turkoz FP, Solak M, Petekkaya I, Keskin O, Kertmen N, Sarici F, et al. Association between common risk factors and molecular subtypes in breast cancer patients. The Breast. 2013;22(3):344–50. doi: 10.1016/j.breast.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Lindelöf B, Krynitz B, Ayoubi S, Martschin C, Wiegleb-Edström D, Wiklund K. Previous extensive sun exposure and subsequent vitamin D production in patients with basal cell carcinoma of the skin, has no protective effect on internal cancers. Eur J Cancer. 2012;48(8):1154–8. doi: 10.1016/j.ejca.2011.06.049. [DOI] [PubMed] [Google Scholar]
- 12.Walker LG, Eremin O. Psychological assessment and intervention: future prospects for women with breast cancer. Semin Surg Oncol. 1996 doi: 10.1002/(SICI)1098-2388(199601/02)12:1<76::AID-SSU11>3.0.CO;2-7. Wiley Online Library. [DOI] [PubMed] [Google Scholar]
- 13.Heng B, Glenn W, Ye Y, Tran B, Delprado W, Lutze-Mann L, et al. Human papilloma virus is associated with breast cancer. Br J Cancer. 2009;101(8):1345–50. doi: 10.1038/sj.bjc.6605282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amrani M, Lalaoui K, El Mzibri M, Lazo P, Belabbas MA. Molecular detection of human papillomavirus in 594 uterine cervix samples from Moroccan women (147 biopsies and 447 swabs) J Clin Virol. 2003;27(3):286–95. doi: 10.1016/s1386-6532(02)00227-5. [DOI] [PubMed] [Google Scholar]
- 15.Damin AP, Karam R, Zettler CG, Caleffi M, Alexandre CO. Evidence for an association of human papillomavirus and breast carcinomas. Breast Cancer Res Treat. 2004;84(2):131–7. doi: 10.1023/B:BREA.0000018411.89667.0d. [DOI] [PubMed] [Google Scholar]
- 16.Ghaffari SR, Sabokbar T, Meshkat Z, Fereidooni F, Dastan J, Rafati M, et al. Tracing Human Papilloma Virus in Breast Tumors of Iranian Breast Cancer Patients. Breast J. 2011;17(2):218–9. doi: 10.1111/j.1524-4741.2010.01053.x. [DOI] [PubMed] [Google Scholar]
- 17.Meshkat Z, Soleimanjahi H, Mirshahabi H, Meshkat M, Kheirandish M, Hassan ZM. Strong immune responses induced by a DNA vaccine containing HPV16 truncated E7 C-terminal linked to HSP70 gene. Iran J Immunol. 2011;8(2):65–75. [PubMed] [Google Scholar]
- 18.Widschwendter A, Brunhuber T, Wiedemair A, Mueller-Holzner E, Marth C. Detection of human papillomavirus DNA in breast cancer of patients with cervical cancer history. J Clin Virol. 2004;31(4):292–7. doi: 10.1016/j.jcv.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Kan C, Iacopetta B, Lawson J, Whitaker N. Identification of human papillomavirus DNA gene sequences in human breast cancer. Br J Cancer. 2005;93(8):946–8. doi: 10.1038/sj.bjc.6602778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan N, Castillo A, Koriyama C, Kijima Y, Umekita Y, Ohi Y, et al. Human papillomavirus detected in female breast carcinomas in Japan. Br J Cancer. 2008;99(3):408–14. doi: 10.1038/sj.bjc.6604502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gumus M, Yumuk P, Salepci T, Aliustaoglu M, Dane F, Ekenel M, et al. HPV DNA frequency and subset analysis in human breast cancer patients' normal and tumoral tissue samples. J Exp Clin Cancer Res. 2006;25(4):515–21. [PubMed] [Google Scholar]
- 22.Kroupis C, Markou A, Vourlidis N, Dionyssiou-Asteriou A, Lianidou ES. Presence of high-risk human papillomavirus sequences in breast cancer tissues and association with histopathological characteristics. Clin Biochem. 2006;39(7):727–31. doi: 10.1016/j.clinbiochem.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Yasmeen A, Bismar TA, Kandouz M, Foulkes WD, Desprez P-Y, Al Moustafa A-E. E6/E7 of HPV type 16 promotes cell invasion and metastasis of human breast cancer cells. Cell Cycle. 2007;6(16):2038–42. doi: 10.4161/cc.6.16.4555. [DOI] [PubMed] [Google Scholar]
- 24.Akil N, Yasmeen A, Kassab A, Ghabreau L, Darnel A, Al Moustafa A. High-risk human papillomavirus infections in breast cancer in Syrian women and their association with Id-1 expression: a tissue microarray study. Br J Cancer. 2008;99(3):404–7. doi: 10.1038/sj.bjc.6604503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de León DC, Montiel DP, Nemcova J, Mykyskova I, Turcios E, Villavicencio V, et al. Human papillomavirus (HPV) in breast tumors: prevalence in a group of Mexican patients. BMC Cancer. 2009;9(1) doi: 10.1186/1471-2407-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawson J, Glenn W, Heng B, Ye Y, Tran B, Lutze-Mann L, et al. Koilocytes indicate a role for human papilloma virus in breast cancer. Br J Cancer. 2009;101(8):1351–6. doi: 10.1038/sj.bjc.6605328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yasmeen A, Bismar TA, Dekhil H, Ricciardi R, Kassab A, Gambacorti-Passerini C, et al. ErbB-2 receptor cooperates with E6/E7 oncoproteins of HPV type 16 in breast tumorigenesis. Cell Cycle. 2007;6(23):2939–43. doi: 10.4161/cc.6.23.4949. [DOI] [PubMed] [Google Scholar]
- 28.Li N, Bi X, Zhang Y, Zhao P, Zheng T, Dai M. Human papillomavirus infection and sporadic breast carcinoma risk: a meta-analysis. Breast Cancer Res Treat. 2011;126(2):515–20. doi: 10.1007/s10549-010-1128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gopalkrishna V, Singh U, Sodhani P, Sharma J, Hedau S, Mandal A, et al. Absence of human papillomavirus DNA in breast cancer as revealed by polymerase chain reaction. Breast Cancer Res Treat. 1996;39(2):197–202. doi: 10.1007/BF01806186. [DOI] [PubMed] [Google Scholar]
- 30.Wrede D, Luqmani Y, Coombes R, Vousden K. Absence of HPV 16 and 18 DNA in breast cancer. Br J Cancer. 1992;65(6):891–4. doi: 10.1038/bjc.1992.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi Y-L, Cho EY, Kim JH, Nam SJ, Oh YL, Song SY, et al. Detection of human papillomavirus DNA by DNA chip in breast carcinomas of Korean women. Tumour Biol. 2008;28(6):327–32. doi: 10.1159/000124238. [DOI] [PubMed] [Google Scholar]
- 32.de Cremoux P, Thioux M, Lebigot I, Sigal-Zafrani B, Salmon R, Sastre-Garau X. No evidence of human papillomavirus DNA sequences in invasive breast carcinoma. Breast Cancer Res Treat. 2008;109(1):55–8. doi: 10.1007/s10549-007-9626-4. [DOI] [PubMed] [Google Scholar]
- 33.Duò D, Ghimenti C, Migliora P, Pavanelli MC, Mastracci L, Angeli G. Identification and characterization of human papillomavirus DNA sequences in Italian breast cancer patients by PCR and line probe assay reverse hybridization. Molecular Med Rep. 2008;1(5):673–7. doi: 10.3892/mmr_00000011. [DOI] [PubMed] [Google Scholar]
- 34.Hachana M, Ziadi S, Amara K, Toumi I, Korbi S, Trimeche M. No evidence of human papillomavirus DNA in breast carcinoma in Tunisian patients. The Breast. 2010;19(6):541–4. doi: 10.1016/j.breast.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Mendizabal-Ruiz A, Morales J, Ramirez-Jirano L, Padilla-Rosas M, Morán-Moguel M, Montoya-Fuentes H. Low frequency of human papillomavirus DNA in breast cancer tissue. Breast Cancer Res Treat. 2009;114(1):189–94. doi: 10.1007/s10549-008-9989-1. [DOI] [PubMed] [Google Scholar]