Abstract
Background. In 2007, Malawi replaced sulfadoxine-pyrimethamine (SP) with an artemisinin-based combination therapy as the first-line treatment for uncomplicated Plasmodium falciparum malaria in response to failing SP efficacy. Here we estimate the effect of reduced SP pressure on the prevalence of SP-resistant parasites and the characteristics of the associated selective sweeps flanking the resistance loci.
Methods. Samples obtained from individuals with clinical malaria during a period of high SP use (1999–2001), a transitional period (2007–2008), and a period of low SP use (2012) were genotyped for resistance markers at pfdhfr-ts codons 51, 59, and 108 and pfdhps codons 437, 540, and 581. Expected heterozygosity was estimated to evaluate the genetic diversity flanking pfdhfr-ts and pfdhps.
Results. An increase in the prevalence of the resistance haplotypes DHFR 51I/59R/108N and DHPS 437G/540E occurred under sustained drug pressure, with no change in haplotype prevalence 5 years after reduction in SP pressure. The DHPS 437G/540E/581G haplotype was observed in 2007 and increased in prevalence during a period of reduced SP pressure. Changes to the sweep characteristics flanking pfdhfr-ts and pfdhps were minimal.
Conclusions. In contrast to the rapid and complete return of chloroquine-susceptible falciparum malaria after chloroquine was withdrawn from Malawi, a reemergence of SP efficacy is unlikely in the near future.
Keywords: malaria, sulfadoxine-pyrimethamine, resistance, selective sweeps, pyrosequencing, DHFR, DHPS
The continued expansion of Plasmodium falciparum resistance to antimalarial chemotherapies is a threat to malaria control and elimination strategies. The return of drug susceptibility to some regions after drug policy changes has renewed interest that previously abandoned drugs, such as chloroquine and sulfadoxine-pyrimethamine (SP), might once again be clinically useful. Malawi was the first African nation to adopt SP as the first-line treatment of uncomplicated malaria, in 1993, in response to high levels of chloroquine resistance in Malawi. Within a decade of the removal of chloroquine pressure, chloroquine-susceptible parasites reemerged and predominated in Malawi [1]. Reemergence was shown, through analysis of selective sweeps, to be due to the expansion of diverse susceptible parasites that had survived chloroquine selective pressure [2, 3]. Within a decade of the switch to SP, treatment failures began to increase in Malawi in association with an increase in the prevalence of resistance polymorphisms within the genes encoding dihydrofolate reductase (DHFR)–thymidylate synthase (encoded by pfdhfr-ts) and dihydropteroate synthase (DHPS; encoded by pfdhps), which are responsible for SP resistance [4]. In 2007, SP was replaced by an artemisinin-based combination therapy (ACT). SP was still used after 2007 for intermittent preventive treatment of malaria in pregnancy (IPTp). Whether SP resistance in Malawi will, similar to chloroquine resistance, decline in the absence of drug pressure remains unknown.
Evidence for directional selection of antimalarial drug resistance polymorphisms has been demonstrated in the form of selective sweeps [5–7]. An allele that provides a fitness advantage will be favored and increase in frequency in a population. As the selected allele increases in frequency, so do flanking neutral markers in linkage with the selected allele, leading to a regional reduction in the heterozygosity of neutral markers flanking the advantageous allele. This phenomenon is referred to as a selective sweep [8]. Aside from providing evidence for positive directional selection of a specific resistance haplotype, the characteristics of a selective sweep can be used to infer the strength of selection and how epidemiological factors affect selection on temporal and spatial scales.
Selective sweeps flanking pfdhfr-ts and pfdhps have been shown in other African populations under SP pressure [9]. In the absence of SP pressure, resistance polymorphisms in pfdhfr-ts and pfdhps may confer a disadvantage to the parasite, allowing them to be outcompeted by susceptible parasites with fully functional DHFR and DHPS enzymes. Polymorphisms in pfdhfr and pfdhps have been shown to confer resistance to SP and to impair enzyme function in a synergistic manner [10, 11]. Under these circumstances, negative selection would begin to reduce the prevalence of the resistant genotypes, while recombination and mutation would begin to increase heterozygosity in markers flanking the resistance-conferring genes, degrading the selective sweep.
The purpose of this study was to estimate how changes in SP pressure affected both the prevalence of SP-resistant haplotypes and the characteristics of the associated selective sweeps. We aimed to test the hypothesis that SP pressure positively selected for SP-resistant haplotypes during a period of high SP use and that a subsequent reduction in SP pressure that occurred following the switch to ACTs in Malawi selected against these highly resistant parasites and allowed for the expansion of SP-susceptible parasites.
METHODS
Study Samples
All samples were collected from Ndirande district within Blantyre, Malawi. Samples from individuals with symptomatic malaria were obtained during 3 intervals. Samples from 1999 to 2001 (n = 689) and 2007–2009 (n = 623) were from individuals with clinical malaria who were participating in therapeutic efficacy studies. Samples from 2012 (n = 968) were from individuals with symptomatic malaria that was identified at the government health center [4, 12]. All samples were collected with informed consent and approval from the University of Maryland Baltimore and local institutional review boards.
Genotyping of Resistance Loci
DNA extraction from filter paper blood cards was performed on all collected samples, using a Qiagen BioRobot (Qiagen, Valencia, California) following the Investigator Bloodcard Protocol. Parasite genotypes at polymorphic sites within pfdhfr-ts and pfdhps were determined via pyrosequencing. Single-nucleotide polymorphisms within codons 51, 59, and 108 of pfdhfr-ts and codons 437, 540, and 581 of pfdhps were genotyped for all samples, using primers and amplification methods adapted from Zhou et al [13]. Pyrosequencing was performed on a PyroMark Q96 MD system (Biotage, Charlotte, North Carolina). Allele frequency was adjusted on the basis of a standard curve [14]. When a polyclonal infection was present, an allele with a relative frequency of ≥80% within a given infection was designated as the predominant allele. Haplotypes (series of pfdhfr-ts or pfdhps alleles) were constructed using only the predominant allele. Samples that were mixed at a single codon were treated as containing both possible haplotypes. Samples without a predominant allele at ≥2 codons were labeled “mixed genotype.” Analyses performed both with and without inclusion of samples that were mixed at 1 codon did not significantly change the estimated prevalence of DHFR and DHPS haplotypes.
Genotyping of Microsatellites
To determine whether reduced heterozygosity around drug resistance genes was the result of selection or demographic processes, we measured expected heterozygosity (He; defined below) in 6 unlinked neutral loci (TA81, TA40, pfPK2, PolyA, TA87, and ARA2) located throughout the P. falciparum genome, using primers and protocols described by Anderson et al [15]. When multiple peaks were identified within the same sample, peaks that were at least one third of the height of the tallest peak were called, and the tallest peak was designated as the predominant allele [16]. Samples without a predominant allele at a given marker were designated as polyclonal. Only samples with a predominant allele were included in estimates of He.
Microsatellites flanking pfdhfr-ts and pfdhps were genotyped for all samples. Eleven polymorphic microsatellites flanking pfdhfr-ts were genotyped using previously described primers and protocols [17, 18]; 6 were upstream (−30 kb, −10 kb, −4.5 kb, −3.8 kb, −1.2 kb, and −0.3 kb), and 5 were downstream (+0.2 kb, +0.52 kb, +1.48 kb, +20 kb, and +50 kb). Eight polymorphic microsatellites flanking pfdhps were genotyped; 4 were upstream (−11.07 kb, −7.49 kb, −2.85 kb, and −0.13 kb), and 4 were downstream (+0.034 kb, +0.51 kb, +1.41 kb, +9.01 kb) of the gene, using primers described by Vinayak et al [19]. PCR cycling conditions were optimized from the original protocols (Supplementary Table 1). Fragment size was visualized using an Applied BioSystems 3730XL high-throughput 96-capillary DNA sequencer. Analysis of electropherograms was performed using GeneMapper software (version 4.0; ABI). A Perl script was used to assign the raw electropherogram scores to an integer allele size based on the expected repeat length and variation seen in the positive controls.
Statistical Analysis
DHFR and DHPS Haplotype Prevalence
The prevalence of each haplotype was estimated as the number of each haplotype observed among the successfully genotyped samples for each resistance gene, divided by the total number of successfully genotyped samples. Only samples with a genotyped allele at all assayed codons were used in the analysis. A SP-resistant haplotype was defined as a haplotype containing any number or combination of resistance polymorphisms (ie, polymorphisms that confer pyrimethamine or sulfadoxine resistance) at the genotyped codons within either of the resistance genes of interest. The SP-susceptible haplotype was defined as the wild type (ie, no mutations at any of the genotyped pfdhfr-ts and pfdhps codons). Samples with mixed-genotype at ≥2 codons within the same gene were excluded because a haplotype could not be determined. DHFR and DHPS haplotypes were treated and reported separately. χ2 tests, with the Yates correction when appropriate, were used to determine whether haplotype prevalence differed significantly between the 3 periods.
He
He, a measure of genetic diversity at each microsatellite locus, was calculated using the following standard equation for He and variance:
He (±1 standard deviation [SD]) was calculated for both pfdhfr-ts and pfdhps during 3 intervals: a period of high SP use, from 1999 to 2001; a transitional period, from 2007 to 2008; and a period of low SP use, during 2012. Samples without a predominant genotype or samples with missing data were excluded from He calculations. Statistical significance was determined via permutation [3]. Diversity ratios were calculated for He (high SP use)/He (transition), He (high SP use)/He (low SP use), and He (transition)/He (low SP use). Calculations for He, standard deviations, and permutations were conducted in R [20].
RESULTS
Haplotype Prevalence
Complete pfdhfr-ts haplotypes were obtained for 394 samples from the period of high SP use, 563 samples from the transitional period, and 549 samples from the period of low SP use. For pfdhps, 550 samples from the period of high SP use produced complete haplotypes, 556 samples from the transitional period produced complete haplotypes, and 546 samples from the period of low SP use produced complete haplotypes. The analysis focused on sweep characteristics flanking DHFR 51I/59R/108N and DHPS 437G/540E haplotypes owing to the small number of double-mutant and single-mutant haplotypes for both pfdhfr-ts and pfdhps in the later periods.
A statistically significant increase in the prevalence of the DHFR 51I/59R/108N was observed between the period of high SP use and the transitional period (87% to 97%; P < .0001; Figure 1A). No change in the DHFR triple-mutant haplotype prevalence over the 5 years between the transitional period and the period of low SP use was observed (P = .94). Concurrent with the increase in the prevalence of the DHFR 51I/59R/108N haplotypes was a decline in the prevalence of the less resistant DHFR 51I/108N and 59R/108N. Unlike the increase in the DHFR 51I/59R/108N prevalence, the decrease in prevalence of the DHFR 51I/108N and 59R/108N haplotypes continued through the period of low SP use. The decrease was significant for both double-mutant haplotypes (P < .0001), although the decrease from the transitional period to the period of low SP use were borderline significant (51I/108N, P = .01; 59R/108N, P = .05). No parasites with susceptible alleles at all DHFR codons were observed in this study. In addition, there was a decrease in the prevalence of the mixed genotype infections (4% to 0%; P < .0001) between the period of high SP use and the transitional period, but there was no significant change in the mixed-genotype prevalence between the transitional period and the period of low SP use.
Figure 1.
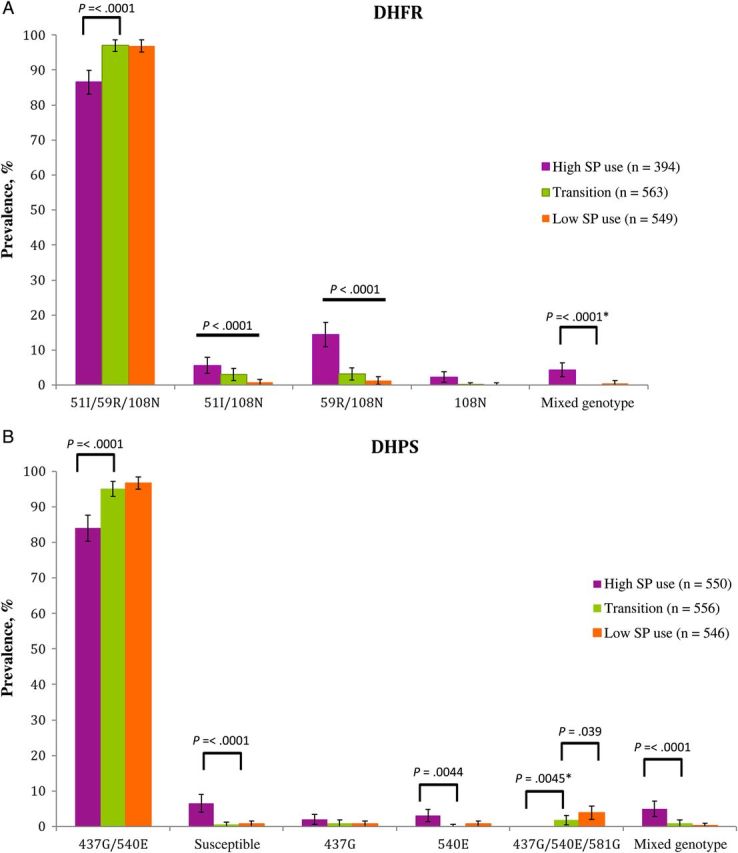
Change in Plasmodium falciparum dihydrofolate reductase (DHFR; A) and dihydropteroate synthase (DHPS; B) haplotype prevalence over 3 periods in the history of sulfadoxine-pyrimethamine (SP) use in Ndirande, Malawi. Prevalence is calculated as the percentage of individuals with a given haplotype. The 3 periods consisted of a period of high SP use (1999–2001), a transitional period (2007–2008), and a period of low SP use (2012). Black bars indicate statistically significant differences. Error bars represent 95% confidence intervals. The asterisks indicate the use of the Yates correction.
The DHPS 437G/540E haplotype prevalence increased from 84% to 95% (437G/540E, P < .0001) between the period of high SP use and the transitional period, but there was no significant change in haplotype prevalence between the transitional period and the period of low SP use (95%–97%; Figure 1B). The DHPS 437G/540E/581G haplotype was not observed in the period of high SP use, but it was present at a low prevalence of 2% during the transitional period (P = .0044, compared with the period of high SP use) and 4% during the period of low SP use (P < .0001, compared with the period of high SP use); the prevalence in the transitional period was significantly different from that in the period of low SP use (P = .039). The prevalence of the haplotype containing all susceptible alleles within DHPS decreased between the period of high SP use and the transitional period (P < .0001), and there was no change between the transitional period and the period of low SP use. No change in haplotype prevalence was observed for the DHPS single-mutant 437G across any of the periods. A reduction in the prevalence of DHPS 540E was seen between the period of high SP use and the transitional period (P = .0044). Similar to the findings for pfdhfr-ts, a decrease in the prevalence of mixed-genotype infections was found between the period of high SP use and the transitional period (P < .0001), although there was no change in haplotype prevalence after the switch from SP to ACTs.
Characteristics of Selective Sweeps
Because of the lack of SP-susceptible parasites in the data set, changes in He at flanking loci were compared to the average He of the unlinked loci, and ±1 SD was used to assess the change in He relative to the average unlinked He for the sample set (Supplementary Figure 1). The average He at unlinked microsatellites was 0.872 for the period of high SP use, 0.897 for the transitional period, and 0.904 for the period of low SP use. These levels were not significantly different from each other (P = .55), though the difference between individual markers was sometimes significant (see Supplementary Figure 1). None of the unlinked loci had He of <0.8, consistent with estimates in other studies [3, 15, 17, 21, 22].
A reduction in He flanking the DHFR 51I/59R/108N (Figure 2A), consistent with a selective sweep, was observed for all periods as compared to the average He of the unlinked neutral microsatellites. At pfdhfr-ts, He returned to that of unlinked markers at 30 kb downstream during the transitional period and the period of low SP use. Comparing the sweep characteristics from the three periods, we observed asymmetrical changes in He across pfdhfr-ts–flanking microsatellites. Upstream changes did not show a consistent trend toward sweep expansion under continued drug pressure, although a reduction in He was observed at the distal downstream edge of the sweep. An increase in He after the reduction of SP pressure occurred at the most distal upstream edge of the sweep. However, no further degradation of the sweep was found between the transitional period and the period of low SP use.
Figure 2.
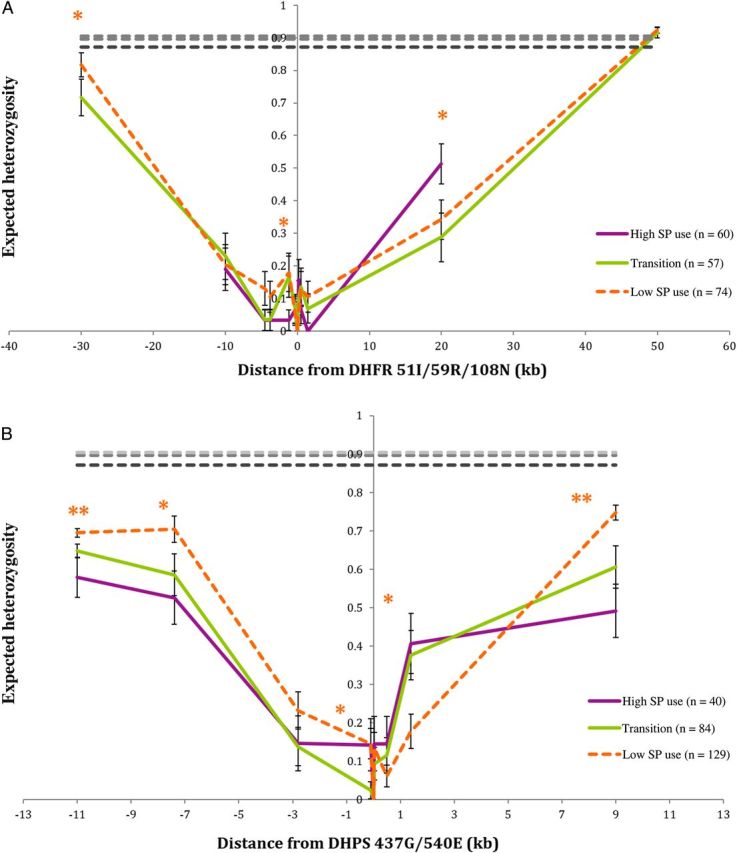
Expected heterozygosity in microsatellite loci flanking the genes encoding Plasmodium falciparum dihydrofolate reductase (DHFR)–thymidylate synthase pfdhfr-ts (A) and dihydropteroate synthase (DHPS) (pfdhps; B). Samples with missing data were excluded. Error bars represent ±1 SD. Dashed lines represent average genomic level expected heterozygosity, based on unlinked loci. The single asterisks indicate a significant difference between 2 periods. The double asterisk indicates a significant difference between all 3 periods, based on permutation. P values of <.05 were considered statistically significant. Abbreviation: SP, sulfadoxine-pyrimethamine.
Analysis of microsatellites flanking pfdhps also indicated the presence of selective sweeps flanking the locus in all 3 periods (Figure 2B). He at the genotyped flanking microsatellites did not return to the average He of unlinked loci. The sweep flanking pfdhps extended at least 10 kb in either direction. Evidence of sweep degradation was found at the extreme edges of the observed sweep. However, overall sweep characteristics did not show a consistent increase or decrease in He between periods.
Haplotype Origins
Mapping of microsatellite haplotypes indicated the pfdhfr-ts and pfdhps haplotypes to be of Southeast Asian origins as compared to V1/S, 3D7, and HB3 control strains (Supplementary Table 2). Analysis of the DHPS 473G/540E/581G haplotype indicated it arose from the DHFR 437G/540E genetic background present in Malawi.
We found that a 37% reduction in the absolute number of samples scored as polyclonal between the periods of high and low SP use (P < .0001). An intermediate reduction in polyclonal samples (33%) was also observed between the period of high SP use and the transitional period (P < .0001). The difference in polyclonal infections between the transitional period and the period of low SP use was not significant (P = .41).
DISCUSSION
Our study demonstrates the persistence of SP-resistant malaria and the presence of selective sweeps 5 years after the removal of SP as the first-line treatment of uncomplicated malaria in Malawi. Our findings suggest little to no fitness cost of the DHFR triple-mutant and DHPS double-mutant haplotypes in the absence of strong SP pressure under current epidemiological conditions. Our findings also present evidence that the DHPS 437G/540E/581G triple-mutant haplotype may be increasing in prevalence despite the switch to ACTs.
As we hypothesized, we have shown an increase in the prevalence of the most highly resistant DHFR and DHPS haplotypes, with a corresponding decrease in the prevalence of less resistant haplotypes and susceptible haplotypes, over the period of persistent high SP use (1999–2007), consistent with selection for the highly resistant haplotypes by SP pressure. However, in contrast with the pattern observed with chloroquine resistance, we did not see a decrease in the prevalence of highly SP-resistant parasites after SP removal as the first-line treatment for uncomplicated malaria. Near fixation of DHFR 51I/59R/108N and DHPS 437G/540E may contribute to the persistence of these highly resistant haplotypes, as few susceptible alleles and no susceptible parasites were found to compete with resistant parasites. Over a greater period, any slight fitness cost to the DHFR 51I/59R/108N and DHPS 437G/540E haplotypes might become more apparent.
Microsatellite analysis was successful in identifying and defining the characteristics of the selective sweeps flanking pfdhfr-ts and pfdhps within the 3 periods, but the changes in sweep characteristics (coupled with prevalence findings) did not suggest a strong fitness cost of the SP-resistant haplotypes. At pfdhfr-ts it appears that selective pressure was maintained and caused a further reduction in He 20 kb downstream during the time of transition from SP to ACTs. After SP pressure was relaxed, in 2007–2008, an increase in He occurred in the period of low SP use in both distal and proximal markers. The increase in He is significant only upstream of pfdhfr-ts. In the absence of a decrease in prevalence of the DHFR triple mutant, it is likely that this increase in He results from the natural decay of the sweep due to recombination over time. The pfdhfr-ts sweep characteristics do not show large-scale degradation of the sweep that would suggest a strong fitness cost of the triple-mutant haplotype in the absence of SP pressure. We cannot however rule out the possibility that compensatory mutations may have arisen on the Malawian genetic background that reduced the fitness cost of DHFR and DHPS resistant haplotypes [23, 24]. Asymmetry in He, as seen flanking pfdhfr-ts and pfdhps, have been reported elsewhere and may be due to differences in mutation rates at each microsatellite locus [9, 17, 25].
The removal of samples with missing data from the analyses reduced sample size and could have biased our findings against rare low-frequency haplotypes and artificially reduced He. However, when sensitivity analyses were performed using samples with varying degrees of missing data, there was no effect on the overall sweep characteristics to suggest that the exclusion of missing data affected the study conclusions. In addition, Hale et al demonstrated that only 25–30 samples are needed to accurately estimate population-level genetic diversity [26].
The decline of the DHFR double-mutant haplotypes without an increase in DHFR susceptible parasites implies continued selection against the DHFR double-mutant haplotypes in favor of the DHFR triple mutant, rather than a fitness cost to the DHFR double mutants, relative to the susceptible parasites. A limitation of estimating haplotype prevalence (as opposed to allele frequency) is the inability to directly attribute an increase in one haplotype to a decrease in another haplotype, as a prevalence estimate is relative to the number of humans sampled and not to the number of parasites sampled. Two recently published methods for determining multiplicity of infection allow for more-accurate assessment of allele/haplotype frequency (as opposed to haplotype prevalence) when assessing resistance allele dynamics [27, 28]. However, the low prevalence of mixed-genotype infections and no change in the proportion of polyclonal infections in 2007–2008 and 2012 suggest that using these methods would be unlikely to have a significant effect on our estimates of haplotype prevalence in this study.
The decline in mixed-genotype infections and the reduction in polyclonal infections in the absence of any change in average He at unlinked loci is consistent with a reduction in malaria transmission in Malawi throughout the 3 study periods. It is likely that sustained drug pressure through 2007, as well as an overall reduction in malaria transmission through malaria control measures and access to effective antimalarial medication, led to the reduction in mixed-genotype and polyclonal infections. A consequence of reducing the amount of malaria in a region may be the extinction of rare haplotypes (such as SP susceptible) due to genetic drift. With little or no selective pressure against the SP-resistant haplotypes, these haplotypes may reach fixation and further reduce the likelihood of a reemergence and expansion of SP-susceptible haplotypes. A minimal metabolic cost to the resistant haplotypes may lead to many decades of persistent SP resistance and prohibit the future clinical use of SP.
Our finding of continued high rates of SP-resistant parasites despite cessation of SP use for the treatment of malaria could also be due in part to antifolate drug pressure that continues from other sources. Use of trimethoprim-sulfamethoxazole (TMP-SMZ) as prophylaxis in people living with HIV and for treatment of acute illnesses and low-level SP pressure through SP-IPTp may be sources of continued selection for SP resistance mutations, because they both act upon the same folate enzymes (DHFR and DHPS) and laboratory evidence suggests cross-resistance [29]. However, to date, there is no evidence that TMP-SMZ has any impact on the prevalence of SP-resistant alleles or that SP resistance influences TMP-SMZ efficacy [30, 31]. In addition, Iriemenam et al found the impact of SP-IPTp on the increase of SP-resistant parasites in pregnant women to be minor, compared with the use of SP for case management in the general population, and therefore it is possible that SP-IPTp may not be the sole cause of the sustained high prevalence of SP resistance in Malawi [32].
Our data confirm the findings by Taylor et al that the Malawian 581G allele arose on the DHPS double-mutant genetic background but indicate that the 581G haplotype emerged by 2007, 3 years earlier than previously reported [33]. The increase in the DHPS 437G/540E/581G prevalence across the 3 periods suggests continued selection for this haplotype. Too few DHPS triple mutants (437G/540E/581G) were found in 2012 to meaningfully assess the extent of recent selection of this haplotype. The increase in the prevalence of the DHPS 437G/540E/581G haplotype could threaten the efficacy of ongoing SP-IPTp in the nation as the presence of 581G allele is associated with SP-IPTp failure [34]. Ongoing molecular monitoring of the 437G/540E/581G haplotype prevalence and SP-IPTp treatment should be performed to determine the influence (if any) this selection could have on IPT efficacy in Malawi.
These data suggest that, in contrast to the rapid and complete return of fully chloroquine-susceptible parasites after cessation of chloroquine use, a similar resurgence of SP susceptibility in Malawi or in similar epidemiological settings is unlikely. Not only did high-level SP resistance persist for 5 years after the removal of SP as the first-line treatment of uncomplicated malaria, but the prevalence of the DHPS 437G/540E/581G haplotype increased. In the face of expanding ACT resistance in Southeast Asia with a risk of spread to Africa, other antimalarial drug classes, perhaps including chloroquine, must be explored, as SP will likely remain an ineffective treatment for uncomplicated malaria in Malawi for the foreseeable future.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Kathy Strauss, Matthew Adams, Sudhaunshu Joshi, Sara Brown, Malathi Vadla, and other laboratory staff of the Center for Vaccine Development Malaria Group, for their contributions to assay optimization and DNA extraction; the University of Maryland School of Medicine sequencing core, where all microsatellite genotyping assays were run; the study participants and their parents, for their participation; and the staff and field teams from the Malaria Alert Center and Blantyre Malaria Project, for their technical assistance.
Disclaimer. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the University of Maryland Baltimore or other associated institutions.
Financial support. This work was supported by the National Institutes of Health (grants U01-AI47858, U19-AI089683, R01-GM084320, R01-AI101713, and NIH U01-AI044824).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Laufer MK, Thesing PC, Eddington ND, et al. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med 2006; 355:1959–66. [DOI] [PubMed] [Google Scholar]
- 2.Wootton JC, Feng X, Ferdig MT, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 2002; 418:320–3. [DOI] [PubMed] [Google Scholar]
- 3.Laufer MK, Takala-Harrison S, Dzinjalamala FK, Stine OC, Taylor TE, Plowe CV. Return of chloroquine-susceptible falciparum malaria in Malawi was a reexpansion of diverse susceptible parasites. J Infect Dis 2010; 202:801–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kublin JG, Dzinjalamala FK, Kamwendo DD, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis 2002; 185:380–8. [DOI] [PubMed] [Google Scholar]
- 5.Triglia T, Menting JG, Wilson C, Cowman AF. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci U S A 1997; 94:13944–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci U S A 1988; 85:9114–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science 2004; 305:1124. [DOI] [PubMed] [Google Scholar]
- 8.Zivkovic D, Stephan W. Analytical results on the neutral non-equilibrium allele frequency spectrum based on diffusion theory. Theor Popul Biol 2011; 79:184–91. [DOI] [PubMed] [Google Scholar]
- 9.McCollum AM, Schneider KA, Griffing SM, et al. Differences in selective pressure on dhps and dhfr drug resistant mutations in western Kenya. Malar J 2012; 11:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuthavong Y. Basis for antifolate action and resistance in malaria. Microbes Infect 2002; 4:175–82. [DOI] [PubMed] [Google Scholar]
- 11.Plowe CV, Kublin JG, Dzinjalamala FK, et al. Sustained clinical efficacy of sulfadoxine-pyrimethamine for uncomplicated falciparum malaria in Malawi after 10 years as first line treatment: five year prospective study. BMJ 2004; 328:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laufer MK, Thesing PC, Dzinjalamala FK, et al. A longitudinal trial comparing chloroquine as monotherapy or in combination with artesunate, azithromycin or atovaquone-proguanil to treat malaria. PLoS One 2012; 7:e42284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z, Poe AC, Limor J, et al. Pyrosequencing, a high-throughput method for detecting single nucleotide polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes of Plasmodium falciparum. J Clin Microbiol 2006; 44:3900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takala SL, Smith DL, Stine OC, et al. A high-throughput method for quantifying alleles and haplotypes of the malaria vaccine candidate Plasmodium falciparum merozoite surface protein-1 19 kDa. Malar J 2006; 5:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology 1999; 119:113–25. [DOI] [PubMed] [Google Scholar]
- 16.Nair S, Williams JT, Brockman A, et al. A selective sweep driven by pyrimethamine treatment in southeast asian malaria parasites. Mol Biol Evol 2003; 20:1526–36. [DOI] [PubMed] [Google Scholar]
- 17.Nash D, Nair S, Mayxay M, et al. Selection strength and hitchhiking around two anti-malarial resistance genes. Proc Biol Sci 2005; 272:1153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roper C, Pearce R, Bredenkamp B, et al. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet 2003; 361:1174–81. [DOI] [PubMed] [Google Scholar]
- 19.Vinayak S, Alam MT, Mixson-Hayden T, et al. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog 2010; 6:e1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, 2008. [Google Scholar]
- 21.Lumb V, Das MK, Singh N, Dev V, Wajihullah, Sharma YD. Characteristics of genetic hitchhiking around dihydrofolate reductase gene associated with pyrimethamine resistance in Plasmodium falciparum isolates from India. Antimicrob Agents Chemother 2009; 53:5173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCollum AM, Mueller K, Villegas L, Udhayakumar V, Escalante AA. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob Agents Chemother 2007; 51:2085–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang H, Patel JJ, Yi M, et al. Genome-wide compensatory changes accompany drug- selected mutations in the Plasmodium falciparum crt gene. PLoS One 2008; 3:e2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown KM, Costanzo MS, Xu W, Roy S, Lozovsky ER, Hartl DL. Compensatory mutations restore fitness during the evolution of dihydrofolate reductase. Mol Biol Evol 2010; 27:2682–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lumb V, Madan R, Das MK, et al. Differential genetic hitchhiking around mutant pfcrt alleles in the Indian Plasmodium falciparum population. J Antimicrob Chemother 2012; 67:600–8. [DOI] [PubMed] [Google Scholar]
- 26.Hale ML, Burg TM, Steeves TE. Sampling for microsatellite-based population genetic studies: 25 to 30 individuals per population is enough to accurately estimate allele frequencies. PLoS One 2012; 7:e45170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider KA, Escalante AA. A likelihood approach to estimate the number of co-infections. PLoS One 2014; 9:e97899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor AR, Flegg JA, Nsobya SL, et al. Estimation of malaria haplotype and genotype frequencies: a statistical approach to overcome the challenge associated with multiclonal infections. Malar J 2014; 13:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer JK, Milhous WK, Cortese JF, Kublin JG, Plowe CV. Plasmodium falciparum cross-resistance between trimethoprim and pyrimethamine. Lancet 2001; 358:1066–7. [DOI] [PubMed] [Google Scholar]
- 30.Gasasira AF, Kamya MR, Ochong EO, et al. Effect of trimethoprim-sulphamethoxazole on the risk of malaria in HIV-infected Ugandan children living in an area of widespread antifolate resistance. Malar J 2010; 9:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thera MA, Sehdev PS, Coulibaly D, et al. Impact of trimethoprim-sulfamethoxazole prophylaxis on falciparum malaria infection and disease. J Infect Dis 2005; 192:1823–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iriemenam NC, Shah M, Gatei W, et al. Temporal trends of sulphadoxine-pyrimethamine (SP) drug-resistance molecular markers in Plasmodium falciparum parasites from pregnant women in western Kenya. Malar J 2012;11:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor SM, Antonia AL, Harrington WE, et al. Independent lineages of highly sulfadoxine-resistant Plasmodium falciparum haplotypes, eastern Africa. Emerg Infect Dis 2014; 20:1140–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gesase S, Gosling RD, Hashim R, et al. High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in northern Tanzania and the emergence of dhps resistance mutation at Codon 581. PLoS One 2009; 4:e4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


