Abstract
Human papillomavirus (HPV) genotype 52 is commonly found in Asian cases of cervical cancer but is rare elsewhere. Analysis of 611 isolates collected worldwide revealed a remarkable geographical distribution, with lineage B predominating in Asia (89.0% vs 0%–5.5%; Pcorrected < .001), whereas lineage A predominated in Africa, the Americas, and Europe. We propose that the name “Asian lineage” be used to denote lineage B, to signify this feature. Preliminary analysis suggested a higher disease risk for lineage B, although ethnogeographical confounders could not be excluded. Further studies are warranted to verify whether the reported high attribution of disease to HPV52 in Asia is due to the high prevalence of lineage B.
Keywords: HPV, cervical cancer, phylogeny, oncogenic risk, Asia
Overall, human papillomavirus genotype 52 (HPV52) ranks as the sixth or seventh most common HPV genotype associated with cervical cancers worldwide [1, 2]. However, studies from East Asia have reported a much higher ranking of HPV52. For instance, HPV52 was the third most common HPV genotype detected in cases of squamous cell carcinoma and the second most common among cases of cervical intraepithelial neoplasia 2 (CIN2) and CIN3 in Hong Kong [3]. Furthermore, HPV52 was the most common genotype in cervical cancer cases in Shanghai [4] and the second most common in cases in Taiwan [5] and Japan [6]. The underlying reason for such a geographical concentration of disease attribution remains obscure. We used a large series of samples collected worldwide to investigate the geographical distribution and risk association of HPV52-variant lineages and thereby improve our knowledge of this non–vaccine-targeted HPV type.
MATERIAL AND METHODS
Study Samples
Cervical and vaginal samples from women or anal samples from men that tested positive for HPV52 were transferred to a central laboratory for sequence analysis. DNA quality was assessed by amplifying a 932-bp fragment of the long-control region (LCR), and HPV genotype was ascertained by demonstrating a nucleotide sequence similarity of >90%, compared with the HPV52 prototype (GenBank accession no. X74481). The local institutional research ethics committee approved the collection of samples. Samples used in this study were sent without inclusion of identifying patient information.
Nucleotide Sequencing
The E6, E7, L1, LCR sequences were amplified by long- or short-fragment polymerase chain reaction (PCR). Long-fragment PCR was performed on good-quality samples, using primers 5′-ATG TCC ATT GAG TCA GGT CC-3′ and 5′-TGC ATT TTC ATC CTC GTC C-3′, and a second PCR was performed using inner primers 5′-GGT CCT GAC ATT CCA TTA CC-3′ and 5′-CCT CTA CTT CAA ACC AGC CT-3′ when necessary. Short-fragment PCR was used when the long-fragment approach failed. E6, E7, L1, and LCR sequences were amplified using primer pairs E6E7 (5′-TGC ACT ACA CGA CCG GTT A-3′ and 5′-CAT CCT CGT CCT CTG AAA TG-3′), L1A (5′-ATG TCC ATT GAG TCA GGT CC-3′ and 5′-GCA CAG GGT CAC CTA AGG TA-3′), L1B (5′-AGG ATG GGG ACA TGG TAG AT-3′ and 5′-CAC AGA CAA TTA CCC AAC AGA C-3′), and LCR (5′-GTC TGC ATC TTT GGA GGA CA-3′ and 5′-TGC GTT AGC TAC ACT GTG TTC-3′), respectively. When necessary, a second PCR, using inner primers E6E7 (5′-TTA CCG TAC CCA CAA CCA CT-3′ and 5′-CCT CTA CTT CAA ACC AGC CT-3′), L1A (5′-GGT CCT GAC ATT CCA TTA CC-3′ and 5′-GGG CAC ATC ACT TTT ACT AGC-3′), L1B (5′-ACA GGA TTT GGT TGC ATG G-3′ and 5′-TTC TTT GTG GAG GTA CGT GG-3′), and LCR (5′-TTT GTT ACA GGC AGG GCT AC-3′ and 5′-CGT TTT CGG TTA CAC CCT A-3′), was performed. The PCR products were sequenced from both directions and were analyzed with SeqScape software (version 2.5, Applied Biosystems). Mutations that occurred only once were confirmed by repeat sequencing from the original sample.
Phylogenetic Tree Construction
The concatenated nucleotide sequences assembled from the E6, E7, L1 and LCR regions were used for phylogenetic tree construction. Representative variants identified previously were incorporated for lineage identification [7]. Maximum-likelihood trees were constructed using the subtree-pruning-regrafting search approach by the Molecular Evolutionary Genetic Analysis software program (version 5.10; available at: http://www.megasoftware.net/) [8]. The data were bootstrap-resampled 1000 times for tree topology evaluation.
Geographical Distribution of Variant Lineages
The detection rate of each variant lineage was compared among regions by the χ2 or Fisher exact test, as appropriate, with correction for multiple comparisons, using the Bonferroni method. Epi Info (version 7.0.8.3; Centers for Disease Control and Prevention, Atlanta, GA) was used to calculate P values. Multivariate analyses were performed to investigate the association between each lineage and disease, with adjustment for age. Subjects with normal cervical cytology findings were used as controls, whereas subjects with histologically confirmed CIN3 or invasive cervical cancer were categorized as cases. Two-tailed P values of <.05 were regarded as statistically significant. SPSS (version 20, IBM) was used for multivariate analysis.
RESULTS
DNA from 611 specimens collected from 14 sites was of sufficient quality for sequencing (Supplementary Table 1). Of these specimens, 73.2% were from Asia, 15.5% were from Europe, 9% were from the Americas, and 2.3% were from Africa. Most samples were from women with normal cervical cytology findings (31.3%) and high-grade lesions (30.1%), including high-grade squamous intraepithelial lesions, CIN2, and CIN3. Twenty-five cervical samples (4.1%) had no associated cytological or histological information, and another 14 (2.3%) were vaginal samples. The mean age (±SD) of study subjects was 41.1 ± 14.0 years (range, 13–88 years).
Lineage Identification
The concatenated E6-E7-L1-LCR sequences derived from referent strains of each sublineage formed distinct branches in the phylogenetic tree, suggesting that these concatenated sequences, composing 40.6% (3226 nt) of the whole HPV52 genome, can be used for lineage identification (Supplementary Figure 1). The tree topology of E6-E7-L1-LCR sequences derived from the 324 unique strains collected in this study revealed 3 closely related but distinct branches, representing lineages A, B, and C, and 1 distantly related branch, representing lineage D.
The phylogenetic trees constructed from L1 or LCR sequences alone showed a topology similar to that of E6-E7-L1-LCR. They were able to identify variants up to the lineage level but could not differentiate sublineages. Signature sequences within the L1 and LCR regions that are useful for lineage/sublineage identification are shown in Supplementary Figure 2. The phylogenetic trees constructed from E6 or E7 sequences alone showed topologies different from that of E6-E7-L1-LCR, and were not useful for lineage identification.
Geographical Distribution of Variant Lineages
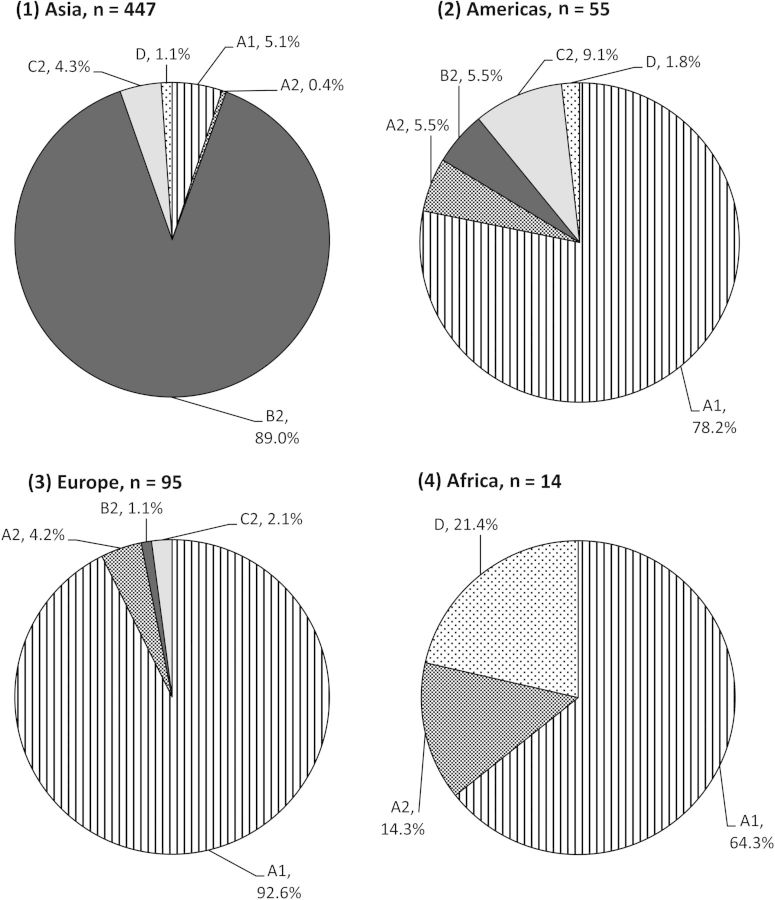
Variation in the geographical distribution of HPV52 lineages was observed (Figure 1). Lineage B was significantly more prevalent in Asia, compared with elsewhere (89.0 vs 0%–5.5%; Pcorrected < .001 for each comparison). In contrast, lineage A predominated in Africa, the Americas, and Europe, accounting for 78.6%–96.8% of the isolates, compared with 5.5% in Asia (Pcorrected < .001 for each comparison). Lineage C was uncommon across all regions (0%–9.1%), with no significant variation in prevalence. Lineage D was rarely detected in the Americas, Asia, and Europe (0%–1.8%) but was found in 3 of 14 samples from Africa, giving a wide 95% confidence interval (CI) of 0%–42.9%.
Figure 1.
Distribution of human papillomavirus genotype 52–variant lineages and sublineages, according to geographical region.
The majority (93.7%) of lineage A variants belonged to sublineage A1, which was consistently observed across regions. All lineage B variants identified in this study were sublineage B2, and all lineage C variants belonged to sublineage C2.
Risk Association of Variant Lineages
The distribution of variant lineages and sublineages according to cervical pathology status is shown in Table 1. Multivariate analyses that adjusted for age were performed to compare subjects with normal cervical cytology findings, as controls, against subjects with histologically confirmed CIN3 or invasive cervical cancer, as cases. Lineage B was found to associate with a significantly higher risk than lineage A (age-adjusted odds ratio [OR], 5.46 [95%CI, 2.28–13.07]). Lineage C was also associated with a significantly higher risk than lineage A (age-adjusted OR, 7.78 [95% CI, 2.26–26.75]. Lineage B appeared to associate with a higher risk than lineage C, but the difference was not statistically significant (age-adjusted OR, 1.42 [95% CI, .56–3.56]. The number of isolates belonging to lineage D was not enough for analysis.
Table 1.
Distribution of Human Papillomavirus Genotype 52–Variant Lineages, According to Cervical Pathology Status
| Lineage, Sublineage | Subjects, No. (%), by Cervical Cytology Findinga |
||
|---|---|---|---|
| Normal (n = 191) | CIN3 (n = 111) | ICC (n = 41) | |
| A (n = 36) | 30 (15.7) | 4 (3.6) | 2 (4.9) |
| A1 (n = 34) | 28 (14.6) | 4 (3.6) | 2 (4.9) |
| A2 (n = 2) | 2 (1.0) | 0 | 0 |
| B (n = 289) | 155 (81.2) | 98 (88.3) | 36 (87.8) |
| B1 (n = 0) | 0 | 0 | 0 |
| B2 (n = 289) | 155 (81.2) | 98 (88.3) | 36 (87.8) |
| C (n = 14) | 5 (2.6) | 6 (5.4) | 3 (7.3) |
| C1 (n = 0) | 0 | 0 | 0 |
| C2 (n = 14) | 5 (2.6) | 6 (5.4) | 3 (7.3) |
| D (n = 4) | 1 (0.5) | 3 (2.7) | 0 |
Abbreviations: CIN3, grade 3 cervical intraepithelial neoplasia; ICC, invasive cervical cancer.
a All CIN3 and ICC cases were diagnosed by histological analysis.
DISCUSSION
Intratypic variants of HPV are divided into lineages based on the topology of phylogenetic tree and a difference of >1% in their full genome sequences [7]. Such classification of variants is important not only for understanding the evolution of HPV, but also because it carries biological implications. HPV52 has evolved into 4 lineages, for which the geographical distribution and risk implication have been uncertain [7] but are addressed in this study. The main strengths of our study are the large number of samples collected from around the world and the ability to restrict risk association analysis to cases with histologically confirmed diagnoses. Nevertheless, this study had limitations: it was not able to account for coinfection with other high-risk HPV genotypes; the number of samples available from some regions, such as Africa, was small; and some samples did not have associated cytological/histological diagnoses. Furthermore, because of colinearity between lineage and geographical distribution, the geographical source of sample could not be one of the covariants in the regression equation. Therefore, we cannot exclude the possibility of ethnogeographical effects on risk association of lineage variants.
To date, only a few studies have investigated the distribution of HPV52 lineages. Chang et al found that, among Taiwanese women, lineage B was the most prevalent (88.2%), followed by lineage C (11.1%), while lineage A was rare (0.7%) [9]. In contrast, lineage A was the most frequently found lineage in Canada, especially among white individuals [10, 11]. Another study examined samples collected from Japan, the Philippines, and Vietnam and reported that lineage B was most prevalent, followed by lineage A [12]. However, that study used E6 and E7 sequences to identify variant lineages, which are suboptimal for such purposes.
Our study assessed the distribution of HPV52 variants based on 611 samples collected from 14 cities across 4 continents, providing the largest data set for assessing geographical distribution. The most remarkable finding was the dominance of lineage B but the rare occurrence of lineage A in Asia. The opposite findings were true in non-Asian regions. Therefore, we propose that the name “Asian (As) lineage” be used to denote lineage B of HPV52 and that “non-Asian (nAs) lineage” be used to denote lineage A, to signify their characteristic geographical distributions.
Results of studies on the risk association of HPV52 variants are limited and inconclusive. Ding et al examined the E6 and E7 sequences of 121 samples from Zhejiang, in eastern China, but could not identify any variant with increased or decreased oncogenic risk [13]. Sun et al analyzed the L1, E6, E7, and LCR sequences of 72 samples from Shengjing, in northeast China [14]. In that study, the variants were not grouped according to the lineage classification system proposed by Chen et al [7], and no significant risk association was observed. Ishizaki et al studied 109 samples from Japan, the Philippines, and Vietnam, and again, no significant association between E6 and E7 sequence variation and abnormal cytology findings was found [12].
Although examination of E6 and E7 sequence variation did not reveal any significant risk association, some interesting findings were observed when lineage classification was taken into account. Chang et al used LCR-E6-E7 sequences to identify the lineage of 280 samples from Taiwan and reported a higher risk of CIN for lineage C variants, compared with lineage B variants [9]. Unfortunately, because lineage A was found in 2 samples only, it was precluded from risk association comparison. Two studies on the risk association of HPV52 variants were available from Canada. Aho et al showed that nonprototypic LCR variant was an independent predictor for viral persistence [11]. The observations from Formentin et al suggested that variant MTL-52-LCR-21, which belongs to sublineage A1, and variant MTL-52-LCR-02, which belongs to sublineage A2, conferred a higher risk. However, most of the isolates available in these Canadian studies were of lineage A, precluding comparison among different lineages. Schiffman et al examined HPV52 samples derived from the Guanacaste Cohort Study and observed that all CIN2+ cases were infected with lineages A/B/C, suggesting a lower risk for lineage D [15]. However, the observation was highly unstable and not statistically significant.
The current study has generated the most comprehensive data for analyzing risk association of HPV52 variant lineages with cervical disease. On the basis of our observations, we propose to classify lineage A as a “low-risk” lineage of HPV52 and lineage B as a “high-risk” lineage. Lineage C is probably “high-risk,” as well. Lineage D is rare and cannot be assigned to a risk category at this stage. Nevertheless, this risk classification should be further evaluated, preferably with assessment on the transforming ability of these variants, using in vitro or in vivo models.
In conclusion, we found that classifying HPV52 variants into lineages carries epidemiological and pathological implications. Lineage B can be regarded as “Asian” and “high-risk” on the basis of its geographical distribution and risk for cervical neoplasia. The reported higher disease attribution of HPV52 in Asia is likely a result of the higher prevalence of lineage B in that region. The unique epidemiological feature of HPV52 in Asia should be considered in the design and evaluation of diagnostic assays and vaccines intended for Asia.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by the Research Fund for the Control of Infectious Diseases, Food and Health Bureau, Hong Kong Special Administrative Region (reference 10090482); the Croatian Ministry of Research, Education, and Sport (to M. G.); the Bill and Melinda Gates Foundation (to K. K. S.-M. and A.-B. M.); the Cancer Society of Canada (to F. C.); the Italian Ministry of Foreign Affairs (to F. De M.); and the Italian Ministry of Health (to F. De M.).
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential conflicts of interest. P. K. S. C. is participating in a clinical trial supported by and has received honoraria as advisory board member and support for attending academic conferences from GlaxoSmithKline. J. P. has received travel support from, received research grant funding from, and served as board member for Merck; has received stock options from Aura Biosciences; has served as board member for Pharmajet; and has received consultation fees (paid to institution) from Qiagen. R. K. has received grants from GlaxoSmithKline and fees from GlaxoSmithKline, Merck Sharp Dome, and Qiagen. K. K. S.-M. is on the scientific and clinical advisory board of and has received stock options from OncoHealth. F. C. has received honoraria from Merck Sharp Dome for lectures on HPV. A.-B. M. has received honoraria from Merck Sharp Dome for being an advisory board member and from Becton Dickinson for serving as a speaker. A. F. has served on advisory boards for and received research grant funding and support to attend HPV-related conferences from GlaxoSmithKline Biologicals and Sanofi Pasteur MSD. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Muñoz N, Bosch FX, Castellsagué X, et al. Against which human papillomavirus types shall we vaccinate and screen? The international perspective. Int J Cancer. 2004;111:278–85. doi: 10.1002/ijc.20244. [DOI] [PubMed] [Google Scholar]
- 2.Smith JS, Lindsay L, Hoots B, et al. Human papillomavirus type distribution in invasive cervical cancer and high-grade cervical lesions: A meta-analysis update. Int J Cancer. 2007;121:621–32. doi: 10.1002/ijc.22527. [DOI] [PubMed] [Google Scholar]
- 3.Chan PK, Cheung TH, Li WH, et al. Attribution of human papillomavirus types to cervical intraepithelial neoplasia and invasive cancers in southern china. Int J Cancer. 2012;131:692–705. doi: 10.1002/ijc.26404. [DOI] [PubMed] [Google Scholar]
- 4.Huang S, Afonina I, Miller BA, Beckmann AM. Human papillomavirus types 52 and 58 are prevalent in cervical cancers from Chinese women. Int J Cancer. 1997;70:408–11. doi: 10.1002/(sici)1097-0215(19970207)70:4<408::aid-ijc6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 5.Ho CM, Chien TY, Huang SH, Lee BH, Chang SF. Integrated human papillomavirus types 52 and 58 are infrequently found in cervical cancer, and high viral loads predict risk of cervical cancer. Gynecol Oncol. 2006;102:54–60. doi: 10.1016/j.ygyno.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 6.Sasagawa T, Basha W, Yamazaki H, Inoue M. High-risk and multiple human papillomavirus infections associated with cervical abnormalities in japanese women. Cancer Epidemiol Biomarkers Prev. 2001;10:45–52. [PubMed] [Google Scholar]
- 7.Chen Z, Schiffman M, Herrero R, et al. Evolution and taxonomic classification of human papillomavirus 16 (HPV16)-related variant genomes: HPV31, HPV33, HPV35, HPV52, HPV58 and HPV67. PLoS One. 2011;6:e20183. doi: 10.1371/journal.pone.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang YJ, Chen HC, Lee BH, et al. Unique variants of human papillomavirus genotypes 52 and 58 and risk of cervical neoplasia. Int J Cancer. 2011;129:965–73. doi: 10.1002/ijc.25724. [DOI] [PubMed] [Google Scholar]
- 10.Formentin A, Archambault J, Koushik A, et al. Human papillomavirus type 52 polymorphism and high-grade lesions of the uterine cervix. Int J Cancer. 2013;132:1821–30. doi: 10.1002/ijc.27874. [DOI] [PubMed] [Google Scholar]
- 11.Aho J, Hankins C, Tremblay C, et al. Genomic polymorphism of human papillomavirus type 52 predisposes toward persistent infection in sexually active women. J Infect Dis. 2004;190:46–52. doi: 10.1086/420787. [DOI] [PubMed] [Google Scholar]
- 12.Ishizaki A, Matsushita K, Hoang HT, et al. E6 and E7 variants of human papillomavirus-16 and -52 in Japan, the Philippines, and Vietnam. J Med Virol. 2013;85:1069–76. doi: 10.1002/jmv.23566. [DOI] [PubMed] [Google Scholar]
- 13.Ding T, Wang X, Ye F, et al. Distribution of human papillomavirus 58 and 52 E6/E7 variants in cervical neoplasia in Chinese women. Gynecol Oncol. 2010;119:436–43. doi: 10.1016/j.ygyno.2010.08.032. [DOI] [PubMed] [Google Scholar]
- 14.Sun Z, Lu Z, Liu J, et al. Genomic polymorphism of human papillomavirus type 52 in women from northeast China. Int J Mol Sci. 2012;13:14962–72. doi: 10.3390/ijms131114962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiffman M, Rodriguez AC, Chen Z, et al. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 2010;70:3159–69. doi: 10.1158/0008-5472.CAN-09-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.