Abstract
Stable isotope-labeled amino acids have long been used to measure the fractional synthesis rate of proteins, although the mass spectrometry platforms used for such analyses have changed throughout the years. More recently, tandem mass spectrometers such as triple quadrupoles have been accepted as the standard platform for enrichment measurement due to their sensitivity and the enhanced specificity offered by multiple reaction monitoring (MRM) experiments. The limit in the utility of such platforms for enrichment analysis occurs when measuring very low levels of enrichment from small amounts of sample, particularly proteins isolated from two-dimensional gel electrophoresis (2D-GE), where interference from contaminant ions impact the sensitivity of the measurement. We therefore applied a high resolution orbitrap mass spectrometer to the analysis of [ring-13C6]-phenylalanine enrichment in individual muscle proteins isolated with 2D-GE. Comparison of samples analyzed on both platforms revealed that the high resolution MS has significantly improved sensitivity relative to the triple quadrupole MS at very low-level enrichments due to its ability to resolve interferences in the m/z dimension. At higher enrichment levels, enrichment measurements from the orbitrap platform showed significant correlation (R2 > 0.5) with those of the triple quadrupole platform. Together, these results indicate that high resolution MS platforms such as the orbitrap are not only as capable of performing isotope enrichment measurements as the more commonly preferred triple quadrupole instruments, but offers unparalleled advantages in terms of mass accuracy and sensitivity in the presence of similar-mass contaminants.
Keywords: Stable isotope, mass spectrometry, high resolution, muscle protein, protein synthesis, fractional synthesis rate, LC-MS/MS
Introduction
The measurement of fractional synthesis rate (FSR) of proteins using stable isotope labeled amino acids as tracers has been performed for over four decades. In the late 1970’s and early 80’s, a 200 mg quadriceps muscle biopsy would yield sufficient protein to measure [1-13C]-leucine enrichment using isotope ratio mass spectrometry (IRMS). In this technique, proteins were precipitated from the muscle homogenate and hydrolyzed to yield individual amino acids. After a cleanup using a cation exchange resin, the leucine methyl ester was cryogenically isolated by preparative gas chromatography. A ninhydrin reaction was then performed to cleave the [1-13C]-leucine, and the liberated CO2 was collected cryogenically prior to distillation into a dual inlet IRMS. Enrichments on the order of 0.005 – 0.02% above natural abundance could be measured by this technique, but 10-20 mg of protein was required [1-3].
In the late 1980’s a newer form of isotope ratio mass spectrometer was introduced which replaced the dual inlet with online sample introduction via a gas chromatograph (GC) and a combustion furnace [4]. The hydrolyzed mixed muscle proteins were converted to a volatile derivative suitable for separation on the GC column. The components were passed sequentially into online oxidation and reduction furnaces which converted them to CO2 and nitrogen, respectively. For 13C-tracers (e.g. L-[1,2-13C]-leucine, [ring-13C6]-phenylalanine) and 15N-tracers (e.g. 15N-phenylalanine), the CO2 and N2 produced, respectively, were analyzed in the mass spectrometer against pulses of reference gas placed before and after the peaks of interest [5-8]. The difference between the online system and the dual inlet approach was that nanograms of material were required for the former and micrograms for the latter. Therefore, it was possible to measure isotope enrichments for fractionated proteins (i.e. mitochondria, sarcoplasmic, and myofibrillar) from a 200 mg muscle sample and determine FSR for each of these muscle protein fractions [9-13].
One disadvantage to the online combustion process was dilution of the 13C label by the large number of 12C carbons present in the derivatized molecule. For example, the n-methyl acetate derivative of [1-13C]-leucine would dilute the 13C label by a factor of nine, which yielded slightly inferior precision measurements relative to the dual inlet procedure. For an FSR experiment where muscle biopsies were taken at 3 and 8 hours, the minimum detectable enrichment difference between the two biopsies was 1.0 delta per mil 13CO2 given a measurement precision of 0.3 delta per mil.
To improve detection limits and reduce the amount of tracer used, the use of multiple stable isotope labeled species (e.g. [ring-13C6]-phenylalanine or [U-13C]-leucine) was introduced. The application of multiple isotope species had been developed by several groups in the mid 1990’s for use with GC/MS systems that ordinarily could not measure such low enrichments with the more commonly used single tracer analogues [14-17]. Using this technique, the m+2 or m+3 vs. m+6 fragments of [ring-13C6]-phenylalanine are monitored rather than m0 vs. m+6. This improves the precision of the measurement greatly because the peak intensities of the two ions are much closer than that of the m0 species.
Further advances in isotope ratio analyses of multiple isotope labeled species were made with the use tandem mass spectrometers, such as triple quadrupole MS, in combination with gas or liquid chromatography (GC/MS/MS or LC/MS/MS, respectively)[18]. These instruments introduced greater specificity by enabling specific fragments of the labeled and unlabeled species to be monitored, subsequently reducing the amount of sample needed for analysis to picograms on column. This allowed the FSR of small amounts of muscle protein samples to be measured with similar precision and accuracy to the GC/C/IR/MS systems [16;18].
The LC/MS/MS method for measuring enrichment has been shown to work well for mixed muscle and plasma protein sub-fractions with relatively high synthesis rates, but several challenges may arise in these types of measurements when applied to measure isotope enrichment in individual muscle proteins with slow synthesis rates. While the use of 2D-GE has added specificity at the individual protein level, contamination of the column with acrylamide monomers causes column degradation over time. Additionally, there is a limit of detection for the isotope label below which the triple quadrupole cannot measure regardless of the sample source or preparation. For FSR measurements that collect biopsies at two or more time points, this detection limit is often observed in the first time point where minimal enrichment may be present. This detection limit is likely a combination of the limit of precision of the system and the noise of the background signal.
We applied high resolution mass spectrometry to overcome the challenges related to the LC/MS/MS approach for measuring isotope enrichment in low abundance individual muscle proteins. The use of higher resolution mass spectrometers for quantification, especially in the pharmaceutical industry, has increased markedly in the past few years with the increase in electronics and detector performance of newer instruments. Linear dynamic ranges spanning 4-5 orders of magnitude are now common for quantitative measurements. The application of high resolution MS to isotope ratio measurements has not been widely reported, but the accurate mass and faster scanning capabilities of such instruments may afford lower detection limits for isotope ratio measurements than is currently possible with unit resolution systems such as triple quadrupole MS. Here we report the application of a high-resolution orbitrap mass spectrometer (orbitrap MS) in combination with ultra-performance liquid chromatography (UPLC) separations for the measurement of [ring-13C6]-phenylalanine enrichment in individual proteins isolated by 2D-PAGE. The results are compared with the current standard for such measurements, a triple quadrupole tandem mass spectrometer, to demonstrate the merits and challenges of using high resolution trapping mass spectrometers for enrichment measurements. This new approach to measure low abundance isotope enrichment has potential for further applications in metabolic flux measurements of very low abundance metabolites.
Methods
Human Studies
We performed studies in healthy study participants in the Mayo Clinic Clinical Research Unit (CRU) after obtaining informed consent as a protocol approved by the Institutional Review Board. In ten healthy human study participants (age = 57.1 ± 20.24 yrs; M:F ratio = 1.5; and BMI = 26.80 ± 3.38 kg/mL), after overnight fast, we infused [ring-13C6]-phenylalanine at a rate of 1 mg/kg FFM/hr for eight hours following a priming dose to achieve an early isotope plateau [19;20]. Percutaneous needle biopsies of the vastus lateralis muscle were performed under local anesthesia at 180 min and 480 min into the infusion [3;21]. The studies were repeated 65-80 days following the first study.
Protein Isolation and Sample Preparation
From the human muscle biopsies, 20-30 mg of quadriceps muscle was homogenized in urea buffer (9.8M urea, 4% CHAPS) and skeletal muscle mitochondria separated using a differential centrifugation [22]. Individual proteins were isolated from the mixture by performing large, high-resolution, 2D-GE [23]. Approximately 200 μg of each protein sample were dissolved in lysis buffer to a final volume of 450 μl. These samples were used to rehydrate 24-cm, pH 4–7 and 6–9, immobilized pH gradient (IPG) strips (Bio-Rad Laboratories, Hercules, CA) in a rehydration tray overnight. The rehydrated IPG strips were subjected to isoelectric focusing in a Protean IEF Cell (Bio-Rad) using a three-step protocol: i) the focusing was achieved with an initial step of 250 V for 15 min; ii) continued with a maximum of 10,000 V increased linearly from 250 V over 6 h; and iii) continued at 10,000 V for 6 h. The cell temperature was kept at 20°C with a maximum current of 50 μA per strip. The IPG strips were then equilibrated for the SDS-PAGE in a two-step equilibration using 5 mL of equilibration buffer per strip (6 M urea, 2% SDS, 0.375 M Tris·HCl, pH 8.8, and 20% glycerol) with 130 mM DTT in the first step and 135 mM iodoacetamide in the second step. The equilibration steps were done in an equilibration tray for 10 min each on a rotary shaker at room temperature. The second-dimension separation by subunit molecular weight was performed by vertical 12%, 24 × 20-cm dimension SDS-PAGE (Ettan DALT system; GE Healthcare Bio-Sciences, Piscataway, NJ). The IPG strips were mounted into the IPG well with molten agarose and then run at 75 V for 24 h or until the dye front reached the bottom of the gel. The protein gel spots were visualized by staining with Coomassie blue (GelCode Blue Stain Reagent; Pierce, Rockford, IL).
Spots were excised from the gel, placed in glass vials, and washed several times with water. An additional 3 mL of HPLC water (Fisher Scientific) was added to each vial, and the gel spot samples were placed on a rocking shaker for 60 min. The proteins were then hydrolyzed at 120 °C for 18 h with 6 M HCl. The following day, the gel spot samples were centrifuged for 5 min at 3,000 rpm and 4 °C, after which 2 mL of water was added to each vial and vortexed. To prepare the AG-50x8 cation exchange column, the resin was rinsed with 4 mL of 4M ammonium hydroxide (NH4OH), followed by 4 rinses with 5 mL water. The column resin was regenerated with 4 mL of 4M HCl and rinsed with 5 mL of 0.1M HCl. The gel spot samples (approx. 2 mL) were transferred to the prepared AG-50 columns, the column was rinsed 4X with 4 mL of HPLC water, and amino acids were eluted into washed glass vials with three 1 mL washes of 4M NH4OH. The eluents were dried overnight in a speed-vac without heat. Amino acids were derivatized with 50 µL of 4M HCl in dry isobutanol at 85 °C for 45 min and dried under nitrogen. For LC-MS/MS analyses, 40 µL of 5% acetonitrile in water was added to each vial, vortexed and transferred to autosampler vials.
Triple Quadrupole Mass Spectrometer
The methodologies used in this approach have been previously published [18]. Briefly, HPLC separation was performed using a Cohesive TX2 multiplexed HPLC system with a Supelco Ascentis Express C18 column (15cm × 2.1 mm, 2.7µm) at a flow rate of 0.2 mL min−1, at room temperature. Solvent A was 99% water (Fisher Scientific), 1% acetonitrile (Fisher Scientific) with 0.1% formic acid (SigmaAldrich), and solvent B was 99.9% acetonitrile and 0.1% formic acid. 10 µL of sample was injected and separated using a gradient of: 20-70% B from 0-5.75 min, 70-95% B from 5.75-6.75 min, 95% B from 6.75-8.75 min, 95-5% B from 8.75-9.75 min, and 5% B from 9.75-13.95 min. The column eluent was passed into a triple quadrupole tandem mass spectrometer (ABSciex API 5000) through the electrospray ionization source operating under the following conditions: ion spray voltage = 5000 V; temperature = 300 °C; curtain gas = 40 a.u.; gas 1 = 30 a.u.; gas 2 = 10 a.u.; declustering potential = 60 V; entrance potential = 10 V; collision energy = 35 V; collision cell exit potential = 13 V; collision gas = 6 a.u..
Analysis of [ring-13C6]-phenylalanine enrichment in gel spot extracts was performed under multiple reaction monitoring (MRM) conditions monitoring the m/z 228.2>127.0 and m/z 224.2>123.0 transitions, which corresponded to the m+6 and m+2 fragments, respectively. The m+6 to m+2 peak area ratios of the unknowns were compared to a [ring-13C6]-phenylalanine calibration curve prepared by mixing known amounts of [ring-13C6]-phenylalanine with a constant amount of natural abundance phenylalanine over a range of 0 – 0.132% enrichment, measured as mole percent enrichment (mpe). FSR calculations were based on the [ring-13C6]-phenylalanine enrichment results for the 2D-GE isolated muscle proteins as well as the enrichment in the corresponding tissue fluids, which were measured on the same platform.
High-resolution Orbitrap Mass Spectrometer
UPLC separations were performed with a Dionex UltiMate 3000 system (ThermoFisher Scientific) and a Zorbax Extended C18 column (Agilent; 5 cm × 2.1 mm, 1.8 µm). Solvent A was 99% water (Fisher Scientific), 1% acetonitrile (Fisher Scientific) with 0.1% formic acid (SigmaAldrich), and solvent B was 99% acetonitrile, 1% water and 0.1% formic acid. The gradient was as follows: 0-6 min: 5-20% B; 6-10 min: 20-95% B; 10-12 min: 95% B; 12-13 min: 95-5% B, at a flow rate of 0.3 mL min−1. An injection volume of 5 µL was used. The UPLC was connected to the ion source through the diverter valve. The HESI ion source was operated with +3.5 kV spray voltage and probe heater temperature of 300 °C. Sheath, auxiliary and spare gas were 47.5, 11.25 and 1.0 (arb. units), respectively. The capillary temperature was maintained at 275 °C.
Analysis of [ring-13C6]-phenylalanine enrichment was performed with a high-resolution orbitrap mass spectrometer (Thermo Q Exactive Plus) using a 14 min targeted MS/MS method. The orbitrap MS was operated at 70,000 resolution, with an AGC target of 5e6 and maximum injection time of 100 ms. An isolation width of 1.0 m/z was applied for isolation of the m+3 and m+6 precursor species at m/z 225.15 and 228.17, respectively, and MS/MS was performed in the HCD cell at 40 V. All resulting fragments within the m/z 50-250 window were detected in the orbitrap.
While multiple reaction monitoring (MRM) experiments cannot be replicated on the orbitrap MS, extracted ion chromatograms (XICs) from the targeted MS/MS experiments were used for quantitative measurements of phenylalanine enrichment. This was achieved by setting a strict 5 ppm window around the m+3 and m+6 product ions (m/z 122.0872 and m/z 126.1010, respectively) which would typically be monitored with Q3 of a triple quadrupole instrument. The area under the curve of the extracted ion chromatograms was then used to calculate enrichment based on the ratio of m+6 to m+3 and the enrichment curve (described above). Data analysis was performed in Xcalibur QuanBrowser.
Results and Discussion
The challenge of detecting low level enrichment from individual proteins with a triple quadrupole (QQQ) MS instrument is demonstrated in Figure 1, which represents the enrichment measurements during the isotope infusion at 180 min biopsy of visit one in Fig. 1a and the 480 min biopsy from visit 2 in Fig. 1b. No enrichment peak is detectable for the first time point, whereas a small peak (shown in red) corresponding to [ring-13C6]-phenylalanine is present at the 480 min time point of visit 2, where enrichment is at the highest level. Given that phenylalanine-specific transitions were monitored in the MRM experiments, the background signal is relatively high in the [12C]-phenylalanine m+2 and the [13C6]-phenylalanine m+6 traces at both time points. SDS-PAGE separations of individual protein extracts require a number of reagents which are not ideal for MS analyses. Among them is polyacrylamide, which is challenging to remove from the gel spots and does have a similar structure to amino acids. We suspected that acrylamide oligomers with m/z values similar to the labeled and unlabeled phenylalanine were the source of the high background signals observed. Regardless of the source of the observed interferences, their presences add further challenges to measuring low level enrichments from individual proteins.
Figure 1.
Chromatographic traces from the triple quadrupole mass spectrometer showing the MRM transitions for the m+2 of [12C]-phenylalanine (m/z 224.2>123.0) in blue and m+6 of [ring 13C6]-phenylalanine (m/z 228.2>127.0) in red. a) Data corresponding to the 180 min biopsy time point for visit 1. b) Data corresponding to the 480 min biopsy time point for visit 2 of the same participant. At 180 min, the signal intensity of [ring 13C6]-phenylalanine is below the baseline noise.
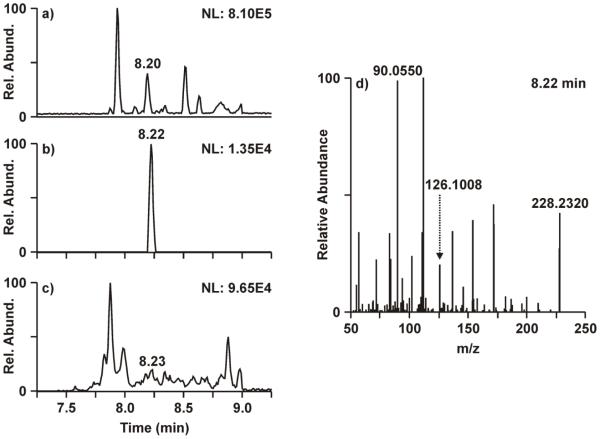
Based on the impediments in measuring low-level enrichment from 2D-PAGE-isolated proteins with the triple quadrupole platform, an orbitrap mass spectrometer was evaluated as an alternative platform for such analyses given its high resolution and accurate mass capabilities. Although the orbitrap MS does not parallel the sensitivity of triple quadrupole platforms when operated at high resolution, it was hypothesized that the enhanced specificity afforded by the high resolution would improve sensitivity relative to the triple quadrupole results by resolving similar-mass contaminant ions from the unlabeled and isotope labeled analytes . Figure 2 demonstrates the advantage of high resolution for resolving the similar-mass chemical interferences. Comparing the base peak chromatogram (Fig. 2a) and the 5 ppm XIC (Fig. 2b), a much larger peak (NL: 3.22E5) elutes at 8.20 min relative to the smaller [ring-13C6]-phenylalanine peak at 8.22 min (NL: 1.35E4). Fragmentation from the m/z 228.17 species at 8.20 min carries over into the MS/MS spectrum at 8.22 min (Fig. 2d), where m/z 90.0550 or m/z 112.1119 alternate as the base peak rather than the m+6 product ion. Opening the extraction window to 500 ppm (Fig. 2c) to mimic the resolution of the triple quadrupole mass analyzer greatly reduces the signal to noise and masks the [ring-13C6]-phenylalanine peak at 8.22 min. The data shown in Figure 2 represent the first time point of the study, where enrichment of [ring-13C6]-phenylalanine is expected to be very low. However, an enrichment measurement could be obtained easily from this sample using the high-resolution orbitrap MS despite the challenges of interfering similar-mass precursors, where no measurement could be made from the triple quadrupole MS (Fig. 1a) data for the same sample.
Figure 2.
Analysis of [13C6]-phenylalanine enrichment with the high-resolution orbitrap MS. Data shown represents the 180 minute time point of visit one. a) Base peak chromatogram from the targeted MS/MS of m/z 228.17, the m+6 precursor ion. b) Extracted ion chromatogram from a) showing m/z 126.1008, the m+6 product ion, at 5 ppm mass accuracy. c) Extracted ion chromatogram from a) showing m/z 126.1008 at 500 ppm mass accuracy. d) Fragmentation spectrum from the targeted MS/MS of m/z 228.17 at 8.22 min.
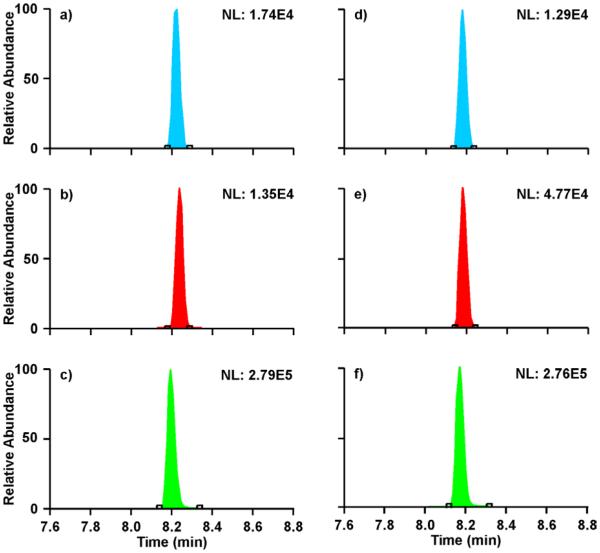
The orbitrap MS results for the first (visit 1-180 min) and last (visit 2-480 min) time points of the study are shown in Figure 3. The XICs in blue (Fig. 3a,d) correspond to the m+3 of the natural abundance species [12C]-phenylalanine, whereas the XICs in red (Fig. 3b,e) correspond to the [ring-13C6]-phenylalanine m+6. Comparing the two time points, one can see that the [ring-13C6]-phenylalanine enrichment increases over time while the [12C]-phenylalanine m+3 remains relatively constant. The XICs in green (Fig. 3c,f) correspond to the contaminant m/z 228.17 species which fragments to m/z 90.0550, the base peak in the MS/MS spectrum in Figure 2d. The abundance of these XICs remains constant over the course of the study, which supports the notion that these species result from the polyacrylamide in the 2D-PAGE process. In the presence of high-abundance similar-mass chemical noise, careful consideration must be made in regards to the injection time and AGC target as it is possible to saturate the c-trap.
Figure 3.
Extracted ion chromatograms from the orbitrap MS for the 180 min biopsy time point of visit 1 (a-c) and the 480 min time point of visit 2 (d-f). The XICs in blue (a and d) correspond to the [12C]-phenylalanine m+3 (m/z 225.15>122.0872). XICs in red (b and e) correspond to the [ring-13C6]-phenylalanine m+6 (m/z 228.17>126.1008). The green XICs (c and f) correspond to m/z 228.17>90.0555, which is representative of the suspected polyacrylamide interference with a similar m/z as [ring-13C6]-phenylalanine.
Table 1 summarizes the results from the QQQ MS and the orbitrap MS for the measurement of [13C6]-phenylalanine enrichment in individual muscle proteins. Results for three different study participants are tabulated across the four time points of the study. In all these participants, no peak (N.P.) was detected in the biopsies taken at 180 and/or 480 minutes in the first visit. These missing peaks result in missing values critical for the calculation of FSR, whereas peaks were detected at both these time points using the high-resolution platform. As the 13C isotope accumulates in the participants’ muscles, the enrichment peaks increased to levels detectable by the QQQ platform and FSR calculations could be performed by the second visit. No clear trend could be observed as to whether the QQQ or orbitrap MS over or underrepresented the enrichment levels.
Table 1.
MPE and FSR results from 13C enrichment measurements performed on both the triple quadrupole (QQQ) and high-resolution (HR) orbitrap MS platforms for three participants.
| Participant 1 | Participant 2 | Participant 3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Visit 1 | Visit 2 | Visit 1 | Visit 2 | Visit 1 | Visit 2 | |||||||
| QQQ | HR | QQQ | HR | QQQ | HR | QQQ | HR | QQQ | HR | QQQ | HR | |
| 180 min MPE | N.P. | 0.004 | 0.038 | 0.019 | 0.010 | 0.012 | 0.048 | 0.046 | N.P. | 0.008 | 0.045 | 0.031 |
| 480 min MPE | 0.018 | 0.009 | 0.046 | 0.040 | N.P. | 0.021 | 0.080 | 0.064 | N.P. | 0.010 | 0.066 | 0.053 |
|
FSR
(%/hr) |
N/A | 0.023 | 0.037 | 0.095 | N/A | 0.040 | 0.125 | 0.071 | N/A | 0.008 | 0.077 | 0.082 |
N.P. = No Peak; N/A = Calculation could not be performed with missing data.
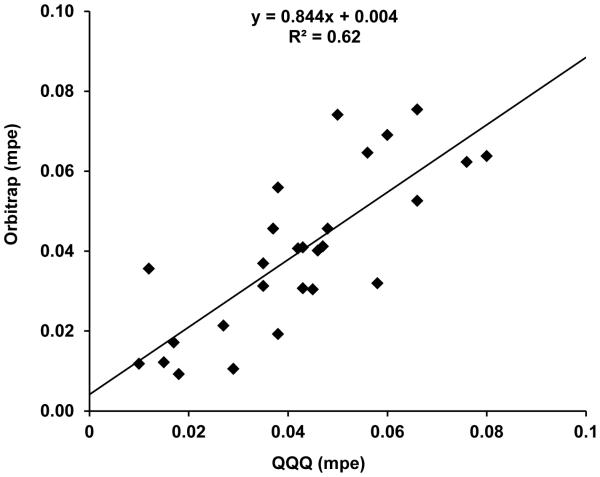
To better compare the two platforms, a correlation analysis was performed using data from the muscle protein gel spot samples that were analyzed on both the triple quadrupole and the orbitrap with the same enrichment curve. Figure 4 shows the matching data points from 10 participants, who contributed data points from either one study day with two time points or two study days with 2-3 time points. This dataset is smaller than that the actual number of biopsies collected as the QQQ MS platform was unable to detect the [ring-13C6]-phenylalanine peak in the first data point for most participants. The correlation between the results from the triple quadrupole and those from the orbitrap mass analyzer is relatively poor, with R2 = 0.62. Generally, the QQQ MS results over-estimated the enrichment level as evidenced by the greater number of data points below the fitted line than above. This could result from the co-eluting contaminant species, as discussed above, contributing to the peak area of the [ring-13C6]-phenylalanine peak. The ability of the high resolution-MS to separate these contaminants from the analyte in the m/z dimension provides more accurate determination of the 13C enrichment.
Figure 4.
Correlation of [ring-13C6]-phenylalanine mpe values determined from the triple quadrupole (QQQ) with those determined from the orbitrap mass spectrometer for ATP synthase. Data points represent the 180 or 480 min time points from one or two study days for 10 participants. The same enrichment curve was used on both platforms.
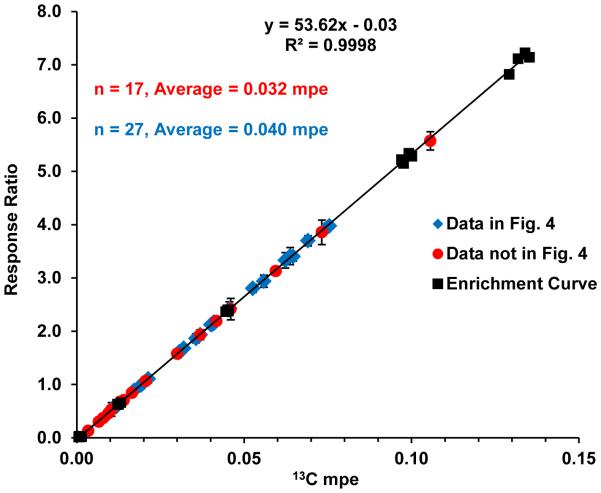
The missing data from the triple quadrupole platform for the earlier time points skews the correlation graph towards higher mole percent enrichment (mpe) values, as those initial time points represent the lowest levels of enrichment. To demonstrate the sensitivity of the orbitrap MS, the corresponding enrichment curve with the results for the 10 participants is shown in Figure 5. The data points which could not be shown in Figure 4 due to missing triple quadrupole data are indicated with red circles (n = 17), whereas the data used in Figure 4 is indicated with blue diamonds (n = 27). The majority of the red data points are clustered near the lower end of the mpe range (average = 0.032, median = 0.021). The blue data points are more widely distributed, but generally have higher mpe values (average = 0.040, median = 0.040) than the red data points. The lowest level of enrichment measured with the high resolution platform was 0.003 mpe in a biopsy taken at the first time point of the first study day. In contrast, the lowest enrichment that was detected with both the orbitrap and the triple quadrupole was three times higher at 0.009 mpe, which represented the second time point of the first study day from the same participant. Collectively, these results demonstrate the disparity between low-level enrichment measurements performed on the triple quadrupole and those performed on the high-resolution orbitrap. The sensitivity gained by using a high resolution mass spectrometer enabled the measurement of the low level enrichments which previously could not be quantified with the triple quadrupole platform.
Figure 5.
Enrichment curve from the orbitrap (black squares) spanning the range from 0-0.132% 13C enrichment. Average mpe results from 10 participants measured with the orbitrap mass spectrometer are overlaid onto the enrichment curve, where the data used to make Fig. 4 is shown as blue diamonds and the data not shown in Fig. 4 is indicated with red circles. The data that lacked corresponding QQQ MS results (red circles; not shown in Figure 4) accounts for 17 of the data points with an average of 0.032 mpe and median of 0.021 mpe. Data used in Figure 4 (blue diamonds) accounts for 27 of the data points, with an average and median mpe of 0.040. Errors bars represent the standard deviation of two replicate measurements.
Overall, the ATP synthase gel spot samples cover a range from 0.003 to 0.106 mpe as determined by the high resolution MS measurements. The dataset presented in Figure 5 (excluding the enrichment curve) was used to determine the mean standard deviations (SD) and coefficients of variation (CV%) for low, medium and high enrichment levels based on two replicate injections per sample. The low enrichment set (0.003 – 0.019, n = 14) had the largest CV% at 6.8%. The CV% generally decreased with increasing enrichment, with 2.8 and 2.6 CV% for medium and high enrichments, respectively. A similar trend was observed in a previous study comparing four GC and LC-based MS and MS/MS platforms, including the same triple quadrupole platform used in this work, for enrichment measurements where CV% increased with decreasing enrichment for all four platforms [18]. The CV% at low and high enrichment from the previous characterization of the triple quadrupole mass spectrometer are smaller (i.e. low mpe: 3.06% CV; high mpe: 1.30% CV) than those observed here for the orbitrap MS, however the range for low enrichment was more narrow (i.e. 0.0091-0.0194 mpe) in comparison with the low enrichment measurement on the orbitrap mass spectrometer (i.e. 0.003-0.019).
In the current study, purified human skeletal muscle ATP synthase β was isolated as previously reported for a mouse study [24]. However, the same level of isotope enrichment observed in mice cannot be achieved in humans due to the larger lean tissue mass and lower rates of protein turnover in humans. Further, skeletal muscle protein turnover rates are relatively slow compared to tissues such as liver and heart, resulting in very low levels of incorporation for stable isotope labeled amino acids. Performing such enrichment measurements by mass spectrometry-based approaches (e.g. GC/C/IRMS or LC/MS/MS) therefore requires both sufficient sensitivity and precision to detect low levels of enrichment. To our best knowledge, we have demonstrated for the first time that low abundance enrichment and fractional synthesis rates of human skeletal muscle ATP synthase can be measured using high resolution orbitrap mass spectrometry. The sensitivity and specificity of this methodology will enable the measurement of synthesis rates within individual muscle proteins under study conditions previously not feasible due to low levels of isotope enrichment. Moreover, the ability to measure low abundance molecular isotope enrichment will offer tremendous opportunities to study metabolite fluxes in humans and animals [25].
Conclusions
Triple quadrupole mass spectrometers have been the standard platform for performing isotope enrichment measurements due to the sensitivity and specificity of multiple reaction monitoring experiments. However, this platform has some limitations for complex samples with low enrichment levels. In particular, the measurement of low-level [ring-13C6]-phenylalanine enrichments from individual muscle proteins isolated from 2D-PAGE has been problematic due to similar-mass contaminants from the 2D-PAGE isolation. These interferences increase the background of the measurement to such an extent that the [ring-13C6]-phenylalanine peak cannot be measured at low enrichments levels. Enrichment of 13C in muscle proteins at multiple time points is critical for the determination of fractional synthesis rates, a measure of muscle protein metabolism. The lack of data from the triple quadrupole platform for the first study time point, where enrichment is at its lowest level, leads to incomplete study results and potentially hinders the broader conclusions that may be drawn about muscle metabolism.
To overcome the challenges presented for enrichment measurement from the protein gel spots, a high resolution orbitrap mass spectrometer was applied to determine its suitability for such measurements as ATP synthase β in human skeletal muscle isolated by 2D-GE. We demonstrated that the high resolution resolved similar-mass interfering chemical species from the [ring-13C6]-phenylalanine, thereby improving signal-to-noise and sensitivity. Although the orbitrap MS cannot perform MRM quantitation in the same manner as triple quadrupole instruments, the results presented here demonstrate that the orbitrap MS is a highly suitable platform for determination of enrichment in individual muscle proteins. Comparison of results for the same sample set analyzed on both the high resolution and triple quadrupole platforms indicated an overestimation of enrichment values from the triple quadrupole mass spectrometer, likely due to contributions from the same-mass chemical interference. Enrichment levels as low as 0.003 mpe were quantifiable using the high resolution platform, whereas the triple quadrupole platform was limited to enrichment levels three times greater (i.e. 0.009 mpe). Moreover, the reproducibility (CV%) of the enrichment measurements from the high-resolution orbitrap MS were comparable to those previously observed for LC/MS/MS, GC/MS/MS and GC/C/IRMS platforms for similar enrichment values. High resolution MS may not be necessary for every level of enrichment measurement, but we have demonstrated the orbitrap MS to be a robust and sensitive platform especially for very low-level enrichment from 2D-GE-isolated muscle proteins.
Acknowledgements
This publication was made possible by Mayo Clinic Metabolomics Resource Core through grant number U24DK100469 from the National Institute of Diabetes and Digestive and Kidney Diseases and originates from the National Institutes of Health Director's Common Fund, UL1 TR000135 from the National Center for Advancing Translational Sciences (NCATS), NIH R01 DK41973, and KL2RR024151.
References
- 1.Halliday D, Ford GC. Mass spectrometry. Walter de Gruyter, Inc.; Hawthorne, NY: 1988. Stable isotopes in clinical investigation. [Google Scholar]
- 2.Halliday D, Read WW. Proceedings of the Nutrition Society. 1981;40:321–334. doi: 10.1079/pns19810048. [DOI] [PubMed] [Google Scholar]
- 3.Nair KS, Halliday D, Griggs RC. Am J Physiol Endocrinol Metab. 1988;254:E208–E213. doi: 10.1152/ajpendo.1988.254.2.E208. [DOI] [PubMed] [Google Scholar]
- 4.Matthews DE, Hayes JM. Anal. Chem. 1978;50:1465–1473. [Google Scholar]
- 5.Balagopal P, Ljungqvist O, Ford GC, Nair KS. Skeletal muscle myosin heavy chain synthesis rate in healthy humans. J Invest Med 44[3] 1996;273A doi: 10.1152/ajpendo.1997.272.1.E45. [DOI] [PubMed] [Google Scholar]
- 6.Yarasheski KE, Smith K, Rennie MJ, Bier DM. Biol Mass Spec. 1992;21:486–490. doi: 10.1002/bms.1200211004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rennie MJ, Edwards RHT, Halliday D, Matthews DE, Wolman SL, Millward DJ. Clin. Sci. 1982;63:519–523. doi: 10.1042/cs0630519. [DOI] [PubMed] [Google Scholar]
- 8.Rennie MJ, Smith K, Watt PW. Am J Physiol Endocrinol Metab. 1994;266:E298–E307. doi: 10.1152/ajpendo.1994.266.3.E298. [DOI] [PubMed] [Google Scholar]
- 9.Rooyackers OE, Balagopal P, Nair KS. Muscle & Nerve. 1997;5(suppl):S93–S96. [PubMed] [Google Scholar]
- 10.Balagopal P, Rooyackers OE, Adey DB, Ades PA, Nair KS. Am J Physiol Endocrinol Metab. 1997;273:E790–E800. doi: 10.1152/ajpendo.1997.273.4.E790. [DOI] [PubMed] [Google Scholar]
- 11.Toth MJ, Matthews DE, Tracy RP, Previs MJ. Am J Physiol Endocrinol Metab. 2005;288:E883–E891. doi: 10.1152/ajpendo.00353.2004. [DOI] [PubMed] [Google Scholar]
- 12.Kumar V, Atherton P, Smith K, Rennie MJ. J Appl Physiol. 2009;1985;106:2026–2039. doi: 10.1152/japplphysiol.91481.2008. [DOI] [PubMed] [Google Scholar]
- 13.Kumar V, Atherton PJ, Selby A, Rankin D, Williams J, Smith K, Hiscock N, Rennie MJ. J Gerontol A Biol Sci Med Sci. 2012;67:1170–1177. doi: 10.1093/gerona/gls141. [DOI] [PubMed] [Google Scholar]
- 14.Patterson BW, Zhang X-J, Chen YP, Klein S, Wolfe RR. Metabolism: Clinical & Experimental. 1997;46:943–948. doi: 10.1016/s0026-0495(97)90084-6. [DOI] [PubMed] [Google Scholar]
- 15.Calder AG, Anderson SE, Grant I, McNurlan MA, Garlick PJ. Rapid Commun. Mass. Spec. 1992;6:421–424. doi: 10.1002/rcm.1290060704. [DOI] [PubMed] [Google Scholar]
- 16.Jaleel A, Short KR, Asmann YW, Klaus KA, Morse DM, Ford GC, Nair KS. Am J Physiol Endocrinol Metab. 2008;295:E1255–E1268. doi: 10.1152/ajpendo.90586.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang XJ, Chinkes DL, Wolfe RR. Am J Physiol Endocrinol Metab. 2002;283:E753–E764. doi: 10.1152/ajpendo.00053.2002. [DOI] [PubMed] [Google Scholar]
- 18.Zabielski P, Ford GC, Persson XM, Jaleel A, Dewey JD, Nair KS. J Mass Spectrom. 2013;48:269–275. doi: 10.1002/jms.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Short KR, Meek SE, Moller N, Ekberg K, Nair KS. Am J Physiol Endocrinol Metab. 1999;276:E1194–E1200. doi: 10.1152/ajpendo.1999.276.6.E1194. [DOI] [PubMed] [Google Scholar]
- 20.Nair KS, Ford GC, Ekberg K, Fernqvist-Forbes E, Wahren J. J Clin Invest. 1995;95:2926–2937. doi: 10.1172/JCI118000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards RH. Lancet. 1971;2:593–5. doi: 10.1016/s0140-6736(71)92165-9. [DOI] [PubMed] [Google Scholar]
- 22.Rooyackers OE, Adey DB, Ades PA, Nair KS. Proc. Natl. Acad. Sci. USA. 1996;93:15364–15369. doi: 10.1073/pnas.93.26.15364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaleel A, Henderson GC, Madden BJ, Klaus KA, Morse DM, Gopala S, Nair KS. Diabetes. 2010;59:2366–2374. doi: 10.2337/db10-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanza IR, Zabielski P, Klaus KA, Morse DM, Heppelmann CJ, Bergen HR, III, Dasari S, Walrand S, Short KR, Johnson ML, Robinson ML, Schimke JC, Jakaitis DR, Asmann YW, Sun Z, Nair KS. Cell Metabolism. 2012;16:777–788. doi: 10.1016/j.cmet.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higashi RM, Fan TW, Lorkiewicz PK, Moseley HN, Lane AN. Methods Mol Biol. 2014;1198:147–167. doi: 10.1007/978-1-4939-1258-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]