Non-muscle myosin IIB plays a major role in applying force on the nucleus to facilitate nuclear translocation through tight spaces during 3D invasive migration, while non-muscle myosin IIA is critical for generating force during active protrusion.
Abstract
Non-muscle myosin II (NMII) is reported to play multiple roles during cell migration and invasion. However, the exact biophysical roles of different NMII isoforms during these processes remain poorly understood. We analyzed the contributions of NMIIA and NMIIB in three-dimensional (3D) migration and in generating the forces required for efficient invasion by mammary gland carcinoma cells. Using traction force microscopy and microfluidic invasion devices, we demonstrated that NMIIA is critical for generating force during active protrusion, and NMIIB plays a major role in applying force on the nucleus to facilitate nuclear translocation through tight spaces. We further demonstrate that the nuclear membrane protein nesprin-2 is a possible linker coupling NMIIB-based force generation to nuclear translocation. Together, these data reveal a central biophysical role for NMIIB in nuclear translocation during 3D invasive migration, a result with relevance not only to cancer metastasis but for 3D migration in other settings such as embryonic cell migration and wound healing.
Introduction
Cellular migration is a crucial aspect of many biological processes, including embryonic development, wound healing, recruitment of immune cells, as well as pathological conditions such as cancer cell invasion and metastasis. Traditionally, examining the mechanics of cell motility has been performed on rigid, 2D substrates, such as glass and plastic. Only in recent years have studies begun to address the roles of cytoskeletal force production during invasive 3D migration (Doyle et al., 2009). For a cell to efficiently migrate through 3D matrices it must overcome obstacles that can inhibit both anterior protrusion and the translocation of the large, bulky nucleus (Wolf et al., 2013; Davidson et al., 2014; Harada et al., 2014). These barriers are absent in 2D migration settings, leaving critical aspects of 3D migration poorly understood (Friedl and Alexander, 2011).
One major player in cellular migration is the motor protein non-muscle myosin II (NMII; Conti and Adelstein, 2008). In mammals, NMII exists as three isoforms (NMIIA, IIB, and IIC) that carry heavy chains encoded by three distinct genes (MYH9, MYH10, and MYH14, respectively). These NMII isoforms are differentially expressed in a cell- and tissue-specific manner and in a developmentally regulated fashion (Wang et al., 2010). Studies in cells and mice have demonstrated that NMII isoforms are capable of both unique and redundant functions (Vicente-Manzanares et al., 2011; Wang et al., 2011). Although it is clear that NMII isoforms contribute to cell polarization and help limit protrusions in 2D and 3D (Fischer et al., 2009; Vicente-Manzanares et al., 2011), little is known about their contribution to nuclear mechanics during 3D migration.
During 2D migration, when a migrating cell forms protrusions, NMIIA stabilizes nascent focal adhesions (Vicente-Manzanares et al., 2007; Choi et al., 2008; Pasapera et al., 2010). As NMIIA filaments move rearward via actin retrograde flow, NMIIB coassembles with the NMIIA filaments and becomes the dominant isoform in the posterior of the cell (Beach et al., 2014). Thus, in persistently migrating cells, NMIIA is enriched anteriorly, whereas NMIIB is enriched in lateral stress fibers and the posterior of the cell (Kolega, 1998; Vicente-Manzanares et al., 2008; Raab et al., 2012). In accordance with the leading edge roles in focal adhesion stabilization, NMIIA has also been shown to be critical for generation of traction force at the leading edge (Jorrisch et al., 2013). Interestingly, although knockdown of NMIIA reduces traction force generation, in most systems this does not significantly reduce migration efficiency along 2D surfaces and in some cases NMIIA knockdown even enhances 2D migration rates (Doyle et al., 2012; Jorrisch et al., 2013). Although there is general consensus that NMIIA has critical roles in focal adhesion dynamics in leading edge protrusions, roles during migration for NMIIB in either 2D or 3D settings are poorly understood.
When mammary gland epithelial cells are driven through an epithelial–mesenchymal transition, NMIIB expression is robustly up-regulated (Beach et al., 2011), suggesting that NMIIB might enhance or contribute to the invasive behavior of mesenchymal-like cells (Taylor et al., 2010; Beach et al., 2011). NMIIB has been shown to play a role in nuclear positioning in cells in 2D. Knockdown of NMIIB in the CHO.K1 cell line results in freely rotating nuclei, indicating a loss of nuclear anchoring (Vicente-Manzanares et al., 2007), and NMIIB knockdown in fibroblasts impairs nuclear repositioning during 2D scratch wound migration (Chang et al., 2013). Blebbistatin treatment, which inhibits all three NMII isoforms, has also been shown to inhibit squeezing of the nucleus during its translocation through restrictive pores in migrating cancer cells (Beadle et al., 2008; Breckenridge et al., 2010). However, the specific NMII isoform responsible for nuclear translocation remains unclear. Given the evidence for NMIIB roles in nuclear positioning in 2D settings, we examine in the current work the contributions of NMIIB in cancer cell migration in 3D settings and identify a role for NMIIB in nuclear translocation through restrictive spaces. We further show that nesprin-2, an outer nuclear membrane–spanning protein that is a major component of the linker of nucleoskeleton and cytoskeleton (LINC) complex (Crisp et al., 2006; Luxton et al., 2010), is a possible mechanistic link for NMIIB-based nuclear translocation during 3D invasion. Our findings support a model where NMIIB-based cytoskeletal elements exhibit an isoform-specific association with the nuclear envelope, thereby enabling the cytoskeleton to exert sufficient forces on the nucleus to squeeze it through tight spaces during 3D invasive migration.
Results
Myosin IIB exerts force on the nucleus
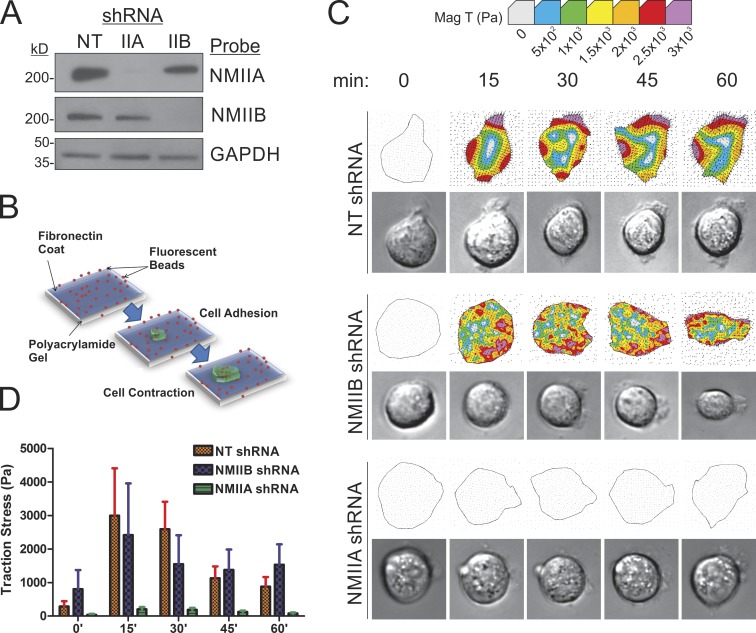
Although NMIIA is recognized to have an important role in exerting forces on nascent adhesions during polarized cell migration in 2D (Jorrisch et al., 2013), it is less clear from previous work whether NMIIB may have roles in generating force relative to matrix adhesions during migration. Recent work demonstrating that NMIIB is critical for mesenchymal cell invasion through collagen gels (Beach et al., 2011) suggests that this isoform may have a previously unrecognized role in force generation during cell migration. We have therefore reexamined contributions of NMIIB to traction force generation in mesenchymal-like breast cancer cells via traction force microscopy (Dembo and Wang, 1999). Mouse mammary gland carcinoma 4T1 cells were subjected to lentiviral-based shRNA depletion of NMIIA or IIB (Fig. 1 A), and then assessed for their ability to exert force on their substrate. Cells were plated on polyacrylamide (PAA) gels coated with 50 µg/ml fibronectin and mean traction stress per cell was measured over 60 min during initial spreading after plating (Fig. 1 B). During this time, control 4T1 cells carrying a nontargeting (NT) control shRNA construct displayed strong traction stress generation, which peaked at ≈15–30 min after adhesion (Fig. 1, C [top] and D [red bars]). NMIIB shRNA had only minimal effect on traction stress generation during active spreading (Fig. 1, C [middle] and D [blue bars]), whereas NMIIA shRNA drastically eliminated traction stress generation (Fig. 1, C [bottom] and D [green bars]).
Figure 1.
NMIIA is essential for traction force generation during initial adhesion and spreading. (A) NMIIA and NMIIB protein expression levels in 4T1 cells stably infected with NT lentiviral constructs or shRNA constructs to deplete NMIIA or NMIIB. (B) Diagram demonstrating TFM. Fluorescent microbeads are embedded within a PAA gel formed on a glass coverslip. The PAA gel is then coated in a continuous layer of 50 µg/ml fibronectin. Plated cells deform the gel during spreading, moving the embedded beads. The cells are then trypsinized, allowing for the relaxation of the gel and return of the beads to their original position. The distance of bead movement is measured to quantify traction stress. (C) 4T1 cell traction force generation during active spreading. Traction stress was measured every 15 min for 1 h after plating. Representative images of control, NMIIA-shRNA, and NMIIB-shRNA cells during the spreading time course are shown. Representative traction stress heat maps (top) and their corresponding phase-contrast image (bottom) are shown. (D) Traction stresses are quantified by averaging the magnitude of all calculated vectors within the area of the cell outline. The mean of the per cell average traction stress was quantified. Error bars are SEM; n = 20 cells.
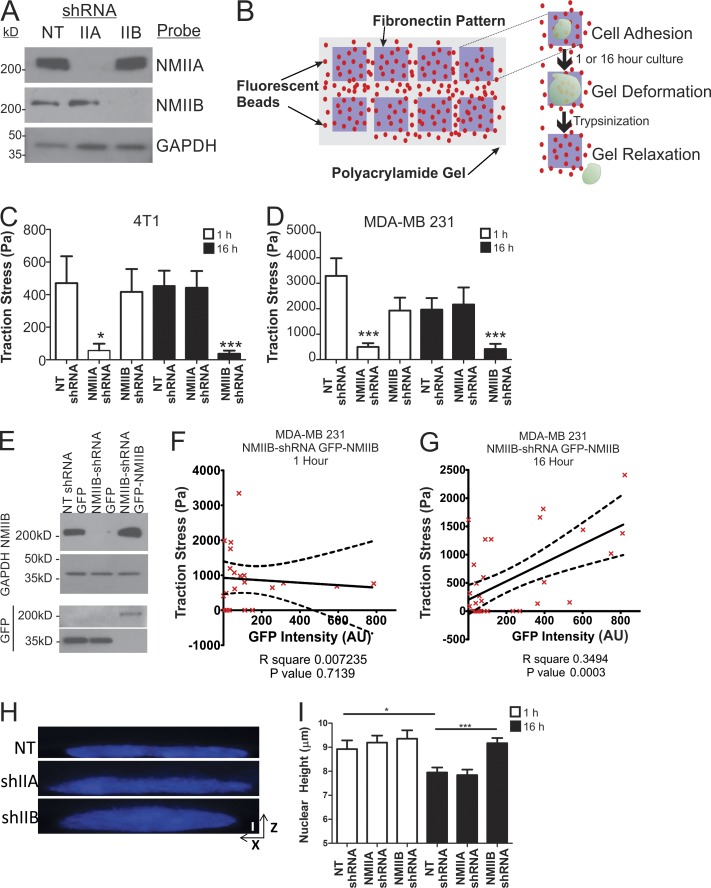
To test whether NMIIA is the only isoform relevant in traction stress generation or whether NMIIB might contribute to traction stresses in settings other than the initial spreading phase, we compared the relative roles of NMIIA versus NMIIB in cells at 1 and 16 h after plating on fibronectin. Experiments were performed with both mouse 4T1 cells and with the human basal-like mammary carcinoma line MDA-MB 231. As with the 4T1 lines, lentiviral-based shRNA was used in MDA-MB 231 cells to deplete NMII isoforms (Fig. 2 A). Cells were plated on constrained micropatterned squares (30 × 30 µm) to eliminate polarized migration and persistent protrusion activity (Fig. 2 B). Similar to the results during cell spreading on unpatterned substrates (Fig. 1), after 1 h of adhesion on patterned surfaces, NMIIA shRNA ablated traction stress in 4T1 and MDA-MB 231 cells (Fig. 2, C and D, open bars). In contrast, NMIIB shRNA had no effect on traction stress in 4T1 cells and only modest effect on traction stress in MDA-MB 231 cell line at this time point (Fig. 2, C and D, open bars). However, when cells were allowed to adhere for 16 h and form steady-state attachments to the matrix, the contributions of NMIIA and NMIIB switched, and NMIIB displayed the dominant contribution to maintaining traction stress (Fig. 2, C and D, shaded bars). These results suggest a critical role for NMIIB in long-term stress generation relative to initial substrate adherence and cell spreading.
Figure 2.
NMIIB is critical for traction force generation in fully spread, nonmigrating cells. (A) NMIIA and NMIIB protein expression levels in MDA-MB231 cells stably infected with lentiviral shRNA constructs to deplete NMIIA or NMIIB. (B) Diagram showing measurement of traction stress, a measure of force generation for cells attached to 30-µm2 patterned squares of fibronectin. Cells were plated and allowed to adhere for 1 or 16 h, and bead positions were imaged before and after trypsin treatment to determine contractile stresses. (C and D) Traction stress measurements were collected on NT shRNA control, NMIIA-shRNA, or NMIIB-shRNA 4T1 cells (C) or MDA-MB-231 cells (D) at 1 h (open bars) and 16 h (closed bars). *, P < 0.01; ***, P < 0.0001; n = 30 cells per condition; error bars represent SEM. (E) MDA-MB 231 were transiently transfected with either a NMIIB-GFP fusion construct or a control-free GFP construct and expression was verified via Western blot. (F and G) MDA-MB 231 NMIIB-shRNA cells were transfected with a GFP-NMIIB fusion construct and traction stress was measured on patterned fibronectin at 1 h (F) or 16 h (G) after plating. GFP intensity of individual cells was quantified and correlated with levels of traction stress. Dashed lines represent 95% confidence interval; n = 21 and 33 for 1 and 16 h time points, respectively. (H and I) MDA-MB 231 cells were plated on fibronectin-coated coverglass for 1 or 16 h before fixation and DAPI staining. They were then analyzed by spinning-disk confocal microscopy to generate x–z projections for nuclear height quantification. (H) Representative x–z projection DAPI images of MDA-MB 231 control, NMIIA shRNA, or NMIIB shRNA at 16-h time point. Vertical bar, 5 µm. (I) Nuclear height among conditions is unchanged at 1 h (open bars). NMIIB knockdown significantly increased nuclear height, whereas NMIIA knockdown had no effect compared with control at 16 h (shaded bars). **, P < 0.01; ***, P < 0.0001; n = 60 cells per condition.
To further explore the contribution of NMIIB in traction stress generation, we rescued NMIIB shRNA cells with GFP-NMIIB (Fig. 2 E) using a GFP-NMIIB fusion construct previously shown to have wild-type NMIIB function both in vitro and in vivo (Bao et al., 2007). GFP-NMIIB expression level, as determined by quantitation of cell-by-cell GFP intensity, was then correlated with traction stress generation. At 1 h after plating on micropatterned fibronectin, there was no correlation between GFP-NMIIB and traction stress (Fig. 2 F), supporting the conclusion that initial traction stress generation during active protrusion is independent of NMIIB. However, a significant positive correlation was observed between GFP-NMIIB expression and traction stress after 16 h of adhesion to micropatterned fibronectin (Fig. 2 G), indicating that NMIIB does contribute to traction stress generation and that this behavior only emerges once cells have had time to develop NMIIB-containing contractile structures. Pearson coefficient analysis confirmed statistical correlation of GFP levels with traction stress at 16 h (P = 0.0003) but not at 1 h (P = 0.7139).
A previous study showed that cytoskeletal forces can control nuclear shape and height (Chancellor et al., 2010). Therefore, we hypothesized that the NMIIB-mediated forces could contribute to nuclear flattening in spread cells under steady-state conditions. As a test of this hypothesis, we measured nuclear height in NT control and NMIIA and NMIIB shRNA-treated MDA-MB 231 cells plated on 50 µg/ml of fibronectin-coated coverglass for either 1 or 16 h (Fig. 2, H and I). No significant difference in nuclear height was observed at 1 h among NT-, NMIIA-, or NMIIB-shRNA–treated cells (Fig. 2 I, open bars). However, NMIIB shRNA caused a significant increase in nuclear height at 16 h, whereas NMIIA shRNA had no effect (Fig. 2 I, shaded bars) and further indicates that NMIIB, and not NMIIA, mediates this nuclear flattening.
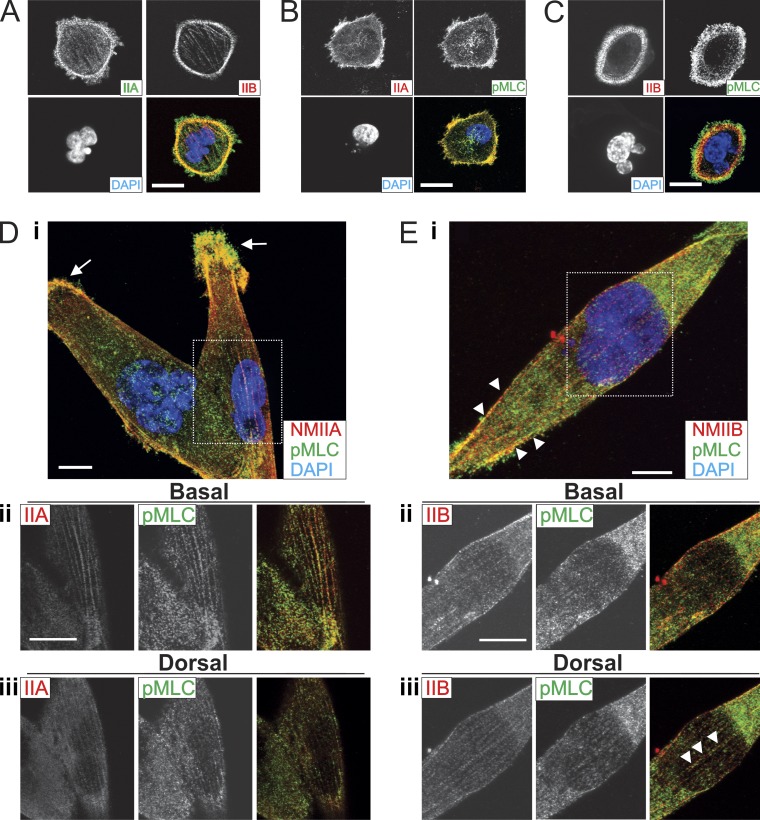
To further explore these isoform-specific contributions, we used immunofluorescence microscopy to observe the localization of the two isoforms in MDA-MB 231 cells 1 and 16 h after plating on fibronectin-coated coverglass. During active spreading (1 h) NMIIA localized in anterior lamellar regions, whereas NMIIB formed circumferential stress fibers just posterior to NMIIA (Fig. 3 A). To identify zones of NMII activation, we costained for phospho-myosin light chain (pMLC). At 1 h after plating, this revealed strong colocalization with NMIIA at the spreading margin (Fig. 3 B). Staining for both NMIIB and pMLC at 1 h revealed a similar pattern, with the strongest pMLC signal being more anterior than the peak NMIIB signal, colocalizing with the NMIIA zone rather than the NMIIB enriched zone (Fig. 3 C). In fully spread cells plated for 16 h, NMIIA localized more robustly to peripheral protrusions (Fig. 3 D, i, arrows) and to basal stress fibers (Fig. 3 D, ii) with modest dorsal fiber formation (Fig. 3 D, iii), whereas NMIIB localized more robustly to lateral fibers (Fig. 3 E, i, arrowheads) and perinuclear fibers that are both basal (Fig. 3 E, ii) and dorsal to the nucleus (Fig. 3 E, iii, arrowheads). These patterns further support our traction force data, demonstrating a dominant role of NMIIA in generation of traction forces in actively spreading cells and robust NMIIB-containing actomyosin fibers present in the perinuclear region in fully spread cells that have had time to develop mature cytoskeletal structures.
Figure 3.
NMIIA is enriched in protrusions and active NMIIB localizes to dorsal perinuclear fibers. MDA-MB 231 cells fixed and immunostained 1 h after plating for NMIIA and NMIIB (A), NMIIA and pMLC (B), or NMIIB and pMLC (C). (D, i) MDA-MB 231 cells fixed and stained for NMIIA and NMIIB 16 h after plating. Dashed box indicates field for ii and iii. NMIIA is enriched in protrusions and NMIIB is enriched in lateral stress fibers (arrows). (D, ii) NMIIA forms basal perinuclear stress fibers. (D, iii) Dorsal localization of NMIIA and NMIIB. (E, i) MDA-MB 231 cell fixed and stained for NMIIB and pMLC 16 h after plating. Dashed box indicates field for ii and iii. NMIIB colocalizes with pMLC in basal perinuclear stress fibers (ii) and in dorsal stress fibers (iii, arrowheads). Bars, 10 µm.
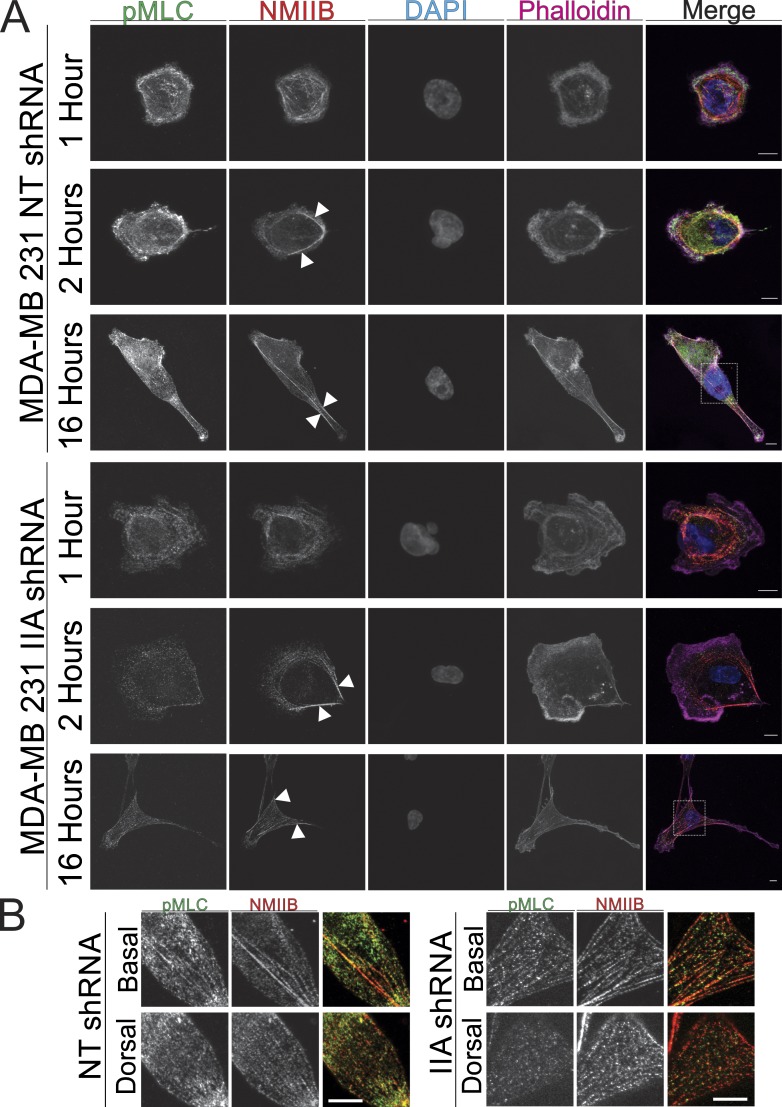
Previous work (Beach et al., 2014) demonstrated that NMIIB coassembles into NMIIA-containing fibers as actomyosin structures mature distal to the leading edge. To determine whether NMIIB’s nuclear association is dependent on the assembly of NMIIA, we assessed NMIIB localization in early stage spreading cells and in fully spread cells in NMIIA shRNA cells. At 1 h both NT control cells and NMIIA shRNA cells exhibit nonpolarized spreading with a strong NMIIB localization interior to the leading edge (Fig. 4 A). As cells begin to polarize at 2 h, NMIIB retains its perinuclear localization in both control and NMIIA shRNA conditions and begins to localize to the rear of the cell (Fig. 4 A, arrowheads). Activated NMIIB, as determined with pMLC colocalization in fibers, formed dorsal stress fibers in NMIIA shRNA cells at the 16-h time point similar to control cells (Fig. 4, A and B). NMIIA shRNA cells exhibit larger protrusions at all time points, relative to control cells, as expected from an earlier study (Vicente-Manzanares et al., 2007). Furthermore, we note that depletion of NMIIA results in a reduced level of detectable pMLC at all time points, presumably because of the reduced overall level of myosin heavy chains and thus myosin II holoenzyme in the NMIIA-depleted cells. In sum, these data show that in the absence of NMIIA NMIIB retains its function of maintaining lateral stress fibers and exerting force on the nucleus.
Figure 4.
Perinuclear localization of NMIIB is not dependent on formation of NMIIA fibers. (A) MDA-MB 231 NT shRNA control or NMIIA shRNA cells were plated on fibronectin-coated coverglass and fixed at 1, 2, or 16 h and stained for NMIIB and pMLC. Arrowheads indicate lateral stress fibers of NMIIB. Dashed boxes indicate region of detail for B. (B) Details of boxes from A. Activated NMIIB fibers in both control and NMIIA shRNA cells were observed below and above the nucleus. All images were acquired with identical parameters, and contrast enhancements were all performed identically. Bars, 10 µm.
NMIIB is critical for nuclear translocation during 3D invasion
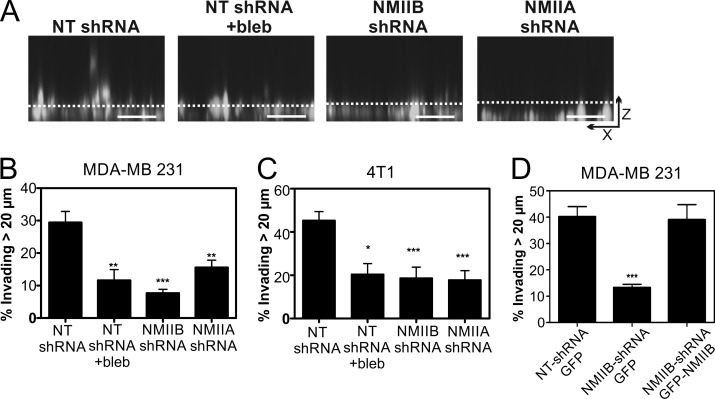
Recent findings suggested that nuclear deformability constitutes a rate-limiting factor in the ability of cells to migrate through dense collagen matrices and that myosin II plays an important role in translocating the nucleus through restrictive pores (Wolf et al., 2013). Therefore, we hypothesized that the NMIIB-dependent forces contributing to traction stress and nuclear flattening in 2D may specifically facilitate nuclear translocation during 3D invasive migration. As an initial test of this hypothesis, we used a bulk population invasion assay. MDA-MB 231 cells were coplated with macrophages in glass-bottom chambers, overlaid with collagen I, and then filled with complete medium to stimulate upward invasive migration, as described previously (Goswami et al., 2005). Global NMII inhibition with blebbistatin significantly reduced invasion (Fig. 5, A and B), as did NMIIA or NMIIB shRNA (Fig. 5, A and B). The same results were observed in 4T1 cells, which displayed robust invasive migration through collagen independent of the presence of macrophages (Fig. 5 C). As a further test of the role of NMIIB in 3D migration, and to alleviate concerns regarding possible off-target effects of NMIIB-shRNA treatment, we performed rescue experiments with transfection of GFP-NMIIB. Expression of GFP-NMIIB in MDA-MB 231 NMIIB shRNA cells rescued 3D collagen invasion to control levels (Fig. 5 D). These results demonstrate that both NMIIA and NMIIB are required for efficient invasion in this setting. However, these studies do not distinguish between redundant versus distinct roles of NMIIA and NMIIB in invasion.
Figure 5.
NMIIA and NMIIB are critical for invasion through 3D collagen gels. (A) Representative x–z projection images of Vybrant DiI fluorescently labeled MDA-MB 231 cells. Dashed line marks 20-µm distance from bottom. Bars, 50 µm. Invasiveness was quantified via confocal z-section analysis, where extent of invasion is scored as the fraction of cells that invade >20 µm into the gel during a 24-h invasion assay. (B) Quantification of invasion of 4T1 cells through 5.8 mg/ml of collagen gel during 24-h incubation. (C) Quantification of invasion of MDA-MB 231 cells through 5.8 mg/ml of collagen gel. MDA-MB-231 cells were co-cultured with nonfluorescent macrophages to enhance invasiveness (Goswami et al., 2005). 4T1 cells were cultured in the absence of macrophages. (D) GFP-NMIIB expression in MDA-MB 231 cells rescued invasion through 3D collagen gels to control levels. Data were compiled from three independent replicates; n = 15–30 fields analyzed per condition. Error bars indicate SEM; *, P < 0.05; **, P < 0.01; ***, P < 0.001 as compared with control.
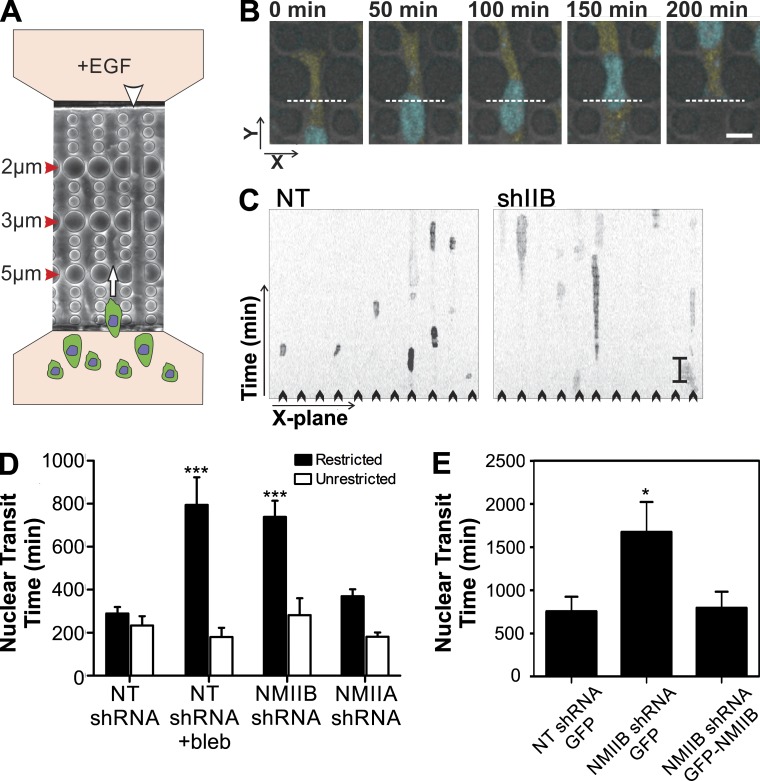
To specifically test NMII isoform roles in nuclear translocation through tight spaces, we used a microfluidic 3D migration device (Davidson et al., 2014) that presents restrictive barriers to nuclear translocation during chemotactic migration, yet does not challenge protrusion formation (Fig. 6, A and B; and Video 1). This device allows high resolution visualization of individual cells and quantification of the rate of nuclear translocation through a series of restrictive polydimethylsiloxane (PDMS) pores ranging from 5 to 2 µm in width (Fig. 6 A, red arrowheads) and 5 µm in height. As an internal control, the chambers also contain 15-µm-wide and 5-µm-high channels in which no restrictive barriers are present (Fig. 6 A, white arrowhead). Kymograph analysis of MDA-MB 231 cells (Fig. 6, B and C) revealed that NMIIB shRNA treatment in MDA-MB 231 cells caused a dramatic increase in the time for cell nuclei required to pass through the 5-µm pores (Fig. 6, C and D, shaded bars). In contrast, NMIIA-shRNA treatment only modestly increased nuclear transit time through the 5-µm pores (Fig. 6 D, shaded bars). Importantly, neither blebbistatin treatment, NMIIA shRNA, nor NMIIB shRNA had any deleterious effect on nuclear transit time through the wide unrestrictive channels in the same chambers (Fig. 6 D, open bars), indicating that NMII activity is not required for unrestricted migration. Expression of GFP-NMIIB in MDA-MB 231 NMIIB shRNA cells rescued parental rates of nuclear translocation in the microfluidic migration device (Fig. 6 E), confirming the specificity of the NMIIB shRNA. Collectively, these data demonstrate a specific role for NMII force production in squeezing the nucleus through tight spaces during chemotactic migration and further show that the NMIIB isoform is the predominant NMII motor protein responsible for this action.
Figure 6.
NMIIB facilitates nuclear translocation through restrictive pores. (A) Schematic showing 3D chemotactic invasion chamber. Cells are seeded in the lower chamber in the diagram, and a chemoattractant gradient is established with 200 ng/ml EGF-containing media in the opposite chamber. Time-lapse images are then collected and the time of nuclear transit through restrictive pores is measured via kymograph analysis. White arrowhead indicates nonrestrictive control channel. White arrow indicates direction of cellular migration. (B) Representative time course images of an MDA-MB 231 cell expressing a Histone2B–CFP nuclear marker passing through a 5 × 5-µm2 pore. The white dashed line in each frame represents the position used for kymograph generation. Bar, 50 µm. (C) Representative kymographs showing time courses of nuclei passing through multiple parallel channels (individual channels denoted by arrowheads) of an invasion chamber. MDA-MB 231 NT shRNA control cells (left) have a shorter nuclear retention in the pore, as compared with NMIIB knockdown cells (right). Vertical bar, 120 min. (D) Quantification of nuclear transit time. *, P < 0.05; ***, P < 0.0001 relative to NT control. Nuclear transit through 5-µm-wide restrictions in shaded bars and unrestricted nuclear transit in open bars. n = 60 to 100 nuclei analyzed per condition. (E) GFP-NMIIB transfection rescues efficient nuclear transit through 3D microfluidic invasion devices to control levels. Error bars indicate SEM; *, P < 0.05.
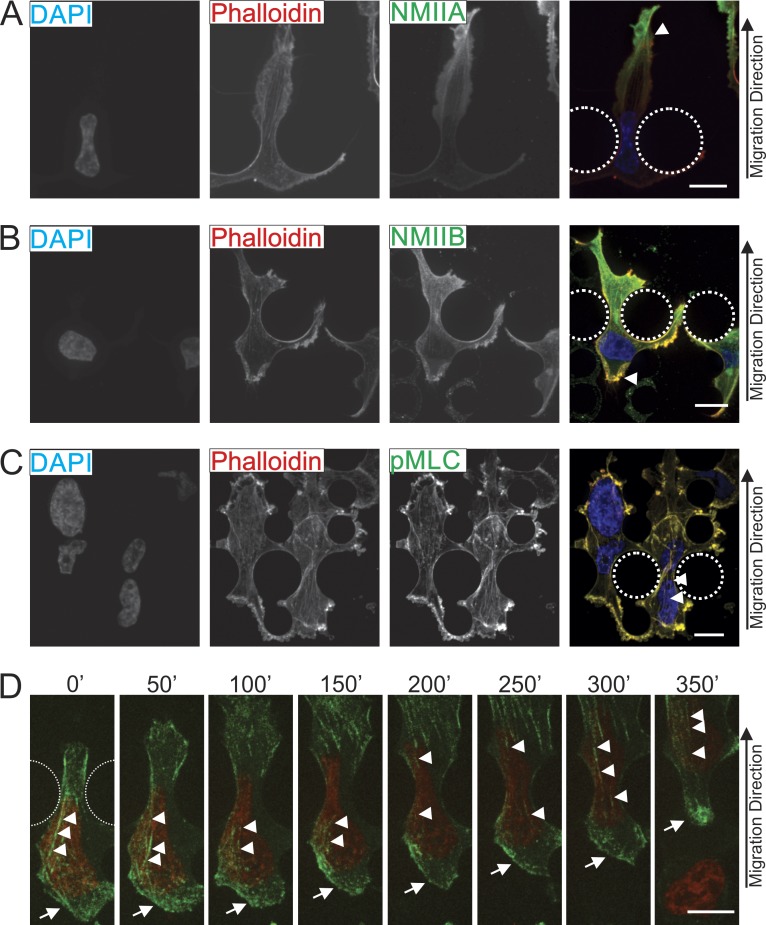
We next asked whether NMIIA and NMIIB displayed differential localization when cells are challenged with restrictive pores in the 3D migration device. As cells translocated through the 5-µm-wide pores, NMIIA consistently exhibited a strong localization in anterior protrusions of the cell (Fig. 7 A, arrowhead), yet did not localize very intensely around the nucleus or in the trailing edge. In contrast, NMIIB was frequently seen throughout the cell with a strong enrichment around the nucleus and in the trailing edge (Fig. 7 B, arrowhead). The actomyosin fibers observed are enriched with active myosin II, shown by the pMLC staining that colocalized strongly with the actin cytoskeleton in both the anterior and posterior regions (Fig. 7 C). These actin fibers can be seen spanning the constricted nucleus in many cases (Fig. 7 C, arrowheads). Similarly, in live-cell imaging of MDA-MB 231 cells expressing GFP-NMIIB, NMIIB accumulated in the perinuclear region and in the cell posterior as cells passed through the 5-µm-wide restriction point in the 3D microfluidic device (Fig. 7 D, arrowheads and arrow, respectively; and Video 2). Collectively, these data support a conserved role for NMIIB in exerting forces on the nucleus in both the nonmigrating state and when the cell is actively squeezing the nucleus through tight spaces during invasive migration.
Figure 7.
NMIIB localizes to the rear and perinuclear region of cells migrating through 3D environments. MDA-MB 231 cells were fixed and stained as they migrated through restriction points of the 3D migration devices. Dotted lines indicate positions of posts that create 5-µm pores. (A) Cells were stained for NMIIA and F-actin (phalloidin). Arrowhead marks leading edge where a high concentration of NMIIA can be observed. (B) Cells are stained for NMIIB and phalloidin. Arrowhead marks NMIIB localization in trailing edge. (C) Cells are stained for pMLC and phalloidin. Arrowheads mark perinuclear stress fibers containing pMLC. (D) Images from time-lapse showing MDA-MB 231 cell expressing nuclear marker H2B-RFP (red) and GFP-NMIIB (green) as it migrates through a 5-µm-wide restriction pore. Arrowheads indicate stress fibers above the nucleus containing GFP-NMIIB and arrows indicate posterior NMIIB accumulation. Bars, 10 µm.
Nesprin-2 provides a possible physical link to the nucleus for NMIIB-based contractile activity
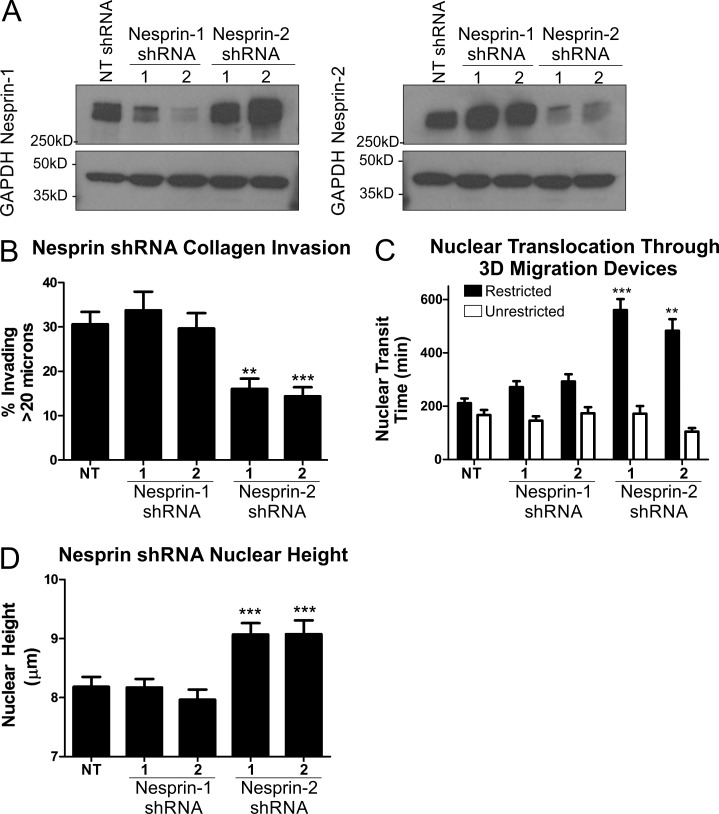
To further explore the mechanism by which NMIIB may exert force on the nucleus, we examined NMIIB interactions with nesprins. Nesprins are a family of large molecular weight proteins that link the cytoskeleton to the nucleus by spanning the outer nuclear membrane and binding the inner nuclear membrane proteins Sun-1/2 (Crisp et al., 2006; Rajgor and Shanahan, 2013). These proteins form the LINC complex, which is a critical component for transmitting forces between the nucleus and cytoskeleton (Lombardi et al., 2011; Isermann and Lammerding, 2013). There are four known isoforms of the nesprin family: nesprin-1 and -2 display calmodulin binding–dependent actin binding, nesprin-3 binds intermediate filaments (via plectin), and nesprin-4 binds microtubules (Rajgor and Shanahan, 2013). Because of their known actin binding activity, we focused our attention on nesprin-1 and -2. Nesprin-1 and -2 protein levels were reduced via stable lentiviral shRNA treatment in MDA-MB 231 cells, as confirmed by Western blot (Fig. 8 A) and immunofluorescence microscopy (Fig. S1) using antibodies provided by MDA Monoclonal Antibody Resource (Randles et al., 2010). We asked whether nesprin depletion had any effect on the efficiency of 3D migration. Knockdown of nesprin-2 but not nesprin-1 conferred a phenotype similar to NMIIB knockdown, with diminished 3D invasion through a collagen gel (Fig. 8 B). Furthermore, similar to NMIIB knockdown, depletion of nesprins resulted in a significant increase in transit time through 5-µm-wide constrictions in the migration device (Fig. 8 C, shaded bars), but had no effect on nuclear translocation through nonrestrictive 15-µm-wide channels (Fig. 8 C, open bars). As seen in NMIIB knockdown cells, depletion of nesprin-2, but not nesprin-1, resulted in increased nuclear height (Fig. 8 D). In conclusion, the nesprin-2 knockdown phenotypes support a model that nesprin-2 is part of the nuclear–cytoskeletal scaffolding system that relays NMIIB-generated force to help translocate the nucleus in cells undergoing 3D migration and to help compress the nucleus in nonmigrating cells in 2D settings.
Figure 8.
Nesprin-2 knockdown recapitulates NMIIB shRNA phenotype. (A) Western blot of nesprin-1 or -2 expression levels. MDA-MB 231 cells were infected with lentivirus expressing shRNA sequences of either NT control, nesprin-1 shRNA targets, or nesprin-2 shRNA targets. To account for off-target effects, two separate constructs were used for nesprin-1 and -2. (B) Collagen invasion of nesprin-1 or -2 shRNA cells (n = 16 fields analyzed). (C) Nuclear translocation time of H2B-RFP–labeled nuclei through 5-µm-wide pores (shaded bars), compared with nonrestrictive control channels (open bars) of 3D migration devices (n = 30–50 nuclei). (D) Nuclear height of nesprin knockdown cells measured from x–z projections of DAPI-stained nuclei (n = 60–70 nuclei). Error bars represent SEM. **, P < 0.01; ***, P < 0.001.
Discussion
Efficient 3D migration requires anterior protrusion, cellular and nuclear translocation, and posterior constriction and retraction. Our data suggests that although both NMIIA and NMIIB isoforms are critical for invasion through a 3D matrix, they serve distinct functions during the overall process. First, we found that NMIIA localizes to anterior lamellar regions and is critical for generating traction stresses during active protrusions. This is consistent with the established role for NMIIA in stabilizing anterior focal adhesions (Vicente-Manzanares et al., 2007; Doyle et al., 2012) and work from Beningo et al. (2001), who reported that nascent anterior focal adhesions in migrating fibroblasts produced strong traction stresses, whereas centrally located and more mature focal adhesions within the same polarized cells produced little traction force. In contrast, NMIIB is dispensable for force generation during anterior protrusion formation, but is critical for long-term force generation in cells confined to patterned squares of fibronectin. This result is consistent with an earlier study in fibroblasts showing that NMIIB, but not NMIIA, drives 3D collagen gel contraction (Meshel et al., 2005). The second process that NMIIA and NMIIB appear to differentially regulate is nuclear deformation and translocation. We observed an NMIIB-specific role in nuclear deformation in both 2D steady-state conditions (i.e., nuclear flattening) and during nuclear translocation in 3D-like invasion assays (squeezing the nucleus through restrictive pores). Recent evidence has identified nuclear deformation to be a major rate-limiting factor in cellular 3D migration (Rowat et al., 2013; Wolf et al., 2013; Davidson et al., 2014; Harada et al., 2014). It has been previously suggested that the higher duty ratio for NMIIB when under load optimizes it for actin cross-linking and prolonged tension generation (Rosenfeld et al., 2003; Kovács et al., 2007). We hypothesize that the sustained force generation necessary to squeeze the nucleus through narrow constrictions in 3D environments is one of the biological processes for which NMIIB has been optimized. Future rescue studies using point mutations to alter NMIIB kinetics independent of perinuclear recruitment might further test this hypothesis.
Consistent with NMIIB being specifically required for these nuclear-based functions, we observed it being recruited to the perinuclear cytoskeleton and cell posterior during 3D pore transmigration. What is the molecular mechanism to accomplish this isoform-specific recruitment? We suggest that these related roles involve a NMIIB-based perinuclear actomyosin network that is present in both nonmigrating and migrating cells. A key question is how such a network would specifically connect to the nuclear envelope. Earlier studies have revealed the presence of a trans-nuclear envelope complex known as the LINC complex that mediates posterior nuclear positioning during fibroblast migration in 2D (Luxton et al., 2011; Chang et al., 2013). Nesprin, a key component of the LINC complex (Luxton et al., 2010), plays a role in nuclear compression in endothelial cells in 2D (Chancellor et al., 2010). We demonstrate here that nesprin-2 mediates nuclear deformation both in a nonmigratory 2D setting and during 3D migration. The phenotypes brought on by the depletion of nesprin-2 expression closely resemble that of NMIIB knockdown, with less nuclear flattening and impaired nuclear translocation through restrictive pores. A functional link between nesprin-2 and NMIIB proteins remains to be demonstrated, but the recapitulation of the NMIIB shRNA phenotype by nesprin-2 shRNA strongly suggests that nesprin-2 contributes to the nuclear anchoring of the NMIIB-associated actomyosin cytoskeleton. Notably, emerin, a protein associated with the LINC complex, has been reported to associate with NMIIB and its depletion in fibroblasts resulted in altered NMIIB organization (Chang et al., 2013), further supporting the concept that NMIB is specifically coupled to the LINC complex in multiple cell types and settings. It is unclear what role nesprin-1 plays in these cells, given the wide range of expression patterns of nesprin-1 and -2 in various tissues and cell lines (Duong et al., 2014). As further evidence of the complex nature of the nesprin family, nesprin-3 has also been shown to play a role in the NMII- and vimentin-dependent forward movement of fibroblast nuclei during 3D lobopodial migration (Petrie et al., 2014). Further studies are needed to address the relative roles of different nesprin isoforms in different settings, as well as the contribution of other members of the LINC complex, such as Sun-1/2 and emerin to nuclear translocation.
Lastly, we note that white blood cells, although highly proficient at amoeboid 3D migration (Lämmermann et al., 2008), do not express NMIIB (Jacobelli et al., 2010). Interestingly, neutrophils also lack many nuclear envelope proteins and have much more deformable nuclei, which may make it unnecessary to have a contractile NMIIB network (Rowat et al., 2013). A recent study demonstrated that under confined conditions, mesenchymal cells can switch to amoeboid modes of migration (Liu et al., 2015), although conditions designed to inhibit nuclear translocation were not a focus of this work. In earlier work, Beach et al. (2011) demonstrated that NMIIB is strongly up-regulated when cells undergo epithelial–mesenchymal transition and that this up-regulation is critical for the acquisition of 3D invasiveness that occurs during epithelial–mesenchymal transition. We suggest that the requirement for NMIIB in 3D invasion may be a hallmark of mesenchymal-type migration (Friedl and Wolf, 2010), perhaps related to mesenchymal-specific nuclear coupling components present in these cells. Further studies are required to address the mechanism of NMIIB association with nesprin-2, and whether or not this mechanocoupling system is present in nontransformed mesenchymal cell types as well as cancer cells.
In conclusion, we present evidence that NMIIB plays a key role in mesenchymal-like cancer cells in applying cytoskeletal forces to the nucleus and that this role is critical for 3D invasive migration both in collagen gels and 3D migration chambers. NMIIB appears to exert this activity via connections to the LINC complex that includes nesprins and Sun proteins.
Materials and methods
Antibodies and reagents
The following antibodies were used: NMIIA for Western blot and immunofluorescence (rabbit pAb PRB-440P; Covance), NMIIB for Western blot (rabbit pAb PRB-445P; Covance), NMIIB for immunofluorescence (mouse mAb ab684; Abcam), GAPDH (rabbit pAb sc-25778; Santa Cruz Biotechnology, Inc.), and GFP (rabbit pAb A6455; Molecular Probes). DAPI and Alexa Fluor 647 Phalloidin were purchased from Life Technologies (D3571 and A22287, respectively) and used according to the manufacturer’s recommendation. Nesprin-1 and -2 antibodies were obtained from MDA Monoclonal Antibody Resource (mouse mAb; MANNES1E and MANNES2G, respectively).
Cell culture and plasmids
Human triple-negative–like breast carcinoma MDA-MB 231 (ATCC) and mouse mammary tumor 4T1 (Aslakson and Miller, 1992) cell lines were cultured in complete RPMI 1640 media containing 10% FBS and 1% penicillin/streptomycin. The SV-40–transformed macrophage cell line BAC 1.2F5 was provided by R. Stanley (Albert Einstein School of Medicine, Bronx, NY) and cultured in MEM α modification media containing 10% FBS, 1% penicillin/streptomycin, and 36 µg/ml of human recombinant CSF-1 (R&D Systems). pLKO.1 shRNA libraries were purchased as bacterial glycerol stocks from Sigma-Aldrich and, after initial screening for effectiveness, the following constructs were used in MDA-MB 231 cells for these studies: NT control, SHC002 (5′-CAACAAGATGAAGAGCACCAA-3′); shIIA, TRCN000029468 (5′-GCCAAGCTCAAGAACAAGCAT-3′); shIIB, TRCN0000123074 (5′-CGGGATTCCTTCCTGAAAGAT-3′). The following constructs were used in 4T1 cells for these studies: shIIA, TRCN0000304377 (5′-TACCCTTTGAGAATCTGATAC-3′); shIIB, TRCN0000110559 (5′-CTTCCAATTTACTCTGAGAA-3′). The shIIB construct for MDA-MB 231 cells targets the 3′ UTR region of the gene: shNesprin1-1, TRCN0000147281 (5′-CGTAGTGATAAGACTGATTT-3′); shNesprin1-2, TRCN0000431083 (5′-CAGAAGTGCTGGTAGCATAAG-3′); shNesprin2-1, TRCN0000303801 (5′-GATGGAAACAATCAATCATA-3′); shNesprin2-2, TRCN0000310545 (5′-CGCTATGAAAGAACGGAGT-3′). Target plasmids were cotransfected with generation 2.0 lentiviral packaging plasmids psPAX.2 and pMD2.G via Lipofectamine 2000 into HEK293t cells. Virus-containing media was then harvested for lentiviral infection to target cells. Positively infected cells were then selected for with 1 or 10 ng/ml puromycin in MDA-MB 231 or 4T1 cells, respectively, for 5 d as determined by kill-curve analysis. LV-RFP and LV-CFP, lentiviral constructs for CFP- or RFP-Histone2B fluorescent tags, were purchased from Addgene and packaged in HEK293t cells, as described for shRNA constructs. Positively infected cells were sorted via flow cytometry. For rescue experiments, pCMV-EGFP control or pCMV-EGFP-MHCIIB fusion plasmids were transfected into MDA-MB 231 cells. Cells were then sorted for GFP-positive cells and selection was maintained in the presence of 400 ng/ml G418.
Western blot analysis
Cells were trypsinized and washed in cold PBS before being lysed in 2× Laemmli sample buffer containing 0.2% β-mercaptoethanol, 1% each of protease inhibitor cocktail, phosphatase inhibitor cocktail 2, and phosphatase inhibitor cocktail 3 (P8340, P5726, P0044, respectively; Sigma-Aldrich). Samples were run on 4–20% Tris-Glycine gels (Life Technologies) and transferred to 0.45-µm PVDF membrane (EMD Millipore). Membranes were blocked in 2% milk solution and probed with the following antibodies: NMIIB (rabbit pAb; 1:1,000; Covance), NMIIA (rabbit pAb; 1:1,000; Covance), GAPDH (rabbit pAb; 1:5,000; Santa Cruz Biotechnology, Inc.), GFP (mouse mAb; 1:3,000; Molecular Probes), Nesprin 1 (MANNES1E; mouse mAB; 1:50; MDA Monoclonal Antibody Resource), Nesprin 2 (MANNES2G; mAb; 1:50; MDA Monoclonal Antibody Resource).
Immunofluorescence imaging
Cells were plated onto 50 µg/ml fibronectin-coated glass coverslips, fixed in 4% paraformaldehyde, and then permeabilized in 0.1% Triton X-100. Cells were then incubated with primary antibody followed by species appropriate Alexa Fluor 488 or 568 secondary antibodies and DAPI and Alexa Fluor 647 Phalloidin (Life Technologies). Samples were then mounted to glass slides using Vectashield (#H-1100; Vector Laboratories). Cells were visualized using an UltraView VOX Spinning Disk Confocal Imaging System (PerkinElmer) with a DM1-6000 SD inverted microscope (Leica) and an electron multiplying charge coupled device C9100-13 camera (63×/NA 1.47 oil; Hamamatsu Photonics) or a TCS SP5 II laser scanning confocal imaging system with five spectral detectors (63×/NA 1.40 oil; Leica). 3D reconstruction and nuclear height measurements were performed with Volocity Software version 6.1.1.
Collagen invasion
Collagen invasion of MDA-MB 231 cells was performed as a macrophage-stimulated (BAC 1.2F5 cells) invasion assay adapted from Goswami et al. (2005). For these experiments, cancer cells were stained with Vybrant-DiI (Life Technologies) and BAC 1.2F5 cells were stained with Vybrant-DiO (Life Technologies). Glass bottom dishes (MatTek Corporation) were treated with 1 N HCl before plating of cells. Cells were plated overnight at a ratio of 2.5:1 BAC 1.2F5/MDA-MB 231 cells. A 5.8-mg/ml acid-solubilized rat tail collagen I gel (#354249; BD) was laid in the well of a glass bottom tissue culture dish and allowed to polymerize at 37°C for 30 min. MEM α modified media containing 10% FBS/1% penicillin/streptomycin and 36 ng/ml human CSF-1 (R&D Systems) was then overlaid above the collagen. Collagen invasion using 4T1 cells was performed in the absence of macrophages, with all other conditions the same as for the MDA-MB 231 experiments. Cells were then incubated overnight at 37°C, fixed with 4% paraformaldehyde, and permeabalized with 0.1% Triton X-100. Cells were visualized using an UltraView VOX Spinning Disk Confocal Imaging System with a DM1-6000 SD inverted microscope (10×/NA 0.7 air). A 100-µm z-stack image at 5-µm increments was obtained. An extended x–z projection was then obtained and invasion was quantified as a percentage of Vybrant DiI signal above 20 µm from the bottom compared with total fluorescence of the field.
Microfluidic 3D migration device
The migration devices were fabricated as described elsewhere (Davidson et al., 2014) using standard lithography techniques to produce SU-8 molds at the Cornell NanoScale Science and Technology Facility and then casting the PDMS device from the SU-8 molds, which were then bonded onto glass slides. In brief, the migration devices contain features of two different heights, produced from an SU-8 mold composed of two layers: one 5-µm-tall layer to form the constrictions channels and one 250-µm-tall layer to create larger chambers directly before and after the constrictions for cell seeding and for cells that have passed through the constrictions. The constriction channels consist of a series of round pillars forming consecutively narrower openings measuring 5, 3, and 2 µm in width, respectively, through which cells pass along a chemotactic gradient established by two large reservoirs on either side of the constrictions (Fig. 6).
Replicas of the SU-8 molds were produced using PDMS. The two components of Sylgard 184 (Dow Corning) were mixed in the 1:10 ratio recommended by the manufacturer; the mixture was degassed to eliminate bubbles and poured over the wafer in a plastic Petri dish. This assembly was then baked at 65°C for 2 h. The resulting PDMS replica was carefully peeled away from the SU-8 structures and individual migration devices were cut out and placed on packing tape to prevent dust from accumulating on the clean surface. Inlets for cells were punched using 1.2-mm biopsy punches, and reservoirs for medium were created using 5-mm biopsy punches (Harris). The newly formed microfluidic devices were baked at 95°C on a hot plate for 5 min, sterilized with 70% ethanol, and then rinsed with sterile water.
Migration chambers were coated in 50 µg/ml fibronectin and cells were loaded into one side using loading ports. Cells were then incubated overnight in RPMI 1640 containing 10% FBS and 1% penicillin/streptomycin. Media was then washed using serum-free media and RPMI 1640 containing 2% FBS was loaded on the side containing cells. RPMI 1640 containing 2% FBS with 200 ng/ml EGF was loaded on the opposite side to the cells. Cells were then analyzed for 48 h using an UltraView VOX Spinning Disk Confocal Imaging System with a DM1-6000 SD inverted microscope (10×/NA 0.7 air; 37°C with 5% CO2) on a C9100-13 camera and analyzed using Volocity Software. To quantify nuclear translocation rate, kymographs were generated transecting the area of the opening of a 5-µm constriction point.
Traction force microscopy
Cell traction stresses were measured as previously described (Zhang et al., 2013). In brief, 5%/0.1% acrylamide/bis-acrylamide gels with a Young’s modulus of 5.8 kPa (Tse and Engler, 2010) were fabricated with 0.1% embedded 0.2-µm Floresbrite-568 (Polysciences) fluorescent microspheres. Gels were polymerized on a 40-mm-diameter PlusOne Bind Silane (GE Healthcare)–coated bottom coverslip. Fibronectin was covalently linked to the surface of the PAA gel by placing a 36-µg/ml sodium-m-periodate (Sigma-Aldrich)–activated fibronectin–coated 25-mm-diameter top coverslip over a 37-µl drop of PAA before polymerization. For analysis of traction stresses during initial spreading, cells were plated on the PAA surfaces and placed in a 37°C microscope chamber. Cells were then identified that were attached, but not yet displaying any spreading. Imaging was initiated on each examined cell at this stage, which was defined as the 0 min time point for presented analysis.
To create patterned fibronectin, a 50-µg/ml activated fibronectin solution was coated onto a PDMS stamp of 30 × 30-µm patterns. The stamp was then pressed against a top coverslip to transfer the square patterned fibronectin, which was in turn used to transfer the fibronectin pattern onto the surface of the PAA gel. After removal of the top coverslip, cells were then plated on the gel for 1 or 16 h. The coverslip with attached cells was then mounted into a 60-mm-diameter microscope chamber for analysis. Cells and microbeads were analyzed with a DMIRE 2100 inverted microscope (40×/NA 0.7; 37°C with 5% CO2; Leica) using an Orca-100 cooled charge coupled device camera (Hamamatsu Photonics). Phase-contrast images of the cells and of the fluorescent microbeads at the surface of the gel were acquired. Cells were then removed by trypsinization and another image of the fluorescent microbeads was obtained. Traction vectors were obtained using DIM software and traction forces were quantified using LIBTRC 2.4 software (Dembo and Wang, 1999).
Online supplemental material
Fig. S1 shows effectiveness of nesprin-1 and -2 shRNA knockdown assessed by immunofluorescence. Two constructs specific for different shRNA target sequences against either nesprin-1 or -2 were used. Expression of both nesprin-1 and -2 were assessed using mouse mAbs from MDA Monoclonal Antibody Resource. Video 1 shows an example of an MDA-MB 231 cell expressing a nuclear H2B-CFP marker and cytoplasmic YFP marker as it migrates through a 5-µm pore in the 3D migration device. Video 2 shows an example of an MDA-MB 231 NMIIB shRNA cell expressing a NMIIB-GFP construct and H2B-RFP nuclear marker as it migrates through a 5-µm pore in the 3D migration device. Perinuclear NMIIB stress fibers and accumulation of NMIIB in the trailing edge should be observed. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201502039/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Micah Dembo for generously sharing the LIBTRC 2.4 image analysis software needed for quantification of traction forces. We thank Dr. Patricia M. Davidson for her contribution to the design of the 3D microfluidic migration devices and the fabrication of the wafers. We thank Dr. Judy Drazba and the Lerner Research Institute Imaging Core for their assistance and guidance with imaging and microscopy.
This work was supported by National Institute of General Medical Sciences grant GM50009 to T.T. Egelhoff. D.G. Thomas was supported by National Institutes of Health Training program in Cancer Pharmacology (R25CA1485052-02). This work was performed in part at the Cornell NanoScale Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation (grant ECCS-0335765). This work was supported by National Institutes of Health awards (R01 HL082792) to J. Lammerding; the Department of Defense Breast Cancer Idea Award (BC102152) to J. Lammerding; a National Science Foundation CAREER award (CBET-1254846) to J. Lammerding; and a Pilot Project Award by the Cornell Center on the Microenvironment and Metastasis through award number U54CA143876 from the National Cancer Institute. This work was also supported by National Cancer Institute grant CA129359 to W.P. Schiemann.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- LINC
- linker of nucleoskeleton and cytoskeleton
- NMII
- non-muscle myosin II
- NT
- nontargeting
- PAA
- polyacrylamide
- PDMS
- polydimethylsiloxane
- pMLC
- phospho-myosin light chain
References
- Aslakson C.J., and Miller F.R.. 1992. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 52:1399–1405. [PubMed] [Google Scholar]
- Bao J., Ma X., Liu C., and Adelstein R.S.. 2007. Replacement of nonmuscle myosin II-B with II-A rescues brain but not cardiac defects in mice. J. Biol. Chem. 282:22102–22111. 10.1074/jbc.M702731200 [DOI] [PubMed] [Google Scholar]
- Beach J.R., Hussey G.S., Miller T.E., Chaudhury A., Patel P., Monslow J., Zheng Q., Keri R.A., Reizes O., Bresnick A.R., et al. 2011. Myosin II isoform switching mediates invasiveness after TGF-β–induced epithelial–mesenchymal transition. Proc. Natl. Acad. Sci. USA. 108:17991–17996. 10.1073/pnas.1106499108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach J.R., Shao L., Remmert K., Li D., Betzig E., and Hammer J.A. III. 2014. Nonmuscle myosin II isoforms coassemble in living cells. Curr. Biol. 24:1160–1166. 10.1016/j.cub.2014.03.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle C., Assanah M.C., Monzo P., Vallee R., Rosenfeld S.S., and Canoll P.. 2008. The role of myosin II in glioma invasion of the brain. Mol. Biol. Cell. 19:3357–3368. 10.1091/mbc.E08-03-0319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beningo K.A., Dembo M., Kaverina I., Small J.V., and Wang Y.L.. 2001. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 153:881–888. 10.1083/jcb.153.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge M.T., Egelhoff T.T., and Baskaran H.. 2010. A microfluidic imaging chamber for the direct observation of chemotactic transmigration. Biomed. Microdevices. 12:543–553. 10.1007/s10544-010-9411-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancellor T.J., Lee J., Thodeti C.K., and Lele T.. 2010. Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys. J. 99:115–123. 10.1016/j.bpj.2010.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W., Folker E.S., Worman H.J., and Gundersen G.G.. 2013. Emerin organizes actin flow for nuclear movement and centrosome orientation in migrating fibroblasts. Mol. Biol. Cell. 24:3869–3880. 10.1091/mbc.E13-06-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.K., Vicente-Manzanares M., Zareno J., Whitmore L.A., Mogilner A., and Horwitz A.R.. 2008. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10:1039–1050. 10.1038/ncb1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti M.A., and Adelstein R.S.. 2008. Nonmuscle myosin II moves in new directions. J. Cell Sci. 121:11–18. 10.1242/jcs.007112 [DOI] [PubMed] [Google Scholar]
- Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., and Hodzic D.. 2006. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 172:41–53. 10.1083/jcb.200509124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson P.M., Denais C., Bakshi M.C., and Lammerding J.. 2014. Nuclear deformability constitutes a rate-limiting step during cell migration in 3-D environments. Cell. Mol. Bioeng. 7:293–306. 10.1007/s12195-014-0342-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo M., and Wang Y.L.. 1999. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 76:2307–2316. 10.1016/S0006-3495(99)77386-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A.D., Wang F.W., Matsumoto K., and Yamada K.M.. 2009. One-dimensional topography underlies three-dimensional fibrillar cell migration. J. Cell Biol. 184:481–490. 10.1083/jcb.200810041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle A.D., Kutys M.L., Conti M.A., Matsumoto K., Adelstein R.S., and Yamada K.M.. 2012. Micro-environmental control of cell migration—myosin IIA is required for efficient migration in fibrillar environments through control of cell adhesion dynamics. J. Cell Sci. 125:2244–2256. 10.1242/jcs.098806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong N.T., Morris G.E., Lam T., Zhang Q., Sewry C.A., Shanahan C.M., and Holt I.. 2014. Nesprins: tissue-specific expression of epsilon and other short isoforms. PLoS ONE. 9:e94380 10.1371/journal.pone.0094380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer R.S., Gardel M., Ma X., Adelstein R.S., and Waterman C.M.. 2009. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr. Biol. 19:260–265. 10.1016/j.cub.2008.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P., and Alexander S.. 2011. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 147:992–1009. 10.1016/j.cell.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Friedl P., and Wolf K.. 2010. Plasticity of cell migration: a multiscale tuning model. J. Cell Biol. 188:11–19. 10.1083/jcb.200909003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami S., Sahai E., Wyckoff J.B., Cammer M., Cox D., Pixley F.J., Stanley E.R., Segall J.E., and Condeelis J.S.. 2005. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 65:5278–5283. 10.1158/0008-5472.CAN-04-1853 [DOI] [PubMed] [Google Scholar]
- Harada T., Swift J., Irianto J., Shin J.W., Spinler K.R., Athirasala A., Diegmiller R., Dingal P.C., Ivanovska I.L., and Discher D.E.. 2014. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell Biol. 204:669–682. 10.1083/jcb.201308029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isermann P., and Lammerding J.. 2013. Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 23:R1113–R1121. 10.1016/j.cub.2013.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobelli J., Friedman R.S., Conti M.A., Lennon-Dumenil A.M., Piel M., Sorensen C.M., Adelstein R.S., and Krummel M.F.. 2010. Confinement-optimized three-dimensional T cell amoeboid motility is modulated via myosin IIA–regulated adhesions. Nat. Immunol. 11:953–961. 10.1038/ni.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorrisch M.H., Shih W., and Yamada S.. 2013. Myosin IIA deficient cells migrate efficiently despite reduced traction forces at cell periphery. Biol. Open. 2:368–372. 10.1242/bio.20133707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J. 1998. Cytoplasmic dynamics of myosin IIA and IIB: spatial ‘sorting’ of isoforms in locomoting cells. J. Cell Sci. 111:2085–2095. [DOI] [PubMed] [Google Scholar]
- Kovács M., Thirumurugan K., Knight P.J., and Sellers J.R.. 2007. Load-dependent mechanism of nonmuscle myosin 2. Proc. Natl. Acad. Sci. USA. 104:9994–9999. 10.1073/pnas.0701181104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmermann T., Bader B.L., Monkley S.J., Worbs T., Wedlich-Söldner R., Hirsch K., Keller M., Förster R., Critchley D.R., Fässler R., and Sixt M.. 2008. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 453:51–55. 10.1038/nature06887 [DOI] [PubMed] [Google Scholar]
- Liu Y.J., Le Berre M., Lautenschlaeger F., Maiuri P., Callan-Jones A., Heuzé M., Takaki T., Voituriez R., and Piel M.. 2015. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 160:659–672. 10.1016/j.cell.2015.01.007 [DOI] [PubMed] [Google Scholar]
- Lombardi M.L., Jaalouk D.E., Shanahan C.M., Burke B., Roux K.J., and Lammerding J.. 2011. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 286:26743–26753. 10.1074/jbc.M111.233700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W., Gomes E.R., Folker E.S., Vintinner E., and Gundersen G.G.. 2010. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 329:956–959. 10.1126/science.1189072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton G.W., Gomes E.R., Folker E.S., Worman H.J., and Gundersen G.G.. 2011. TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus. 2:173–181. 10.4161/nucl.2.3.16243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshel A.S., Wei Q., Adelstein R.S., and Sheetz M.P.. 2005. Basic mechanism of three-dimensional collagen fibre transport by fibroblasts. Nat. Cell Biol. 7:157–164. 10.1038/ncb1216 [DOI] [PubMed] [Google Scholar]
- Pasapera A.M., Schneider I.C., Rericha E., Schlaepfer D.D., and Waterman C.M.. 2010. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 188:877–890. 10.1083/jcb.200906012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie R.J., Koo H., and Yamada K.M.. 2014. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 345:1062–1065. 10.1126/science.1256965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Swift J., Dingal P.C.D.P., Shah P., Shin J.-W., and Discher D.E.. 2012. Crawling from soft to stiff matrix polarizes the cytoskeleton and phosphoregulates myosin-II heavy chain. J. Cell Biol. 199:669–683. 10.1083/jcb.201205056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D., and Shanahan C.M.. 2013. Nesprins: from the nuclear envelope and beyond. Expert Rev. Mol. Med. 15:e5 10.1017/erm.2013.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randles K.N., Lam T., Sewry C.A., Puckelwartz M., Furling D., Wehnert M., McNally E.M., and Morris G.E.. 2010. Nesprins, but not sun proteins, switch isoforms at the nuclear envelope during muscle development. Dev. Dyn. 239:998–1009. 10.1002/dvdy.22229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld S.S., Xing J., Chen L.Q., and Sweeney H.L.. 2003. Myosin IIb is unconventionally conventional. J. Biol. Chem. 278:27449–27455. 10.1074/jbc.M302555200 [DOI] [PubMed] [Google Scholar]
- Rowat A.C., Jaalouk D.E., Zwerger M., Ung W.L., Eydelnant I.A., Olins D.E., Olins A.L., Herrmann H., Weitz D.A., and Lammerding J.. 2013. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J. Biol. Chem. 288:8610–8618. 10.1074/jbc.M112.441535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.A., Parvani J.G., and Schiemann W.P.. 2010. The pathophysiology of epithelial-mesenchymal transition induced by transforming growth factor-β in normal and malignant mammary epithelial cells. J. Mammary Gland Biol. Neoplasia. 15:169–190. 10.1007/s10911-010-9181-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse J.R., and Engler A.J.. 2010. Preparation of hydrogel substrates with tunable mechanical properties. Curr. Protoc. Cell Biol. Chapter 10:10.16. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Zareno J., Whitmore L., Choi C.K., and Horwitz A.F.. 2007. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 176:573–580. 10.1083/jcb.200612043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Koach M.A., Whitmore L., Lamers M.L., and Horwitz A.F.. 2008. Segregation and activation of myosin IIB creates a rear in migrating cells. J. Cell Biol. 183:543–554. 10.1083/jcb.200806030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Newell-Litwa K., Bachir A.I., Whitmore L.A., and Horwitz A.R.. 2011. Myosin IIA/IIB restrict adhesive and protrusive signaling to generate front–back polarity in migrating cells. J. Cell Biol. 193:381–396. 10.1083/jcb.201012159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Ma X., Conti M.A., Liu C., Kawamoto S., and Adelstein R.S.. 2010. Nonmuscle myosin II isoform and domain specificity during early mouse development. Proc. Natl. Acad. Sci. USA. 107:14645–14650. 10.1073/pnas.1004023107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Ma X., Conti M.A., and Adelstein R.S.. 2011. Distinct and redundant roles of the non-muscle myosin II isoforms and functional domains. Biochem. Soc. Trans. 39:1131–1135. 10.1042/BST0391131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., Te Lindert M., Krause M., Alexander S., Te Riet J., Willis A.L., Hoffman R.M., Figdor C.G., Weiss S.J., and Friedl P.. 2013. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 201:1069–1084. 10.1083/jcb.201210152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Guo W.H., Rape A., and Wang Y.L.. 2013. Micropatterning cell adhesion on polyacrylamide hydrogels. Methods Mol. Biol. 1066:147–156. 10.1007/978-1-62703-604-7_13 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.