The shootin1–cortactin interaction participates in netrin-1–induced F-actin–adhesion coupling and in the promotion of traction forces for axon outgrowth.
Abstract
Motile cells transduce environmental chemical signals into mechanical forces to achieve properly controlled migration. This signal–force transduction is thought to require regulated mechanical coupling between actin filaments (F-actins), which undergo retrograde flow at the cellular leading edge, and cell adhesions via linker “clutch” molecules. However, the molecular machinery mediating this regulatory coupling remains unclear. Here we show that the F-actin binding molecule cortactin directly interacts with a clutch molecule, shootin1, in axonal growth cones, thereby mediating the linkage between F-actin retrograde flow and cell adhesions through L1-CAM. Shootin1–cortactin interaction was enhanced by shootin1 phosphorylation by Pak1, which is activated by the axonal chemoattractant netrin-1. We provide evidence that shootin1–cortactin interaction participates in netrin-1–induced F-actin adhesion coupling and in the promotion of traction forces for axon outgrowth. Under cell signaling, this regulatory F-actin adhesion coupling in growth cones cooperates with actin polymerization for efficient cellular motility.
Introduction
Throughout life, directional cell migration underlies various physiological processes, including gastrulation, neuronal network formation, tissue development, immune responses, and wound healing. To achieve properly controlled migration, motile cells sense environmental chemical signals and transduce them into protrusive activity (Xu et al., 2003; Van Haastert and Devreotes, 2004). Actin filaments (F-actins) polymerize at the leading edge of motile cells and depolymerize proximally (Pollard and Borisy, 2003; Le Clainche and Carlier, 2008), which, in conjunction with myosin II activity, induces retrograde flow of F-actins (Forscher and Smith, 1988; Katoh et al., 1999; Medeiros et al., 2006). Modulation of mechanical coupling between F-actin retrograde flow and cell adhesions by “clutch” molecules is thought to play a key role in the signal-force transduction required for regulatory cell migration (Mitchison and Kirschner, 1988; Suter and Forscher, 2000; Le Clainche and Carlier, 2008; Toriyama et al., 2013). An increase in the coupling efficiency produces traction forces on extracellular substrates and concurrently reduces the speed of the F-actin retrograde flow, thereby converting actin polymerization into force that pushes the leading edge membrane. However, the molecular machinery that executes the regulatory coupling remains unknown despite considerable efforts to identify clutch molecules (Bard et al., 2008; Shimada et al., 2008; Giannone et al., 2009; Lowery and Van Vactor, 2009; Thievessen et al., 2013).
Shootin1 is a key molecule involved in neuronal polarization and axon outgrowth (Toriyama et al., 2006, 2010; Inagaki et al., 2011; Sapir et al., 2013). It accumulates at the leading edge of axonal growth cones, and mediates the mechanical coupling between F-actin retrograde flow and the cell adhesion molecule (CAM) L1-CAM (Kamiguchi et al., 1998) as a clutch molecule (Shimada et al., 2008). Pak1 is a downstream kinase of Cdc42 and Rac1, and is also involved in axon outgrowth and cell migration (Jacobs et al., 2007; Delorme-Walker et al., 2011). Recently, we reported that the attractive axon guidance molecule netrin-1 (Serafini et al., 1994; Li et al., 2008) induces Pak1-mediated shootin1 phosphorylation in axonal growth cones (Toriyama et al., 2013). This in turn enhances the coupling between F-actins and shootin1, thereby promoting the traction forces for axon outgrowth. However, the molecular basis of this regulatory coupling was not elucidated (Shimada et al., 2008).
The F-actin binding protein cortactin (Weed and Parsons, 2001) accumulates at sites of dynamic actin assembly, such as the lamellipodia and filopodia of axonal growth cones and migrating cells (Wu and Parsons, 1993; Weed et al., 2000; Decourt et al., 2009; Kurklinsky et al., 2011), and the invadopodia of cancer cells (MacGrath and Koleske, 2012b). It induces membrane protrusion, lamellipodia persistence, and formation of filopodia and invadopodia (Kinley et al., 2003; Bryce et al., 2005; Mingorance-Le Meur and O’Connor, 2009; Spillane et al., 2012), and is thought to play a key role in motility of diverse cell types (Cheng et al., 2000; Vidal et al., 2002; Bryce et al., 2005; Kirkbride et al., 2011; MacGrath and Koleske, 2012b).
We have analyzed the molecular basis of the coupling between F-actin retrograde flow and cell adhesions in axonal growth cones, and show here that cortactin directly mediates the linkage between F-actin retrograde flow and shootin1 as a clutch molecule. Our data further suggest that the shootin1–cortactin interaction serves as a crucial regulatory interface for signal–force transduction in axon outgrowth.
Results
Cortactin directly interacts with shootin1 in axonal growth cones
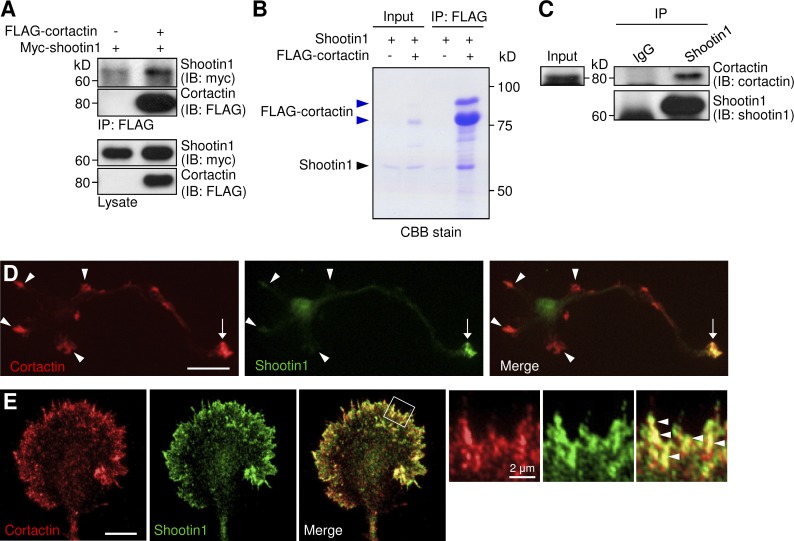
To address the missing link between shootin1 and F-actins in the clutch machinery (Shimada et al., 2008), we searched for actin-binding proteins that interact with shootin1 using coimmunoprecipitation assays. Among the seven actin binding proteins examined (fascin, VASP, p21-ARC, Esp8, XAC2, capping protein β1, and cortactin), we detected an interaction between cortactin and shootin1 expressed in COS7 cells (Fig. 1 A). A binding assay using purified shootin1 and FLAG-cortactin confirmed their direct interaction in vitro (Fig. 1 B), and, furthermore, cortactin was immunoprecipitated with shootin1 in P6 rat brain lysates (Fig. 1 C). Fig. 1 (D and E) shows the subcellular localization of cortactin and shootin1 in cultured rat hippocampal neurons. As previously reported (Du et al., 1998; Toriyama et al., 2006), cortactin was localized in growth cones of axons and immature dendrites of cultured hippocampal neurons (Fig. 1 D, arrows and arrowheads), whereas shootin1 accumulated preferentially in axonal growth cones (arrows). Cortactin was highly colocalized with shootin1 in the peripheral region of axonal growth cones (Fig. 1 E, arrowheads). Together, these results indicate that cortactin interacts directly with shootin1 in axonal growth cones.
Figure 1.
Cortactin directly interacts with shootin1 in axonal growth cones. (A) Coimmunoprecipitation of shootin1 and cortactin in COS7 cells. Cells were cotransfected with myc-shootin1 and FLAG-cortactin, and lysates were incubated with anti-FLAG antibody. The immunoprecipitates were immunoblotted with anti-FLAG or anti-myc antibody. (B) In vitro binding assay using purified shootin1 (2 µM) and purified FLAG-cortactin (2 µM). Proteins were incubated with anti-FLAG antibody, and the immunoprecipitates were then analyzed by SDS-PAGE and CBB staining. 0.125% of the input proteins were also analyzed. As reported previously (Wu and Parsons, 1993; MacGrath and Koleske, 2012a), purified cortactin is composed of a single major band at 80 kD and an additional upper band (blue arrowheads). (C) Coimmunoprecipitation of endogenous shootin1 and cortactin from rat (postnatal day 6) brain lysates. Brain lysates were incubated with anti-shootin1 antibody or control IgG. The immunoprecipitates were immunoblotted with anti-shootin1 or anti-cortactin antibody. (D) Rat hippocampal neurons (3 d in vitro) costained with anti-cortactin (red) and anti-shootin1 (green) antibodies. Arrows and arrowheads denote an axonal growth cone and minor process growth cones, respectively. (E) Fluorescence images of an axonal growth cone labeled with anti-cortactin (red) and anti-shootin1 (green) antibodies. Enlarged views of the filopodia in the white square are shown to the right. Arrowheads indicate cortactin colocalized with shootin1. Bars: (D) 20 µm; (E, main panels) 10 µm.
Cortactin interacts with F-actin retrograde flow in axonal growth cones
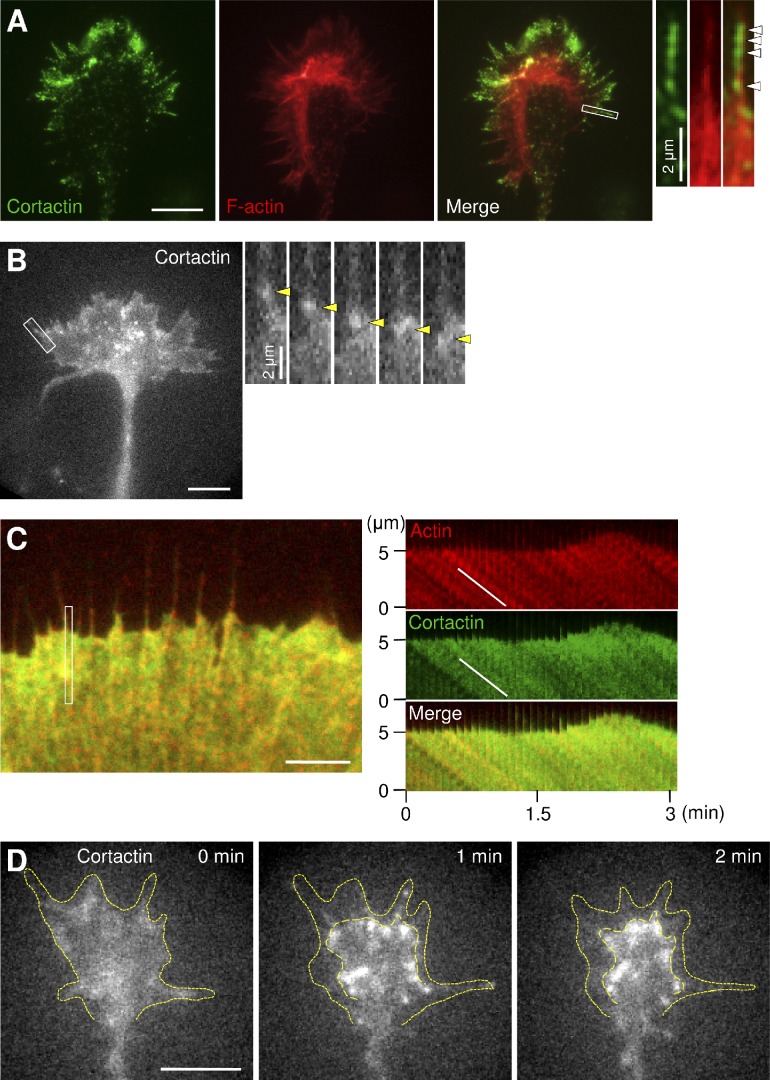
Cortactin also colocalized with F-actins in the growth cone peripheral domain (Fig. 2 A), where F-actins undergo directional retrograde flow. Punctate staining of cortactin preferentially colocalized with F-actins in the distal parts of filopodia and lamellipodia (Fig. 2 A, arrowheads), as reported previously (Yamada et al., 2013). This is consistent with the previous observations that actin molecules are incorporated into F-actins at distal parts (Watanabe and Mitchison, 2002) and that cortactin binds preferentially to newly polymerized ATP/ADP-Pi-F-actin (Bryce et al., 2005). To analyze the interaction between cortactin and F-actin retrograde flow at axonal growth cones, we expressed EGFP-cortactin in hippocampal neurons and performed live-cell fluorescence microscopy (Watanabe and Mitchison, 2002; Shimada et al., 2008). EGFP-cortactin displayed fluorescent features that moved retrogradely at the peripheral domain of growth cones (Fig. 2 B, arrowheads; and Video 1). To compare the movement of the fluorescent features of cortactin directly with that of F-actin retrograde flow, we coexpressed EGFP-cortactin and mCherry-actin in XTC fibroblasts, which are suitable for live-cell fluorescence microscopy of actin in lamellipodia (Watanabe and Mitchison, 2002). The signals of EGFP-cortactin and mCherry-actin showed retrograde movement at similar speeds (EGFP-cortactin, 5.21 ± 0.14 µm/min, n = 30; mCherry-actin, 5.29 ± 0.11 µm/min, n = 30; Fig. 2 C and Video 2). To confirm the interaction between cortactin and F-actin retrograde flow, we inhibited actin polymerization with cytochalasin D. Treatment of XTC fibroblasts with 1 µM cytochalasin D led to disruption of F-actin flow at the leading edge of lamellipodia, as previously reported (Shimada et al., 2008); this is accompanied by similar redistribution of EGFP-cortactin signals (Fig. S1 and Video 3). Similar disruption of EGFP-cortactin signals was induced by cytochalasin D in axonal growth cones (Fig. 2 D and Video 4), indicating that cortactin interacts with F-actin retrograde flow at axonal growth cones.
Figure 2.
Cortactin interacts with F-actin retrograde flow. (A) Fluorescent images of an axonal growth cone labeled with anti-cortactin antibody (green) and phalloidin for F-actin (red). Enlarged views of the filopodium in the white rectangle are shown to the right. Arrowheads indicate cortactin accumulation in a filopodium. (B) A fluorescent feature image of EGFP-cortactin in an axonal growth cone (left) and a time series of the boxed area at 5-s intervals (right). See Video 1. Yellow arrowheads denote a fluorescent feature of EGFP-cortactin moving retrogradely. (C) A fluorescent feature image of mCherry–β-actin (red) and EGFP-cortactin (green) coexpressed in an XTC fibroblast (left). The kymographs (right) of the peripheral region indicated by the rectangle in the left image show that the fluorescent features of EGFP-cortactin and those of actin moved at similar speeds (white lines). See Video 2. (D) Time-lapse fluorescent feature images of EGFP-cortactin in an axonal growth cone treated at 0 min with 1 µM cytochalasin D. See Video 4. Dotted lines indicate the leading edge of the growth cone and boundary of fluorescent features. Bars: (A, B, and D) 10 µm; (C) 5 µm.
Cortactin functions as a clutch molecule, mediating the linkage between F-actin retrograde flow and L1-CAM
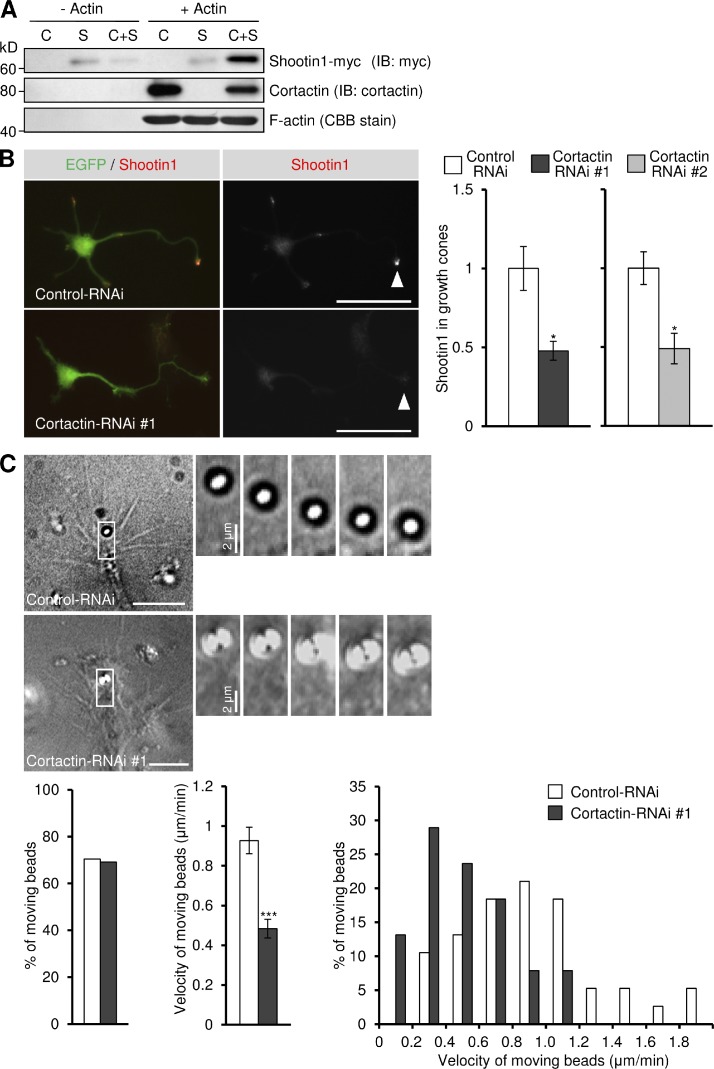
To determine whether cortactin directly mediates the linkage between F-actin and shootin1, we performed an F-actin cosedimentation assay. As shown in Fig. 3 A, purified cortactin associated with F-actin as reported (Wu and Parsons, 1993), whereas no direct interaction between F-actin and shootin1 was detectable. However, shootin1 cosedimented with F-actin in the presence of cortactin, indicating that cortactin mediates directly the linkage between F-actin and shootin1 in vitro. To further examine whether cortactin mediates the linkage between F-actins and shootin1 in growth cones, we quantified shootin1 immunoreactivity in neurons cultured on polylysine-coated coverslips (see Materials and methods). Reduction of cortactin expression by RNAi (Fig. S2, A and B) significantly decreased the localization of endogenous shootin1 to axonal growth cones (Fig. 3 B) without affecting the amount of F-actin (Fig. S2 C), thereby suggesting its dissociation from F-actins.
Figure 3.
Cortactin mediates the linkage between F-actin and shootin1 as a clutch molecule. (A) Cosedimentation of shootin1 with F-actin in the presence of cortactin. Polymerized actin (5 µM) was incubated with purified cortactin (1.5 µM; C), purified shootin1-myc (1.5 µM; S), or both (C+S). After centrifugation, the pellets were immunoblotted with anti-cortactin or anti-myc antibody. F-actin was detected by Coomassie brilliant blue (CBB) staining. (B) Hippocampal neurons were transfected with miRNA against cortactin (#1 or #2) or control miRNA, and cultured on polylysine-coated coverslips for 48 h. They were then fixed and immunostained with anti-shootin1 antibody. The vectors for the miRNAs are designed to coexpress EGFP. Arrowheads indicate an axonal growth cone. Relative fluorescence intensities of shootin1 in growth cones are shown on the right (n = 289 neurons). (C) DIC micrographs showing retrograde movement of L1-CAM-Fc–coated beads on axonal growth cones (3 d in vitro) expressing a control miRNA or cortactin miRNA, and a time series of the indicated areas at 30-s intervals (right). See Videos 5 and 6. The bottom panel illustrates the percentage of beads that showed retrograde flow on growth cones expressing control miRNA (n = 38) or cortactin miRNA (n = 38; left), the mean velocity of moving beads (middle), and the percentage of moving beads with the indicated velocities (right). Data represent means ± SEM (error bars); *, P < 0.05; ***, P < 0.001. Bars: (B) 50 µm; (C) 10 µm.
To determine whether cortactin, by linking shootin1 to F-actins, mediates the mechanical coupling between F-actin retrograde flow and L1-CAM, we performed bead-tracking experiments. Microbeads coated with L1-CAM-Fc were placed on axonal growth cones using laser optical tweezers. As the L1-CAM molecules on the bead bind homophilically to L1-CAM molecules expressed in the growth cone plasma membrane, the bead can monitor the movement of L1-CAM in the membrane (Shimada et al., 2008). On control growth cones, 70% of the beads showed retrograde movement (Fig. 3 C and Video 5) at a velocity of 0.93 ± 0.07 µm/min (mean ± SE, n = 38), as reported previously (Shimada et al., 2008). However, suppression of cortactin by RNAi significantly reduced the flow velocity of the beads (0.48 ± 0.05 µm/min, n = 38; Fig. 3 C and Video 6), indicating a reduction of the linkage between F-actin retrograde flow and L1-CAM. We conclude from these experiments that cortactin functions as a clutch molecule mediating the linkage between F-actin retrograde flow and L1-CAM in growth cones.
Cortactin is involved in shootin1-mediated and L1-CAM–dependent axon outgrowth
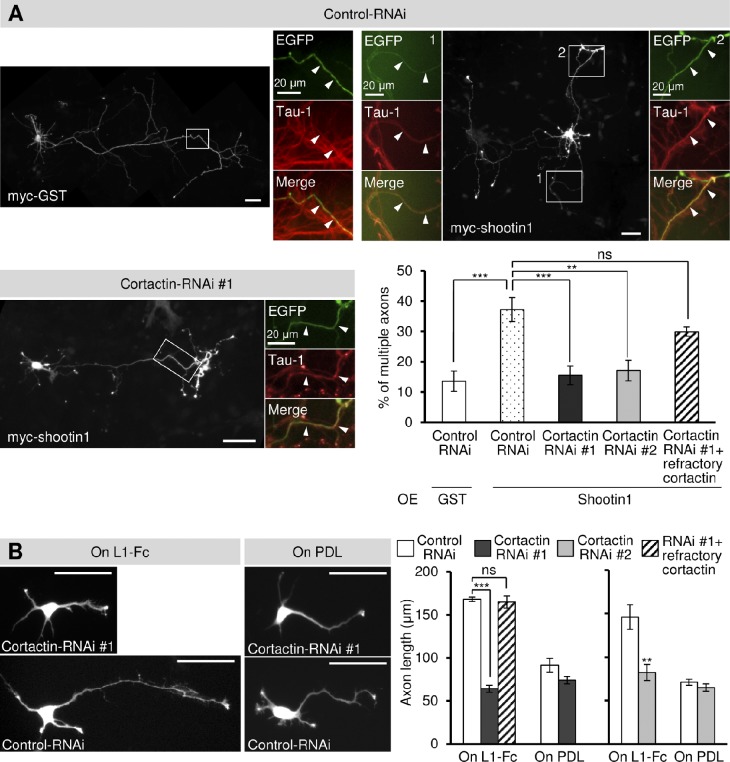
We next investigated the role of cortactin in axon outgrowth. Consistent with a previous study (Toriyama et al., 2006), overexpression of shootin1 in hippocampal neurons cultured on coverslips coated with L1-CAM-Fc promoted formation of multiple axons (Fig. 4 A). However, this shootin1-mediated axon formation was inhibited by reducing the level of cortactin by RNAi, and this inhibition was rescued by expression of an RNAi-refractory cortactin (Fig. 4 A). As reported previously (Shimada et al., 2008), hippocampal neurons cultured on L1-CAM-Fc developed longer axons than those on a control substrate, polylysine (Fig. 4 B). Suppression of cortactin expression by RNAi resulted in a significant decrease in axonal length on L1-CAM-Fc but not on polylysine (Fig. 4 B). In addition, expression of an RNAi-refractory cortactin rescued the effect of cortactin RNAi. Our previous work also showed that shootin1 knockdown selectively inhibits axonal outgrowth on L1-CAM–coated substrates (Shimada et al., 2008). Thus, consistent with the idea that cortactin constitutes a clutch system involving shootin1 and L1-CAM, shootin1-mediated and L1-CAM–dependent axon outgrowth is also cortactin-dependent.
Figure 4.
Cortactin is involved in shootin1-mediated and L1-CAM–dependent axon outgrowth. (A) Hippocampal neurons overexpressing myc-GST (control) or myc-shootin1 were cultured on coverslips coated with L1-CAM-Fc for 6 d and then immunostained with anti-myc and tau-1 antibodies. The neurons were also cotransfected with a vector to express a control miRNA or cortactin miRNA (#1 or #2), which also coexpresses EGFP. RNAi-refractory cortactin was also coexpressed in the indicated experiments. Arrowheads in the enlargements of the boxed areas of the main images indicate axons labeled by the axon-specific marker tau-1 antibody. The graph shows the percentage of neurons with multiple axons. A total of 509 neurons were examined in three independent experiments. (B) Hippocampal neurons were transfected with a control miRNA or cortactin miRNA (#1 or #2) and cultured on coverslips coated with L1-CAM-Fc (L1-Fc) or polylysine (PDL) for 48 h. RNAi-refractory cortactin was also coexpressed in the indicated experiments. The graphs show axon length. A total of 1,111 neurons were examined in three independent experiments. Data represent means ± SEM (error bars); ***, P < 0.01; **, P < 0.02. Bars, 50 µm.
The shootin1–cortactin interaction is enhanced by Pak1-mediated shootin1 phosphorylation
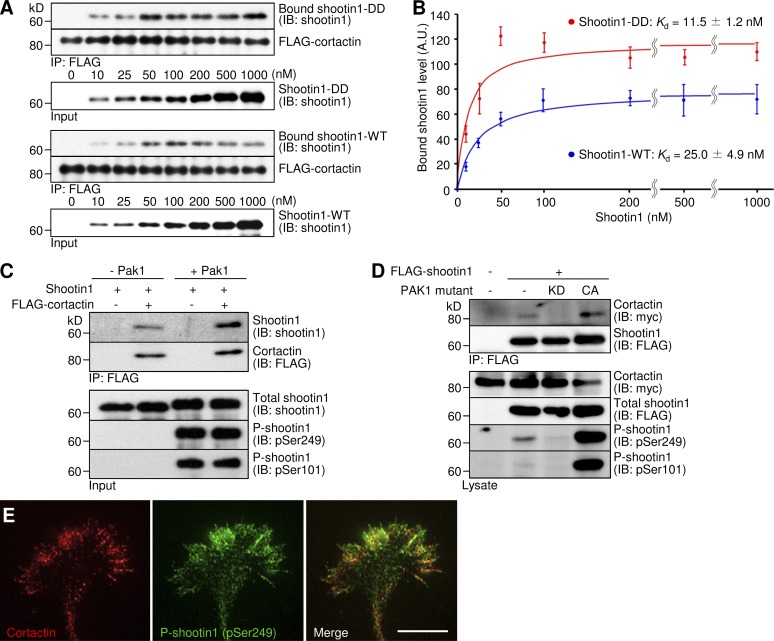
Netrin-1 induces Pak1-mediated shootin1 phosphorylation at Ser101 and Ser249, thereby enhancing the interaction between F-actin and shootin1 (Toriyama et al., 2013). To determine whether the shootin1–cortactin interaction functions as a key interface for the netrin-induced F-actin adhesion coupling, we analyzed the interaction between the phosphorylated shootin1 and cortactin. An in vitro binding assay detected the interaction of cortactin with phosphomimic shootin1 (shootin1-DD), in which Ser101 and Ser249 were replaced by aspartate (Figs. 5 A and S3 A). Importantly, the interaction of cortactin with shootin1-DD was stronger than that with shootin1 wild type (shootin1-WT). This corresponded to a 2.2-fold decrease (P < 0.05, n = 4) in the apparent dissociation constant for shootin1-DD (Kd = 11.5 ± 1.2 nM) compared with shootin1-WT (Kd = 25.0 ± 4.9 nM; Fig. 5 B). Consistently, shootin1 phosphorylated by Pak1 showed a stronger interaction with cortactin than did unphosphorylated shootin1 (Fig. 5 C). Coimmunoprecipitation assays detected cortactin–shootin1–DD and cortactin–shootin1–WT interactions in COS7 cells (Fig. S3 B). However, we could not detect an interaction between cortactin and unphosphorylated shootin1 (shootin1-AA) where Ser101 and Ser249 were replaced by alanine. Consistent with this, ectopic expression of a constitutively active PAK1 increased phosphorylation of FLAG-shootin1 at Ser101 and Ser249 in COS cells; this in turn promoted the shootin1–cortactin interaction (Fig. 5 D). Conversely, expression of a dominant-negative PAK1 decreased shootin1 phosphorylation and inhibited the interaction (Fig. 5 D). In axonal growth cones, phosphorylated shootin1 was highly colocalized with cortactin (Fig. 5 E), strengthening our conclusion that shootin1 phosphorylation by Pak1 promotes the shootin1–cortactin interaction.
Figure 5.
Pak1-mediated shootin1 phosphorylation enhances shootin1–cortactin interaction. (A and B) In vitro binding assay using purified shootin1 and purified FLAG-cortactin. Shootin1-DD or shootin1-WT at increasing concentrations were incubated with FLAG-cortactin and anti-FLAG antibody. The immunoprecipitates were immunoblotted with anti-shootin1 or anti-FLAG antibody (A), and the bound shootin1-DD and -WT were then quantified (B). Data represent means ± SEM (error bars; n = 4). (C) In vitro binding assay using purified Pak1-phosphorylated shootin1 and purified FLAG-cortactin. Shootin1-WT (80 nM) or Pak1-phosphorylated shootin1-WT (80 nM) were incubated with FLAG-cortactin (80 nM) and anti-FLAG antibody. The immunoprecipitates were immunoblotted with anti-shootin1, anti-pSer249-shootin1, anti–pSer101-shootin1, or anti-FLAG antibody. (D) Coimmunoprecipitation of shootin1 and cortactin in COS7 cells. Cells were transfected with vectors to express FLAG-shootin1 and myc-cortactin; some of them were also cotransfected with a vector to express dominant-negative Pak1 (KD) or constitutively active Pak1 (CA) as indicated. Cell lysates were then incubated with anti-FLAG antibody. The immunoprecipitates were immunoblotted with anti-myc, anti-FLAG, anti-pSer249-shootin1, or anti–pSer101-shootin1 antibody. (E) Fluorescence images of an axonal growth cone costained with anti-cortactin (red) and anti–pSer249-shootin1 (green) antibodies. Bar, 5 µm.
Cortactin mediates netrin-1–induced F-actin-substrate coupling and the promotion of traction force for axon outgrowth
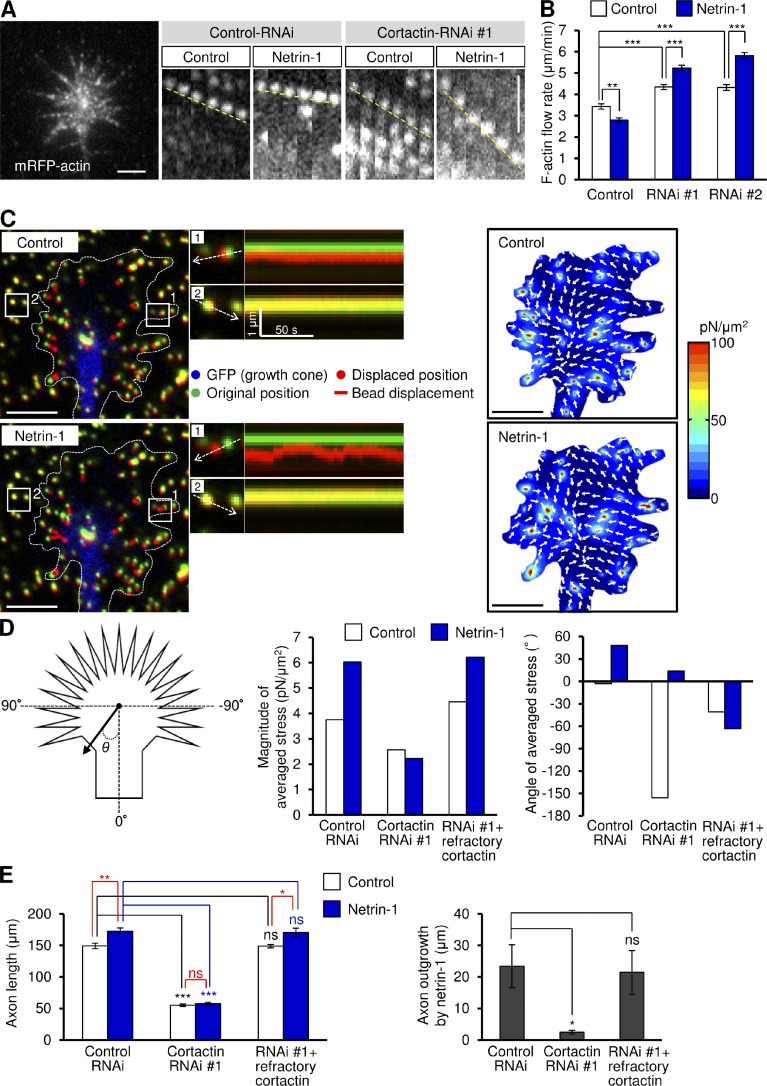
We further examined the role of cortactin in netrin-1–induced F-actin–substrate coupling by monitoring the F-actin flow in growth cones. Hippocampal neurons expressing mRFP-actin were cultured on coverslips coated with L1-CAM-Fc, and F-actin flow in the growth cone was monitored by live-cell fluorescence microscopy (Fig. 6 A). Coupling between F-actins and substrate reduces the speed of F-actin flow (Suter et al., 1998; Le Clainche and Carlier, 2008; Toriyama et al., 2013). In control growth cones, the fluorescent features of mRFP-actin moved retrogradely at 3.4 ± 0.1 µm/min (mean ± SE, n = 40), as previously reported (Toriyama et al., 2013). Consistent with the role of cortactin as a clutch molecule, a reduction of cortactin expression by RNAi increased the velocity of F-actin flow (Fig. 6, A and B), reflecting increased slippage of F-actin–substrate coupling (Le Clainche and Carlier, 2008; Lowery and Van Vactor, 2009). However, netrin-1 stimulation significantly decreased the velocity of F-actin flow, but increased it when cortactin expression was reduced (Fig. 6, A and B), suggesting that netrin-1–mediated F-actin–substrate coupling is also cortactin dependent.
Figure 6.
Cortactin is involved in netrin-1–induced F-actin–substrate coupling and promotion of traction force for axon outgrowth. (A) Fluorescent feature images of mRFP-actin at axonal growth cones expressing control miRNA or cortactin miRNA in the presence or absence of 300 ng/ml netrin-1. Kymographs of the fluorescent features of mRFP-actin in filopodia at 5-s intervals are shown (F-actin flows are indicated by broken yellow lines). (B) F-actin retrograde flow rate measured from the kymograph analysis in A. 200 fluorescent features were analyzed. (C) Fluorescence images (left) showing an axonal growth cone of a 2 d in vitro neuron expressing EGFP and cultured on L1-CAM–coated polyacrylamide gel embedded with 200-nm fluorescent beads. The pictures show representative images from time-lapse series taken every 3 s for 150 s before (control) and 60 min after netrin-1 (300 ng/ml) stimulation (see Video 7). The original and displaced positions of the beads in the gel are indicated by green and red colors, respectively. The bead displacements are also indicated by red bars. Broken lines indicate the boundary of the growth cone. The kymographs (middle) along the axis of bead displacement (white broken arrows) at the indicated areas 1 and 2 of the growth cone show movement of beads recorded every 3 s. The bead in area 2 is a reference bead. The right panel shows the stress maps during the initial 30-s observations. (D) Statistical analyses (right) of the magnitude and angle (θ) of the traction forces under axonal growth cones expressing control miRNA or miRNA against cortactin before or after netrin-1 stimulation. RNAi-refractory cortactin was also coexpressed in the indicated experiments. n = 32 growth cones. (E) 3 h after plating, neurons expressing control miRNA or cortactin miRNA #1 were incubated with BSA (control) or 300 ng/ml netrin-1 for 40 h. RNAi-refractory cortactin was also coexpressed in the indicated experiments. Axon length was then analyzed. Data represent means ± SEM; ***, P < 0.01; **, P < 0.02; *, P < 0.05; ns, not significant. Bars: (A) 2 µm; (C) 5 µm. n = 543 neurons.
To determine whether cortactin is involved in generating traction force for axon outgrowth, we performed traction force microscopy. Hippocampal neurons were cultured on L1-CAM-Fc–coated polyacrylamide gels embedded with 200-nm fluorescent beads. Traction forces under the growth cones were monitored by visualizing force-induced deformation of the elastic substrate, which is reflected by displacement of the beads from their original positions. As reported previously (Chan and Odde, 2008; Toriyama et al., 2013), the reporter beads under the growth cones moved dynamically toward the center in a “load-and-fail” manner (Fig. 6 C and Video 7) and the net force was oriented toward the rear of the growth cones (Fig. 6 D). The magnitude of the traction force increased markedly after netrin-1 treatment, which is consistent with our observation that netrin-1 promotes clutch engagement (Toriyama et al., 2013). Suppressing cortactin expression by RNAi decreased both the basal level of traction forces and netrin-1–mediated promotion of traction forces at growth cones (Fig. 6 D). Consistently, cortactin RNAi also decreased axon length and inhibited netrin-1–induced axon outgrowth (Fig. 6 E). In addition, reexpression of an RNAi-refractory cortactin rescued the effects of cortactin RNAi (Fig. 6, D and E). The present data for cortactin knockdown are similar to those obtained previously by shootin1 RNAi (Toriyama et al., 2013), and suggest that cortactin is involved in netrin-1–induced generation of traction force for axon outgrowth.
The shootin1–cortactin interaction mediates netrin-1–induced generation of traction force and axon outgrowth
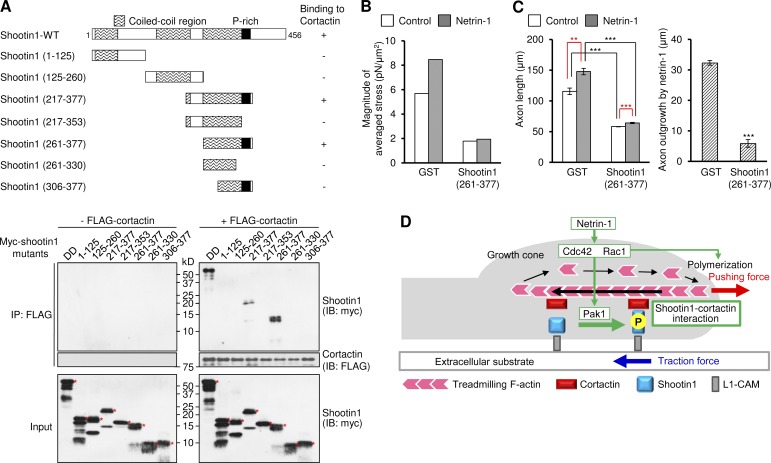
Finally, we asked whether the interaction between shootin1 and cortactin is involved in netrin-1–induced generation of traction force and axon outgrowth. To address this, we analyzed the shootin1 region that interacts with cortactin, and the cortactin domain that binds to shootin1-DD, using deletion mutants. An in vitro binding assay showed that residues 261–377 of shootin1 (shootin1 (261–377)) were essential and sufficient to bind to cortactin (Fig. 7 A), whereas the cortactin repeats region (residues 80–287) interacted with shootin1-DD (Fig. S4 A). To examine whether shootin1 (261–377) disturbs the interaction between full-length shootin1 and cortactin, we overexpressed shootin1 (261–377) in XTC fibroblasts. We fused the nuclear export signal (NES) LSLKLAGLDL (Fukuda et al., 1996) to shootin1 (261–377), as shootin1 (261–377) unexpectedly accumulated in the neuronal nucleus (unpublished data). Overexpression of a control protein myc-GST in hippocampal neurons did not decrease the number of EGFP-shootin1 signals that displayed retrograde movement (Fig. S4 B and Video 8). However, overexpression of myc-NES-shootin1 (261–377) substantially decreased the number of EGFP-shootin1 signals that displayed retrograde movement, without affecting the retrograde flow of mRFP-actin signals (Fig. S4 B and Video 9), thereby suggesting that myc-NES-shootin1 (261–377) disturbs the interaction between shootin1 and cortactin. When expressed in hippocampal neurons, Myc-NES-shootin1 (261–377) was highly colocalized with cortactin in axonal growth cones (Fig. S4 C). As in the case of cortactin RNAi, overexpression of myc-NES-shootin1 (261–377) in hippocampal neurons led to a decrease in both the basal level of traction forces and netrin-1–induced generation of traction forces at growth cones (Fig. 7 B). Overexpression of myc-NES-shootin1 (261–377) also decreased axon length and inhibited netrin-1–induced axon outgrowth (Fig. 7 C). Together, these data suggest that the shootin1–cortactin interaction mediates netrin-1–induced generation of traction force and concomitant axon outgrowth.
Figure 7.
Shootin1–cortactin interaction mediates netrin-1–induced generation of traction force and axon outgrowth. (A, top) Schematic representation of WT and shootin1 deletion mutants, and their abilities to interact with cortactin. (A, bottom) In vitro binding assay using purified myc-tagged shootin1 mutants and purified FLAG-cortactin. Myc-shootin1 mutants (80 nM) were incubated with FLAG-cortactin (80 nM) and anti-FLAG antibodies. The immunoprecipitates were immunoblotted with anti-myc or anti-FLAG antibody. Asterisks denote myc-tagged shootin1 mutants. (B) Statistical analyses of the magnitude of the traction forces under axonal growth cones overexpressing myc-GST or myc-NES-shootin1 (261–377) before or after netrin-1 stimulation. n = 19 growth cones. (C) 3 h after plating, hippocampal neurons overexpressing myc-GST or myc-NES-shootin1 (261–377) were incubated with BSA (control) or 300 ng/ml netrin-1 for 40 h. The graph shows axon length. A total of 544 neurons were examined in three independent experiments. (D) A model for signal–force transduction in axon outgrowth. Netrin-1 induces Pak1-mediated shootin1 phosphorylation, which in turn promotes the interaction between shootin1 and cortactin. The shootin1–cortactin interaction couples F-actin retrograde flow with cell adhesion, thereby transmitting the force of F-actin retrograde flow (black arrow) onto the extracellular substrate (blue arrow). This also reduces the speed of the F-actin flow, thereby converting actin polymerization into force that pushes the leading edge membrane (red arrow). F-actin adhesion coupling and actin polymerization, under the activation of Cdc42 and Rac1, cooperate complementarily for efficient promotion of protrusive forces. Data represent means ± SEM (error bars); ***, P < 0.01; *, P < 0.05; ns, nonsignificant.
Discussion
Molecular machinery to regulate F-actin–adhesion coupling
Coupling between F-actin retrograde flow and adhesion by clutch machinery has long been implicated in the regulation of cellular protrusion and migration. The present study reveals that the shootin1–cortactin complex serves as a core regulatory component of the clutch machinery in the axonal growth cone. Here, Pak1-mediated shootin1 phosphorylation enhances the shootin1–cortactin interaction, thereby executing signal–force transduction in axon outgrowth (Fig. 7 D). The identified clutch machinery uses L1-CAM as a CAM, and the shootin1–L1-CAM interaction is the missing link at present: whether shootin1 interacts directly with L1-CAM as well as whether this interaction is regulated by signaling are unclear. Previous studies reported that Pak1 and other protein kinases phosphorylate cortactin (Vidal et al., 2002; Grassart et al., 2010; MacGrath and Koleske, 2012b), raising the possibility that cortactin phosphorylation may regulate the shootin1–cortactin interaction. In addition, modification of the interaction between F-actin and cortactin (Zhang et al., 2007; MacGrath and Koleske, 2012a) would also influence F-actin adhesion coupling.
In nonneuronal cells, the integrin-based focal adhesion (FA) is thought to play a key role in F-actin–substrate coupling at the cellular front, and candidate clutch components such as vinculin and talin have been identified (Le Clainche and Carlier, 2008; Zhang et al., 2008; Thievessen et al., 2013). However, the FA is a complex structure composed of a large number of different proteins, and the molecular machinery that regulates F-actin–FA coupling is unclear. Cortactin is expressed in nonneuronal cells (Weed and Parsons, 2001; MacGrath and Koleske, 2012b), and although shootin1 analyzed here is a brain-specific protein (Toriyama et al., 2006), we have found that an alternative splicing variant of shootin1 is expressed in peripheral tissues (unpublished data). A shootin–cortactin interaction in nonneuronal cells is an intriguing issue for future analyses.
Cortactin as a clutch molecule
Cortactin interacts with a large number of signaling, cytoskeletal, and membrane trafficking proteins, and multiple mechanisms for cortactin-mediated cell motility have been proposed (Kirkbride et al., 2011; MacGrath and Koleske, 2012b). Cortactin binds to the Arp2/3 complex (Weed et al., 2000), which is essential for F-actin nucleation (Pollard and Borisy, 2003), and serves as a cofactor for its activation (Uruno et al., 2001; Weaver et al., 2001). Cortactin stabilizes Arp2/3-mediated F-actin branches (Weaver et al., 2001), and is also reported to play a role in endocytic processes (Cao et al., 2003; Sauvonnet et al., 2005) and secretion of extracellular matrix (Sung et al., 2011).
Our data present a previously unknown mechanism for cortactin-mediated cell motility. Reduction of endogenous cortactin did not significantly decrease the F-actin content in the growth cones (Fig. S2 C), suggesting that cortactin-mediated F-actin nucleation and stabilization through Arp2/3 complex do not have a dominant effect. This is further supported by the finding that cortactin RNAi led to an increase in the rate of F-actin flow in growth cones (Fig. 6, A and B). If cortactin contributed to axon outgrowth by activating actin polymerization through Arp2/3 complex, cortactin RNAi should lead to a decrease in the F-actin flow rate; however, the inverse effect was observed. We therefore conclude that cortactin promotes traction force and L1-CAM–dependent axon outgrowth mainly by coupling F-actins with cell adhesions.
A recent study reported that cortactin can bind and stabilize F-actins (Courtemanche et al., 2015); it also forms a ring complex together with dynamin 1 that mechanically stabilizes F-actin bundles in growth cone filopodia (Yamada et al., 2013). The cortactin-dependent force generation may cooperate with the F-actin stabilizing activity of cortactin in the formation of growth cone filopodia and axon outgrowth. In addition, the present mechanism may explain the observation of Decourt et al. (2009) that cortactin accumulates at IgCAM adhesion sites together with F-actins in Aplysia californica growth cones. Interestingly, another recent study showed that inhibition of Arp2/3 complex, which interacts with cortactin, led to delocalization of Arp2/3 complex from the leading edge of A. californica growth cones and an increase in F-actin flow rate (Yang et al., 2012). The effect on the F-actin flow rate appears difficult to explain in terms of the Arp2/3 complex activity that nucleates F-actins to generate actin networks; thus, it would be interesting to examine whether inhibition of Arp2/3 complex also leads to a delocalization of cortactin from the leading edge, which may inhibit F-actin adhesion coupling and subsequently increase the F-actin flow rate.
Cooperation between F-actin adhesion coupling and actin polymerization in protrusive activity
Stimulation of actin polymerization also plays a key role in cellular motility (Pollard and Borisy, 2003; Lowery and Van Vactor, 2009). The regulatory clutch machinery identified here, in conjunction with actin polymerization, provides an integrated view of how forces for cellular motility are generated under cell signaling. As shown here, an increase in the coupling efficiency produces traction force on the extracellular substrate (Fig. 7 D, blue arrow). This also reduces the speed of F-actin retrograde flow, thereby converting actin polymerization into force that pushes the leading edge membrane (Fig. 7 D, red arrow). However, promotion of actin polymerization at the leading edge not only produces force to push the front (Fig. 7 D, red arrow) but also increases the F-actin flow rate by counteracting force (Pollard and Borisy, 2003; Medeiros et al., 2006); this in turn increases the traction force on the extracellular substrate (Fig. 7 D, blue arrow). In this regard, F-actin adhesion coupling and actin polymerization play complementary roles in triggering protrusive forces.
Activation of Cdc42 and Rac1 promotes not only Pak1-mediated shootin1 phosphorylation but also actin polymerization (Rohatgi et al., 1999; Miki et al., 2000; Peng et al., 2003; Pollard and Borisy, 2003; Le Clainche and Carlier, 2008; Fig. 7 D). Stimulation of axonal growth cones with netrin-1, which activates Cdc42 and Rac1 (Shekarabi et al., 2005; Li et al., 2008; Toriyama et al., 2013), led to an increase in the F-actin flow rate when promotion of F-actin adhesion coupling was inhibited by cortactin RNAi (Fig. 6, A and B). We believe that this effect reflects increased actin polymerization (Pollard and Borisy, 2003; Medeiros et al., 2006) induced by the activation of Cdc42 and Rac1. In this case, in contrast to the control experiment, netrin-1 stimulation did not promote formation of traction force or axon outgrowth (Fig. 6, D and E). Similar results were obtained when F-actin adhesion coupling was blocked by shootin1 RNAi (Toriyama et al., 2013). Together, these data underscore the role of F-actin adhesion coupling in linking increased actin polymerization to forces for protrusive activity.
In conclusion, the present study identifies a molecular complex that regulates F-actin adhesion coupling, and thus paves the way for acquiring molecular-level and structural insights underlying it. Intracellular asymmetric activation of F-actin adhesion coupling would contribute to directional axon formation and guidance (Li et al., 2008; Mai et al., 2009). The system involving polymerizing F-actins, the molecular clutch, and adhesion molecules (Fig. 7 D) would integrate not only diffusible chemical signals but also adhesive and mechanical cues (Esch et al., 1999; Chan and Odde, 2008) to regulate protrusive activity. A comprehensive elucidation of this system will lead to a profound understanding of cell migration, development, and various pathological processes.
Materials and methods
Cultures and transfection
Hippocampal neurons prepared from embryonic day 18 rat embryos were cultured on coverslips coated with L1-CAM-Fc or polylysine as described previously (Shimada et al., 2008). For preparation of L1-CAM-Fc, COS-7 cells were transfected with pCAGGS-L1-CAM-Fc vector using the calcium phosphate method. The culture media were conditioned for 48 h, and the L1-CAM-Fc were purified from the culture supernatant by a recombinant protein A–Sepharose column (GE Healthcare). All experiments except for the measurement of forces were performed on L1-CAM–coated or polylysine-coated glass surfaces. The neurons were transfected with vectors using Nucleofector (Lonza) before plating. We previously reported that shootin1 accumulation at the growth cone is influenced by the length of neurite (Toriyama et al., 2010). Therefore, to examine the effects of cortactin RNAi on shootin1 localization in axonal growth cones (Fig. 3 B), we cultured neurons on polylysine-coated coverslips because suppression of cortactin expression by RNAi did not result in a decrease in axonal length on polylysine (Fig. 4 B). XTC cells were cultured as described previously (Higashida et al., 2004) and transfected with vectors using FuGENE 6 (Roche). COS7 cells were cultured in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Japan Bio Serum) and transfected with vectors by the calcium phosphate method.
DNA constructs
Rat cortactin cDNA was amplified by PCR from a rat brain cDNA library (Takara Bio Inc.) with the primers 5′-CTGGATCCATGTGGAAAGCTTCTGCAGGC-3′ and 5′-CTGGATCCCTACTGCCGCAGCTCCACATAG-3′. cDNA fragments of cortactin and shootin1 deletion mutants were amplified by PCR and subcloned into pGEX-6P-1 (fused to N-terminal GST-tag, tac promoter; GE Healthcare), pCAGGS-myc (fused to N-terminal myc-tag, β-actin promoter; Niwa et al., 1991), pCMV-myc (fused to N-terminal myc-tag, cytomegalovirus [CMV] promoter; Agilent Technologies), or pEGFP-C1 (fused to N-terminal EGFP-tag, CMV promoter; Takara Bio Inc.) vector. The generation of unphosphorylated (S101A/S249A: AA) and phosphomimetic (S101D/S249D: DD) mutants of shootin1 has been described previously (Toriyama et al., 2013). pCAGGS-myc was used to overexpress proteins under the β-actin promoter as described previously (Toriyama et al., 2006). Unexpectedly, myc-shootin1 (261–377) preferentially accumulated in the neuronal nucleus, whereas it was mainly localized in the cytoplasm of XTC cells. To express myc-shootin1 (261–377) in the cytoplasm of neurons, we fused the NES LSLKLAGLDL (Fukuda et al., 1996) to the N terminus of myc-shootin1 (261–377).
RNAi
For RNAi experiments, we used a Block-iT Pol II miR RNAi expression kit (Invitrogen). The targeting miRNA sequences of cortactin miRNA #1 (5′-GCAGCACAGCATGTCCTTGTA-3′) and cortactin miRNA #2 (5′-TACATCGCGTCTGCGTGTGTT-3′) correspond to nucleotides 2708–2728 and nucleotides 1719–1739 in the 3′ UTR region of rat cortactin mRNA, respectively. The control vector pcDNA 6.2-GW/EmGFP-miR-neg encodes an miRNA (5′-GAAATGTACTGCGCGTGGAGACGTTTTGGCCACTGACTGACGTCTCCACGCAGTACATTT-3′) that targets no known vertebrate gene. To ensure a high level of expression of cortactin miRNA before axon outgrowth, neurons transfected with miRNA were cultured in suspension on noncoated polystyrene dishes for 24 h (Toriyama et al., 2006), and then collected and seeded onto coated-glass coverslips, polyacrylamide gels, or glass-bottom dishes. Reduction of cortactin levels in neurons was confirmed by immunoblotting and immunocytochemistry, using anti-cortactin antibody (Fig. S2, A and B). As cortactin miRNA #1 and cortactin miRNA #2 target 3′ UTR regions of cortactin mRNA, cortactin exogenously expressed by the pCMV-myc vector functioned as an RNAi-refractory cortactin.
Protein preparation and in vitro kinase assay
Recombinant proteins were expressed in Escherichia coli as GST fusion proteins and purified on Glutathione Sepharose columns (GE Healthcare), after which GST was removed from them by PreScission protease (GE Healthcare). L1-CAM-Fc was prepared as described previously (Shimada et al., 2008). Kinase reactions were performed in 20 µl kinase buffer (50 mM Hepes, pH 7.5, 10 mM MgCl2, 2 mM MnCl2, 1 mM DTT, and 125 µM ATP, in the presence or absence of 10 µCi γ-[32P]ATP) containing 250 ng active GST-Pak1 (Invitrogen) and 2.1 µg purified shootin1.
In vitro binding assay and F-actin sedimentation assay
Purified FLAG-cortactin and shootin1 were incubated overnight at 4°C in reaction buffer (0.3% CHAPS, 20 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 1 mM DTT) and centrifuged for 15 min at 17,400 g at 4°C. The supernatants were incubated with 15 µl (bed volume) of anti-FLAG M2 gel (Sigma-Aldrich) for 2 h at 4°C. Anti-FLAG M2 gels were washed three times with wash buffer (reaction buffer + 150 mM NaCl) and once with TED buffer (20 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 1 mM DTT). For elution, the gels were incubated with 25 µl of FLAG peptide (400 µg/ml; Sigma-Aldrich) for 1 h at 4°C. The supernatants were analyzed by immunoblotting. Apparent dissociation constants were calculated by nonlinear regression using GraphPad Prism 6 (GraphPad Software).
Human nonmuscle actin was obtained from Cytoskeleton, Inc. F-actin sedimentation assays were performed according to the manufacturer’s protocol with slight modifications. We used actin stored in general actin buffer (5 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2) supplemented with 0.2 mM ATP. Actin was polymerized in polymerization buffer (50 mM KCl, 2 mM MgCl2, and 1 mM ATP), and polymerized actin was centrifuged for 1 h at 100,000 g at 4°C. After removal of the supernatant, the pellet was resuspended in buffer A (20 mM Tris-HCl, pH 8.0, 50 mM KCl, 2 mM MgCl2, 1 mM EDTA, 0.5 mM DTT, and 0.5 mM ATP). To perform the F-actin sedimentation assay, 1.5 µM of purified protein was incubated with 7.5 µg of polymerized actin for 1 h at room temperature. 25 µl of the mixture was overlaid on 50 µl of buffer A containing 10% sucrose, and centrifuged for 1 h at 100,000 g at 4°C. The pellet fractions were washed three times with buffer A and analyzed by immunoblotting.
Immunocytochemistry, microscopy, immunoprecipitation, and immunoblotting
Cultured neurons were fixed with 3.7% formaldehyde in Krebs buffer for 10 min at room temperature, followed by treatment for 15 min with 0.05% Triton X-100 in PBS on ice and 10% fetal bovine serum in PBS for 1 h at room temperature. They were then stained with antibodies (mouse anti-cortactin, mouse anti-Tau1, rabbit anti-shootin1, and rabbit anti-myc antibodies) and Alexa Fluor 350 phalloidin (Invitrogen), as described previously (Shimada et al., 2008). Secondary antibodies and phalloidin were conjugated with Alexa Fluor 488 (Invitrogen) or Alexa Fluor 594 (Invitrogen). We used 50% glycerol and 50% PBS as the mounting medium. Fluorescence and phase-contrast images of neurons were acquired using a fluorescence microscope (Axioplan2; Carl Zeiss) equipped with a plan-Neofluar 20× 0.50 NA or 63× oil 1.40 NA objective lens (Carl Zeiss), a charge-coupled device camera (AxioCam MRm; Carl Zeiss), and imaging software (Axiovision3; Carl Zeiss). Axon length and the relative fluorescence intensity of shootin1 or F-actin were measured using Multi Gauge software (Fujifilm). Immunoprecipitation and immunoblotting were performed as described previously (Shimada et al., 2008). For immunoprecipitation, supernatants of brain lysates or cell lysates were incubated with antibodies overnight at 4°C, and immunocomplexes were then precipitated with protein G–Sepharose 4B (GE Healthcare). After washing out beads with RIPA buffer, immunocomplexes were analyzed by immunoblotting.
Bead tracking, live-cell fluorescence microscopy, and traction force microscopy
Bead tracking and live-cell fluorescence microscopy were performed as described previously (Shimada et al., 2008). In brief, 1-µm-diameter polystyrene beads (Polysciences, Inc.) were coated with L1-CAM-Fc. We used the laser optical trap system to place beads on growth cones cultured in Leibovitz’s L-15 medium (Invitrogen). Differential interference contrast (DIC) images of beads were acquired at room temperature using a fluorescence microscope (Eclipse TE2000-U; Nikon) equipped with a plan-Apochromat 60× water immersion 1.20 NA objective lens (Nikon) and a digital video camera (DCR-SR100; Sony). Bead velocity was quantified using Multi Gauge software. The live-cell fluorescence imaging data in Fig. 6 A were obtained using neurons cultured on coverslips coated with L1-CAM-Fc, whereas the other data for live-cell imaging were obtained with neurons or XTC cells cultured on polylysine-coated coverslips.
Traction force microscopy was performed as described previously (Toriyama et al., 2013). In brief, neurons were cultured on L1-CAM-Fc–coated polyacrylamide gels embedded with fluorescent microspheres (200 nm diameter; Invitrogen). Time-lapse imaging of fluorescent beads and growth cones was performed at 37°C using a confocal microscope (LSM710; Carl Zeiss) equipped with a C-Apochromat 63×/1.2 W Corr objective lens. The growth cone area was determined from EGFP fluorescence or DIC images. Traction forces under the growth cones were monitored by visualizing force-induced deformation of the elastic substrate, which is reflected by displacement of the beads from their original positions, and expressed as vectors (Toriyama et al., 2013). The force vectors detected by the beads under individual growth cones were then averaged, and were expressed as vectors composed of magnitude and angle (θ; Fig. 6 D, left). To compare the forces under different conditions, the force vectors of individual growth cones under each condition were then averaged, and the magnitude and angle of the averaged force vector were obtained (Fig. 6 D, right two panels).
Statistical analysis
Significance was determined by the unpaired Student’s t test in most cases. For multiple comparisons, we used one-way analysis of variance (ANOVA) with Schaffer’s post hoc test.
Materials
Preparations of anti-shootin1, anti-pSer101-shootin1, and anti-pSer249-shootin1 antibodies have been described elsewhere (Toriyama et al., 2006, 2013). Antibodies against cortactin (4F11) and actin were obtained from EMD Millipore. Antibodies against Tau-1, myc, and FLAG were obtained from EMD Millipore, MBL, and Sigma-Aldrich, respectively. Cytochalasin D and polylysine were obtained from EMD Millipore and Sigma-Aldrich, respectively. Alexa Fluor 594 phalloidin and Netrin-1 were obtained from Invitrogen and R&D Systems, respectively. The constructs of fascin, VASP, p21-ARC, Esp8, XAC2 and capping protein β1 were provided by N. Watanabe (Kyoto University, Kyoto, Japan).
Online supplemental material
Fig. S1 shows the effect of cytochalasin D on retrograde movement of the fluorescent features of EGFP-cortactin in XTC fibroblasts. Fig. S2 shows suppression of cortactin by RNAi and the effect of cortactin suppression on the F-actin level in axonal growth cones. Fig. S3 shows an in vitro binding assay and coimmunoprecipitation assay, demonstrating that Pak1-mediated shootin1 phosphorylation enhances the interaction between shootin1 and cortactin. Fig. S4 shows the cortactin repeats region that interacts with shootin1-DD, and demonstrates that shootin1 (261–377) disrupts shootin1–cortactin interaction and colocalizes with F-actins in axonal growth cones. Video 1 is a time-lapse video showing movement of the fluorescent features of EGFP-cortactin in an axonal growth cone as described in Fig. 2 B. Video 2 is a time-lapse video showing movement of the fluorescent features of EGFP-cortactin and mCherry-actin in an XTC fibroblast as described in Fig. 2 C. Video 3 is a time-lapse video showing the effect of cytochalasin D on retrograde movement of EGFP-cortactin and mCherry-actin signals in XTC fibroblasts as described in Fig. S1. Video 4 is a time-lapse video showing the effect of cytochalasin D on retrograde movement of the fluorescent features of EGFP-cortactin in an axonal growth cone as described in Fig. 2 D. Video 5 is a time-lapse video showing retrograde movement of an L1-CAM-Fc–coated bead on an axonal growth cone expressing control miRNA as described in Fig. 3 C. Video 6 is a time-lapse video showing retrograde movement of an L1-CAM-Fc–coated bead on an axonal growth cone expressing cortactin miRNA as described in Fig. 3 C. Video 7 is a time-lapse video showing netrin-1–induced promotion of traction force under an axon outgrowth cone as described in Fig. 6 C. Video 8 is a time-lapse video showing movement of the fluorescent features of EGFP-shootin1 and mRFP-actin in an axonal growth cone overexpressing Myc-NES-GST as described in Fig. S4 B. Video 9 is a time-lapse video showing movement of the fluorescent features of EGFP-shootin1 and mRFP-actin in an axonal growth cone overexpressing Myc-NES-shootin1 (261–377) as described in Fig. S4 B. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201505011/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201505011.dv.
Supplementary Material
Acknowledgements
We thank N. Watanabe for providing constructs of fascin, VASP, p21-ARC, Esp8, XAC2, and capping protein β1; Y. Sakumura for discussion; and T. Hakoshima, A. Urasaki and H. Katsuno for reviewing the manuscript.
This research was supported by a Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research on Innovative Areas (25102010), JSPS KAKENHI (23370088), the Global COE Program at Nara Institute of Science and Technology (NAIST; MEXT), the Osaka Medical Research Foundation for Incurable Diseases, and the NAIST Interdisciplinary Promotion Project.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CAM
- cell adhesion molecule
- CMV
- cytomegalovirus
- DIC
- differential interference contrast
- FA
- focal adhesion
- NES
- nuclear export signal
- WT
- wild type
References
- Bard L., Boscher C., Lambert M., Mège R.M., Choquet D., and Thoumine O.. 2008. A molecular clutch between the actin flow and N-cadherin adhesions drives growth cone migration. J. Neurosci. 28:5879–5890. 10.1523/JNEUROSCI.5331-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce N.S., Clark E.S., Leysath J.L., Currie J.D., Webb D.J., and Weaver A.M.. 2005. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr. Biol. 15:1276–1285. 10.1016/j.cub.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Cao H., Orth J.D., Chen J., Weller S.G., Heuser J.E., and McNiven M.A.. 2003. Cortactin is a component of clathrin-coated pits and participates in receptor-mediated endocytosis. Mol. Cell. Biol. 23:2162–2170. 10.1128/MCB.23.6.2162-2170.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.E., and Odde D.J.. 2008. Traction dynamics of filopodia on compliant substrates. Science. 322:1687–1691. 10.1126/science.1163595 [DOI] [PubMed] [Google Scholar]
- Cheng Y., Leung S., and Mangoura D.. 2000. Transient suppression of cortactin ectopically induces large telencephalic neurons towards a GABAergic phenotype. J. Cell Sci. 113:3161–3172. [DOI] [PubMed] [Google Scholar]
- Courtemanche N., Gifford S.M., Simpson M.A., Pollard T.D., and Koleske A.J.. 2015. Abl2/Abl-related gene stabilizes actin filaments, stimulates actin branching by actin-related protein 2/3 complex, and promotes actin filament severing by cofilin. J. Biol. Chem. 290:4038–4046. 10.1074/jbc.M114.608117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decourt B., Munnamalai V., Lee A.C., Sanchez L., and Suter D.M.. 2009. Cortactin colocalizes with filopodial actin and accumulates at IgCAM adhesion sites in Aplysia growth cones. J. Neurosci. Res. 87:1057–1068. 10.1002/jnr.21937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Walker V.D., Peterson J.R., Chernoff J., Waterman C.M., Danuser G., DerMardirossian C., and Bokoch G.M.. 2011. Pak1 regulates focal adhesion strength, myosin IIA distribution, and actin dynamics to optimize cell migration. J. Cell Biol. 193:1289–1303. 10.1083/jcb.201010059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Weed S.A., Xiong W.C., Marshall T.D., and Parsons J.T.. 1998. Identification of a novel cortactin SH3 domain-binding protein and its localization to growth cones of cultured neurons. Mol. Cell. Biol. 18:5838–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch T., Lemmon V., and Banker G.. 1999. Local presentation of substrate molecules directs axon specification by cultured hippocampal neurons. J. Neurosci. 19:6417–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forscher P., and Smith S.J.. 1988. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J. Cell Biol. 107:1505–1516. 10.1083/jcb.107.4.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Gotoh I., Gotoh Y., and Nishida E.. 1996. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J. Biol. Chem. 271:20024–20028. 10.1074/jbc.271.33.20024 [DOI] [PubMed] [Google Scholar]
- Giannone G., Mège R.M., and Thoumine O.. 2009. Multi-level molecular clutches in motile cell processes. Trends Cell Biol. 19:475–486. 10.1016/j.tcb.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Grassart A., Meas-Yedid V., Dufour A., Olivo-Marin J.C., Dautry-Varsat A., and Sauvonnet N.. 2010. Pak1 phosphorylation enhances cortactin-N-WASP interaction in clathrin-caveolin-independent endocytosis. Traffic. 11:1079–1091. 10.1111/j.1600-0854.2010.01075.x [DOI] [PubMed] [Google Scholar]
- Higashida C., Miyoshi T., Fujita A., Oceguera-Yanez F., Monypenny J., Andou Y., Narumiya S., and Watanabe N.. 2004. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 303:2007–2010. 10.1126/science.1093923 [DOI] [PubMed] [Google Scholar]
- Inagaki N., Toriyama M., and Sakumura Y.. 2011. Systems biology of symmetry breaking during neuronal polarity formation. Dev. Neurobiol. 71:584–593. 10.1002/dneu.20837 [DOI] [PubMed] [Google Scholar]
- Jacobs T., Causeret F., Nishimura Y.V., Terao M., Norman A., Hoshino M., and Nikolić M.. 2007. Localized activation of p21-activated kinase controls neuronal polarity and morphology. J. Neurosci. 27:8604–8615. 10.1523/JNEUROSCI.0765-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiguchi H., Hlavin M.L., Yamasaki M., and Lemmon V.. 1998. Adhesion molecules and inherited diseases of the human nervous system. Annu. Rev. Neurosci. 21:97–125. 10.1146/annurev.neuro.21.1.97 [DOI] [PubMed] [Google Scholar]
- Katoh K., Hammar K., Smith P.J., and Oldenbourg R.. 1999. Birefringence imaging directly reveals architectural dynamics of filamentous actin in living growth cones. Mol. Biol. Cell. 10:197–210. 10.1091/mbc.10.1.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinley A.W., Weed S.A., Weaver A.M., Karginov A.V., Bissonette E., Cooper J.A., and Parsons J.T.. 2003. Cortactin interacts with WIP in regulating Arp2/3 activation and membrane protrusion. Curr. Biol. 13:384–393. 10.1016/S0960-9822(03)00107-6 [DOI] [PubMed] [Google Scholar]
- Kirkbride K.C., Sung B.H., Sinha S., and Weaver A.M.. 2011. Cortactin: a multifunctional regulator of cellular invasiveness. Cell Adhes. Migr. 5:187–198. 10.4161/cam.5.2.14773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurklinsky S., Chen J., and McNiven M.A.. 2011. Growth cone morphology and spreading are regulated by a dynamin-cortactin complex at point contacts in hippocampal neurons. J. Neurochem. 117:48–60. 10.1111/j.1471-4159.2011.07169.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C., and Carlier M.F.. 2008. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 88:489–513. 10.1152/physrev.00021.2007 [DOI] [PubMed] [Google Scholar]
- Li X., Gao X., Liu G., Xiong W., Wu J., and Rao Y.. 2008. Netrin signal transduction and the guanine nucleotide exchange factor DOCK180 in attractive signaling. Nat. Neurosci. 11:28–35. 10.1038/nn2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery L.A., and Van Vactor D.. 2009. The trip of the tip: understanding the growth cone machinery. Nat. Rev. Mol. Cell Biol. 10:332–343. 10.1038/nrm2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGrath S.M., and Koleske A.J.. 2012a Arg/Abl2 modulates the affinity and stoichiometry of binding of cortactin to F-actin. Biochemistry. 51:6644–6653. 10.1021/bi300722t [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGrath S.M., and Koleske A.J.. 2012b Cortactin in cell migration and cancer at a glance. J. Cell Sci. 125:1621–1626. 10.1242/jcs.093781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai J., Fok L., Gao H., Zhang X., and Poo M.M.. 2009. Axon initiation and growth cone turning on bound protein gradients. J. Neurosci. 29:7450–7458. 10.1523/JNEUROSCI.1121-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros N.A., Burnette D.T., and Forscher P.. 2006. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat. Cell Biol. 8:216–226. 10.1038/ncb1367 [DOI] [PubMed] [Google Scholar]
- Miki H., Yamaguchi H., Suetsugu S., and Takenawa T.. 2000. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature. 408:732–735. 10.1038/35047107 [DOI] [PubMed] [Google Scholar]
- Mingorance-Le Meur A., and O’Connor T.P.. 2009. Neurite consolidation is an active process requiring constant repression of protrusive activity. EMBO J. 28:248–260. 10.1038/emboj.2008.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., and Kirschner M.. 1988. Cytoskeletal dynamics and nerve growth. Neuron. 1:761–772. 10.1016/0896-6273(88)90124-9 [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., and Miyazaki J.. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 108:193–199. 10.1016/0378-1119(91)90434-D [DOI] [PubMed] [Google Scholar]
- Peng J., Wallar B.J., Flanders A., Swiatek P.J., and Alberts A.S.. 2003. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr. Biol. 13:534–545. 10.1016/S0960-9822(03)00170-2 [DOI] [PubMed] [Google Scholar]
- Pollard T.D., and Borisy G.G.. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., and Kirschner M.W.. 1999. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 97:221–231. 10.1016/S0092-8674(00)80732-1 [DOI] [PubMed] [Google Scholar]
- Sapir T., Levy T., Sakakibara A., Rabinkov A., Miyata T., and Reiner O.. 2013. Shootin1 acts in concert with KIF20B to promote polarization of migrating neurons. J. Neurosci. 33:11932–11948. 10.1523/JNEUROSCI.5425-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvonnet N., Dujeancourt A., and Dautry-Varsat A.. 2005. Cortactin and dynamin are required for the clathrin-independent endocytosis of γc cytokine receptor. J. Cell Biol. 168:155–163. 10.1083/jcb.200406174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini T., Kennedy T.E., Galko M.J., Mirzayan C., Jessell T.M., and Tessier-Lavigne M.. 1994. The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell. 78:409–424. 10.1016/0092-8674(94)90420-0 [DOI] [PubMed] [Google Scholar]
- Shekarabi M., Moore S.W., Tritsch N.X., Morris S.J., Bouchard J.F., and Kennedy T.E.. 2005. Deleted in colorectal cancer binding netrin-1 mediates cell substrate adhesion and recruits Cdc42, Rac1, Pak1, and N-WASP into an intracellular signaling complex that promotes growth cone expansion. J. Neurosci. 25:3132–3141. 10.1523/JNEUROSCI.1920-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Toriyama M., Uemura K., Kamiguchi H., Sugiura T., Watanabe N., and Inagaki N.. 2008. Shootin1 interacts with actin retrograde flow and L1-CAM to promote axon outgrowth. J. Cell Biol. 181:817–829. 10.1083/jcb.200712138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane M., Ketschek A., Donnelly C.J., Pacheco A., Twiss J.L., and Gallo G.. 2012. Nerve growth factor-induced formation of axonal filopodia and collateral branches involves the intra-axonal synthesis of regulators of the actin-nucleating Arp2/3 complex. J. Neurosci. 32:17671–17689. 10.1523/JNEUROSCI.1079-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung B.H., Zhu X., Kaverina I., and Weaver A.M.. 2011. Cortactin controls cell motility and lamellipodial dynamics by regulating ECM secretion. Curr. Biol. 21:1460–1469. 10.1016/j.cub.2011.06.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter D.M., and Forscher P.. 2000. Substrate-cytoskeletal coupling as a mechanism for the regulation of growth cone motility and guidance. J. Neurobiol. 44:97–113. [DOI] [PubMed] [Google Scholar]
- Suter D.M., Errante L.D., Belotserkovsky V., and Forscher P.. 1998. The Ig superfamily cell adhesion molecule, apCAM, mediates growth cone steering by substrate-cytoskeletal coupling. J. Cell Biol. 141:227–240. 10.1083/jcb.141.1.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thievessen I., Thompson P.M., Berlemont S., Plevock K.M., Plotnikov S.V., Zemljic-Harpf A., Ross R.S., Davidson M.W., Danuser G., Campbell S.L., and Waterman C.M.. 2013. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202:163–177. 10.1083/jcb.201303129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama M., Shimada T., Kim K.B., Mitsuba M., Nomura E., Katsuta K., Sakumura Y., Roepstorff P., and Inagaki N.. 2006. Shootin1: A protein involved in the organization of an asymmetric signal for neuronal polarization. J. Cell Biol. 175:147–157. 10.1083/jcb.200604160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama M., Sakumura Y., Shimada T., Ishii S., and Inagaki N.. 2010. A diffusion-based neurite length-sensing mechanism involved in neuronal symmetry breaking. Mol. Syst. Biol. 6:394 10.1038/msb.2010.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama M., Kozawa S., Sakumura Y., and Inagaki N.. 2013. Conversion of a signal into forces for axon outgrowth through Pak1-mediated shootin1 phosphorylation. Curr. Biol. 23:529–534. 10.1016/j.cub.2013.02.017 [DOI] [PubMed] [Google Scholar]
- Uruno T., Liu J., Zhang P., Fan Yx C., Egile R., Li S.C., Mueller, and Zhan X.. 2001. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat. Cell Biol. 3:259–266. 10.1038/35060051 [DOI] [PubMed] [Google Scholar]
- Van Haastert P.J., and Devreotes P.N.. 2004. Chemotaxis: signalling the way forward. Nat. Rev. Mol. Cell Biol. 5:626–634. 10.1038/nrm1435 [DOI] [PubMed] [Google Scholar]
- Vidal C., Geny B., Melle J., Jandrot-Perrus M., and Fontenay-Roupie M.. 2002. Cdc42/Rac1-dependent activation of the p21-activated kinase (PAK) regulates human platelet lamellipodia spreading: implication of the cortical-actin binding protein cortactin. Blood. 100:4462–4469. 10.1182/blood.V100.13.4462 [DOI] [PubMed] [Google Scholar]
- Watanabe N., and Mitchison T.J.. 2002. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 295:1083–1086. 10.1126/science.1067470 [DOI] [PubMed] [Google Scholar]
- Weaver A.M., Karginov A.V., Kinley A.W., Weed S.A., Li Y., Parsons J.T., and Cooper J.A.. 2001. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11:370–374. 10.1016/S0960-9822(01)00098-7 [DOI] [PubMed] [Google Scholar]
- Weed S.A., and Parsons J.T.. 2001. Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene. 20:6418–6434. 10.1038/sj.onc.1204783 [DOI] [PubMed] [Google Scholar]
- Weed S.A., Karginov A.V., Schafer D.A., Weaver A.M., Kinley A.W., Cooper J.A., and Parsons J.T.. 2000. Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J. Cell Biol. 151:29–40. 10.1083/jcb.151.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., and Parsons J.T.. 1993. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J. Cell Biol. 120:1417–1426. 10.1083/jcb.120.6.1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Wang F., Van Keymeulen A., Herzmark P., Straight A., Kelly K., Takuwa Y., Sugimoto N., Mitchison T., and Bourne H.R.. 2003. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 114:201–214. 10.1016/S0092-8674(03)00555-5 [DOI] [PubMed] [Google Scholar]
- Yamada H., Abe T., Satoh A., Okazaki N., Tago S., Kobayashi K., Yoshida Y., Oda Y., Watanabe M., Tomizawa K., et al. 2013. Stabilization of actin bundles by a dynamin 1/cortactin ring complex is necessary for growth cone filopodia. J. Neurosci. 33:4514–4526. 10.1523/JNEUROSCI.2762-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Zhang X.F., Pollard T.D., and Forscher P.. 2012. Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J. Cell Biol. 197:939–956. 10.1083/jcb.201111052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yuan Z., Zhang Y., Yong S., Salas-Burgos A., Koomen J., Olashaw N., Parsons J.T., Yang X.J., Dent S.R., et al. 2007. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol. Cell. 27:197–213. 10.1016/j.molcel.2007.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Jiang G., Cai Y., Monkley S.J., Critchley D.R., and Sheetz M.P.. 2008. Talin depletion reveals independence of initial cell spreading from integrin activation and traction. Nat. Cell Biol. 10:1062–1068. 10.1038/ncb1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.