Abstract
This Data Descriptor announces the submission to public repositories of the PNNL Biodiversity Library, a large collection of global proteomics data for 112 bacterial and archaeal organisms. The data comprises 35,162 tandem mass spectrometry (MS/MS) datasets from ~10 years of research. All data has been searched, annotated and organized in a consistent manner to promote reuse by the community. Protein identifications were cross-referenced with KEGG functional annotations which allows for pathway oriented investigation. We present the data as a freely available community resource. A variety of data re-use options are described for computational modelling, proteomics assay design and bioengineering. Instrument data and analysis files are available at ProteomeXchange via the MassIVE partner repository under the identifiers PXD001860 and MSV000079053.
Subject terms: Environmental microbiology, Pathogens, Proteome informatics, Proteomics
Background & Summary
Global measurements of -omic molecular data (genome, transcriptome, proteome, metabolome, etc.) are changing the way we research and think about biological systems. Computational biology research, which attempts to identify novel biological phenomena using these large-scale global measurements, depends on publically available data for training and testing new algorithms. Repositories like GEO1 were vital to the development of robust computational methods for analyzing microarray and other genomics technologies. Therefore, depositing complementary proteomics data for a large number of organisms is a similarly valuable public resource.
Researchers at the Pacific Northwest National Laboratory have participated in hundreds of collaborative projects that have involved mass spectrometry-based proteomic analysis of more than 300 species or distinct environmental communities. A portion of this data has been freely available through our website (omics.pnl.gov) for almost a decade, while metadata is maintained by our in-house LIMS systems2. In addition to the numerous project specific publications, meta-analyses of this massive corpus have advanced both computational algorithms3–5 and biological discovery6–8. The size of the library, however, has precluded broad distribution due to a lack of public repositories large enough to host the data. Recently, the ProteomeXchange9 repository system enabled accommodation of significantly larger data volumes.
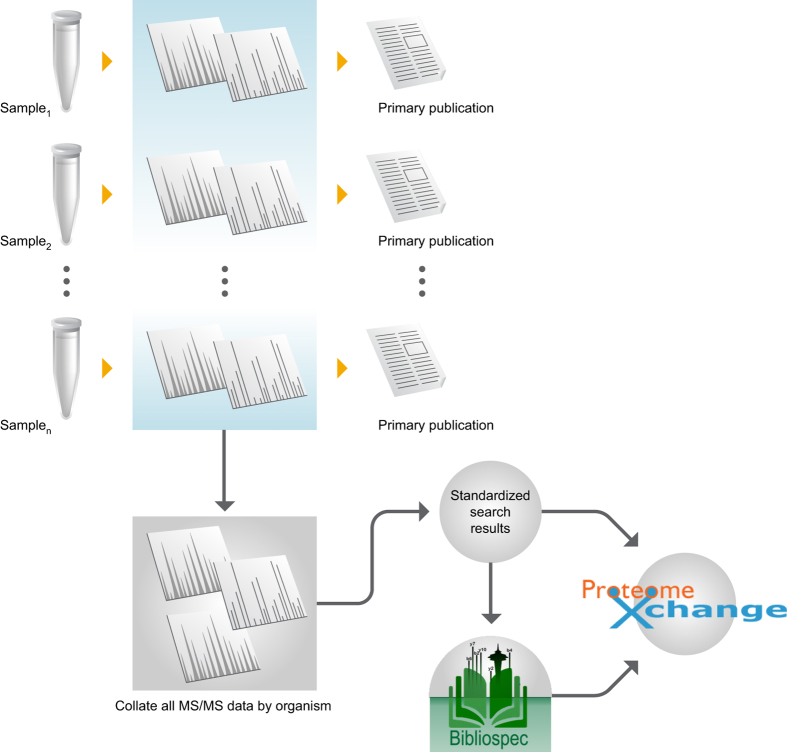
The purpose of this Data Descriptor is to announce the deposition of proteomics data from 112 microbial organisms representing 15 phyla into public 3rd party repositories (Table 1 (available online only)). All the data has been prepared, parsed and organized in a uniform manner to facilitate analysis and reuse (Fig. 1). The combined data deposited is 13 TB (compressed) from 35,162 mass spectrometry files and their associated analysis files. In total, the library contains >70 million spectra identified at q<0.0001, with 3 million peptides from 230,000 proteins. The median number of observed proteins per organism is 2154, or roughly half of the annotated proteins in the proteome. By releasing this data, we hope to promote open science. In this manuscript, we describe a variety of re-uses for mass spectrometry, algorithmic computation and basic biology.
Table 1. Overview of the organisms included in the PNNL Biodiversity Library.
| Organism Name | # datasets | Proteins | Peptides | Spectra | False Spectra |
|---|---|---|---|---|---|
| Each organism is listed along with information about how many datasets, proteins, peptides and spectra were included in the Library creation. The number of false spectra identifications, as estimated by the decoy search strategy, is also included to help understand the quality of the data in the Library. | |||||
| Acidiphilium_cryptum_JF-5 | 40 | 1454 | 7306 | 123,944 | 0 |
| Actinosynnema_mirum_DSM_43827 | 85 | 2083 | 13975 | 80,974 | 0 |
| Anabaena_variabilis | 341 | 3324 | 35814 | 456,600 | 13 |
| Anaeromyxobacter_dehalogenans | 20 | 1250 | 7685 | 54,092 | 0 |
| Anaplasma_phagocytophilium | 129 | 690 | 3066 | 8,363 | 0 |
| Arthrobacter_sp_FB24 | 353 | 3279 | 46122 | 663,861 | 29 |
| Bacillus_anthracis_Ames | 20 | 849 | 6487 | 36,014 | 0 |
| Bacillus_anthracis_Sterne | 158 | 2506 | 26165 | 152,339 | 0 |
| Bacillus_subtilis_168 | 242 | 2435 | 39415 | 709,179 | 0 |
| Bartonella_henselae_Houston-1 | 160 | 1245 | 36347 | 508,803 | 1 |
| Borrelia_burdorferi_B31 | 104 | 877 | 19060 | 193,743 | 1 |
| Brachybacterium_faecium_DSM_4810 | 78 | 2088 | 29248 | 121,914 | 0 |
| Burkholderia_mallei | 189 | 1970 | 33824 | 270,609 | 2 |
| Candidatus_chloracidobacterium_thermophilum | 111 | 563 | 2360 | 6,037 | 7 |
| Caulobacter_crescentus_CB15 | 1016 | 2971 | 63773 | 2,320,212 | 41 |
| Cellulomonas_flavigena_DSM_20109 | 103 | 2382 | 38033 | 175,148 | 0 |
| Cenarchaeum_symbiosum | 108 | 358 | 757 | 4,172 | 0 |
| Chlorobaculum_tepidum_WT | 94 | 1584 | 26561 | 137,150 | 0 |
| Chloroflexus_aurantiacus | 225 | 2736 | 44200 | 323,036 | 16 |
| Clostridium_thermocellum | 408 | 2110 | 61618 | 1,140,251 | 5 |
| Cryptobacterium_curtum_DSM_15641 | 78 | 1115 | 17888 | 75,903 | 0 |
| Cyanobacterium_synechocystis_PCC6803 | 279 | 2251 | 27351 | 344,473 | 0 |
| Cyanothece_sp_ATCC51142 | 1404 | 3309 | 57219 | 1,879,239 | 0 |
| Cyanothece_strain_ATCC51472 | 93 | 2146 | 20702 | 134,879 | 0 |
| Cyanothece_strain_PCC7424 | 109 | 2182 | 20763 | 91,389 | 0 |
| Cyanothece_strain_PCC7425 | 104 | 2590 | 27429 | 114,498 | 0 |
| Cyanothece_strain_PCC7822 | 166 | 3230 | 37493 | 336,052 | 0 |
| Cyanothece_strain_PCC8801 | 90 | 2399 | 23313 | 145,714 | 0 |
| Cyanothece_strain_PCC8802 | 81 | 679 | 3958 | 22,785 | 0 |
| Dehalococcoides_ethenogenes | 437 | 1025 | 25378 | 262,644 | 0 |
| Deinococcus_radiodurans_R1 | 368 | 2212 | 43409 | 1,002,931 | 0 |
| Delta_proteobacterium_NaphS2 | 158 | 2210 | 13007 | 94,378 | 0 |
| Desulfovibrio_desulfuricans_G20 | 463 | 2564 | 48057 | 916,183 | 28 |
| Desulfovibrio_sp_ND132 | 92 | 2573 | 42717 | 225,546 | 0 |
| Desulfovibrio_vulgaris_Hildenborough | 218 | 2397 | 36518 | 451,518 | 0 |
| Dethiosulfovibrio_peptidovorans_DSM_11002 | 78 | 1778 | 25287 | 95,940 | 0 |
| Ehrlichia_chaffeensis | 127 | 532 | 4045 | 12,545 | 0 |
| Enterobacter_cloacae_SCF1 | 58 | 1793 | 19194 | 235,721 | 0 |
| Escherichia_coli_BL21 | 191 | 1564 | 20792 | 190,645 | 0 |
| Escherichia_coli_K-12 | 3925 | 3324 | 122114 | 11,592,291 | 461 |
| Escherichia_coli_RK4353 | 31 | 1766 | 17089 | 53,334 | 0 |
| Fibrobacter_succinogenes_S85 | 122 | 1505 | 17408 | 265,219 | 0 |
| Geobacter_bemidjiensis_Bem_T | 780 | 2566 | 33313 | 682,897 | 0 |
| Geobacter_metallireducens_GS-15 | 155 | 2455 | 27442 | 245,691 | 0 |
| Geobacter_sulfurreducens_PCA | 1122 | 2770 | 45298 | 2,597,915 | 0 |
| Geobacter_uraniumreducens | 287 | 2779 | 42812 | 401,586 | 0 |
| Haloferax_volcanii | 34 | 1463 | 9502 | 56,669 | 0 |
| Halogeometricum_borinquense_DSM_11551 | 83 | 2216 | 18776 | 56,225 | 0 |
| Halorhabdus_utahensis_DSM_12940 | 84 | 2082 | 22987 | 89,820 | 0 |
| Heliobacterium_modesticaldum | 77 | 1549 | 22105 | 107,100 | 0 |
| Kineococcus_radiotolerans_SRS30216 | 192 | 2742 | 38694 | 480,705 | 0 |
| Kosmotoga_olearia_TBF_19-5-1 | 25 | 878 | 7219 | 29,080 | 0 |
| Methanosarcina_barkeri | 89 | 1852 | 24889 | 116,224 | 0 |
| Methanospirillum_hungatei_JF-1 | 78 | 1722 | 23041 | 60,300 | 0 |
| Methylophilales_HTCC2181 | 61 | 1172 | 20615 | 104,603 | 2 |
| Mycobacterium_tuberculosis | 647 | 2714 | 35206 | 1,296,018 | 7 |
| Nakamurella_multipartita_DSM_44233 | 78 | 2171 | 19782 | 57,582 | 0 |
| Nocardiopsis_dassonvillei_DSM_43111 | 80 | 1965 | 16507 | 76,404 | 0 |
| Novosphingobium_aromaticivorans_F199 | 23 | 1563 | 12783 | 75,850 | 1 |
| Opitutaceae_bacterium_TAV2 | 125 | 2229 | 20659 | 292,644 | 0 |
| Pelagibacter_ubique_HTC1062 | 426 | 1201 | 30536 | 1,294,945 | 58 |
| Pelobacter_carbinolicus_DSM_2380 | 22 | 830 | 4168 | 19,078 | 0 |
| Prochlorococcus | 121 | 1285 | 19310 | 158,225 | 0 |
| Pseudomonas_aerunginosa | 40 | 3054 | 24670 | 109,854 | 1 |
| Pseudomonas_fluorescens_PfO-1 | 397 | 3437 | 44819 | 535,883 | 0 |
| Pseudonocardia_sp | 48 | 188 | 448 | 1,406 | 0 |
| Ralstonia_pickettii | 194 | 5,786 | 0 | ||
| Rhodobacter_capsulatus_SB1003 | 495 | 2761 | 58894 | 1,316,844 | 76 |
| Rhodobacter_sphaeroides_2.4.1 | 1639 | 3362 | 72977 | 3,982,596 | 47 |
| Rhodopseudomonas_palustris | 587 | 2743 | 25366 | 247,373 | 0 |
| Roseiflexus_castenholzii | 76 | 2537 | 28414 | 91,063 | 0 |
| Saccharomonospora_viridis_DSM_43017 | 78 | 1918 | 15458 | 39,384 | 0 |
| Salmonella_typhi_TY2 | 499 | 2729 | 50292 | 9,832,722 | 385 |
| Salmonella_typhimurium_ATCC_14028 | 3701 | 3779 | 91496 | 1,555,617 | 28 |
| Salmonella_typhimurium_LT2 | 823 | 3214 | 62399 | 1,051,565 | 3 |
| Sanguibacter_keddieii_DSM_10542 | 78 | 2213 | 20489 | 89,427 | 0 |
| Shewanella_amazonensis_SB2B | 114 | 2079 | 27295 | 469,397 | 32 |
| Shewanella_baltica_OS155 | 631 | 2988 | 47725 | 1,874,615 | 112 |
| Shewanella_baltica_OS185 | 190 | 2853 | 41613 | 395,867 | 4 |
| Shewanella_baltica_OS195 | 65 | 2193 | 23560 | 175,237 | 5 |
| Shewanella_baltica_OS223 | 75 | 2158 | 22391 | 92,424 | 0 |
| Shewanella_denitrificans_OS217 | 91 | 2313 | 25045 | 240,069 | 0 |
| Shewanella_frigidimarina_NCIMB_400 | 66 | 2286 | 26819 | 192,758 | 7 |
| Shewanella_loihica_PV-4 | 68 | 2082 | 27260 | 181,814 | 5 |
| Shewanella_oneidensis_MR-1 | 3954 | 3307 | 94195 | 5,561,459 | 442 |
| Shewanella_putrefaciens_200 | 64 | 2200 | 25524 | 191,946 | 2 |
| Shewanella_putrefaciens_CN-32 | 78 | 2338 | 29135 | 254,187 | 14 |
| Shewanella_putrefaciens_W3-18-1 | 80 | 1975 | 18900 | 273,705 | 12 |
| Shewanella_sp_ANA-3 | 62 | 2154 | 25870 | 145,474 | 4 |
| Shewanella_sp_MR-4 | 65 | 2052 | 25724 | 195,547 | 6 |
| Shewanella_sp_MR-7 | 64 | 2217 | 29196 | 178,776 | 16 |
| Sinorhizobium_medicae | 40 | 2557 | 17954 | 164,088 | 2 |
| Sinorhizobium_meliloti_1021 | 137 | 2547 | 21631 | 581,060 | 2 |
| Slackia_heliotrinireducens_DSM_20476 | 79 | 1645 | 22440 | 105,667 | 1 |
| Stackebrandtia_nassauensis_DSM_44728 | 104 | 2158 | 22213 | 138,735 | 0 |
| Sulfolobus_acidocaldarius_DSM_639 | 33 | 1380 | 16093 | 92,402 | 0 |
| Synechococcus_sp_PCC7002 | 1050 | 2606 | 52469 | 1,842,452 | 1 |
| Syntrophobacter_fumaroxidans | 80 | 1893 | 18846 | 71,287 | 0 |
| Thermobispora_bispora_DSM_43833 | 78 | 1470 | 9193 | 23,861 | 0 |
| Thermosynechococcus_elongatus_BP-1 | 137 | 1266 | 8161 | 182,698 | 0 |
| Thermosynechococcus_sp_NAK55 | 47 | 1106 | 9587 | 87,550 | 0 |
| Thermotoga_maritima | 281 | 1526 | 52050 | 544,795 | 6 |
| Thiocapsa_marina_DSM_5653T | 130 | 1632 | 8577 | 26,649 | 0 |
| Verrucomicrobium_sp_TAV1 | 32 | 2288 | 15059 | 79,896 | 0 |
| Verrucomicrobium_sp_TAV5 | 33 | 2116 | 12893 | 87,443 | 2 |
| Xylanimonas_cellulosilytica_DSM_15894 | 200 | 2487 | 41090 | 319,030 | 0 |
| Yersinia_enterocolitica | 86 | 1622 | 11983 | 107,731 | 2 |
| Yersinia_pestis_CO92 | 210 | 2302 | 33289 | 993,714 | 19 |
| Yersinia_pestis_KIM | 240 | 1371 | 11084 | 98,780 | 0 |
| Yersinia_pestis_Pestoides_F | 99 | 2153 | 22980 | 523,779 | 16 |
| Yersinia_pseudotuberculosis_IP_32953 | 400 | 2151 | 24690 | 301,175 | 11 |
| Yersinia_pseudotuberculosis_PB1_Plus | 99 | 2029 | 20729 | 470,575 | 16 |
| 35162 | 229,537 | 3,147,576 | 70,455,991 | 1951 |
Figure 1. Workflow for library creation.
Biological samples are used to create MS/MS data as part of an experiment and primary publication. All of this data is stored on our servers for re-analysis. Historical data was collated by organism and researched for release in the Biodiversity Library.
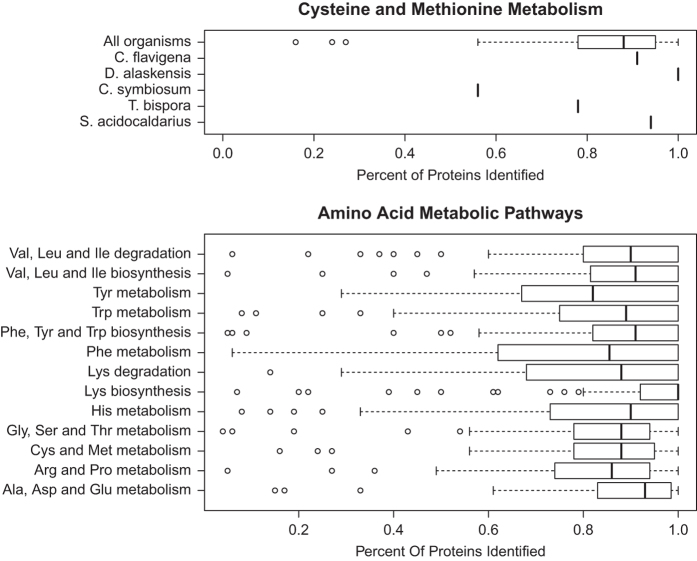
As part of the analysis, we have cross referenced protein identifications to KEGG functional annotation where possible. Nine of the 112 organisms are not processed by KEGG, and therefore were excluded from this additional analysis. When viewing the Library as a whole, annotated biological pathways are broadly covered by the identified proteins. For example, the reference ‘cysteine and methionine metabolism pathway’ as defined by KEGG consists of 81 orthologous genes participating in 73 reactions. As expected, not all orthologs are annotated in every genome, e.g., Cellulomonas flavigena has only 23 of the 81 genes. By searching all MS/MS data with standard RefSeq databases, we can easily identify that 21 of the 23 Cellulomonas genes were observed in MS/MS data, or 91%. When considering all organisms in the Library, the median coverage of the cysteine and methionine metabolism pathway is 89%. A summary of the coverage of every KEGG pathway for each organism is presented in Supplementary Table 1. Using KEGG pathway categories, we determined the median coverage of all functionally classified proteins (Fig. 2). For example, in all 13 pathways for amino acid metabolism, the median coverage across the entire library is 89%. This high coverage is seen for most KEGG pathway categories: 82% for lipid metabolism, 83% for vitamin and cofactor metabolism, etc.
Figure 2. Kegg pathway coverage.
(top) Using the pathway classifications provided by KEGG, we can determine how many annotated proteins were identified in mass spectrometry data. The cysteine and methionine metabolism pathway is provided as an example. For each organism, we calculate the percentage of identified proteins. C. flavigena has 23 proteins in the pathway, 21 of which were observed (91%). The box plot shows average coverage of the 103 organisms that KEGG has annotated. Circles depict outliers. (bottom) Pathway coverage for all 13 amino acid metabolic pathways is shown.
Methods
As the library encompasses 35,162 mass spectrometry files from 10+ years of research, it is impossible to fully describe the evolving and diverse protocols for experimental sample preparation or data acquisition. In Supplementary Table 2, we have provided data from our LIMS system2 about each sample data file (called a dataset). Below is a set of descriptions that represent a large fraction of the methods applied to generate the released datasets.
Either an established or optimized protein extraction protocol was applied to each sample7. In brief, a typical experimental approach included global (total), insoluble, and soluble protein extractions from lysed cell cultures that were then washed and suspended in 100 mM NH4HCO3, pH 8.4 buffer.
Global protein extracts were denatured and reduced by adding urea, thiourea, and dithiothreitol (DTT) followed by incubation at ~60 °C for ~30 min. Following incubation, the global protein samples were diluted to reduce salt concentration and then proteolytic digested, at 37 °C for ~4 h, using sequencing grade trypsin (Roche, Indianapolis, IN) at a ratio of 1 unit per 50 units of protein (1 unit=~1 μg of protein). Following incubation, digested samples were desalted using an appropriately sized C-18 SPE column (Supelco, St Louis, MO) and a vacuum manifold. The collected peptides were concentrated to a final volume ranging from 50 to 100 μl and measured using the BCA assay (Pierce Chemical Co., Rockfort, IL) according to the manufacturer's instructions.
Insoluble protein extracts were produced by ultracentrifuging the cell lysate at 4 °C and 100,000 rpm for 10 min. The resulting supernatant that contained soluble proteins was separated from the pellet and retained for digestion as previously described for the global extraction. The pellet was washed by suspending it in 100 mM NH4HCO3, pH 7.8, using mild sonication and then ultracentrifuged at 100,000 rpm for 5 min, again at 4 °C. Following centrifugation, the pellet was resuspended in a solubilizing solution that contained urea, thiourea, 1% CHAPS in 50 mM NH4HCO3, pH 7.8. An aliquot of 50 mM DTT solution was also added to final concentration of 5 mM. The insoluble protein sample was then incubated and digested as described above with the exception that a 50 mM NH4HCO3, pH 7.8 buffer was used for the dilution step. Following proteolytic digestion, the pH of the sample was slowly lowered to <4.0 by adding small volumes (1 to 2 μl) of 20% formic acid. Removal of salts and detergent was performed using either an appropriately sized strong cation exchange (SCX) or solid phase extraction column (Supelco, St Louis, MO) and vacuum manifold. Peptides were then concentrated and their concentration measured as described above.
The HPLCs used to run the samples were built in-house utilizing various commercial pumps, valves, and auto samplers, all of which were coordinated by a custom software package called LCMSnet. The data sets analyzed for this paper were run using LC columns that were 75 μm inner diameter, and either 30 or 65 cm in length. These LC columns were packed in house with Phenomenex Jupiter C18 3 μm porous beads. The flow rate was 300 nl/min. Mobile phase A is 0.1% formic acid in water and mobile phase B is 0.1% formic acid in acetonitrile. The 100 min gradient was delivered by starting at 5% mobile phase B and advancing to 8, 12, 35, 60, and 75% at times (in minutes) 2, 20, 75, 97, 100 respectively. Typically 2.5 μg of peptides were loaded to the head of the column or to a trapping column. Although operating conditions varied by capabilities of each instrument, typical conditions for each are as follows. The LTQ was run in data-dependent MS-MS mode, selecting the top 10 parent ions from each survey scan. The LTQ-Orbitrap and the Velos-Orbitrap instruments were typically set to have a high resolution survey scan of 60,000 resolution followed by the top 6 or 10 data-dependent MS-MS scans, respectively. Because of the diversity of data sets presented in this work, this is not a comprehensive list of conditions. Instrumentation details can be found in the raw data files (.RAW or.mzML).
Code availability
Software used in the generation of this project is largely third party software as described in the Data Records section, i.e., MSGF+ and Bibliospec. The only remaining software was to link protein identifications to KEGG functional assignments. This was done via custom parsing of the files and cross-referencing the KEGG database. This code is trivially reproducible.
Data Records
To maximize the utility and ease of access, the data described in this publication have been uploaded to the ProteomeXchange9 with accession PXD001860 via MassIVE (Data Citation 1). On MassIVE (identifier MSV000079053), each organism’s data is located in a separate folder, with both raw and processed data as described below. Data is organized around a tandem mass spectrometry file that represents one run of the instrument on a biological sample. In our terminology this is called a dataset. Each dataset has the following associated files.
Mass spectrometry data
Each dataset is available in the original vendor format and the community standard open format mzML10. These files contain the raw mass spectra. Mass spectrometry data is a combination of MS and MS/MS data showing both the detection of all analytes at a particular time in chromatography (MS data) and the fragmentation of a particular analyte (MS/MS data). See the review by Aebersold and Mann for a basic primer of proteomic mass spectrometry data11.
Peptide identifications
Each dataset is associated with a file describing the peptides that were identified via the spectra. This file was created using the MSGF+ algorithm12 version v9979. All 35,162 datasets were analyzed with a consistent set of parameters. Searches included oxidized methionine as an optional post-translational modification, and specified partial trypsin specificity. For experiments that utilized iodoacetimide as an alkylation agent, the static modification (C+57) was also added. Precursor and fragment mass tolerances were set according to the resolving power of the mass analyzer. The output of MSGF+ is stored in the community standard mzIdentML format13, which describes the peptide/spectrum match (PSM), search parameters and scoring details.
The one caveat for peptide identification was that three organisms did not have a RefSeq proteome set derived from a publically available genome sequence. Escherichia coli RK4353 did not have a sequence genome at NCBI, so we used the relative BW2952 strain. Cyanothece strain ATCC51472 also lacks a sequence at NCBI; we substituted strain 8801. Thiocapsa marina DSM_5653T lacks a RefSeq genome; the Genbank submission was used instead.
Metadata
Data acquired at PNNL has been tracked using an in-house LIMS system since 2000. Each dataset is recorded with a variety of details including: acquisition date and time, instrument, chromatography details, organism, etc. These metadata are presented in Supplementary Table 2 with this publication.
Spectrum library
A spectrum library is a condensed collection of annotated tandem mass spectra. In addition to serving as an efficient storage format for very large datasets, these libraries are also utilized for annotating new datasets14,15. With this deposition, we created a spectrum library for each microbial organism using Bibliospec16. Peptide/spectrum matches were filtered for high quality matches (MSGF+’s q-value <0.0001). When viewed in aggregate, the 112 organisms had 70,455,991 spectra passing this cutoff (with 1951 false hits and an estimated FDR of 2e-5). This strict filtering is necessary to control false-positives when creating very large libraries. The libraries, stored as.blib files, are also available on the MassIVE repository.
Technical Validation
When releasing the Library, we took a conservative stance on spectral quality. Considering the large number of spectra, even a 1% false-positive rate would mean polluting the resource with nearly one million false-positive spectral identifications. Moreover, a well-known problem in proteomics is that aggregating numerous datasets leads to the inflation of false-positives when considering the entire group. This is especially true when rolling results up to a peptide or protein level as many true spectra are associated with a single true protein, whereas false proteins are typically represented by very few false-positive spectra identifications. The primary method to reduce false-positive peptide and protein identifications is to be more stringent on spectrum quality.
When aggregating 35,162 datasets into the Library, using a typical qvalue cutoff of 0.01 on each individual dataset was insufficient to ensure high quality of the library as a whole (Fig. 3). Although the spectral false discovery was indeed 1%, the protein level false discovery was an astonishing 37%. We applied a qvalue cutoff of 0.0001, or two orders of magnitude more stringent than common practice. In this filtering process, 23 million true-positive spectra are removed. Although this may seem overly conservative, the more stringent filter also removed 600,000 false-positive peptides and 200,000 false-positive protein identifications. This allowed for a permissible false-discovery rate at spectrum, peptide, and protein levels (0.00002, 0.00009 and 0.001 respectively).
Figure 3. False Discovery Rate.
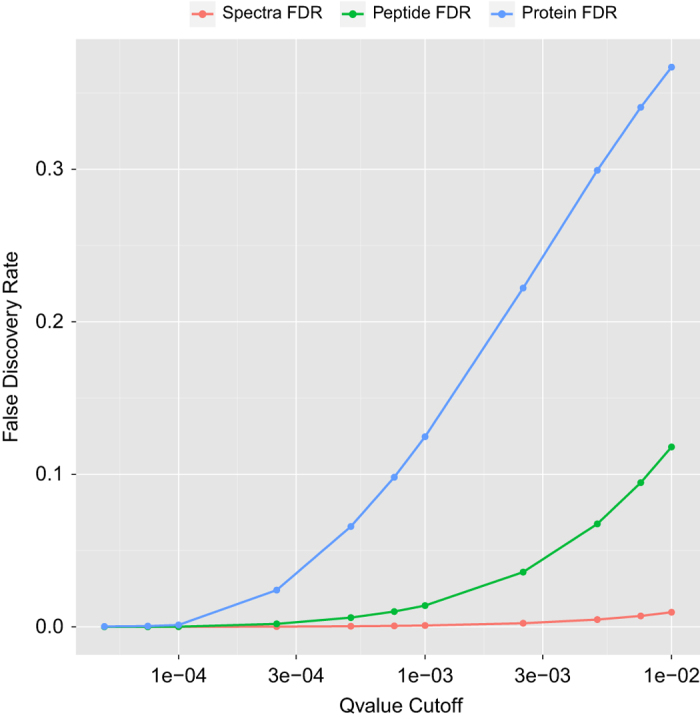
Due the large nature of the Library, the false-discovery rate of the aggregated data can inflate significantly, especially when rolled up to protein and peptide level. Data is shown for the FDR of the entire Library when using a specified qvalue cutoff of PSMs from the MSGF+ results. When using a loose PSM filter of qvalue<0.01, the protein and peptide FDR rates are unacceptably high. We choose the cutoff qvalue<0.0001, which produces high data quality at spectra, peptide and protein levels.
Usage Notes
Our purpose in depositing such a large corpus of data is to promote reuse and open science. The richness of the PNNL Biodiversity Library is seen in both the breadth and depth of coverage for proteins and phylogeny. Besides sheer size, a unique feature of the Library is the pairs of spectra that come from similar peptides; one million peptides in the Library are one mutation away from another peptide (edit distance=1). These pairs originate from orthologues, where the proteins share significant sequence identity (Fig. 4). Indeed, 21,721 peptides have four or more ‘one mutation’ neighbours. This vast web of sequence related spectra can be productively mined for a wide variety of bioinformatics and fundamental mass spectrometry research.
Figure 4. Peptide observation across taxa.

This is a multiple sequence alignment of a section of an ABC transporter (accessions and organism given), with observed peptides from the PNNL Biodiversity Library in blue. For simplicity, we displayed sequences from the Actinobacteria phylum, with Roseiflexus as an out group. The right side of the alignment shows consistent discovery in proteomics data across the phylum and in the out group. The left side of the alignment is only observed in the proteomics data for Arthrobacter sp. FB24.
Ion fragmentation
Exploring the fundamentals of fragmentation is typically done working with purified peptides in low throughput17,18. With the Biodiversity library, however, pairs of related spectra could easily be mined to understand the effect of residue changes on the intensity of fragment ions. For example, there are 2,854 peptides where sequences only differ in that an alanine residue is changed to a serine residue. Additionally, many peptides are repeatedly identified. Indeed 53,828 peptides have over 200 spectra. Replicate spectra for a peptide are often used in understanding and modelling fragmentation patterns. However, in the library we note that 30,672 peptides with over 200 spectra are from conserved regions of proteins found in multiple organisms. Thus they contain distinct background and noise in the MS/MS spectra, aiding in the identification of novel fragment peaks.
Proteotypic peptides
Computational prediction of which peptides are discoverable in experimental conditions is a valuable tool in proteomics workflows19. Such machine learning efforts will undoubtedly improve with the 3 million peptides provided by the PNNL Biodiversity Library. Yet the related sequences mentioned above provide a truly distinct perspective on peptide observability. Several important features of orthology can be utilized to improve the quality of machine learning predictions. First, as seen in Fig. 4, there are regions of a protein sequence which are fundamentally observable. In many orthologs spanning a large phylogeny, these regions are consistently observed. The sequence variation present in these regions can be leveraged to identify the physiochemical factors that govern mass spectrometry identification. Also seen are regions that are rarely observed. These could provide valuable negative training data for machine learning approaches.
Library search of MS/MS data
Spectrum annotation via library search is both faster and more sensitive than database search algorithms20. Due to a lack of data, library search has previously not been practical except for the most commonly used model systems (e.g., human and yeast). Since the Biodiversity Library contains data for nearly every model system, including numerous environmentally and medically relevant microbes, peptide identification via spectrum library matching becomes an attractive alternative to database searching.
Novel scoring functions
Bioinformatics algorithms to identify peptides from mass spectrometry data are constantly being developed and refined. For these, having free access to a large pool of training data is essential12,21–23. With data presented on different classes of instruments and multiple fragmentation modalities, the PNNL Biodiversity Library is an ideal source of data to test new scoring functions.
Unidentified spectra
Another application that we envision is the investigation of unidentified or unattributed spectra. With tens of thousands of LC-MS/MS data sets, there are literally hundreds of millions of fragmentation spectra for which there is not a confident identification using the current search tool and parameters. Of those unidentified species, many are fragmented in multiple data sets; spectrum averaging or other methods could be utilized to obtain a confident identification.
Novel post-translational modifications
For simplicity and sensitivity, only the most common post-translational modification (oxidized methionine) was included in the database search parameters. However, numerous post-translational modifications are observable in proteomics mass spectrometry24. Some modifications are rare, and therefore not commonly included in database searches. We recently uncovered a novel PTM switch in Salmonella for S-thiolation25 and believe that many such unexpected post-translational modifications exist. Identifying which observed PTMs are functionally relevant is a difficult task, but observing it across different taxa and showing evolutionary conservation provides a valuable filter for high-priority targets26,27.
Proteogenomics
The process of using peptides from mass spectrometry to assist genome annotation, or proteogenomics, has been very successful in identifying both false-negative omissions in a genome’s protein list, and also false-positives. To date most of the work in this area has been focused on a single genome, or a group of closely related genomes4,28,29. With the Biodiversity Library, one can now attempt to leverage identifications across an entire phylum, or perhaps the entire tree of life.
Additional Information
How to cite this article: Payne, S. H. et al. The Pacific Northwest National Laboratory library of bacterial and archaeal proteomic biodiversity. Sci. Data 2:150041 doi: 10.1038/sdata.2015.41 (2015).
Supplementary Material
Acknowledgments
We gratefully acknowledge the productive collaborations and teams that have generated the data used. Acknowledgements include, but are not limited to, those named here: Lucy Shapiro, Himadri Pakrasi, Bernhard Palsson, Stephen Giovannoni, Kim Lewis, Fred Heffron, Vladimir Motin, Tim Donohue, Andrew Lang, Derek Lovely, Carrie Harwood, Jorge Rodrigues, Donald Bryant, Ann Summers, Robert Landick, Mike Daly, John Battista, Jim Fredrickson, and Margie Romine. SHP was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Early Career Research Program. Portions of this research were supported by the Department of Energy Office of Biological and Environmental Research Genome Sciences Program under the Pan-omics Program, and by the NIH National Institute of General Medical Sciences (GM103493), National Institute of Allergy and Infectious Diseases (Y1-AI-8401), the Laboratory Directed Research and Development Program (LDRD) at Pacific Northwest National Laboratory (PNNL). Work was performed in the Environmental Molecular Science Laboratory, a U.S. Department of Energy (DOE) national scientific user facility at Pacific Northwest National Laboratory (PNNL) in Richland, WA. Battelle operates PNNL for the DOE under contract DE-AC05-76RLO01830.
Footnotes
The authors declare no competing financial interests.
Data Citations
- Payne S. H. 2015. ProteomeXchange. PXD001860
References
- Edgar R., Domrachev M. & Lash A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebel G. R. et al. PRISM: a data management system for high-throughput proteomics. Proteomics 6, 1783–1790 (2006). [DOI] [PubMed] [Google Scholar]
- Frank A. M. et al. Spectral archives: extending spectral libraries to analyze both identified and unidentified spectra. Nature methods 8, 587–591 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N. et al. Comparative proteogenomics: combining mass spectrometry and comparative genomics to analyze multiple genomes. Genome research 18, 1133–1142 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb-Robertson B. J. et al. A support vector machine model for the prediction of proteotypic peptides for accurate mass and time proteomics. Bioinformatics (Oxford, England) 26, 1677–1683 (2010). [DOI] [PubMed] [Google Scholar]
- Venter E., Smith R. D. & Payne S. H. Proteogenomic analysis of bacteria and archaea: a 46 organism case study. PloS ONE 6, e27587 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callister S. J. et al. Comparative bacterial proteomics: analysis of the core genome concept. PloS ONE 3, e1542 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J., Gupta N., Smith R. D. & Pevzner P. A. Does trypsin cut before proline? Journal of proteome research 7, 300–305 (2008). [DOI] [PubMed] [Google Scholar]
- Vizcaino J. A. et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic acids research 41, D1063–1069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens L. et al. mzML--a community standard for mass spectrometry data. Molecular & cellular proteomics: MCP 10, R110.000133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebersold R. & Mann M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003). [DOI] [PubMed] [Google Scholar]
- Kim S. et al. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Molecular & cellular proteomics: MCP 9, 2840–2852 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. R. et al. The mzIdentML data standard for mass spectrometry-based proteomics results. Molecular & cellular proteomics: MCP 11, M111.014381 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S. E. Estimating probabilities of correct identification from results of mass spectral library searches. Journal of the American Society for Mass Spectrometry 5, 316–323 (1994). [DOI] [PubMed] [Google Scholar]
- Lam H. Building and searching tandem mass spectral libraries for peptide identification. Molecular & cellular proteomics: MCP 10, R111.008565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frewen B. & MacCoss M. J. Using BiblioSpec for creating and searching tandem MS peptide libraries. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis... [et al.] Chapter 13, Unit 13.17 (2007). [DOI] [PubMed] [Google Scholar]
- Dongré A. R., Jones J. L., Somogyi Á. & Wysocki V. H. Influence of Peptide Composition, Gas-Phase Basicity, and Chemical Modification on Fragmentation Efficiency: Evidence for the Mobile Proton Model. Journal of the American Chemical Society 118, 8365–8374 (1996). [Google Scholar]
- Savitski M. M., Falth M., Fung Y. M., Adams C. M. & Zubarev R. A. Bifurcating fragmentation behavior of gas-phase tryptic peptide dications in collisional activation. Journal of the American Society for Mass Spectrometry 19, 1755–1763 (2008). [DOI] [PubMed] [Google Scholar]
- Mallick P. et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nature biotechnology 25, 125–131 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang X., Li Y., Shao W. & Lam H. Understanding the improved sensitivity of spectral library searching over sequence database searching in proteomics data analysis. Proteomics 11, 1075–1085 (2011). [DOI] [PubMed] [Google Scholar]
- Geer L. Y. et al. Open mass spectrometry search algorithm. Journal of proteome research 3, 958–964 (2004). [DOI] [PubMed] [Google Scholar]
- Tabb D. L., Fernando C. G. & Chambers M. C. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. Journal of proteome research 6, 654–661 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne S. H. et al. Phosphorylation-specific MS/MS scoring for rapid and accurate phosphoproteome analysis. Journal of proteome research 7, 3373–3381 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witze E. S., Old W. M., Resing K. A. & Ahn N. G. Mapping protein post-translational modifications with mass spectrometry. Nature methods 4, 798–806 (2007). [DOI] [PubMed] [Google Scholar]
- Ansong C. et al. Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proceedings of the National Academy of Sciences of the United States of America 110, 10153–10158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner S. et al. Accurate annotation of peptide modifications through unrestrictive database search. Journal of proteome research 7, 170–181 (2008). [DOI] [PubMed] [Google Scholar]
- Macek B. et al. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Molecular & cellular proteomics 7, 299–307 (2008). [DOI] [PubMed] [Google Scholar]
- Payne S. H., Huang S. T. & Pieper R. A proteogenomic update to Yersinia: enhancing genome annotation. BMC genomics 11, 460 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallien S. et al. Ortho-proteogenomics: multiple proteomes investigation through orthology and a new MS-based protocol. Genome research 19, 128–135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Payne S. H. 2015. ProteomeXchange. PXD001860