Abstract
The mechanisms that regulate skeletal muscle differentiation, fiber type diversity and muscle regeneration are incompletely defined. Forkhead transcription factors are critical regulators of cellular fate determination, proliferation, and differentiation. We identified a forkhead/winged helix transcription factor, Foxj3, which was expressed in embryonic and adult skeletal muscle. To define the functional role of Foxj3, we examined Foxj3 mutant mice. Foxj3 mutant mice are viable but have significantly fewer Type I slow-twitch myofibers and have impaired skeletal muscle contractile function compared to their wild type controls. In response to a severe injury, Foxj3 mutant mice have impaired muscle regeneration. Foxj3 mutant myogenic progenitor cells have perturbed cell cycle kinetics and decreased expression of Mef2c. Examination of the skeletal muscle 5′ upstream enhancer of the Mef2c gene revealed an evolutionary conserved forkhead binding site (FBS). Transcriptional assays in C2C12 myoblasts revealed that Foxj3 transcriptionally activates the Mef2c gene in a dose dependent fashion and binds to the conserved FBS. Together, these studies support the hypothesis that Foxj3 is an important regulator of myofiber identity and muscle regeneration through the transcriptional activation of the Mef2c gene.
Keywords: forkhead, Foxj3, Myocyte enhancer factor, Mef2c, Muscle fiber type, Muscle regeneration, Myogenesis, Gene disruption technologies
Introduction
The molecular networks that govern muscle differentiation, fiber diversity and muscle regeneration remain incompletely defined. Previous studies have uncovered an essential role for the basic helix loop helix (bHLH) MyoD family members (i.e., MyoD, myf5, MRF4 and myogenin) and myocyte enhancer factor 2 (Mef2) factors, but the repertoire of interacting factors and upstream regulators that coordinately regulate skeletal muscle differentiation and fiber type specification remain an area of intense interest. Studies have demonstrated that the bHLH MyoD factors interact with Mef2 factors to synergistically coactivate the myogenic program. Global elimination of Mef2c results in early embryonic lethality due to cardiovascular defects (Lin et al., 1997). Conditional elimination of Mef2c in skeletal muscle revealed a role in fiber type specification as there was a significant decrease in Type I oxidative slow myofibers (Potthoff et al., 2007). While efforts have focused on downstream targets of Mef2c, few direct upstream regulators of Mef2c have been identified in the skeletal muscle lineage.
Forkhead/winged helix transcription factors perform an array of functions including fate specification, pattern formation, cellular proliferation, cellular differentiation and organogenesis (Lehmann et al., 2003). Members of the forkhead/winged helix transcription factors function through a DNA-binding dependent mechanism or alternatively through protein–protein interactions to regulate gene expression (Wijchers et al., 2006). For example, the forkhead/winged helix factor, Foxk1, is expressed in myogenic progenitor cells and functions as a cell cycle regulator by regulating the cyclindependent kinase inhibitor, p21KIP(Garry et al., 2000). Mice lacking Foxk1 have perturbed skeletal muscle regeneration due to impaired activation/proliferation of the myogenic progenitor cell population (Hawke et al., 2003a). In addition to Foxk1, other forkhead factors including FoxO factors modulate cell signaling pathways, growth and atrophy of adult skeletal muscle (Sandri et al., 2004). These studies suggest that forkhead factors have critical roles in the regulation of skeletal muscle development and regeneration.
To further identify forkhead factors expressed in the myogenic lineages, we undertook a candidate based screen. Using this strategy, we identified Foxj3 as being significantly upregulated in differentiating myoblasts. To further explore the functional role of Foxj3 in vivo, we obtained a Foxj3 gene-targeted ES cell line and generated mutant mice that have a β-geo cassette flanked by two splice acceptors inserted into the Foxj3 locus. The resulting mutant Foxj3 allele produces a transcript that encodes exons 1–5 (1–176 amino acids) of the Foxj3 protein that lacks a transcriptional activation domain. Mice homozygous for both the mutant alleles (referred to as Foxj3m/m) are viable, but have impaired skeletal muscle contractility and decreased Type I oxidative myofibers compared to their wild type controls. In addition, Foxj3m/m mice have impaired skeletal muscle regeneration following injury, impaired cell cycle kinetics of the myogenic progenitor cell population and decreased expression of Mef2c. Examination of the 5′ upstream skeletal muscle enhancer of Mef2c revealed a highly conserved forkhead binding site (FBS). Transcriptional assays in C2C12 myoblasts demonstrated that Foxj3 activates a Mef2c-luciferase reporter and mutagenesis of the FBS in the Mef2c skeletal muscle enhancer ablates this transcriptional activation. Together these studies reveal that Foxj3 is a transcriptional activator of Mef2c, and is an important regulator for adult muscle fiber type identity and skeletal muscle regeneration.
Results
We have utilized an array of techniques to define the expression of forkhead family members in the skeletal muscle lineage during development and regeneration. To complement these studies, we utilized a C2C12 myoblast differentiation assay to analyze gene expression during discrete phases of myogenesis (Shi and Garry, 2006). We identified a novel member of the forkhead/winged helix family, Foxj3, which was dynamically expressed during C2C12 myogenic differentiation using semi-quantitative RT-PCR (Fig. 1 and Supplemental Figure S1). This differentiation assay revealed that Foxj3 and Mef2c preceded myoglobin expression during myogenesis. These studies support the hypothesis that Foxj3 is expressed and regulated during myogenesis.
Fig. 1.
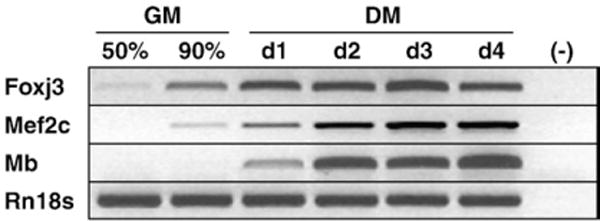
Foxj3 is expressed in C2C12 myoblasts. Semiquantitative RT-PCR analysis of transcript expression during C2C12 differentiation from myoblasts (50% or 90% confluency, cultured in growth medium or GM) to myotubes (day 1 or d1 to day 4 or d4, cultured in differentiation medium or DM) demonstrates that Foxj3 is expressed in myoblasts prior to myoglobin (Mb) expression. Rn18s was used as a loading control. (−) represents a negative control (lacking reverse transcriptase).
Generation of Foxj3m/m mice
To define the functional role of Foxj3, we generated Foxj3 mutant mice (referred to as Foxj3m/m) from a BayGenomics© ES cell “trapped” line (Stryke et al., 2003). The Foxj3 trapped ES cell line contained an insertion of a β-galactosidase/neomyocin (β-geo) cassette with two separate splice acceptors (Fig. 2A). The resulting mRNA predicted transcript encodes exons 1 through 5 and contains all but the last six residues of the forkhead DNA-binding domain. No transcripts were detected following exon 6 of the native Foxj3 gene (Figs. 2A–C). Previous studies using Gal4 transcriptional assays demonstrated that the carboxy-terminus harbors the activation domain of Foxj proteins and is essential for Foxj family of proteins functional activity (Pérez-Sánchez et al., 2000). Thus, we hypothesized that the fusion protein encoded from Foxj3m/m mice would result in a nonfunctional, transcriptionally inactive fusion protein. Additionally, we predicted that the Foxj3-β-gal fusion protein would recapitulate Foxj3 endogenous expression patterns.
Fig. 2.
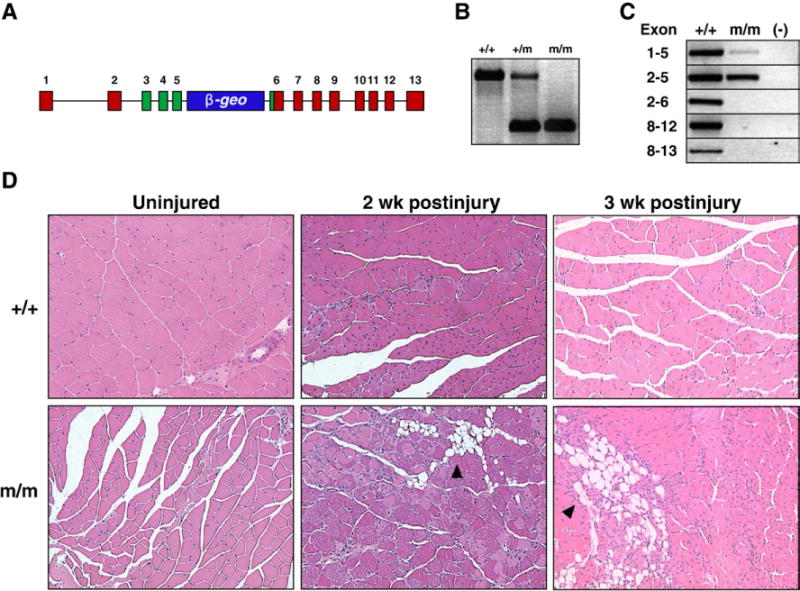
Foxj3m/m mice have impaired skeletal muscle regeneration. (A) Schematic outlying the Foxj3-β-geo “gene trapped” locus. (B) PCR genotyping of tails from Foxj3 mice. (C) The Foxj3 mutant transcript in Foxj3m/m gastrocnemius cDNA does not encode beyond exon 5 using RT-PCR techniques. Note that the transcript is amplified up to exon 5 in the Foxj3m/m muscle; however, only the wild type transcript can be amplified from exon 6 and beyond. (D) Histological sections of WT (+/+) and Foxj3m/m gastrocnemius skeletal muscle uninjured, 2 weeks, and 3 weeks post-cardiotoxin injury from 3-month-old male littermates. Note that the wild type skeletal muscle is fully regenerated with restoration of skeletal muscle architecture within two weeks (note central nuclei representing regenerated myofibers). Conversely, theFoxj3m/m cardiotoxin injured skeletal muscle fails to fully regenerate up to 3 weeks postinjury. Black arrowheads mark areas of necrosis and adipogenesis that form and replace myofibers following injury in the Foxj3m/m mice.
Foxj3 mutant mice are viable
Mice homozygous for the Foxj3-β-geo targeted allele were generated from matings of heterozygote mice. Initially, our studies utilized the C57/B6J:129OlaP2Hsd mixed strain; however, we have backcrossed the original chimeric mice with inbred C57/B6J and 129OlaP2Hsd strains separately and over six generations and we observed no strain variances. Mice homozygous with the Foxj3 mutant alleles, generated from heterozygote matings, were viable and born at normal Mendelian ratios (Supplemental Figure S2). Semiquantitative RT-PCR of gastrocnemius skeletal muscle from wildtype and homozygous Foxj3 mutant mice revealed that expression of the Foxj3 mRNA transcript was as predicted limited to exons 1–5, and that the carboxy-terminal activation domain in exons 8–12 were not transcribed (Fig. 2C).
Foxj3m/m mice have impaired skeletal muscle regeneration following injury
As previously described (Landgren and Carlsson, 2004), Foxj3 was expressed in the myogenic lineage during embyogenesis (Supplemental Figure S3) and in the in vitro myogenic differentiation assay (Fig. 1 and Supplemental Figure S1). We then examined the ability of Foxj3m/m mice to regenerate their skeletal muscle following a severe myonecrotic injury which destroys approximately 90% of the adult skeletal muscle. In response to this cardiotoxin-induced injury, wild type skeletal muscle regenerates within a 2-week period and has restoration of its architecture. In contrast, Foxj3 mutant mice had a severe regenerative impairment that was evident at 2 and 3 weeks following delivery of cardiotoxin. We observed persistent myonecrosis and widespread replacement of the myofibers with adipocytes (Fig. 2D). Using electron microscopy, we observed the presence of myogenic progenitor cells (i.e., satellite cells) in the unperturbed Foxj3m/m skeletal muscle and there were no significant differences in their number compared to the gender and age matched control mice (Supplemental Figure S4 and data not shown).
Foxj3 mutant myogenic progenitor cells have an increased proliferative capacity
To determine the proliferative capacity of the mutant and wild type myogenic progenitor cell population, we performed cell cycle kinetics assay and an in situ proliferation assay. We isolated myogenic progenitor cells from wild type and mutant skeletal muscle and analyzed the cell cycle kinetics in asynchronized proliferating cells. The mutant myogenic progenitor cells had a significant decrease in the G0/G1 phase and a significant increase in the G2/M phase as assayed by flow cytometry (p<0.05; n=3 littermate pairs) (Figs. 3A and B). These cell cycle results were further supported using the proliferation marker Ki67 (which labels all proliferating cells) in regenerating Foxj3 mutant vs. control skeletal muscle performed 5 days following cardiotoxin induced injury. These results demonstrate an increased proliferative capacity, as measured by Ki67 expression (Fig. 3C) that represented a 65% increase compared to the wild type control (p<0.05; n=3) (Fig. 3D). Additionally, most of the Ki67-positive cells were also positive for MyoD expression, which supports the notion that the impaired regenerative defect may also be due in part to increased proliferation of the mutant myogenic progenitor cells (Fig. 3E). Importantly, we observed no significant differences in Ki67 expression in unperturbed Foxj3 mutant and wild type control skeletal muscle (data not shown). These results, in combination with the studies revealing no significant differences in the number of myogenic progenitor cells using electron microscopy or TUNEL-positive cells in Foxj3m/m vs. control unperturbed skeletal muscle further support the conclusion that Foxj3 is important in cell cycle regulation as opposed to having a role in the maintenance of the myogenic progenitor cell population (Supplemental Figure S4).
Fig. 3.
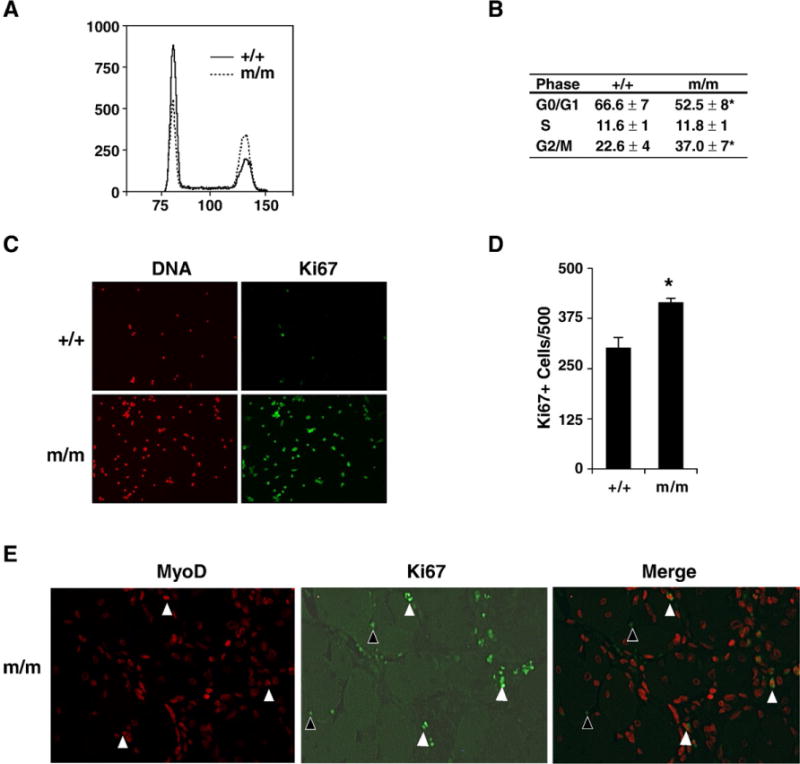
Foxj3m/m MPCs have perturbed cell cycle kinetics. (A) Flow cytometry profile of wild type and Foxj3m/m MPCs reveals that mutant MPCs have decreased number of cells in the G0/G1 stage and increased cells in the G2/M stage of the cell cycle. (B) Table outlines quantitative results of experiments undertaken in panel A representing the average cell cycle kinetics of WT and Foxj3m/m MPCs (n=3 separate experiments; *p<0.003). (C) Ki67 immunostaining of WT and mutant injured skeletal muscle reveals increased proliferation in the absence of Foxj3 following injury. (D) Quantitation of the Ki67 immunostaining reveals approximately a 65% increase in cellular proliferation in the Foxj3m/m injured skeletal muscle (*p<0.05). (E) Immunohistochemical staining of MyoD (red) and Ki67 (green) expression following injury. White arrowheads indicate that a population Ki67 positive cells are also positive for MyoD expression. A sub-population of cells are also positive for Ki67 (black arrowheads) but not myogenic (MyoD negative) in regenerating skeletal muscle.
shRNA knockdown of endogenous Foxj3 results in increased myoblast proliferation
To validate the in vivo increased myoblast proliferation phenotype observed in the injured Foxj3m/m mice, we undertook shRNA knockdown of endogenous Foxj3 in C2C12 myoblasts. Using four separate shRNA and an empty vector control, we observed knockdown of Foxj3 transcript in two out of the four shRNA constructs examined (Fig. 4A). Knockdown of endogenous Foxj3 in C2C12 myoblasts resulted in increased proliferation of Foxj3 myoblasts and an induction of cells in the proliferative phase of the cell cycle (Figs. 4B and C). No evidence of cellular death was observed in any of the samples (n=3 replicates, performed in three separate experiments).
Fig. 4.
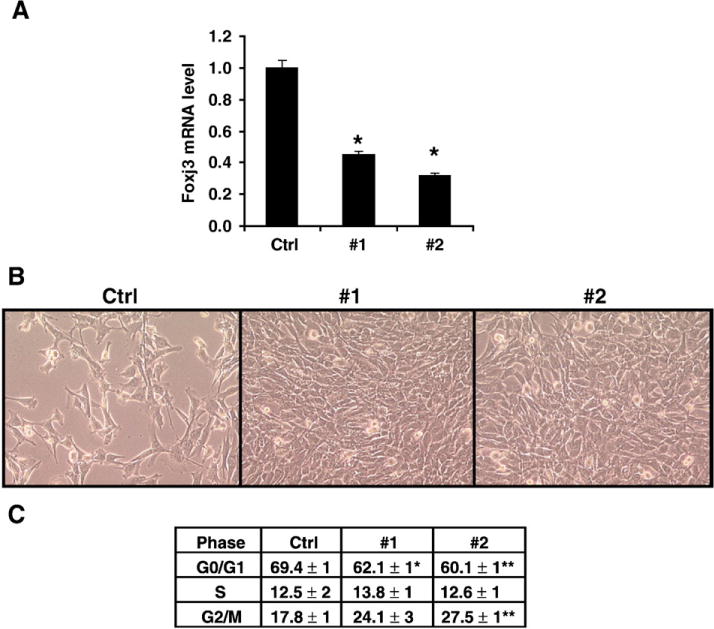
shRNA knockdown of endogenous Foxj3 results in increased myoblast proliferation. (A) Real-time PCR confirming knockdown of endogenous Foxj3 transcript in cells transfected with shRNA directed against Foxj3 (n=3 replicates; *p<0.05). (B) Phase microscopy of C2C12 myoblasts 48 h post-transfection of both control (pRS; empty vector) and shRNA directed against mouse Foxj3; shRNA #1 and #2. Note increased number of myoblasts with knockdown of Foxj3. (C) Table summarizing the results of the FACS histogram profiles of the C2C12 myoblasts transfected with control and shRNA Foxj3 vectors (n=3 replicates; *p<0.03; **p<0.003).
Foxj3m/m skeletal muscles have decreased Type I fibers and impaired contractility
We next examined the fiber type diversity of the oxidative, slow twitch soleus muscle and the glycolytic fast twitch EDL muscle in adult Foxj3m/m male mice using metachromatic ATPase fiber type analysis. We observed that both the soleus and EDL muscles of the Foxj3m/m mice had decreased numbers of oxidative Type I fibers and significantly increased numbers of Type IIa/b fibers compared to their wild type controls (Figs. 5A–C; n=3; *, p<0.05).
Fig. 5.
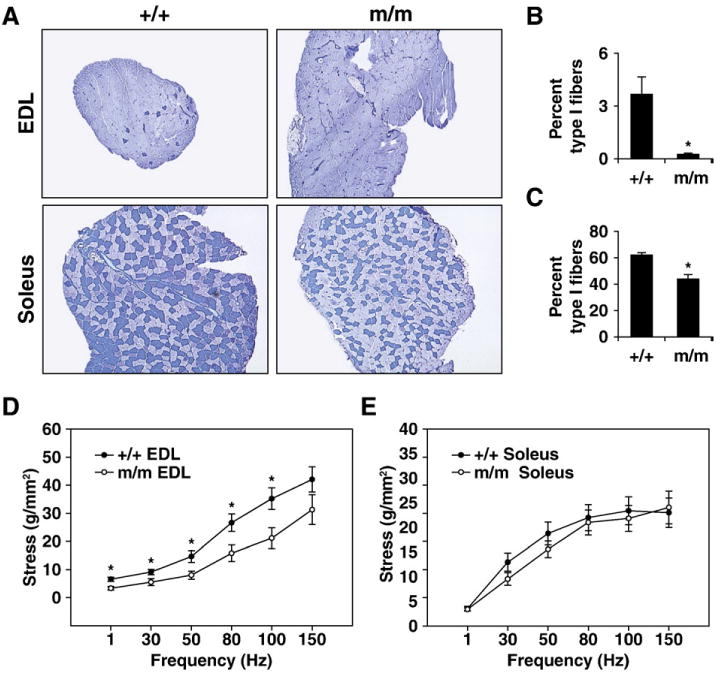
Foxj3 is essential for the determination of adult myofiber identity. (A) Metachromatic ATPase staining of EDL and soleus skeletal muscles of WT and Foxj3m/m mice. Foxj3m/m muscles have decreased numbers of Type 1 oxidative slow-twitch fibers and increased numbers of Type IIa/b fast twitch fibers. (B) Quantitation of Type 1 fibers in wild type and Foxj3m/m EDL muscles (n=4 separate muscles; 3 littermate pairs; *p<0.05) reveals decreased Type 1 fibers compared to WT control. (C) Quantitation of Type 1 fibers in wild type and Foxj3m/m soleus muscles (n=4 separate muscles; 3 littermate pairs; *p<0.05) reveals decreased Type 1 fibers compared to WT control. (D and E) Isolated wild type and Foxj3m/m EDL (D) and soleus (E) skeletal muscle stress-frequency responses. Both the EDL and soleus muscles of the Foxj3 mutant mice showed a right shift in the stress–frequency relationship compared to their wild type littermates (n=4 separate muscles; 3 littermate pairs; *p<0.05).
We next assessed the ability of the muscles to produce stress (force normalized to muscle cross sectional area; Grange et al., 2002) to determine if any functional deficits existed in the Foxj3m/m muscles resulting from the shift in fiber types. Compared to WT controls, Foxj3m/m EDL muscles (Fig. 5D) had decreased stress (with increased stimulation frequency compared to the wild type control; n=6; p<0.05), but stress was minimally effected in the soleus (Fig. 5E). These results establish that Foxj3 is important for fiber type diversity and physiological function of skeletal muscle.
Foxj3 functions as a transcriptional activator of gene expression
We next undertook a series of transcriptional assays to evaluate the functional role of Foxj3 as a modulator of gene transcription. We utilized an in vitro Gal4-UAS-luciferase reporter-based assay in C2C12 myoblasts (Chang et al., 2005). The Foxj3 full length construct (containing the entire open reading frame) and the Foxj31–176 aa mutant transcript were fused to the Gal4-DNA binding domain and transfected with a UAS-luciferase reporter into C2C12 myoblasts. Overexpression of the full-length Gal4-Foxj3 and the truncated transcripts were expressed in the nuclear compartment (Supplemental Figure S5A and S5B). Overexpression of the full length Gal4-Foxj3 construct was capable of activating the reporter 12-fold in a dose-dependant fashion (Supplemental Figure S5C; n=3;*, p<0.05) whereas the Foxj31–176 aa mutant transcript had no transcriptional activity)(Supplemental Figure S5D). These results support the hypothesis that Foxj3 functions as a transcriptional activator of gene expression.
Mef2c is a direct downstream target gene of Foxj3 in myoblasts
To identify potential downstream target genes of Foxj3, we utilized oligonucleotide microarray technology to examine dysregulated genes in Foxj3m/m myogenic progenitor cells. We identified over 900 genes that were significantly dysregulated (p<0.005) greater than two-fold and validated several of the most dysregulated transcripts by semiquantitative RT-PCR (Fig. 6A). Analysis of the Foxj3 dysregulated transcripts suggested that this forkhead protein regulated a Mef2c-dependent pathway as a number of the downregulated transcripts contained Mef2c-binding elements within their promoters. Recent studies have validated a number of these downregulated transcripts (troponins I and T, myomesin2) as direct Mef2c downstream target genes (Blais et al., 2005). Myocyte enhancer factor 2C (Mef2c) was downregulated more than 10-fold in the Foxj3 mutant myoblasts compared to the wild type controls. In addition, we observed that Mef2c target genes were also downregulated including Tnni1 and Tnnt2 (Fig. 6A). Upon examination of the 5′ upstream promoter of Mef2c, we identified a highly conserved forkhead binding site within the skeletal muscle enhancer region (−1.1 kb /+77 bp, Wang et al., 2001) (Fig. 6B). Using transgenic technology, Wang and colleagues demonstrated that this 1.2 kb skeletal muscle enhancer region directs lacZ reporter expression exclusively in embryonic and adult skeletal muscle lineages. To confirm our hypothesis that Foxj3 was able to bind to the evolutionary conserved FBS and was a direct regulator of Mef2c in vivo, we performed a chromatin immunoprecipitation (ChIP) assay and PCR with primers specific for the FBS located in the Mef2c 5′ upstream skeletal enhancer region. When we overexpressed the Foxj3-GFP fusion construct in C2C12 myoblasts, and purified the bound chromatin, the segment containing the FBS in the 1.2 kb Mef2c skeletal muscle enhancer was amplified, but not in the negative IgG control (Fig. 6C). These studies support the conclusion that Foxj3 binds to the Mef2c promoter in vivo.
Fig. 6.
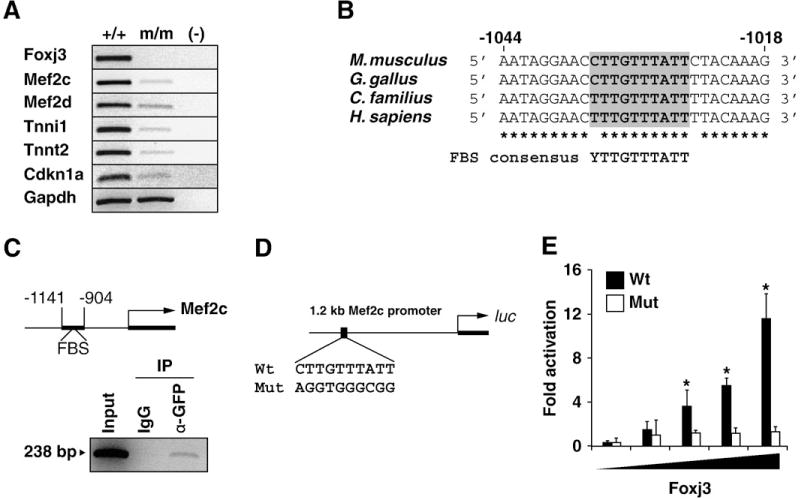
Mef2c is a direct downstream target of Foxj3 in skeletal muscle. (A) Semi-quantitative RT-PCR analysis of WT and Foxj3m/m MPCs reveals decreased expression of Mef2c and Mef2c-dependent target genes in the absence of Foxj3. Decreased expression of Cdkn1a or p21KIP corresponds to the increased proliferative capacity of the mutant MPCs as demonstrated in Fig. 3A. The housekeeping gene Gapdh is used as the loading control. (B) Homology alignment of the 5′ upstream Mef2c promoter reveals an evolutionarily conserved forkhead binding site or FBS. (C) Chromatin immunoprecipitation (ChIP) of Foxj3 using primers for the FBS in the Mef2c skeletal muscle enhancer indicate that Foxj3 directly binds to the FBS. IgG and 10% total chromatin (Input) are shown as controls. (D) Schematic outlining the Mef2c upstream fragment that harbors the FBS (and corresponding mutant construct) fused to the luciferase (luc) reporter constructs that were used in the transcriptional assays in panel E. (E) Foxj3 transcriptionally activates the Mef2c-luc skeletal muscle enhancer reporter in a dose dependent fashion in C2C12 myoblasts (n=3; *p < 0.005). Mutagenesis of the FBS ablates the ability of Foxj3 to transcriptionally activate the Mef2c-luc reporter.
Based on the results of the ChIP assay and the evolutionary conserved region of the Mef2c promoter, we fused the −1.1 kb/+77bp (referred to as 1.2 kb Mef2c) to a luciferase reporter and transfected C2C12 myoblasts with increasing amounts of Foxj3. Using these transcriptional assays, we observed that Foxj3 trancriptionally activated the 1.2 kb Mef2c-luc up to 12-fold in a dose dependant fashion (Fig. 6D and E). The Foxj31–176 aa mutant transcript that was produced in the Foxj3m/m mice had essentially no transcriptional activity (Supplemental Figure S5E and S5H). Using site-directed mutagenesis, we mutated the forkhead binding site (FBS) in the 1.2 kb Mef2c-luc reporter, and noted that overexpression of Foxj3 in C2C12 myoblasts lacked any transcriptional activity (Figs. 6D and E). Collectively, these studies support the conclusion that Foxj3 is a direct upstream regulator of the Mef2c gene.
Discussion
Molecular networks coordinately regulate skeletal myogenesis during embryogenesis and regeneration of adult skeletal muscle. Many of these transcriptional networks (MyoD family members, Mef2 factors, Pax3, Pax7, Foxk1, etc.) have been defined using gene disruption strategies that confirmed or uncovered novel essential roles in developing and regenerating muscle (reviewed by Shi and Garry, 2006). While intense interest has focused on downstream pathways that impact muscle differentiation and fiber type specification, the upstream regulators are incompletely defined. In the present study, we have begun to further decipher the regulators of the myogenic program by making three principal findings. First, we utilized molecular technologies to generate Foxj3 mutant mice (not Foxj3 null mice) that were viable. Importantly, we recognize that this genetic model may produce a truncated functional Foxj3 protein. As Foxj3 is expressed in skeletal muscle, we examined the skeletal muscle regenerative capacity in Foxj3 mutant mice. We observed that mutant Foxj3 mice had impaired muscle regeneration.
Another forkhead transcription factor, Foxk1, has been implicated as essential for normal skeletal muscle regeneration (Garry et al., 2000). However, the Foxj3m/m regenerative phenotype exhibits considerable differences with the Foxk1 null phenotype. Foxk1 is restricted to the MPC population and mice lacking Foxk1 have perturbed skeletal muscle regeneration due to decreased MPCs and an inability to efficiently activate the MPC population (Garry et al., 2000). In contrast, these studies emphasize that while the absence of either forkhead factor results in impaired muscle regeneration, the underlying mechanism is distinct for both Foxj3 and Foxk1.
A second major finding of the present study is that Foxj3 is an important cell cycle regulator. While no proliferative changes were observed in the unperturbed Foxj3m/m skeletal muscle, our in vivo studies (following cardiotoxin injury) and in vitro studies (isolation and activation of the MPC population) demonstrate that Foxj3 is an antiproliferative factor. While the mechanistic detail regarding Foxj3′s antiproliferative role is incompletely defined, one possible mechanism involves decreased Mef2c expression. Previous studies have demonstrated that Mef2c interacts with the RNA helicase, CHAMP (Mov10l1), which induces the cyclin dependent kinase inhibitor p21KIP (cdkn1a) in striated muscle to promote cellular quiescence (Liu and Olson, 2002). Many other forkhead factors have repressive and permissive (Foxk1) influences on cell cycle kinetics and therefore have important regulatory roles during embryogenesis, growth and regeneration. Our studies have further defined a novel forkhead transcription factor that regulates cell cycle progression in the muscle lineage.
A third major finding of this study is that Foxj3 is a transcriptional activator of Mef2c. The Fox transcription factor family has a number of members such as the FoxO family that directly regulate gene expression and impact muscle growth and function. For example, Foxo3 has been shown to transcriptionally activate the ubiquitin ligase, atrogin-1 gene that results in atrophy of the myofibers (Sandri et al., 2004). In the present study, we utilized transcriptome analysis, transcriptional assays, mutagenesis and binding studies to support the conclusion that Foxj3 is a direct upstream activator of the Mef2c gene.
Mice lacking Mef2c are embryonic lethal at E9.5 due to cardiovascular defects (Lin et al., 1997). While the role of Mef2c in the heart has been elucidated in the past few years, the exact role of Mef2c in skeletal myogenesis is unclear (reviewed by Berkes and Tapscott, 2005). A recent study utilizing a temperature sensitive Drosophila Mef2 orthologue (dMEF2), surprisingly revealed that dMEF2 was not essential for adult myogenesis (Baker et al., 2005). While flies lacking dMEF2 had severe neurological deficits and an inability to fly, they had normal patterning of skeletal muscle. However, studies in vertebrates support a role for Mef2c in skeletal myogenesis.
Recent studies undertaken by Hughes and colleagues demonstrated that simultaneous morpholino knockdown of zebrafish Mef2c and Mef2d resulted in a loss of thick filament proteins and the disruption of sarcomeric structure (Hinits and Hughes, 2007). In addition, it has been conclusively demonstrated that Mef2c is an essential upstream transcriptional activator of troponins in skeletal muscle (Di Lisi et al., 1998; Blais et al., 2005) and myofiber identity (Wu et al., 2000). Conditional transgenic technologies have revealed a broader role for Mef2c in cellular maintenance in various lineages. The conditional deletion of Mef2c in skeletal muscle lineages using a MCK-Cre transgenic line resulted in a severe decrease of type 1 fibers (Potthoff et al., 2007). Potthoff and colleages clearly demonstrated that HDAC-mediated inhibition of Mef2c was the essential regulatory step in the conversion of oxidative fibers to slow-twitch, non-oxidative, fasttwitch fibers. The authors further demonstrated that the conditional loss of Mef2c in the skeletal muscle lineage, results in decreased expression of structural Mef2c target genes, including troponins I and T and myomesin2.
In the Foxj3m/m skeletal muscle, we observed decreased expression of Mef2c as well as decreased expression of the Mef2c downstream target genes. The decreased expression of Mef2c and its downstream targets resulted in perturbed fiber type specification and impaired contractility. The decrease in overall maximum stress output in Foxj3 mutant mice could be the result of the decreased levels of troponins as a consequence of Mef2c downregulation. This could lead to decreased activation of the contractile apparatus. For reasons that are not clear, the decrease in stress was greater in the EDL than the soleus of the Foxj3 mutant mice. Together these results establish Foxj3 is a modulator of myofiber identity through a likely MEF2-dependent pathway.
Collectively, these studies have identified a novel forkhead transcriptional regulator, Foxj3 for Mef2c gene expression. Moreover, Foxj3 mutant mice demonstrate a phenotype consistent with decreased Mef2c expression and emphasize the role of Foxj3 in skeletal muscle biology including cell cycle kinetics, fiber type diversity and muscle regeneration.
Methods
Knockout mice
Foxj3 mutant mice were generated from the BayGenomics Foxj3-trapped ES-cell line (XL913) of the129P2OlaHsd strain (Stryke et al., 2003). After ES cell expansion and sequence confirmation, the Foxj3-trapped ES cells were injected into blastocysts, and chimeric mice were born to foster mothers. Progeny that were greater than 90% chimeric were then mated to both C56/B6J (Jackson Laboratory) and the 129P2OlaHsd (Harlan) mice to ensure germline transmission. Unless specified, wild type littermates from Foxj3 heterozygote crosses were used for all experiments (See Supplemental Materials and Methods for genotyping primers).
Mice were kept on a 4% fat irradiated chow (Harlan) diet and exposed to standard 12-h light/dark cycles. Mouse weights were recorded on a Mettler PE 200 scale. Mice were housed in a sterile, pathogen-free animal facility. All animal protocols were approved by the Institutional Animal Care and Use Committees at the University of Texas Southwestern Medical Center at Dallas.
Immunohistochemistry/histochemistry of histological sections
All tissues were perfusion fixed using 4% paraformaldehyde at 4 C and washed in 1 PBS before sectioning. Sections were stained with hemotoxylin and eosin, nuclear fast red, and other antisera as previously described (Meeson et al., 2007).
Skeletal muscle tissues/sections for fiber type analysis were performed following cryoembedding of tissues in OCT (Tissue Tek) and Gum Tragacanth (Sigma) mix as previously described (Meeson et al., 2007). The protocol for metachromatic ATPase fibertyping has been previously described (Garry et al., 1996).
Primers
All primers used in this study were from Operon, and RTPCR primer sequences can be found in the Supplemental Materials and Methods section.
Plasmids
The murine Foxj3 gene (NM_172699; NCBI Gene Bank) consisting of a 1869 base pair open reading frame was cloned into the pCDNA3.1-NT-GFP (Invitrogen), pM1 (Gal4 vector) and the pCDNA3.1-N-myc plasmids using standard techniques. Foxj3 constructs that contained the amino terminus and the DNA-binding domain (Foxj31–176aa) were cloned in frame in the same expression plasmids. The Gal4-UAS-luciferase system has been previously described (McKinsey et al., 2006). More detailed information about the plasmids used in these studies is available upon request.
The Mef2c skeletal muscle-specific promoter (−1.1 kb/+77 bp) containing the forkhead binding site has been previously described (Wang et al., 2001). The promoter region was amplified from genomic DNA and cloned into the pGL3-basic promoter with a minimal TATA promoter (pGL3-TATA). Mutagenesis of the forkhead binding site in the Mef2c-luciferase promoter construct was performed using the QuikChange II Site-Directed Mutagenesis Kit (Strategene). The Mef2c FBS was mutated using the mutant primer 5′ TAATAGGAACAGGTGGGCGGCTACAAAGC 3′ and its reverse compliment.
Cell culture and transfection assays
Unless otherwise noted, all transfections were performed using C2C12 myoblasts grown in 20% fetal calf serum/DMEM (Gibco). Transfections of 105 C2C12 myoblasts were performed in six-well plates using a Lipofectamine (Invitrogen) kit, and total protein was harvested 48 h post-transfection. Up to 2 μg of plasmid DNA was transfected in serum-free Optimem media (Gibco). Luciferase assays were performed using a Luciferase Assay System (Promega) following the manufacturer’s instructions. All fold changes were normalized to a lacZ loading control, as previously described (Meeson et al., 2007). Myotube differentiation was promoted by exposing 80% confluent myoblast cultures to differentiation medium (DMEM supplemented with 2% heat inactivated horse serum, antibiotics, insulin and transferrin) as previously described (Hawke et al., 2003b).
Antisera
The following antisera were used in these studies: mouse monoclonal anti-Ki67 serum (Nova Castra; 1:1,000 dilution), rabbit anti-GFP serum (Molecular Probes; 1:1,000 dilution) and mouse monoclonal anti-MyoD serum (Nova Castra, 1:500 dilution). The secondary antisera utilized in these studies included a FITC conjugated goat anti-mouse serum (1:50 dilution; Jackson ImmunoResearch) and a rhodamine conjugated goat anti-rabbit serum (1:50 dilution; Jackson ImmunoResearch).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using the ChIP Kit-M for mammalian cells (Progeneron, Inc). C2C12 myoblasts (2×106) were transfected with 4 μg Foxj3-GFP, and harvested for nuclear protein 48 h post-transfection. Nuclear lysates were sonicated using a 550 Sonic Dismembrator (Fisher Scientific) sonicator and immunoprecipitated with 6 μg of anti-GFP serum (Molecular Probes) overnight at 4 C. Selected samples were incubated with IgG (Sigma) as an internal control. Lysates were then bound on UltralLink Immobilized Protein A/G beads (Pierce), and chromatin was then purified using Tripure (Roche) before elution in sterile water. 6 μL of five-fold diluted input and antibody-bound chromatin were used in the PCR reaction with primers specific for the region in the Mef2c skeletal muscle enhancer containing the forkhead binding site (see Supplemental Materials and Methods).
Primary MPC isolation, culture and immunohistochemical analyses
As previously described (Meeson et al., 2007), primary MPC cultures were established following the isolation of MPCs from hindlimb skeletal muscle of 2-day-old neonatal mice. Cells were preplated and cultured in F-10 growth medium (Gibco/Invitrogen) supplemented with 25 ng/ml fibroblast growth factor (Invitrogen). WT and Foxj3m/m MPCs were isolated from postnatal day 2 littermate pups generated from Foxj3 heterozygote breeders. Asynchronously dividing WT and Foxj3m/m MPCs were fixed for 10 min with 4% paraformaldehyde and immunostained as previously described (Meeson et al., 2007).
Electron microscopy and satellite cell quantitation
As previously described (Meeson et al., 2007), tibialis anterior muscles from 3 to 4 month old male adult WT and Foxj3m/m mice were harvested, perfusion fixed with 3% gluteraldehyde and postfixed with buffered 1% osmium tetroxide. Samples were then dehydreated with ethanol, embedded in Spurr resin, polymerized, sectioned and placed on copper grids. Sections were examined using a JEOL 1200 EXII TEM. MPCs were quantified using criteria as outlined previously (Garry et al., 1997).
Muscle physiology protocol
Both extensor digitorum longus (EDL) and soleus muscles were dissected from anesthetized WT control and Foxj3m/m mice aged 130–190 days (2 mg xylazine–20 mg ketamine per 100 g of body mass i.p.). Muscles were incubated at 30 °C in an oxygenated (95% O2–5% CO2) physiological salt solution (PSS; pH 7.6) containing (in mM):120.5 NaCl, 4.8 KCl, 1.2 MgSO4, 20.4 NaHCO3, 1.6 CaCl2, 1.2 NaH2PO4, 10.0 dextrose, and 1.0 pyruvate. Muscles were prepared for isometric contractions using a dual-mode servomotor system (300B, Aurora Scientific) as previously described (Wolff et al., 2006). Resting muscle force was set at 1.00 ± 0.05 g, (i.e., L0, muscle length at which twitch force was maximal) that was automatically maintained by a stepper motor (Wolff et al., 2006). The stepper motor was temporarily inactive during isometric contractions. Dynamic Muscle Control software (DMC Version 4.1.6, Aurora Scientific) was used to record force output in response to electrical activation of the muscles via closely flanking platinum electrodes. Each EDL and soleus muscle was subjected to a force-frequency (F-F; 1, 30, 50, 80, 100, 150 Hz) protocol. Forces for the F-F protocol were converted to stress, or force normalized to muscle cross sectional area, as described (Grange et al., 2002). Physiological data were analyzed using a one-way ANOVA. (JMP 7.0).
Semiquantitative RT-PCR
Total RNA was extracted from cells and homogenized tissues using Tripure (Roche). 1 μg of total RNA was reverse transcribed using the SuperScript First Strand Synthesis kit (Invitrogen). cDNA was then diluted ten-fold in water, and PCR amplified using Go Taq Master Mix (Promega), and visualized on 1% agarose gels stained with ethidium bromide.
shRNA experiments
Four separate sets of short hairpin RNA (shRNA; HuSH-29 kit, Origene) directed against murine Foxj3 were cloned into the pRS plasmid under the U6 promoter and used for transfection assays in C2C12 myoblasts. The empty vector (pRS) and a nonsense (pRS-GFP) plasmid were used as negative controls. Effectiveness of the Foxj3 knockdown was tested in C2C12 myoblasts using both semiquantitative RT-PCR for endogenous Foxj3. 0.5 μg of shFoxj3 or vector was transfected into C2C12 myoblasts using lipofectamine (Invitrogen) in serum free Optimem (Gibco) media for 4 h. After 4 h, the transfection media was replaced with growth media containing 20% FBS in DMEM (Gibco). Cells were harvested 48 h post transfection for RNA, protein, and cellular content in a manner previously described (Meeson et al., 2007). Real-time PCR of Foxj3 was run following reverse transcription of 1 μg of total RNA (Superscript II kit/Invitrogen) using SYBRGreen Master Mix (Applied Biosystems). Samples were run on an ABI Prism 7900 light cycler and data were analyzed as previously described (Meeson et al., 2007).
cDNA microarray data
Total RNA was harvested from wild type and Foxj3m/m myoblasts, and labeled for Affymetrix arrays as previously described (Goetsch et al., 2003). cDNA microarray data can be accessed at the NCBI Gene Expression Omnibus (accession number GSE10628).
Cardiotoxin-induced muscle regeneration
Cardiotoxin (Calbiochem) was delivered intramuscularly into the gastrocnemius muscle of adult transgenic mice as previously described (Meeson et al., 2007).
Statistical analysis
All non-physiological p-values were calculated using Student’s t-test (two-tailed) analysis.
Supplementary Material
Acknowledgments
The Gal4-UAS plasmids and the original −1.1 kb/+77 bp Mef2c skeletal muscle enhancer reporter plasmids were provided by Eric Olson. Additionally, the authors wish to thank Dr. James A. Richardson and John M. Shelton for assistance with the histological analyses and Nan Jiang for assistance with the myoblast isolations.
Funding support was obtained from the National Institutes of Health (AR47850), the Muscular Dystrophy Association and the American Heart Association. M.S.A. is a NIH pre-doctoral fellow, and D.J.G. is an Established Investigator of the AHA.
Nonstandard abbreviations
- Fox
Forkhead box
- FBS
forkhead binding site
- Mef2c
Myocyte enhancer factor 2c
- luc
luciferase
- β-gal
β-galactosidase
- bHLH
basic Helix-Loop-Helix
- H&E
hemotoxylin and eosin
- NFR
nuclear fast red
- WT
wild type
- GFP
green fluorescent protein
- DMEM
Dulbecco’s modification of eagle media
- ES
embryonic stem
- ChIP
chromatin immunoprecipitation
- EDL
extensor digitorum longus
- TA
tibialis anterior
- MCK
muscle creatine kinase
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2009.11.015.
Footnotes
Conflicts of interest
The authors declare no conflicts of interests.
References
- Baker PW, Tanaka KK, Kiltgord N, Cripps RM. Adult myogenesis in Drosophila melanogaster can proceed independently of myocyte enhancer factor-2. Genetics. 2005;170(4):1747–1759. doi: 10.1534/genetics.105.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16(4–5):585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Blais A, Tsikitis M, Acosta-Alvear D, Sharan R, Klugar Y, Dynlacht BD. An initial blueprint for myogenic differentiation. Genes Dev. 2005;19(5):553–569. doi: 10.1101/gad.1281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Bezprozvannaya S, Li S, Olson EN. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc Natl Acad Sci. 2005;102(23):8120–8125. doi: 10.1073/pnas.0503275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lisi R, Millino C, Altruda F, Schiaffino S, Ausoni S. Combinatorial cis-acting elements control tissue-specific activation of the cardiac troponin I gene in vitro and in vivo. J Biol Chem. 1998;273(39):25371–25380. doi: 10.1074/jbc.273.39.25371. [DOI] [PubMed] [Google Scholar]
- Garry DJ, Bassel-Duby RS, Richardson JA, Grayson J, Neufer PD, Williams RS. Postnatal development and plasticity of specialized muscle fiber characteristics in the hindlimb. Dev Genet. 1996;19(2):146–156. doi: 10.1002/(SICI)1520-6408(1996)19:2<146::AID-DVG6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Garry DJ, Yang Q, Bassel-Duby RS, Williams RS. Persistent expression of MNF identifies myogenic stem cells in postnatal muscles. Dev Biol. 1997;188(2):280–294. doi: 10.1006/dbio.1997.8657. [DOI] [PubMed] [Google Scholar]
- Garry DJ, Meeson AP, Elterman J, Zhao Y, Yang P, Bassel-Duby RS, Williams RS. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc Natl Acad Sci. 2000;97(10):5416–5421. doi: 10.1073/pnas.100501197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetsch SC, Hawke TJ, Gallardo TD, Richardson JA, Garry DJ. Transcriptional profiling and regulation of the extracellular matrix during muscle regeneration. Physiol Genomics. 2003;14(3):261–271. doi: 10.1152/physiolgenomics.00056.2003. [DOI] [PubMed] [Google Scholar]
- Grange RW, Gainer TG, Marschner KM, Talmadge RJ, Stull JT. Fast-twitch skeletal muscles of dystrophic mouse pups are resistant to injury from acute mechanical stress. Am J Physiol Cell Physiol. 2002;283(4):1090–1101. doi: 10.1152/ajpcell.00450.2001. [DOI] [PubMed] [Google Scholar]
- Hawke TJ, Jiang N, Garry DJ. Absence of p21CIP rescues myogenic progenitor cell proliferative and regenerative capacity in Foxk1 null mice. J Biol Chem. 2003a;278(6):4015–4020. doi: 10.1074/jbc.M209200200. [DOI] [PubMed] [Google Scholar]
- Hawke TJ, Meeson AP, Jiang N, Graham S, Hutcheson K, DiMaio JM, Garry DJ. p21 is essential for normal myogenic progenitor cell function in regenerating skeletal muscle. Am J Physiol Cell Physiol. 2003b;285(5):C1019–C1027. doi: 10.1152/ajpcell.00055.2003. [DOI] [PubMed] [Google Scholar]
- Hinits Y, Hughes SM. Mef2s are required for thick filament formation in nascent muscle fibres. Development. 2007;134(13):2511–2519. doi: 10.1242/dev.007088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgren H, Carlsson P. FoxJ3, a novel mammalian forkhead gene expressed in neuroectoderm, neural crest, and myotome. Dev Dyn. 2004;231(2):396–401. doi: 10.1002/dvdy.20131. [DOI] [PubMed] [Google Scholar]
- Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox’s in development and disease. Trends Genet. 2003;19(6):339–344. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276(5317):1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZP, Olson EN. Suppression of proliferation and cardiomyocyte hypertrophy by CHAMP, a cardiac-specific RNA helicase. Proc Natl Acad Sci. 2002;99(4):2043–2048. doi: 10.1073/pnas.261708699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Kuwahara K, Bezprozvannaya S, Olson EN. Class II histone deacetylases confer signal responsiveness to the ankyrin-repeat proteins ANKRA2 and RFXANK. Mol Biol Cell. 2006;17(1):438–447. doi: 10.1091/mbc.E05-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeson AP, Shi X, Alexander MS, Williams RS, Allen RE, Jiang N, Adham IM, Goetsch SC, Hammer RE, Garry DJ. Sox15 and Fhl3 transcriptionally coactivate Foxk1 and regulate myogenic progenitor cells. EMBO J. 2007;26(7):1902–1912. doi: 10.1038/sj.emboj.7601635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Sánchez C, Arias-de-la-Fuente C, Gómez-Ferreria MA, Granadino B, Rey-Campos J. FHX.L and FHX.S, two isoforms of the human fork-head factor FHX (FOXJ2) with differential activity. J Mol Biol. 2000;301(4):795–806. doi: 10.1006/jmbi.2000.3999. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Wu H, Arnold MA, Shelton JM, Backs J, McAnally J, Qi X, Bassel-Duby RD, Olson EN. Modulation of myofiber identity and function by histone deactylase degradatioin and MEF2 activation. J Clin Invest. 2007;117(9):2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophyrelated ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117(3):399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Garry DJ. Muscle stem cells in development, regeneration, and disease. Genes Dev. 2006;20(13):1692–1708. doi: 10.1101/gad.1419406. [DOI] [PubMed] [Google Scholar]
- Stryke D, Kawamoto M, Huang CC, Johns SJ, King LA, Harper CA, Meng EC, Lee RE, Yee A, L’Italien L, et al. BayGenomics: a resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 2003;31(1):278–281. doi: 10.1093/nar/gkg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DZ, Valdez MR, McAnally J, Richardson JA, Olson EN. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development. 2001;128(22):4623–4633. doi: 10.1242/dev.128.22.4623. [DOI] [PubMed] [Google Scholar]
- Wijchers PJ, Burbach JP, Smidt MP. In control of biology: of mice, men and Foxes. Biochem J. 2006;397(2):233–246. doi: 10.1042/BJ20060387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff AV, Niday AK, Voekler KA, Call JA, Evans NP, Granata KP, Grange RW. Passive mechanical properties of maturing extensor digitorum longus are not affected by lack of dystrophin. Muscle Nerve. 2006;34(3):304–312. doi: 10.1002/mus.20588. [DOI] [PubMed] [Google Scholar]
- Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, Simard AR, Michel RN, Bassel-Duby RD, Olson EN, et al. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 2000;19(9):1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


