Abstract
Uncertainty about the phase of strings of single nucleotide polymorphisms (SNPs) creates complications in genetic analysis although methods have been developed for phasing population-based samples. However, these methods can only phase a small number of SNPs effectively, and become unreliable when applied to SNPs spanning many linkage disequilibrium (LD) blocks. Here we show how to phase more than one thousand SNPs simultaneously for a large fraction of the 35,528 Icelanders genotyped by Illumina chips. Moreover, haplotypes that are identical by descent (IBD) between close and distant relatives, e.g. those separated by 10 meioses or more, can often be reliably detected. This method is particularly powerful in studies of the inheritance of recurrent mutations and fine-scale recombinations in large sample sets. A further extension of the method allows us to impute long haplotypes for individuals who are not genotyped.
The availability of high density SNP arrays has revolutionized genetic studies. However, genotypes of SNPs from these arrays are not phased. For many genetic analyses, it would be empowering if the uncertainty about phase could be eliminated. Many statistical methods have been proposed to phase SNPs for a set of individuals sampled from a population1-5. These methods, that we call local phasing, exploit strong correlations of SNP alleles within LD blocks. Some can be very slow computationally, but the main limitation of these methods is that SNPs that are separated by many LD blocks cannot be reliably phased.
Family data provide a simple way to phasing. When both parents of the proband are genotyped, SNPs that are not triply heterozygous, i.e. heterozygous for both parents and child, can be phased. The method presented here, that we call long-range phasing (LRP), is based on the same principle as phasing with parents, the difference being that we can often perform the task even when the parents are not genotyped. When the parents are genotyped, our method can phase many of the SNPs that are triply heterozygous.
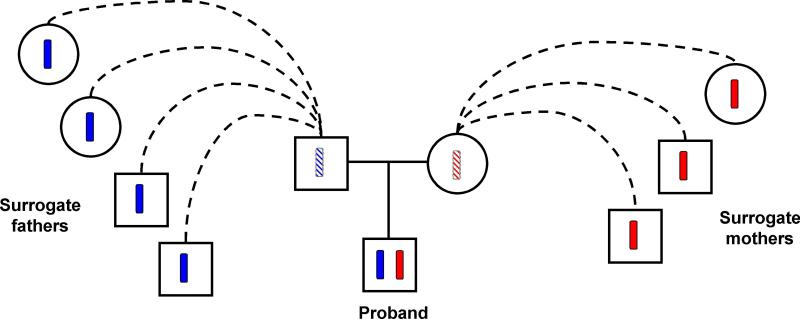
To understand the method, some knowledge of the Icelandic population is necessary. A genealogy database constructed by deCode includes 740,033 individuals with 410,551 born at or after 1900, and about 316,000 now living. In particular, the part of the genealogy after 1650 is rather complete and accurate. Using the latter to perform simulations, we studied the population characteristics of IBD sharing among the 35,528 Icelanders we have genotyped using Illumina chips. For a random proband in this set and for a particular genomic locus, for his/her paternal and maternal chromosome respectively, there is on average 17.6 and 18.1 other individuals in the genotyped set who have inherited the locus IBD (Table 1). This means that the average kinship coefficient between the genotyped individuals is approximately (17.6+18.1)/(4 ×35,528) ~ 2.5 × 10−4. This is not particularly high as Icelanders are not inbred, but is nonetheless high enough that a substantial number of the genotyped people are expected to share a region IBD. This has important implications. Consider two individuals who are nth-degree cousins separated by 2 × (n + 1) meioses. Their chance of sharing a locus IBD is 2−2n . This chance is small if n is larger than 2 or 3, but given that they do share a locus IBD, they are expected to share a region on average 200/(2n + 2) centiMorgans (cM) in genetic length. If n is 9, the shared region is on average 10 cM in width. With data for about 300,000 SNPs, they would share a haplotype that on average includes about 1,000 SNPs. When two people share a haplotype, for each SNP making up the haplotype, they would have at least one allele identical by state (IBS), and IBS ≥ 1 for 1,000 or more SNPs consecutively would usually be above the noise level. For more closely related individuals, the expected width of the shared region is larger and even easier to detect. Once a relative is shown to share a region IBD with the proband, he/she can be used to phase the proband just like a parent; the relative functions as a surrogate father if he carries the paternal haplotype of the proband, and a surrogate mother if he carries the maternal haplotype of the proband (Fig. 1). Note however that (a) a surrogate father (mother) is not necessarily male (female), (b) there can be multiple surrogate fathers and surrogate mothers, and (c) for the same proband the surrogate fathers and mothers change from locus to locus. The phase of a heterozygous SNP in the proband is determined if any of the parents, real or surrogate, is homozygous. Actually, it is better than that. Note that surrogate parenthood is a non-directional relationship, i.e. if B is a surrogate parent of A, then A is a surrogate parent of B. Also, if B and C are respectively a surrogate father and surrogate mother of A, B can assist the phasing of C through the phasing of A even though B and C do not share a haplotype. The key concepts are best captured by a graph where the nodes correspond to typed individuals and edges are put between pairs who are surrogate parents of each other. Two individuals are surrogate-relatives if they are connected, directly or indirectly, with respect to this haplotype-sharing graph, and the distance between them is the length of the shortest path linking them. We refer to this as the Erdös distance as it is a clear analogue to the Erdös number defined for co-authorships6. Thus surrogate parents are surrogate relatives with Erdös distance 1. Surrogate relatives with Erdös distance 2 or above by definition do not share a haplotype at the locus and hence may not be related at all. However, what gives LRP extraordinary power is that a SNP that is heterozygous in the proband can be phased if one of the surrogate relatives, regardless of Erdös distance, is homozygous (Methods). With our data, many of the typed individuals have more than 30,000 surrogate relatives, including some who have only one surrogate parent. Often every SNP is phased.
Table 1.
Population characteristics of typed and untyped individuals in Iceland.
| Expected Number of Typed Individuals Sharing a Specific Locus IBD |
|||||||
|---|---|---|---|---|---|---|---|
| All Relatives | Descendants | Legacy Coefficient | |||||
| Proband | Ave. YOB | Count | Paternal Allele | Maternal Allele | Paternal/Maternal | Average | Sum |
| chip typed | 1947 | 35528 | 17.6 | 18.1 | 0.243 | 0.176 | 6259 |
| untyped, yob ≥ 1900 | 1963 | 375032 | 16 | 16.4 | 0.082 | 0.054 | 20130 |
| untyped, 1850 ≤ yob < 1900 | 1873 | 101599 | 14.8 | 14.8 | 0.525 | 0.168 | 17071 |
YOB = year of birth. Legacy coefficient is the probability that the paternal/maternal haplotype of an individual is transmitted to at least one typed child or grandchild.
Fig. 1.
The concept of surrogate parenthood. Typed relatives who share either the paternal or maternal haplotypes of the proband can be used to phase the proband as though they are actual parents. These relatives are referred to as surrogate fathers and surrogate mothers respectively. A surrogate father does not have to be a male and a surrogate mother does not have to be a female. Surrogate parenthood is locus specific. A sibling can be a surrogate father for one locus and a surrogate mother for another locus. However, for a locus where the sibling shares both haplotypes with the proband, the sibling is like a twin and cannot be used to phase the proband.
Some useful applications of the method are presented below.
Phasing
Our first target is a 10Mb region on chromosome 6 (NCBI Build 36: 26.5Mb to 36.5Mb) which includes the MHC region (~ 29.7 Mb to 33.3Mb). A total of 2187 SNPs, a subset of 290,449 SNPs in the genome that satisfy various quality and yield criteria (Methods), covering an extended region of approximately 15Mb (24.3Mb to 39.1Mb) are used for phasing the 1469 SNPs in the target region. Genetically, the target region is approximately 6 cM7 and the extended region is approximately 10cM.
Applying LRP in a simple and conservative manner, i.e. only individuals who share IBS ≥ 1 for the entire extended region with a proband are considered as potential surrogate parents, 1995 (5.6%) of the 35,528 typed individuals were not phased at all either because no surrogate parents were identified for them initially or putative surrogate parents identified were eliminated later in the phasing process due to incompatibilities (Methods). Among the others, 30,954 (87.1% of the total) were phased for every SNP, and 2579 (7.3%) were phased for 90.4% of the heterozygous genotypes (Table 2). Overall, counting all 35,528 individuals, the proportion of heterozygous SNPs phased (yield) was 93.7% (16,201,012 out of 17,287,391). There are 2839 father-mother-offspring trios among the typed individuals. To empirically evaluate the accuracy of our phasing method, we removed the parents (3826 individuals) in these trios from the list of typed individuals. Because some of the removed parents are themselves offspring in typed trios, 2718 offspring were left. We phased these 2718 offspring/probands by (1) applying LRP to the reduced list of 31,702 individuals, and compared the results to (2) phasing the offspring using data from their parents only. From (1), 200 (7.4%) individuals could not be phased. Among the others, 2299 were phased for all SNPs, and 219 were partially phased (yield = 84.5%). The overall yield including the unphased individuals was 91.4%. Using (2), all probands were partially phased with a yield of 80.6%. Among the 978,802 heterozygous genotypes phased by both methods, there were 845 (0.086%) discrepancies (Supplementary Table 1a online). Individually, there were no discrepancy between LRP and trio phasing for 2456 (97.5%) of the 2518 offspring phased by LRP. Among the 62 probands with discrepancies, 43 had a discrepancy for only a single SNP. Considering that nearly one million phased genotypes were compared, many of these discrepancies could be attributed to miscalled genotypes in the parents, i.e. the LRP result could often be correct. Ten offspring had more than 3 discrepancies. Three of these 10 are siblings and they account for over 50% (468/845) of the total genotype discrepancies. One sib has IBS ≥ 1 with each of the other two sibs, who shared both haplotypes in this region, but the sharing does not result from sharing one haplotype for the entire target region, but rather sharing the paternal haplotype for part of the region and the maternal haplotype for another part with overlap. This complication contributed directly to the phasing mistake and we expect that future adjustments to the algorithm will reduce such errors (Supplementary Note online).
Table 2.
Phasing results for three genomic regions.
| All Genotyped Persons (N = 35528) | Offspring in Genotyped Trios (N = 2718) | ||
|---|---|---|---|
| LRP | LRP without parents | trio data only | |
| MHC (10 Mb): | |||
| Fully Phased | 30954 | 2299 | 0 |
| Partially Phased (yield) | 2579 (90.4%) | 219 (84.5%) | 2718 (80.6%) |
| Unphased | 1995 | 200 | 0 |
| Overall Yield | 93.7% | 91.4% | 80.6% |
| 15q25 (10 Mb): | |||
| Fully Phased | 31401 | 2345 | 0 |
| Partially Phased (yield) | 2094 (86.4%) | 173 (86.9%) | 2718 (80.2%) |
| Unphased | 2033 | 200 | 0 |
| Overall Yield | 93.5% | 91.7% | 80.2% |
| 15q25 (6.4 Mb) | |||
| Fully Phased | 32627 | 2464 | 0 |
| Partially Phased (yield) | 1333 (87.4%) | 98 (80.4%) | 2718 (79.9%) |
| Unphased | 1568 | 156 | 0 |
| Overall Yield | 95.2% | 93.6% | 79.9% |
Yield = proportion of heterozygous SNPs phased
The phased MHC region has lower recombination rate than the genome-wide average while the density of typed SNPs is substantially higher than average. We applied the same phasing algorithm to a location on 15q25 where the recombination rate is approximately 1 cM/Mb and where we have about 90 typed SNPs per Mb, both close to the genome average. We considered two regions, one longer (~ 10.0 Mb, 895 SNPs) and one shorter (~ 6.4 Mb, 574 SNPs) (Supplementary Results online). For the longer region, the results matched the MHC closely; overall yield of 93.5% for the run with 35,528 individuals and a discrepancy rate of 0.080% in the trio test. Results for the shorter region were better; overall yield of 95.2% with a discrepancy rate of 0.022% (Table 2 and Supplementary Table 1a). For all three regions investigated, as part of the trio test with parents removed when applying LRP, we found that those offspring without siblings genotyped (1249 out of 2718) have yield that are approximately 1% to 2% lower than the overall, but there is little difference in the discrepancy rates (Supplementary Table 1b online).
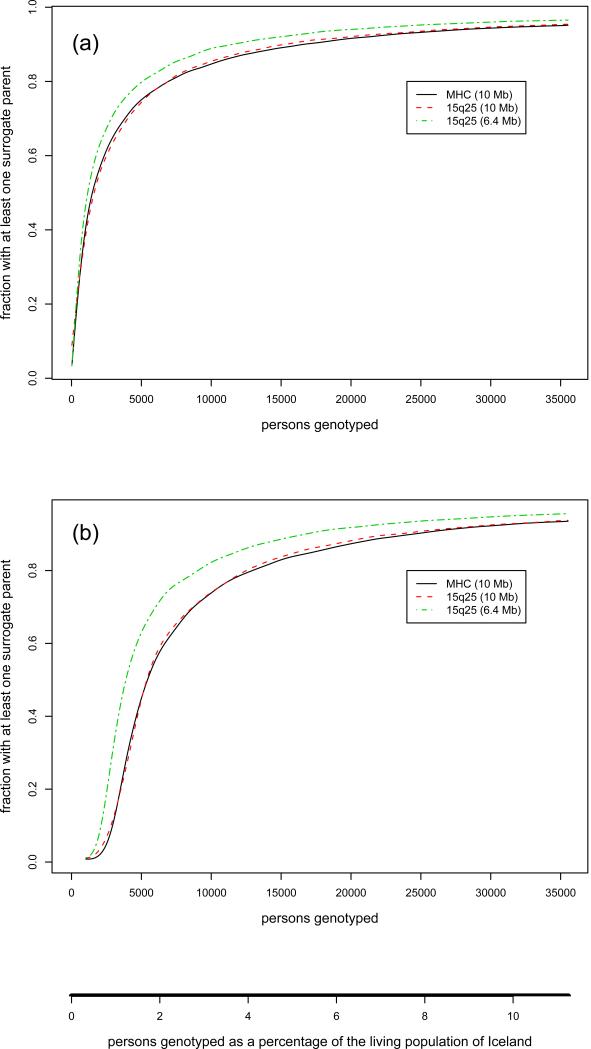
To further understand the workings of LRP, we randomly ordered the 35,528 typed individuals, and removed 50 of them at a time from the haplotype-sharing graph. Every time individuals were removed, we recomputed the fraction of individuals who had at least one surrogate parent (Fig. 2a), and the fraction of individuals belonging to the main, i.e. largest connected, cluster in the haplotype-sharing graph (Fig. 2b). The results were similar for the MHC region and the 10 Mb 15q25 region, and higher for the 15q25 6.4 Mb region. Specifically, with as little as 2% of the living population (~ 6300) typed, for the MHC and the 10 Mb 15q25 regions, about 78% of the individuals would have at least one surrogate parent and about 59% of the individuals belong to the main cluster. Similar results were achieved for the shorter 6.4 Mb 15q25 region by having about 1.5% (~ 4700) of the living population typed. With improved analytical tools (see below), and in some cases focusing on smaller regions, useful results could possibly be obtained with only 1% of the living population genotyped. Based on the fraction of the population typed, we believe that these results would apply, albeit crudely, to populations of various sizes, including large out-bred populations (see Supplementary Note for a discussion of how LRP might work with non-Icelandic data).
Fig. 2.
As a function of sample size, in absolute number and as a fraction of the size of the living population in Iceland (316,000), (a) the fraction of typed individuals with at least one surrogate parent and (b) the fraction of individuals in the largest connected cluster in the haplotype sharing graph. A person with one or more surrogate parents can at least be partially phased. Individuals in the main cluster have a large number of surrogate relatives, and often every SNP can be phased.
The yield of LRP presented here is high, but it would likely be even higher if not for the current algorithm, which is a first attempt at implementing a procedure that can process data in mass and give reliable results, and is both conservative and inefficient. Much information is not utilized, and we expect that by incorporating a number of refinements (Supplementary Note) to the procedure, the yield would increase without elevating the error rate. Currently, yield is limited by the criterion that an individual is considered a surrogate parent only if he is IBS ≥ 1 with the proband for the entire extended region. This could not only rule out true surrogate parents due to a single genotyping error, but also eliminates individuals who share a very long haplotype with the proband, part of it extending beyond the extended region on one side, but the shared haplotype does not cover the entire target or extended region. Utilizing these individuals in a proper manner is crucial to our ultimate goal of phasing chromosomes in their entirety, achievable by stitching together phasing results from overlapping target regions.
Comparisons were made with PHASE2 and fastPHASE5. PHASE is too slow for meaningful comparisons. For the shorter 15q25 region, fastPHASE and LRP processed 10,000 typed individuals in 92 hours and 16 minutes respectively. Based on the trio test, the discrepancy rate is 30.4% for fastPHASE and 0.066% for LRP. For individuals/SNPs it can phase, we estimate that LPR can phase a region 800 times longer than fastPHASE with a similar probability of not making any errors (Supplementary Results and Supplementary Table 2 online). These results reinforce the point that local phasing methods are not designed to phase long regions on their own. However, improvements to LRP could be achieved through incorporating local phasing ideas (Supplementary Note).
Studying Fine-scale Recombination with Increased Sample Size
There is much interest in recombination hotspots and their evolution8-10. Apart from methods that estimate historic recombination rates based on linkage disequilibrium (LD) patterns, a recent study investigated recombination events directly utilising high-density SNP data from 725 related individuals falling into 82 nuclear families11. A number of interesting observations were made even though there was only information on 728 meioses.
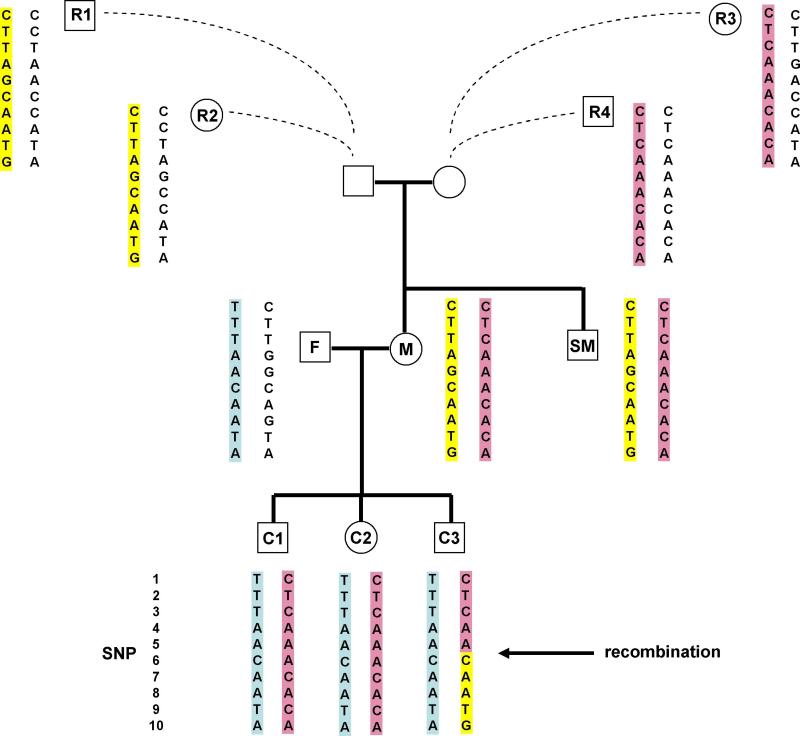
The main difficulty with studying recombination events in chromosomes transmitted to the children is that the parents need to be phased. But phasing the parents directly requires genotyping the grandparents, a serious limiting factor. An alternative is to utilize nuclear families with 3 or more children genotyped. Here, in effect, the children are used to phase the parents. Specifically, if both parents and two children are genotyped, one can detect a recombination event, but it would be impossible to tell in which child the event occurred. With more than two children, by assuming that the chance that more than one child has gone through a recombination event in a small region is negligible, the uncertainty in phase is resolved by the majority rule. Fig. 3 displays an example of a recombination observed at a known hotspot in the MHC region. The recombination event between SNPs rs2532924 and rs3095089 (SNP5 and SNP6) could be deduced from data of the two parents and the three children (C1 to C3). It is clear that all three children share the maternal allele IBD for rs2532924. Since C1 and C2 also share the maternal allele for rs3095089 IBD, but not C3, one can deduce that a recombination event occurred between the two SNPs for the maternal meiosis of C3. If the data for C1 were not available, one could still deduce that a recombination event occurred in either C2 or C3, but assigning it to a specific child would not be possible. If neither C1 nor C2 were genotyped, the recombination event could not be inferred using traditional approaches without genotyping the maternal grandparents.
Fig. 3.
Applying long-range phasing to determine a recombination event. The results from phasing a 10Mb region including the MHC were used, although only the 10 SNPs around the recombination event are highlighted. By phasing M using relatives R1 to R4, the recombination event in C3 could be deduced based on data from the trio F, M and C3 only, without the need of data from C1 and C2, or data from the parents of M. Having R2 and R4 could actually be better than having the two parents of M. A SNP informative for recombination in the children has to be heterozygous in M. Here both SNP5 and SNP6 are. To phase M, one of her parents (if typed) or surrogate relatives needs to be homozygous. In this case, R2 and R4 are each homozygous for both SNP5 and SNP6, so having one of them would be sufficient to deduce the precise location of the recombination. By contrast, R1 is homozygous at SNP6, but heterozygous at SNP5. With R1 only, we could deduce that a recombination in C3 occurred between SNP3 (the closest marker on the left that is heterozygous for M and homozygous for R1) and SNP6, but some resolution would be lost. The same could happen if one or both parents of M were typed. Surrogate relatives who are not surrogate parents of M can also help. E.g. the uncertain phase of SNP 5 in R1 can be resolved by surrogate parents of his sharing the other haplotype. Surrogate parents of R1 are surrogate relatives of M with Erdös distance 2.
Applying LRP, we were able to phase the mother using her more distant relatives. This phasing information together with the data on the father and C3 alone allowed us to infer the same recombination event that was deduced by also using the data of C1 and C2. As detailed in the figure legend (Fig. 3), having relatives R2 to R4 could actually provide better resolution of the recombination event than having the parents of M genotyped. As noted earlier, a substantial fraction of the heterozygous SNPs in M could remain unphased even with data on parents, but having a large number of surrogate parents and surrogate relatives could have every SNP phased. Also worth noting is that a sibling of M, SM, is genotyped, but because he shares both haplotypes with M he is actually not useful for phasing M at this locus. By contrast, R2 and R4, the key individuals, are separated from M by 7 meioses and hence not close relatives.
By phasing the parents with LRP, recombination events can be estimated from trio data. In addition to 194 nuclear families with both parents and three or more children genotyped, the 35,528 chip-typed individuals include 1257 and 475 nuclear families with respectively one and two children genotyped. The latter families alone could provide up to 4414 = 2 × (1257 + 2 × 475) meioses for the study of fine-scale recombination.
Studying the Inheritance of a Recurrent Deletion
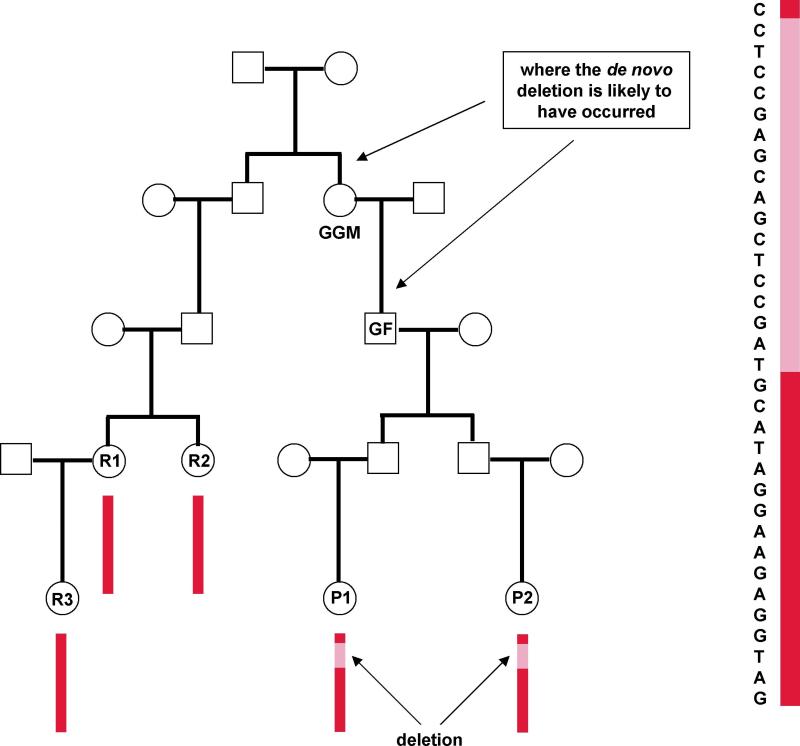
Recent studies support the notion that recurrent structural mutations may contribute substantially to the risks of psychiatric disorders such as autism12 and schizophrenia13. Recently, we found evidence suggesting that a recurrent deletion at 15q11.2 is associated with schizophrenia14. Assuming that association is real, the penetrance is not very high. In Iceland, a total of 63 carriers were observed: 4 in 648 schizophrenics, one in a parent of a schizophrenic, and 58 in 32,442 controls (OR ~ 3.5). Here we explore the inheritance of this deletion independent of its putative association with schizophrenia. Out of the 63 chromosomes with the deletion, 14 of the parents of origin were chip-typed. Twelve of these 14 parents also carry the deletion (i.e. part of the 63) and one does not, the latter points to a de novo event for the proband. Given that many of the deletion carriers do not have a ‘first-generation’ de novo mutation, we investigated the relationship between the 63 chromosomes with the deletion through long-range phasing. The deletion resides in a difficult region. Chromosome 15 has a very short p-arm. Among the SNPs used, only two are on the left of the deletion (51 are inside), meaning that the information in determining whether two chromosomes are IBD comes mostly from SNPs on the right side. This region also has a high recombination rate so that less SNPs than average are included for a specific genetic distance. Nonetheless, substantial progress was made and Fig. 4 displays some of the results. Probands 1 and 2 (P1 and P2) carry the deletion IBD (sharing a haplotype ~ 1500 SNPs in length not including the 51 SNPs in the deleted region). Relatives 1 to 3 (R1, R2 and R3) also share a long haplotype with the probands (over 4000 SNPs with P1 and ~ 1500 SNPs with P2), but they do not have the deletion. One can deduce from the family relationship that the grandfather (GF) has the deletion, and the mutation event occurred either at the meiosis of GF or GGM. By phasing R1 to R3, the haplotype of the 51 SNPs that were deleted in P1 and P2 could be reconstructed together with the haplotype background of over 4000 SNPs.
Fig. 4.
The inheritance of a chromosome associated with a deletion. Typed are P1-P2 and R1-R3. Long-range phasing revealed that they all share a haplotype with over 1000 SNPs, although only P1 and P2 carry the deletion. Displayed are alleles of every 3rd of the first 100 SNPs on chromosome 15, including 17 of the 51 SNPs deleted. It can be inferred from the family structure that the shared region was transmitted to P1 and P2 through GGM and GF. Note: with only two typed SNPs (one shown) on the left of the deletion, the first two SNPs might only be IBS and not IBD between R1-R3 and P1-P2 as it could not be ruled out that a recombination event close by had taken place at one of the intermediary meioses, particularly since it is known that a recombination often accompanies a deletion event23.
Overall, by studying the haplotype backgrounds, we deduced that the 63 deletions correspond to approximately 31 separate mutation/deletion events (Supplementary Results). A rather complex pattern of inheritance was indicated. Firstly, carriers of these deletions are not completely infertile and, moreover, could pass on the deletion to their children (one carrier with 5 children overall passed on the deletion to all 4 of the chip-typed children). However, the probability that the carriers could pass on the deletion to a child appears to be substantially lower than that under a model of neutrality. The many haplotype backgrounds observed for the chromosomes with the deletion indicate that the deletion occurs rather frequently as a de novo event. This is in sharp contrast to other rare variants such as the BRAC2 mutation in Iceland where all chromosomes with the mutation appear to have a single founder and share a haplotype background15,16. If the deletions were inherited neutrally, they would be expected to have a much higher frequency in the population than observed. Hence, this novel approach provides support to the notion that the deletions are under negative selection, but statistical methods still need to be developed to test for negative selection formally and to estimate its magnitude.
Imputing Haplotypes into Untyped Individuals
In Fig. 4, it can be inferred that GGM and GF, who are not genotyped, both carry the haplotype shared by R1-R3 and P1-P2. In general, a haplotype can be imputed into an untyped proband if two genotyped relatives share a long haplotype IBD and the genealogy indicates that the path of IBD sharing goes through the proband. Standard conditions for that are:
One of the chip-typed relatives is a descendant, preferably a child or a grandchild.
With some exceptions (see 3), the other chip-typed relative should not be a descendant. Unless the mate of the proband is chip-typed, this relative is preferably substantially more closely related to the proband than the mate.
The other genotyped relative could also be a descendant if either the mate of the proband is genotyped, or the two chip-typed descendants are from different mates (e.g. half-sibs).
Condition 1 is required because one could only reliably deduce an untyped proband carry a haplotype if a descendant inherited it. Conditions 2 and 3 ensure that the chance that the shared haplotype was transmitted to the descendant through the mate instead of the proband is small. If the descendant is a grandchild, in addition to the mate of the proband, data are needed to show that the sharing of the haplotype did not go through the other two grandparents instead.
Condition 1 is often the limiting factor. As indicated in Table 1, for untyped probands, the average number chip-typed relatives expected to share the paternal/maternal chromosome IBD exceeds 15. By contrast, for the un-typed probands born at or after 1900, the expected number of typed descendants carrying the paternal/maternal allele IBD is only 0.082. We define the legacy coefficient of an individual as the probability that the paternal/maternal haplotype is transmitted to at least one typed child or grandchild. For example, the legacy coefficient is 0.5 if exactly one child of the proband is chip-typed, and 0.25 if exactly one grandchild is typed (Supplementary Methods). The legacy coefficient, with some discount, should be about the probability that the paternal/maternal haplotype of an untyped proband can be imputed.
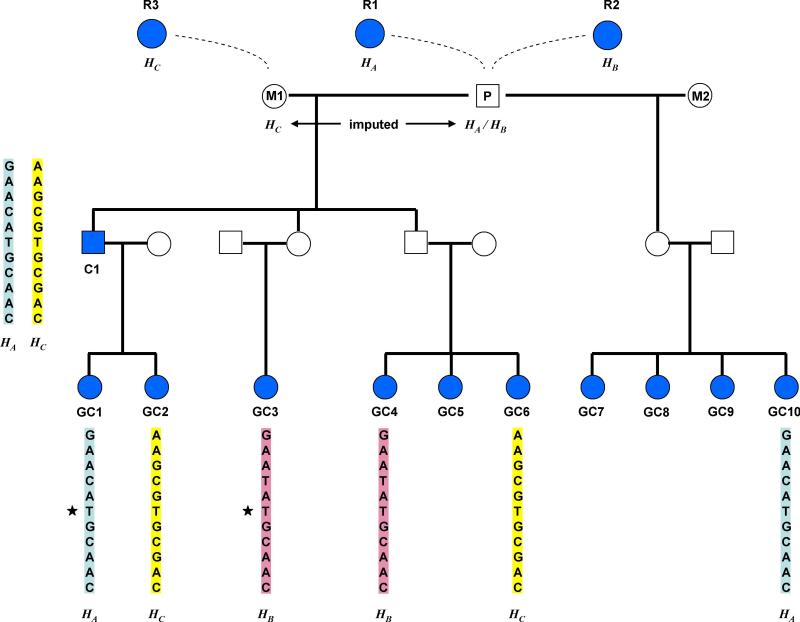
In practice, haplotype imputation is done in two steps. The first step involves using LRP to simultaneously phase typed individuals and to identify haplotypes that are IBD. The second step overlays this information on the pedigree. Based on how individuals sharing a haplotype IBD are related, it can then be inferred that some untyped individuals must also carry the haplotype. The following example (Fig. 5) highlights how the second step works. The proband (P) is a deceased lung cancer patient. One of his children (C1) and 10 of his grandchildren (GC1 to GC10) are chip-typed. His legacy coefficient is 0.89. Here we focus on a region around the nicotinic acetylcholine receptor genes on 15q25 where variants, including allele T of SNP rs1051730, were recently shown to associate with smoking behaviour, lung cancer, and peripheral arterial disease17-20. As detailed in the figure legend, there are many sources of information from the pedigree that allowed us to impute HA and HB, two phased haplotypes composed of 1001 consecutive SNPs centred at rs1051730, into P. In particular, we know that P is homozygous TT for rs1051730.
Fig 5.
Imputing haplotypes into an untyped proband P. One of his children (C1) and 10 of his grandchildren (GC1 to GC10) are chip-typed (in blue). A region on 15q25 with 1001 typed SNPs centred at rs1051730 was investigated. All typed individuals were phased although only three haplotypes, HA , HB and HC, are highlighted. Haplotype HA could be imputed into P because C1 and GC10, descendants of P with different mates share HA IBD, satisfying Conditions 1 and 3. R2 shares HB IBD with GC3 and GC4, satisfying Conditions 1 and 2, and allow us to impute HB into P. However, as an exception to Conditions 2 and 3, HB can actually be imputed into P in an alternative way that does not require R2 and only employs the data from the descendants. Given that GC3 and GC4 share HB, it must be carried by either P or M1. The same with HC since it is shared by C1 and GC6. Given that GC4 and GC6 are related to P and M1 in the same way, HB and HC cannot both originate from M1. Since C1 has both HA and HC, and HA is established to be from P, HC must be from M1. This highlights that there could be extra information in addition to what can be deduced from the pair-wise sharing of relatives. Because R1 is related to P on his father side and R2 is a relative on his mother side, we can deduce that HA is the paternal haplotype of P and HB is the maternal haplotype, information useful for an imprinting model. While GC5, GC7, GC8 and GC9 do not play a role in the imputation of P here, they do contribute to the imputation of P for other regions in the genome. If C1 was not genotyped, GC1 and GC2 could be used to impute C1 and P.
Given that C1 is typed, his children, GC1 and GC2, are not crucial for imputing P. However, as a proof of principle, suppose C1 is not typed. At this locus, because GC1 and GC2 carry HA and HC respectively, they could be used to impute C1. The two haplotypes inferred this way agree completely with the actual genotypes of C1.
For untyped probands born after 1900, the average legacy coefficient is only 0.054, much less than the 0.176 of typed probands. This is because the untyped individuals are on average younger and there is some clustering in people chip-typed. Still, with 375,032 probands in this category, approximately 20,000 paternal and maternal haplotypes each could be imputed. For the 101,599 untyped individuals born between 1850 and 1900, the average legacy coefficient is 0.168. This corresponds to another 17,000 paternal and maternal haplotypes each. Another approximately 2000 paternal and haplotypes each could be deduced for individuals born before 1850. Overall, this corresponds to about 76,000 haplotypes being potentially imputable. The emphasis is on haplotypes instead of individuals because often only one haplotype of a person could be determined (e.g. M1 in Fig. 5). Also, while the 76,000 is an average number that applies to all locations in the genome with good coverage by the SNPs, who and which haplotype could be imputed vary from locus to locus. Finally, even when a haplotype was passed on to a typed child or grandchild, there are still instances where the haplotype cannot be reliably imputed. We believe that the chance of this is around 10%.
Discussion
We demonstrated that when a substantial fraction of a population is genotyped with a high-density SNP array, there is much more information in the data than what lies on the surface. Compared to existing approaches that either focus on close relatives, i.e. family-based analyses, or very distantly related (‘unrelated’) individuals, i.e. LD-based analyses, the conceptual leap here lies in the recognition of the utility of moderately to distantly related individuals, e.g. those separated by 3 to 20 or more meioses. As the number of meioses increases, the decrease in the probability of IBD sharing for any particular relative is compensated for by the exponential increase in the number of such relatives. While it is generally recognized among investigators familiar with linkage analysis that IBD sharing can often be detected without precise knowledge of the pedigree structure, it still came as a surprise when we realized that the information could be exploited in this systematic and extensive manner.
To utilize the data fully, statistical methods need to be developed on two fronts. Many obvious methodological refinements could increase yield and reduce error rates. Most importantly, even though this new method can be applied to tasks that were previously impossible, its power can be further enhanced by incorporating ideas behind existing methods. In our phasing examples, LRP on its own was performing very well, but for more difficult regions or with a smaller sample typed, local phasing, which in effect could generate more informative markers than bi-allelic ones, could assist in both phasing and the detection of IBD sharing of shorter regions. Apart from being the first procedure that can systematically impute haplotypes into completely untyped individuals, our method can also be used in conjunction with previous methods for the imputation of untyped variants into individuals with other markers typed21,22. In another front, we emphasize that proper, and sometimes sophisticated, statistical methods are needed in situations where valid estimates and measures of statistical significance are required.
The method presented should be transferable to settings other than that in Iceland if certain conditions are met and with proper adjustments (Supplementary Note). Long-range phasing and IBD detection do not require explicit knowledge of the genealogy. However, the number of individuals genotyped has to be above a certain threshold. While many factors play a role, we speculate that having as little as 1% of a population genotyped may be adequate for the method to yield useful results. This would still correspond to a very big sample size for a large population, but it may be attainable in less than a decade given the fast pace in technological advance. This could be achievable in the near future, or already achieved, for smaller populations, including isolated regions within a large nation. Indeed, the results shown here are particularly relevant for the planning of biobanks. The genealogy plays an important role in the imputation of untyped individuals. Still, even without it, haplotypes could be imputed into a proband if the mate and at least one child are genotyped, with the other relatives used to assist in resolving phase uncertainties, e.g. when both mate and child are heterozygous. With high density SNP data, close relationships could often be detected and it may be possible to reconstruct small families. It remains to be seen, with further methodological development, whether information on the mitochondria, the sex chromosomes and knowledge of ancestries/ethnicities of the proband and mate could assist in haplotype imputation. Recently, as a consequence of the unprecedented success of case-control genome-wide association studies, family-based studies have faded into the background. The results presented here are a reminder of the fact that genetics is ultimately a study of inheritance. Familial relations always play an important role and sometimes in unexpected ways.
Methods
Individuals selected for the phasing project
The genotypes of 35,528 persons were used in this study. These individuals are participants in a large number of on-going genome-wide association studies being conducted in-house. All participants signed an informed consent. Four different Illumina chip-based arrays were used over the course of data collection, with a common core of over 300,000 SNPs. For inclusion in the study, the genotype yield of a person on that person's chip had to exceed 98% (1561 individuals were excluded due to this criterion).
SNPs selected for phasing
A total of 290,449 SNPs were used for this phasing study. Each SNP had a genotyping yield greater than 95% on all four different chip types that were used to generate the genotypes and had a Hardy-Weinberg statistic within and across chip types that was not significant at the P = 0.0001 level. A few additional markers were excluded because they were monomorphic or having allele frequencies that vary by over 2% across the chip types. In all, 6.5% (n=20,220) of the 310,669 SNPs that were common to all the chip types were excluded from the study.
Identifying individuals with a deletion
The microdeletion at 15q11.2 was identified in the course of a study of the association of copy number variations (CNV) with schizophrenia. An Icelandic population based sample of 2160 trios and 5558 parent offspring pairs who had been genotyped on one of the Illumina chips was used to identify de novo deletion and duplication regions using dose (a probe-based intensity measure) and loss of heterozygosity analysis. The 15q11.2 deletion region was one of the 66 regions identified and subsequently investigated for association. In Iceland, 4 out of 646 schizophrenia patients and 58 out of 32,442 controls were shown to carry the deletion. In addition, one parent of a schizophrenia patient also carried the deletion. Each of the 63 persons with a deletion in this region met the criteria for inclusion in this phasing study.
General principles behind long-range phasing
For a proband A, (putative) surrogate parents are identified based on IBS sharing. Ideally, the surrogate parents can be separated into two groups, which correspond to surrogate fathers and surrogate mothers. However, determining which group is which is not necessary for the purpose of phasing. Group 1 shares haplotype H1 with the proband while Group 2 shares H2. For a SNP that is heterozygous in A, phase is determined if at least one of the surrogate parents is homozygous. For example, if a surrogate parent in Group 1 is homozygous for the major allele, then H1 has the major allele and H2 has the minor allele. Consider a SNP that is heterozygous for A and all of his surrogate parents. Its phase can still be determined as long as one of his surrogate relatives with Erdös distance higher than 1 is homozygous. For example, let B be a member of Group 1 and apart from H1, let H3 be the other haplotype she carries. Treating B as the proband, one group of her surrogate parents includes everyone in Group 1 but her plus A, while the other group includes individuals carrying H3. Suppose a member of this latter group is homozygous for the major allele, this would imply that H3 has the major allele which in turn implies that H1 has the minor allele. In general, consider a SNP that is heterozygous (1,2) for A and all of his surrogate relatives with a Erdös distance of K or less. If a surrogate relative C with Erdös distance K+1 is homozygous (1,1) , then the haplotype of A through which she is linked to C by the shortest path has allele 2 if K is odd, and allele 1 if K is even.
Incompatibilities and Error Detection
If the putative surrogate parents identified for a proband are true, it should be possible to classify them all as a single group (when only surrogate mothers or fathers exist) or to divide them into two groups. However, any attempt to group the surrogate parents could result in incompatibilities. Specifically, for a SNP that is heterozygous in the proband, an incompatibility occurs when two surrogate parents in the same group have different homozygous genotypes or if two surrogate parents in separate groups have the same homozygous genotype. Incompatibilities could also result from genotypes of more distant surrogate relatives. This happens when a surrogate parent is heterozygous, but data from her other surrogate relatives nonetheless suggest that the allele she shared with the proband has to be, say, the minor allele, which happens to be in contradiction with the data of the other surrogate parents. Indeed, the fact that any surrogate relatives, regardless of Erdös distance, could contribute to the phasing of each other also means that for individuals belonging to the main cluster in the haplotype-sharing graph, even a single genotyping error for one of the SNPs can often be detected because of resulting incompatibilities. While this means that the data have extraordinary power in the detection of irregularities, the challenge is in the determination of what is the cause of an incompatibility. Incompatibilities could result from one of the following:
Simple genotyping errors.
Misinterpretation of the data due to the presence of structural mutations such as deletions and duplications in some of the individuals. While this could be considered as a form of genotyping errors, the difference from I is that these errors are systematic rather than random, and in some cases may affect the calling of a long sequence of SNPs.
A putative surrogate parent does not actually share a haplotype with the proband at the region at all, or more frequently the surrogate parent only shares a haplotype with the proband for part of the region (some recombination event has cut-off the IBD sharing at a certain location, but by chance the IBS sharing continues).
With so many SNPs and individuals studied simultaneously, I is unavoidable. While practical phasing procedures will have to allow for some errors, the importance of high quality genotypes cannot be overstated. Based on 50 individuals who were typed twice, for individuals and SNPs that passed our quality control criterion, we put the error rate at around 0.01% or lower, which is consistent with the discrepancies we observed for the trio test. Given proper treatment, isolated genotyping errors which impact SNPs individually do not pose a substantial problem. With II, the best approach is to identify them in advance based on extra information such as probe intensities and SNPs that are specifically designed to capture CNVs, with special attention paid to known locations harbouring such variants. Our study of the deletion on 15q11.2 is one such example. There phasing is performed two ways, with and without SNPs in the deletion region (Supplementary Results). The impact of III is discussed below using the MHC region as an example.
The current phasing algorithm and the phasing of the 10 Mb MHC region
LRP was performed in two rounds of three steps each. At Round 1, Step 1 identified putative surrogate parents for each proband. To minimize the impact of III described above, only those individuals who had IBS ≥ 1 for all 2187 SNPs in the extended 14Mb (10 cM) region were selected. This increased the chance that a putative surrogate parent actually shares a haplotype IBD with the proband for the entire 10 Mb target region. Missing genotypes were treated as wild cards and considered to be consistent with sharing. Even though we plan to do that in the future, imputation was not attempted. At Step 2, for each proband, surrogate parents were first checked for incompatibilities. A proband was phased at this step only if no incompatibilities were observed for any of the SNPs. Because genotypes phased at this step would contribute to the next phasing step, this ensured that only very high quality results would be carried over. Note that at Step 2, data of surrogate parents entered the processing of a proband as unphased. But every surrogate parent was himself a proband. At Step 3, surrogate parents carried with them the phasing information obtained from Step 2. This in effect utilized surrogate relatives with Erdös distance 2. Probands who were partially phased at Step 2 could now have more of their heterozygous genotypes being phased. Most of the probands who were not phased at all before due to incompatibilities were also phased here. With additional information provided by some of the putative surrogate parents who were now partially phased, a reasonable but ad-hoc, i.e. rule-based instead of model-based, procedure (Supplementary Methods) was used to resolve the incompatibilities. Sometimes, for a proband successfully phased for most of the SNPs, an individual SNP could be declared unphasable because of incompatibilities that resulted from the genotypes of many surrogate parents. Often the cause could be a genotyping error in the proband. Nevertheless, genotype correction was not attempted. Note that probands were processed one at a time, but the updated information for one proband was not applied to the phasing of the others until the next step/iteration, and thus the ordering of the probands had no impact on the results. After each proband had been processed, Step 3 was then repeated, and the successive iterations brought in information contributed by surrogate relatives with Erdös distance 3 and higher. Round 1 was completed when the results of the iterations stabilized. At Step 1 of Round 2, a review was performed to identify surrogate parents who were part of multiple incompatibilities (Supplementary Methods). They were then removed from the surrogate parent list, i.e. even genotypes of theirs that did not lead to incompatibilities would no longer be used. Steps 2 and 3 were then repeated.
Note that since missing or possibly wrong genotypes were not imputed or corrected, the final phase result was fully compatible with the orgininal genotyping information that entered the algorithm.
Supplementary Material
References
- 1.Hawley ME, Kidd KK. HAPLO: a program using the EM algorithm to estimate the frequencies of multi-site haplotypes. J Hered. 1995;86:409–11. doi: 10.1093/oxfordjournals.jhered.a111613. [DOI] [PubMed] [Google Scholar]
- 2.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halperin E, Eskin E. Haplotype reconstruction from genotype data using Imperfect Phylogeny. Bioinformatics. 2004;20:1842–9. doi: 10.1093/bioinformatics/bth149. [DOI] [PubMed] [Google Scholar]
- 4.Marchini J, et al. A comparison of phasing algorithms for trios and unrelated individuals. Am J Hum Genet. 2006;78:437–50. doi: 10.1086/500808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–44. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goffman C. And what is your Erdos number? American Mathematical Monthly. 1969;76:791. [Google Scholar]
- 7.Kong A, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–7. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 8.Winckler W, et al. Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–11. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- 9.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–4. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 10.Jeffreys AJ, Neumann R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum Mol Genet. 2005;14:2277–87. doi: 10.1093/hmg/ddi232. [DOI] [PubMed] [Google Scholar]
- 11.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–8. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 12.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 13.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–43. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 14.Stefansson H. Large recurrent microdeletios associated with schizophrenia. Nature. 2008 doi: 10.1038/nature07229. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thorlacius S, et al. A single BRCA2 mutation in male and female breast cancer families from Iceland with varied cancer phenotypes. Nat Genet. 1996;13:117–9. doi: 10.1038/ng0596-117. [DOI] [PubMed] [Google Scholar]
- 16.Gudmundsson J, et al. Frequent occurrence of BRCA2 linkage in Icelandic breast cancer families and segregation of a common BRCA2 haplotype. Am J Hum Genet. 1996;58:749–56. [PMC free article] [PubMed] [Google Scholar]
- 17.Saccone SF, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 20.Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burdick JT, Chen WM, Abecasis GR, Cheung VG. In silico method for inferring genotypes in pedigrees. Nat Genet. 2006;38:1002–4. doi: 10.1038/ng1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 23.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–47. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.