Abstract
Mongolians have played a significant role in modern human evolution, especially after the rise of Genghis Khan (1162[?]–1227). Although the social cultural impacts of Genghis Khan and the Mongolian population have been well documented, explorations of their genome structure and genetic imprints on other human populations have been lacking. We here present the genome of a Mongolian male individual. The genome was de novo assembled using a total of 130.8-fold genomic data produced from massively parallel whole-genome sequencing. We identified high-confidence variation sets, including 3.7 million single nucleotide polymorphisms (SNPs) and 756,234 short insertions and deletions. Functional SNP analysis predicted that the individual has a pathogenic risk for carnitine deficiency. We located the patrilineal inheritance of the Mongolian genome to the lineage D3a through Y haplogroup analysis and inferred that the individual has a common patrilineal ancestor with Tibeto-Burman populations and is likely to be the progeny of the earliest settlers in East Asia. We finally investigated the genetic imprints of Mongolians on other human populations using different approaches. We found varying degrees of gene flows between Mongolians and populations living in Europe, South/Central Asia, and the Indian subcontinent. The analyses demonstrate that the genetic impacts of Mongolians likely resulted from the expansion of the Mongolian Empire in the 13th century. The genome will be of great help in further explorations of modern human evolution and genetic causes of diseases/traits specific to Mongolians.
Keywords: Mongolian genome, de novo assembly, genetic variations, patrilineal origin, genetic imprints
Introduction
The Mongolian ethnic group, a population of East Asia, has approximately 10 million individuals. They primarily reside in China, Mongolia, Russia, the Republic of Kazakhstan, and other countries. The ethnogenesis of Mongolians is vaguely known. It was first recorded during the Tang Dynasty as “Mongol” or “Meng-wu,” a tribe of the Shih-wei (Twitchett and Fairbank 1994). The group is broadly considered to be a founding population of the New World (Kolman et al. 1996; Merriwether et al. 1996; Starikovskaya et al. 2005; Reich et al. 2012). The rise of the Mongolian Empire and conquests of the Eurasia continent (from the 13th to 19th centuries) (Twitchett and Fairbank 1994; Weatherford 2005) under Genghis Khan and his successors have played a major role in the last 1,000 years of human evolution. Known as a typical nomadic people, Mongolians have evolved into a modern day ethnic group with their own culture, language, life style (Komatsu et al. 2006, 2008, 2009), and phenotypic and physiological traits (Zheng et al. 2002) through recent adaptation to characteristic environments.
Next-generation sequencing technologies made the sequencing of the 1,000 genomes (1000 Genomes Project Consortium 2010, 2012) a reality and facilitated genome-based, personal medicine. Representative genomes of increasing numbers of human populations have been sequenced to dissect the structure and history, including Indian (Reich et al. 2009), American (Reich et al. 2012), and Jewish (Behar et al. 2010). In addition, genome-wide genetic variation maps have been compiled for population-specific genetics research, such as Dutch (Genome of the Netherlands Consortium 2014) and British (http://www.uk10k.org). However, Mongolian population history has only been explored through Y haplogroup (Zhong et al. 2010) and M haplogroup (Derenko et al. 2007), and the studies of genetics and diseases of Mongolians are still at a rudimentary level (Svobodova et al. 2007; Tsunoda et al. 2012). A Mongolian reference genome and population data are lacking. They are increasingly necessary to explore characteristics of population evolution, disease, and personal healthcare.
In this study, we sequenced the genome of a representative Mongolian male individual with high coverage (>100×) by using the next sequencing technology. We then presented a high-quality Mongolian genome draft produced from hierarchical de novo assembly strategy. Based on human reference genome (GRCh37/hg19), we constructed a high-resolution Mongolian personal genetic variation map, including single nucleotide polymorphisms (SNPs), short insertions and deletions (indels), structural variations (SVs), and novel sequences and haplotypes. We also predicted Mendelian diseases risk for the individual by analyzing potential functional SNPs. Through the haplogroup analyses of Y chromosome and mitochondria genome, we traced the patrilineal and matrilineal transmissions of the Mongolian genome. Based on the sequence of Mongolian genome, we investigated the genetic imprints of Mongolians on global ethnic groups through different approaches. Broadly, the Mongolian genome data and analyses will be of value to future researches on the origin and evolution of Euro-Asian-America populations and Mongolian characteristic traits and diseases.
Materials and Methods
Ethics Statement
The sample donor has signed the written informed consent. According to the related items of informed consent form (supplementary fig. S1, Supplementary Material online), the donor has agreed that his genomic data can be used for genetic studies and can be freely released to the public for future studies. The study has been approved by the Institutional Review Board on Bioethics and Biosafety.
Libraries Preparation and Sequencing
Genomic DNA was extracted from the peripheral blood of sample donor. DNA libraries with multiple insert sizes (200, 500, 800 bp, 2, 5, 10, 20, and 40 kb) were constructed according to the protocol of Illumina sequencing platform. For libraries with short insert size (200, 500, and 800 bp), 3 µg of DNA for each library was fragmented to the expected insert sizes, repaired the ends, and ligated to Illumina standard paired-end adaptors. Ligated fragments were size selected for 200, 500, and 800 bp on agarose gel and were purified by polymerase chain reaction (PCR) amplification to produce the corresponding libraries. For the mate-pair libraries with large insert sizes (2, 5, 10, 20, and 40 kb), 60 µg of genomic DNA was needed for each library. We cyclized genomic DNA, digested linear DNA, fragmented cyclized DNA, and purified biotinylated DNA and then performed adaptor ligation. Finally, all libraries were sequenced on the Illumina HiSeq 2000 sequencing platform.
De Novo Assembly of the Genome
De novo assembly of the Mongolian genome was performed by using short oligonucleotide analysis package SOAPdenovov2 (Luo et al. 2012). We applied a hierarchical assembly strategy to construct the genome sequence from contigs to scaffolds, in which we added paired-end reads step by step from short insert size to long insert size. The reads, due to PCR duplication and adaptor contamination, plus the low quality ones, were filtered out. Read pairs from the libraries with short insert size (<1 kb) were then assembled into distinct contigs based on the K-mer overlap information. Next, read pairs derived from long insert-size libraries (>1 kb) were aligned to the contig sequences, and the paired information was used to construct the scaffolds. For the final step of gap filling, we used the read pairs that had one read anchored on a contig and the mate read located within the gap region to perform local assembly.
We anchored the scaffolds onto the chromosomes of the human reference genome by following several steps. First, we extracted seed sequences (20 kb, without N) from each scaffold. For small scaffolds (<40 kb), only one effective sequence was randomly selected as the seed sequence. For large scaffolds (≥40 kb), multiple seeds (20 kb) were extracted from each one. We then aligned the seed sequences onto the human reference genome. The cutoff value of similarity was set at 90%. We ultimately obtained the chromosome sequences for the Mongolian genome.
Assembly Evaluation
In this study, we used several approaches to evaluate the Mongolian genome assembly. A total of 7,974 expressed sequence tags (ESTs) and 42 bacterial artificial chromosome sequences (BAC) were downloaded from the National Center for Biotechnology Information (NCBI) database. In the evaluation, BLAT software (Kent 2002) was applied to align the ESTs to the assembled genome. Nucmer (V3) (Kurtz et al. 2004) and BLASTN were used to map the BAC to the scaffolds. Additionally, we calculated GC content of every 500-bp (with a 250 bp of step size) sliding window along the genome draft and compared the GC content with the human reference genome and YH genome to assess assembly quality.
Assembly Annotation
We here also carried out the annotation of the genome draft using the pipeline developed by BGI (Li, Fan, et al. 2010). For repeats annotation, we used the RepeatModeler (1.0.5) to construct the de novo repeat library. We also used the RepeatMasker (version 3.3.0) (http://repeatmasker.org) with the repeat library (RepBase 16.01) to predict known translocation elements and applied the Tandem Repeat Finder to identify tandem repeats.
We further predicted a high-quality gene set for the Mongolian genome by applying a combined strategy of integrating homology-based and de novo approaches. The prediction was based on the repeat-masked assembly using Augustus (Stanke et al. 2006) and GENSCAN (Burge and Karlin 1997). The size of each gene was required to be at least 150 bp. Homology-based gene prediction was then carried out as follows: 1) Rough alignment of sequences to the protein sequences of Chimpanzee (Pan troglodytes) and humans, 2) precise mapping using GeneWise (Birney et al. 2004), 3) transcripts clustering, 4) building the gene-scaffold, 5) filtering pseudogenes, and 6) Untranslated region attachment. We ultimately manually integrated de novo prediction gene set and synteny alignment prediction gene set into a final consensus gene set.
Annotation of gene function was finally performed based on alignment of the genes to the SwissProt and Translated EMBL Nucleotide Sequence Data Library databases (Bairoch and Apweiler 2000). The motifs and domains were predicted by scanning the InterProScan (Zdobnov and Apweiler 2001) of the sequences against publicly available databases, including Pfam (Finn et al. 2010), PRINTS (Attwood et al. 1994), PROSITE (Hulo et al. 2006), ProDom (Bru et al. 2005), and SMART (Letunic et al. 2012). We also aligned annotated genes to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Ogata et al. 1999) and identified the best matched pathways for each gene.
Identification of SNPs and Short Indels
To build the personal genetic variation map for the Mongolian genome, we first aligned selected high-quality short reads onto the human reference genome by using the Burrows–Wheeler Aligner program (BWA, v 0.6.2) (Li and Durbin, 2009).
We then adopted a multialgorithm supporting strategy (fig. 1B) to identify the high confidence SNPs and short indels. Based on the alignment of selected short reads, each of SOAPsnp (Wang et al. 2008; Li R, Li Y, et al. 2009), GATK (McKenna et al. 2010; DePristo et al. 2011), and SAMtools (Li, Handsaker, et al. 2009) was used to detect SNPs of the Mongolian genome, respectively. Rigorous quality control of three raw SNP sets was then carried out. For the set derived from SOAPsnp, we used the following criteria: 1) The lowest quality value of genotype was set to 13 (P > 0.95), 2) each allele of every candidate SNP had to be supported by at least three uniquely mapped reads, 3) the number of total covered reads did not exceed 100 and uniquely mapped reads were not less than 50% of the total reads, and 4) the average copy number of each site (mapped hit number/mapped reads number) was not larger than 2. For the SNP sets from GATK and SAMtools, we refined the SNPs by using the same threshold value of genotype quality (≥13) and read coverage (≥6 and ≤100). Subsequently, three SNP sets were integrated into a final SNP set by selecting the sites that were supported by at least two of three approaches.
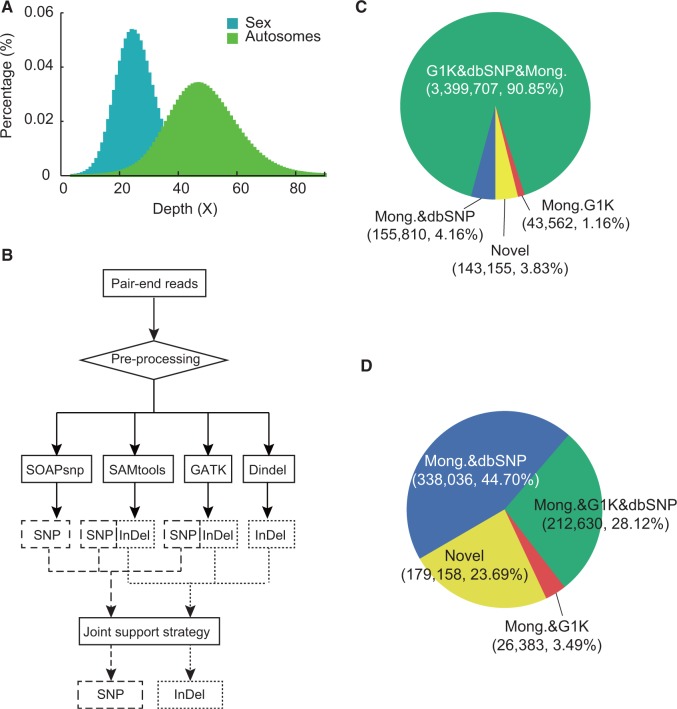
Fig. 1.—
Mapping depth, detection strategy, and variant composition (SNP and short indel) in construction of genetic variation map. (A) Depth distribution based on the alignment. (B) The strategy of SNP and short indel identification. (C, D) Composition of SNP and short indel sets of the Mongolian genome compared with variant sets of dbSNP and 1000 genomes project.
Similarly, we applied three methods, GATK, SAMtools, and Dindel (Albers et al. 2011) to identify short indels (fig. 1B). For each raw short indel set, we required every candidate to be covered by ≥6 reads and call quality to be ≥13. The sites supported by at least two methods were also integrated into a final short indel set.
Evaluation of SNPs and Short Indels
To evaluate the accuracy of our final SNP set and short indel set in this study, we genotyped the Mongolian genome using HumanOmni2.5S Beadchip (Illumina). We then compared the genotyping data derived from the chip with the calls predicted by sequencing to assess the accuracy of our SNP calling. In addition, we designed PCR and carried out Sanger sequencing for dozens of SNPs and short indels to evaluate the accuracy of our identified SNPs and indels.
SVs Detection
In this work, we used the assembly-based SOAP detection pipeline (Li et al. 2011) to identify the high-quality SV set of the Mongolian genome. The pipeline includes the following several steps: 1) Alignment, 2) SV candidate calling, and 3) SV validation (supplementary fig. S6, Supplementary Material online). First, we aligned the assembled scaffolds to the human reference genome using the BLAT program. The mapped scaffolds were then aligned to the human reference genome again by using the LASTZ program (Harris 2007) for precise mapping. SV candidates were called by using the SOAPsv program (http://soap.genomics.org.cn). Large insertions/deletions (>100 bp), the largest proportion of the SV candidates, were finally filtered by S/P ratio (number of single-end mapped reads/number of paired-end mapped reads) (Li et al. 2011). Length of the majority of candidates ranges from 50 bp to 100 kb.
Novel Sequence Detection
We defined novel sequence of the Mongolian genome using unmapped reads with respect to the reference human genome. The reads unmapped onto the human reference genome were collected and regarded as the primary candidates of novel sequences. We further mapped these primary candidates to YH genome (Luo et al. 2012) and collected the remaining unmapped reads. Subsequently, we aligned these unmapped reads again onto the Mongolian assembled draft by using rigorous parameters. The reads that had no mismatch and uniquely mapped ones were retained. Finally, the sequences with length ≥100 and covered by at least three uniquely mapped reads were defined as the novel sequences of the Mongolian genome.
Haplotype Block Prediction
We here predicted the haplotype blocks of the Mongolian genome based on the genotype data of the Human Genome Diversity Project (HGDP) (Li et al. 2008). In brief, using the Haploview software (Barrett et al. 2005), we first combined genotypes of ten Mongolian individuals of HGDP and the studied individual to infer the haplotype blocks for the small population (R2 > 0.8). Then, through scanning the SNP sites of the individual which overlapped with predicted haplotype blocks, we ultimately obtained the haplotype blocks of the Mongolian genome.
Functional SNPs and Diseases Risk
To predict the Mendelian diseases risks of the individual, we scanned an in-house human mutation database for each SNP of the Mongolian genome. An SNP is considered disease related if it meets all of following criteria: 1) Is functionally related (synonymous, missense, stop gain, stop loss, splicing error, and frame shift), 2) has low mutated allele frequency in the human population (<0.05), and 3) is reported to be related to a disease in at least two cases. According to recommendations from the American College of Medical Genetics and Genomics (ACMG), we finally divided all targeted mutations into five categories: Pathogenic, likely pathogenic, uncertain, likely benign, and benign.
Haplogroup Analysis of Y Chromosome and Mitochondrial Genome
We performed inference of patrilineal inheritance of the individual through Y haplogroup analysis. We checked alleles of all markers released in the Y haplogroup phylogenetic tree (Karafet et al. 2008) and corrected inconsistent ones when compared with the genotypes of dbSNP database (build 135) and synteny analysis between Chimpanzee and human reference genomes. We reassigned the ancestral alleles and strand information to all markers (SNPs) (supplementary table S17, Supplementary Material online). In total, 537 of 571 sites were finally confirmed and used for the subsequent Y haplogroup analysis. We compared the alleles of these markers with the phylogenetic tree and traced the patrilineal transmission pathway of the studied individual. Based on the located lineage, we inferred the ancestor of the male.
Similarly, we also traced the matrilineal inheritance of the individual by comparing the ancestral/derived alleles of all markers against the released phylogenetic trees derived from two mitochondria genome versions, namely, RSRS (Behar et al. 2012) and rCRS (Anderson et al. 1981; Andrews et al. 1999). When multiple lineages share derived alleles of the same markers, the one with the largest number of contiguous markers is considered the proper matrilineal transmission pathway.
Ancestral Proportion Analysis
In this study, we used a resource-efficient computing program ADMIXTURE (Alexander et al. 2009). The method is based on an algorithm of maximum-likelihood estimation to account for an assumed ancestral proportion of the Mongolian genome. We utilized genotype data of 1,042 individuals of HGDP from 50 global populations (plus the studied Mongolian sample) as the genetic reference to estimate the ancestral proportion. To avoid potential bias, we conducted a linkage disequilibrium analysis using PLINK program (v1.07) (Purcell et al. 2007) to filter out closely linked sites (r2 > 0.4). In the final analysis, we set the assumed ancestral populations from K = 2 to 20.
D Test (ABBABABA Test)
We performed D tests (ABBABABA test) (Green et al. 2010; Reich et al. 2010; Rasmussen et al. 2011) to evaluate gene flows between Mongolians and other modern human populations. We calculated standard errors and Z-scores using the bootstrap method to estimate significance of the D values.
We carried out the D tests using the different population model for different block sizes (1, 2, 5, 8, and 10 Mb) (supplementary table S17, Supplementary Material online) and chose to use the one with the smallest standard error in the subsequent bootstrap analyses. In the D-test analysis, sampling size was set at 10% of D-value pool and bootstrap time was set to 50,000 for each population combination.
We used each of Chimpanzee genome and Yoruba genome (Bentley et al. 2008) as an outgroup for every comparison.
Gene Flows Based on the Analysis of Haplotype Blocks
Next, we used the haplotype blocks to investigate the genetic imprints of Mongolians on other human populations. We assume that two populations have greater degree of gene flows in evolutionary history when they share more haplotype block compared with another two (Pearson χ2 test, P = 0.05). We constructed the cursory haplotype blocks from the HGDP genotypes of several groups using the Haploview program, including samples of Yoruba, Mongolian (ten of HGDP plus the individual of this study), Russian, Han, Caucasian (Adygei), Maya, French, Brahui, and Palestinian. In the analysis, the same number of individuals was selected for each group. To eliminate genetic noises caused by relatively shorter haplotypes of Africans, we excluded the shared haplotype blocks with Africans from each group. The shared haplotypes of different population pairs were compared against each other and Pearson chi-square tests were calculated to determine the significance of differences.
Data Access
All genome sequenced reads have been deposited in the NCBI short read archive database (access number: SRA105951). The assembled genome, annotation results, SNPs, short indels, and SVs produced in the study have been deposited in our Mongolian genome database, http://dx.doi.org/10.5524/100104. They also can be obtained from another ftp site: ftp://public.genomics.org.cn/BGI/MongolianGenome.
Results
Sampling and Genome Sequencing
We chose a Mongolian male living in the Inner Mongolia Autonomous Region of China as the study sample. The donor signed the informed consent form for use of his genomic data in the study and agreed to release of the data to the public prior to sampling. The genomic DNA was extracted from the peripheral blood of the individual.
We sequenced genomic DNA of the individual using the whole-genome shotgun strategy on Illumina HiSeq 2000 sequencing platform. A total of 5.6 billion paired-end reads were generated from sequencing of libraries with different fragment sizes (200, 500, 800 bp, 2, 5, 10, 20, and 40 kb) and the coverage reached 130.8-folds of the genome. Total of 392.37-Gb genomic data were produced (supplementary table S1, Supplementary Material online).
De Novo Assembly and Annotation
De novo assembly yielded over 2,000 contigs/scaffolds and a draft genome with a total length of 2.9 Gb. The N50 sizes of contigs and scaffolds have reached 56.2 kb and 7.6 Mb, respectively (table 1). Approximately 90% of the genome draft was covered by 431 scaffolds with each having size larger than 1.4 Mb, of which the largest scaffold spans 36 Mb. 98.69% of the draft was covered by more than 20-folds (supplementary fig. S2, Supplementary Material online), which presents high assembly accuracy at the nucleotide level. The assembly covered 96.68% of 42 BAC and 98.3% of 7,974 ESTs derived from NCBI database, indicating high coverage of the genome including genic regions (supplementary tables S2 and S3, Supplementary Material online). Distribution of GC content is similar to that of the human reference genome and YH genome (Luo et al. 2012) (supplementary fig. S3, Supplementary Material online) indicating the high quality of the genome draft. Furthermore, we were able to align 96% of the scaffolds onto chromosomes by mapping the representative seed sequences (20 kb) to the human reference genome.
Table 1.
Summary of the Mongolian Genome Assembly
| Contig |
Scaffold |
|||
|---|---|---|---|---|
| Size (bp) | Number | Size (bp) | Number | |
| N90 | 13,773 | 52,725 | 1,461,661 | 431 |
| N80 | 24,225 | 37,548 | 2,880,171 | 292 |
| N70 | 34,160 | 27,774 | 4,350,155 | 210 |
| N60 | 44,637 | 20,547 | 5,856,852 | 154 |
| N50 | 56,244 | 14,915 | 7,632,466 | 111 |
| Longest | 517,634 | — | 35,963,476 | — |
| Total size | 2,823,488, 473 | — | 2,881,945, 563 | — |
| Total number (≥100 bp) | — | 320,927 | — | 221,013 |
| Total number (≥2 kb) | — | 84,214 | — | 3,251 |
Using the integrated strategy of de novo prediction and homology-based alignment, the assembled genome was annotated for repeat elements, coding genes, and other noncoding RNAs. A total of 1,362-Mb repetitive sequences were predicted, accounting for 47.3% of the genome draft. The largest proportion of the repetitive sequences is LINE, contributing 31.3% of the genome (supplementary table S4, Supplementary Material online). Similarly, using the integrated strategy, we identified 21,264 protein-coding genes, with an average size of 41 kb for mRNA, 1.5 kb for coding sequences (CDS), 177 bp for exon, and 5.5 kb for intron (supplementary table S5, Supplementary Material online), which is similar to that of the human reference genome (47, 1.7, 173, and 5.3 kb, respectively). Scanning the public protein databases revealed that 91.7% of gene models having at least one matched hits, showing the high-confidence gene set of the genome assembly.
Genetic Variations Pattern
To build the genetic variations pattern of the Mongolian genome, we picked four lanes of genomic data generated from four libraries with small insert size (∼500 bp) for subsequent analyses. A total of 1.39 billion reads (139.20 Gb) were obtained and the proportion of high quality data (Q20) reached as high as 93.8% (supplementary table S6, Supplementary Material online).
All selected short reads were then mapped onto the human reference genome. 96.76% of the reads were mapped to the reference, accounting for 46.5-fold effective genome coverage (without N regions) (fig. 1A) (supplementary table S6, Supplementary Material online). In total, 99.74% and 94.42% of the effective genome was covered by at least 1 read and 20 reads, respectively (supplementary table S7, Supplementary Material online). 0.48% of all bases in the uniquely mapped reads mismatched the human reference genome.
Using a combined variation calling strategy that integrates multiple methods simultaneously (fig. 1B, see Materials and Methods), we identified a total of 3,742,234 high-confidence SNPs and 756,234 short indels (<50 bp) (supplementary table S10, Supplementary Material online), which are distributed across the genome with varying density (fig. 2). To evaluate the variations, we first compared SNP set with the Illumina 2.5M genotyping data of the Mongolian genome. After excluding missing sites and sites with low quality from the chip genotyping data, the genotype concordance rate between the SNP call set and the chip genotyping set (both position and two alleles) reached 98.48% in a total of 2,356,566 sites. Additional 24,941 sites (1.06%) in two data sets have one matching allele (supplementary table S8A, Supplementary Material online). For 685,036 SNPs, the two-allele and one-allele matching rates are 99.67% and 0.32%, respectively (supplementary table S8B, Supplementary Material online). Second, we used PCR and Sanger sequencing to validate dozens of SNPs and short indels. Out of 34 SNPs tested, both alleles of 31 SNPs (all 12 homozygous and 19 heterozygous) were verified completely. For the remaining three heterozygous SNPs, only one allele had been validated. From a different point of view, 17 of 18 exonic SNPs were confirmed in the experimental validation. In total, 51 (Exon, 26; Intron, 8; Intergenic, 17) of 54 short indels were validated completely (supplementary table S9, Supplementary Material online).
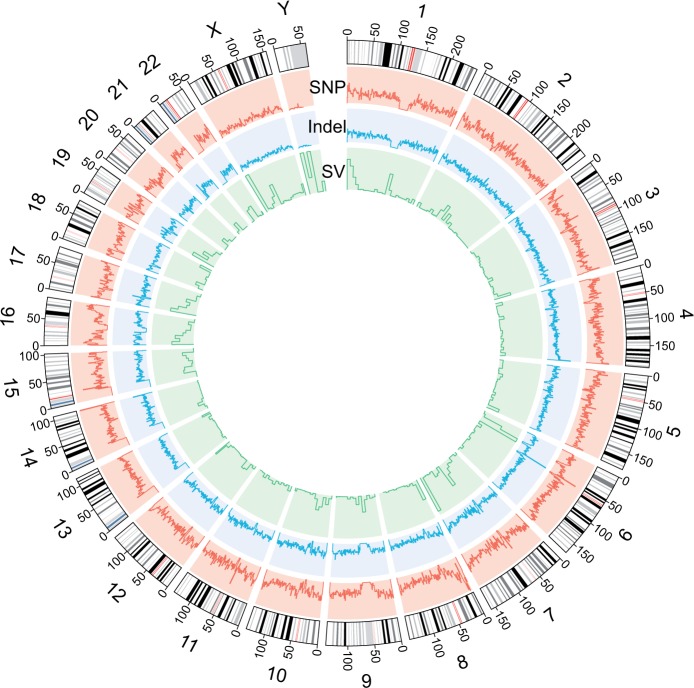
Fig. 2.—
Genetic variation pattern of Mongolian genome. Distribution of genetic variations by chromosomes, including SNP, short indel, and SV. The sizes of window and sliding step are 1 and 0.5 Mb for SNP and short indel, 2 and 1 Mb for SV.
Based on the SNP and short indel sets of the Mongolian genome, we investigated the distribution of these two types of variation in the public databases, including dbSNP (build 135) and the 1000 genomes project (released in May 2011) (supplementary fig. S4, Supplementary Material online). For SNPs, a total of 3,399,707 (90.85%) were found in both databases; 155,810 (4.16%) and 43,562 (1.16%) were identified in dbSNP set and 1000 genomes set, respectively; and 143,155 (3.83%) were novel (fig. 1C). For short indels, 212,630 (28.12%) were found in both sets; 338,038 (44.7%) and 26,383 (3.49%) were only included in dbSNP or 1000 genomes, respectively; and 179,158 (23.69%) were novel (fig. 1D).
ANNOVAR software (Wang et al. 2010) was used to annotate SNPs and short indels (supplementary table S10, Supplementary Material online). A majority of SNPs and short indels were located in the intergenic regions (60.58% of SNPs, 58.25% of indels) and introns (34.36% of SNPs, 36.85% of indels). A total of 43,014 (1.15%) SNPs and 10,131 (1.34%) short indels were located in potential regulatory regions (2-kb flanking regions of each gene). Only 21,233 (0.57%) of SNPs and 528 indels (0.07%) were found in CDS, indicating higher conservation of coding regions. Additionally, as many as 532 SNPs and 514 indels were found, involving 535 and 468 genes, respectively, which give rise to large functional effects, such as missense, stop gain, stop loss, splicing error, and frame shift in gene expression (supplementary table S11, Supplementary Material online). Distribution of short indels in CDS (without those of 1 bp) shows that 3n-bp indels are much more frequent than non-3n-bp indels (supplementary fig. S5, Supplementary Material online).
To identify the SVs of the Mongolian genome, the assembly-based SV detection strategy was applied. We ultimately obtained 15,088 large indels (>50 bp) and 1,210 candidate inversions (from 8 bp to 45 kb) (fig. 2 and supplementary table S12, Supplementary Material online). The length distribution of the SVs in the Mongolian genome is very similar to that of YH genome (Li et al. 2011), including a peak at the size of about 300 bp, potentially caused by abundance of Alu elements (supplementary fig. S7, Supplementary Material online). However, the comparative analysis also shows that the Mongolian genome has fewer short SVs (<100 bp) and more large SVs (>100 bp) than YH genome.
Novel sequences possess typical individual and population specificity (Li R, Li Y, et al. 2010) and here were regarded as a type of genetic variation. By comparing the Mongolian genome with the human reference genome and YH genome, a total of 26.9-Mb novel sequences were obtained, including 15,198 scaffolds. Scanning the annotated gene set of the Mongolian genome, we found 24 genes in these novel sequences. Fourteen of them exist in the databases of KEGG, Trembl (Bairoch and Apweiler 2000), SwissProt, and InterProScan (supplementary table S13, Supplementary Material online).
In this study, we introduced the genotype data of Mongolian individuals from the HGDP to predict the haplotype blocks of Mongolian genome. A total of 8,592 haplotype blocks were first obtained based on the genotype data of 11 Mongolian individuals (ten of HGDP and Mongolian genome), which cover 9.1% of human reference genome. In these Mongolian haplotype blocks, we found that 2,310 with an average size of 16.4 kb are in Mongolian genome (supplementary table S14 and fig. S8, Supplementary Material online).
Functional SNPs and Mendelian Diseases
Based on an in-house human mutation database, we obtained 16 heterozygous SNPs (table 2) after filtering with a series of criteria (see Materials and Methods). From further analyses of these potential diseased related mutations, only one was identified as pathogenic, one is likely pathogenic, two are benign, nine are likely benign, and remaining three SNPs are not supported by current medical data. The pathogenic mutation (rs60376624, c.1400C>G) located in gene SLC22A5 was proposed to be a causative mutation for systemic primary carnitine deficiency (CDSP) (Koizumi et al. 1999; Yoon et al. 2012). CDSP is an autosomal recessive disorder of the carnitine cycle and patients with CDSP have defects in the ability to transform fat to energy during periods of stress and fasting (Longo et al. 2006). The likely pathogenic mutation (rs17102999, c.2825C>T) in MLH3, an Asian-specific mutation, was reported to be present at low frequency in endometrial cancer patients (Tylor et al. 2006).
Table 2.
Risks of Mendelian Diseases Based on Functional SNPs
| Disease | Gene | SNP ID | Mutation | Pathogenicitya |
|---|---|---|---|---|
| Systemic primary Carnitine deficiency | SLC22A5 | rs60376624 | c.1400C > G (p.Ser467Cys) | Pathogenic |
| Endometrial cancer | MLH3 | rs17102999 | c.2825C > T (p.Thr942Ile) | Likely pathogenic |
| Crohn’s disease | IL23R | rs76418789 | c.445G > A (p.Gly149Arg) | Benign |
| Parkinson disease | PARK7 | rs71653619 | c.293G > A (p.Arg98Gln) | Benign |
| Hypertension | XDH | rs45523133 | c.514G > A (p.Gly172Arg) | Likely benign |
| Colorectal cancer | MSH6 | rs63750252 | c.3488A > T (p.Glu1163Val) | Likely benign |
| Glaucoma 1 | OPTN | rs75654767 | c.1634G > A (p.Arg545Gln) | Likely benign |
| Carnitine palmitoyltransferase 1 deficiency | CPT1A | rs2229738 | c.823G > A (p.Ala275Thr) | Likely benign |
| Hemolytic uremic syndrome | CFH | rs62625015 | c.3226C > G (p.Gln1076Glu) | Likely benign |
| Macular dystrophy | ABCA4 | rs76258939 | c.3626T > C (p.Met1209Thr) | Likely benign |
| Hyperoxaluria | AGXT | rs34664134 | c.590G > A (p.Arg197Gln) | Likely benign |
| Idiopathic generalized epilepsy (IGE) | CLCN2 | rs111656822 | c.2063G > A (p.Arg688Gln) | Likely benign |
| Fuchs endothelial cornea dystrophy (FECD) | COL8A2 | rs75864656 | c.464G > A (p.Arg155Gln) | Likely benign |
| Nephrotic syndrome | NPHS1 | rs114849139 | c.2869G > C (p.Val957Leu) | Uncertain |
| Cardiomyopathy | MYBPC3 | rs193068692 | c.478C > T(p.Arg160Trp) | Uncertain |
| Ectopia lentis | ADAMTSL4 | rs76075180 | c.926G > A (p.Arg309Gln) | Uncertain |
aThe pathogenicity is annotated based on the recommendations of the ACMG.
Haplogroup Analysis
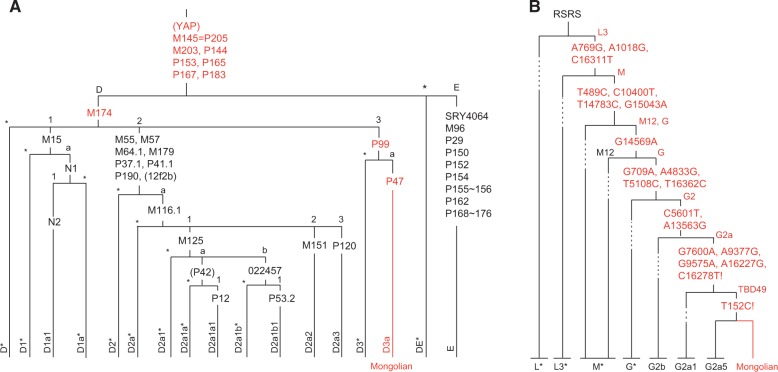
Based on the released markers reported in the previous Y haplogroup study (Karafet et al. 2008), we used a confirmed set of 537 markers to trace the patrilineal transmission of the Mongolian individual (supplementary table S15, Supplementary Material online). By scanning the ancestral/derived alleles of the selected markers in the released Y haplogroup tree, we found a consecutive patrilineal transmission pathway and assigned the patrilineal ancestor of the individual to the lineage D3a (fig. 3A and supplementary fig. S9, Supplementary Material online). This inference supports the frequent presence of lineage D (D3) in Mongolians as widely reported in previous studies (Deng et al. 2004; Katoh et al. 2005; Shi et al. 2008; Zhong et al. 2010).
Fig. 3.—
Haplogroup of Y chromosome and mitochondria genome. (A) Patrilineal transmission of Mongolian genome based on Y haplogroup. Red markers are validated as derived genotypes in the Mongolian genome and remaining ones (black) are ancestral type. The bracketed markers were not validated in the Mongolian genome. (B) Matrilineal transmission of the Mongolian genome based on mitochondria haplogroup. Red markers were confirmed to be derived in the Mongolian genome. The mutations labeled with an exclamation point are reported in the mitochondria sequence of rCRS version. L* represents all L lineages but L3, including L0, L0, L2, L4, L5, and L6; L3* represents all L3 lineages but M, including L3a to L3f, L3h, L3i, L3k, L3x, and N; M* represents all lineages prefixed “M,” D, and Q, but G; G* represents G1, G3, and G4.
We then traced the matrilineal transmission of the individual through Mt haplogroup analysis. Similarly, by scanning the ancestral/derived alleles of all markers in the Mt haplogroup database, we discarded one aberrant mutation (C182T) and located the matrilineal ancestor of the Mongolian genome in a novel sublineage under the lineage G2a (T152C) in the RSRS tree as well as the sublineage G2a2 (T711C and C8943T) under the lineage G2a (T152C) in rCRS tree (fig. 3B and supplementary fig. S10 and table S16, Supplementary Material online). The lineage occurs frequently in populations of Northeast Asia, including Mongolians, Daur, Buryat, and Yakut (Tanaka et al. 2004; Derenko et al. 2007).
Genetic Imprints of Mongolians
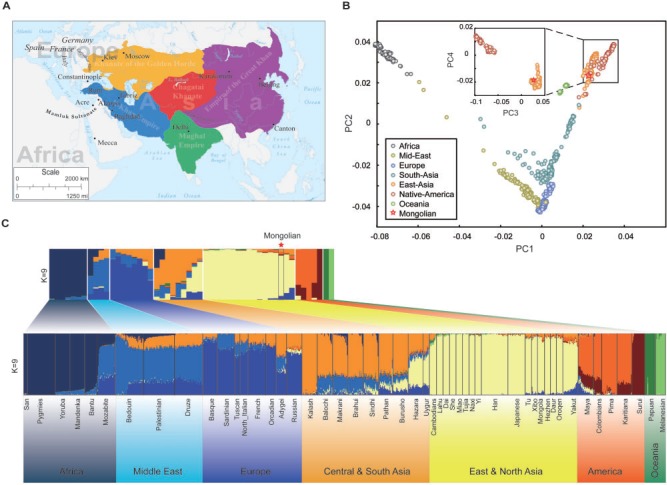
After formation, the Mongol Empire expanded into the largest contiguous empire in human history under Genghis Khan and his successors. The Mughal Empire expanded the reign of Mongolian lineage to the Indian subcontinent (Richards 1993) (fig. 4A and supplementary fig. S11, Supplementary Material online). Although sociocultural impacts of Mongolians have been well documented, the evidences of molecular genetics are limited and attract great attention. We introduced here the genotype data of HGDP and Indian ethnic groups (Reich et al. 2009) to investigate the genetic imprints of Mongolians on other modern populations.
Fig. 4.—
History and population genetic structure. (A) World regions once occupied by Mongolians from the 13th century to 19th century. (B) PCA plots of 1,042 individuals from 50 global ethnic groups. (C) The ancestry proportion plot of 1,042 individuals using the ADMIXTURE with K = 9. The Mongolian genome is marked by a red star.
Principal component analysis (PCA) was performed to confirm the position of the sample in genetic maps of human populations. As we expected, the analysis placed the Mongolian genome close to the populations of East Asian and American (fig. 4B, major plot), especially in the northern, East Asian populations such as the Daur, Oroqen, Han, and Japanese (fig. 4B, minor plot). We further estimated the ancestral proportions of the Mongolian genome with the ADMIXTURE program, assuming the ancestor groups from K = 2 to 20 (supplementary fig. S12, Supplementary Material online). The geographically representative estimation of K = 9 (fig. 4C) presented the ancestral proportions of Mongolian genome. The analysis indicates that Mongolians mainly possess four ancestral proportions, including East Asians, South/Central Asians, Europeans, and from the Americas. The proportion of Americans supports that Mongolians might have contributed to the foundation of the New World as reported (Kolman et al. 1996; Merriwether et al. 1996). The part of North/East Asians might have been resulted from the recent common ancestor before moving into East Asia and gene flows after divergence from groups in other continents. The remaining two ancestral proportions of South/Central Asians and Europeans likely reflect gene flows between Mongolians and those populations.
We then applied the D test (ABBABABA test) to estimate the gene flows between the Mongolians and other human populations. In the analysis, the bootstrap method using a tested block size of 5 Mb (supplementary table S17, Supplementary Material online) was employed to calculate the statistic D value (see Materials and Methods). Using Chimpanzee as the outgroup, the fewest shared ancestral alleles with Africans and adjacent populations (Middle East) (|Z| >> 7 or P << 10−12) were found, indicating no evidence of gene flows between Mongolians and Africans after the ancestors of Mongolians leaving Africa (supplementary table S18, Supplementary Material online). We then used Yoruba genome as the outgroup to capture the gene flows between Mongolians and other non-Africans. Consistent with the inferences from the PCA and ADMIXTURE, the groups of East Asian (Daur, Oroqen, Hezhen, Xibo, Tu, Han, and Japanese) shared more ancestral alleles than groups of other regions (table 3 part 1). Given the geographic isolation, the relatively large amount of ancestral alleles shared with Native Americans (Maya) most likely have resulted from the Mongolians’ contribution to peopling of the Americas, but not from any recent gene flow (table 3 part 2). The fact that groups in Central/South Asia and Europe shared more ancestral alleles with Mongolians than the groups in Middle East and less with the groups in East Asia and America (table 3 part 3) is probably a result from the traces of early human migration and/or the expansion of Mongolians. However, in each geographic region, such as Middle East, Central/South Asia, and Europe, different populations possess significantly different amount of shared ancestral alleles (Middle East: Druze > Palestinian > Bedouin; European: Russian/Adygei >French/North_Italian/Tuscan/Orcadian > Sardinian/Basque in France/other groups; Central/South Asian: Hazara/Uygur >Kalash/Burusho/Pathan > Makrani) (table 3 parts 4 and 5 and supplementary table S18, Supplementary Material online). This observation approximately matches the route of the Mongol Empire expansion in the 13th century (fig. 4A and supplementary fig. S11, Supplementary Material online).
Table 3.
D Test between Mongolians and Other Modern Humans
| Group 1 | Group 2 | Yoruba |
Chimp |
|||||
|---|---|---|---|---|---|---|---|---|
| D (%) | SE (%) | Z | D (%) | SE (%) | Z | |||
| East Asian to other (Part 1) | Han | Maya | −3.4 | 0.34 | −10 | −4.4 | 1.20 | −3.6 |
| French | Han | 13.7 | 0.49 | 28.2 | 13.3 | 1.08 | 12.3 | |
| Daur | Russian | −11.9 | 0.21 | −56.8 | −12.1 | 0.11 | −106.7 | |
| Brahui | Daur | 14.2 | 0.35 | 40.8 | 13.3 | 0.14 | 94.8 | |
| Hezhen | Sindhi | −13.6 | 0.48 | −28.6 | −13.2 | 0.31 | −42.3 | |
| Japanese | North Italian | −14.2 | 0.44 | −32.5 | −13.1 | 0.31 | −42.1 | |
| Han | Palestinian | −16.5 | 0.56 | −29.2 | −16.5 | 0.71 | −23.3 | |
| American to other (Part 2) | Maya | Russian | −9.3 | 0.57 | −16.2 | −7.8 | 0.85 | −9.2 |
| Maya | Papuan | −8.3 | 0.15 | −55.4 | −8.8 | 0.25 | −35.7 | |
| Maya | North Italian | −10.9 | 0.40 | −27.1 | −10.1 | 0.31 | −33.1 | |
| Maya | Palestinian | −13.7 | 0.58 | −23.5 | −13.3 | 0.61 | −22.0 | |
| Bedouin | Maya | 32 | 0.25 | 128.3 | 31.7 | 0.43 | 73.0 | |
| Brahui | Maya | 11.3 | 0.91 | 12.4 | 10.1 | 0.66 | 15.4 | |
| Middle East groups to other (Part 3) | Bedouin | Brahui | 23.1 | 0.82 | 28.2 | 24.2 | 0.27 | 90.3 |
| Druze | French | 1.1 | 0.23 | 4.7 | 1.6 | 0.24 | 6.7 | |
| Palestinian | Russian | 5.1 | 0.28 | 18 | 5.1 | 0.51 | 10.1 | |
| Palestinian | Sindhi | 4.4 | 0.47 | 9.4 | 4.3 | 0.38 | 11.4 | |
| In European (Part 4) | Adygei | Russian | 0.2 | 0.19 | 1.2* | −0.7 | 0.26 | −2.8* |
| French | North Italian | −0.2 | 0.17 | −1.2* | −0.3 | 0.15 | −2.2* | |
| French | Tuscan | 0 | 0.40 | −0.1* | −0.7 | 0.40 | −1.8* | |
| French | Orcadian | −0.4 | 0.24 | −1.5* | 0.7 | 0.63 | 1.2* | |
| Basque | Sardinian | −0.6 | 0.24 | −0.3* | −0.8 | 0.80 | −1.0* | |
| Adygei | French | −1.9 | 0.32 | −5.8 | −2.2 | 0.94 | −2.3 | |
| French | Sardinian | −0.3 | 0.08 | −3.8 | −1.1 | 0.23 | −4.9 | |
| In South/Central Asian (Part 5) | Hazara | Uygur | 0 | 0.26 | 0.2* | 0.1 | 0.64 | 0.1* |
| Kalash | Burusho | 0.2 | 0.47 | 0.5* | 0.6 | 0.78 | 0.8* | |
| Pathan | Burusho | 0.6 | 0.27 | 2.1* | 0.1 | 0.25 | 0.2* | |
| Balochi | Brahui | 0.2 | 0.30 | 0.7* | −0.2 | 0.10 | −1.9* | |
| Balochi | Sindhi | −1.5 | 0.99 | −1.5* | −1.6 | 0.86 | −1.9* | |
| Makrani | Sindhi | 0.5 | 0.51 | 1.0* | 0.3 | 0.24 | 1.4* | |
| Hazara | Burusho | −5.3 | 0.25 | −21.2 | −5.5 | 0.24 | −22.6 | |
| Burusho | Brahui | −2.4 | 0.23 | −10.2 | −2.3 | 0.46 | −5.1 | |
| In Indian (Part 6) | Aonaga | Siddi | 27.9 | 1.15 | 24.2 | 23.1 | 0.26 | 89.1 |
| Kashmiri Pandit | Siddi | 20.1 | 1.37 | 14.7 | 16.7 | 0.23 | 71.6 | |
| Meghawal | Siddi | 19.1 | 0.33 | 58 | 17.4 | 0.78 | 22.3 | |
| Siddi | Vysya | −19.2 | 0.39 | −48.7 | −17.4 | 0.45 | −38.8 | |
| Indian to other (Part 7) | Brahui | Siddi | −5 | 0.87 | −5.7 | −5.8 | 5.08 | −11.5 |
| Balochi | Siddi | −4.2 | 0.34 | −12.3 | −5.3 | 0.37 | −14.3 | |
| French | Siddi | −3.5 | 0.57 | −6.2 | −4.4 | 0.54 | −8.2 | |
| North Italian | Siddi | −4.5 | 0.77 | −5.9 | −4.6 | 0.34 | −13.5 | |
Note.—D, the ABBABABA test statistic value; Z, Z-score. *, Mongolians possess statistically equivalent shared alleles with group1 and group 2 in the analyses of using two out-groups simultaneously. A tested stable block size 5 Mb (supplementary table S19, Supplementary Material online) was selected to calculate the D value and standard error by using the bootstrap method. The threshold values is |Z| = 3.
We also used the genotype data of Indian populations to investigate the genetic imprints of Mongolian lineage on the Indians. The results of D test showed that the Indian groups share different amounts of ancestral alleles with Mongolians (supplementary table S19, Supplementary Material online). We also found the people of Siddi, a subgroup of Dravidian who live on the southwest coast, have been proposed to be the closest group to Africans (Reich et al. 2009) possess the most shared ancestral alleles with Mongolians compared with other Indian groups. This commonality might have been introduced during the time of Mughal Empire (table 3 part 6). Although Indians have a certain amount of shared ancestral alleles, comparative analysis shows that the shared ancestral alleles with Indians are significantly fewer than that with Europeans and Central/South Asians, such as French, Italian, Balochi, and Brahui (|Z| >> 7 or P << 10−12) (table 3 part 7 and supplementary table S20, Supplementary Material online). It indicates that the shared ancestral alleles in Indians might have been introduced by historically later introduction of Mongolian linage, which is consistent with the records about the ancestry of Mughal Empire founders (Richards 1993).
We additionally used reconstructed haplotype blocks of representative human populations (Yoruba, French, Russian, Caucasian [Adygei], Brahui, Mongolian, Han, and Maya) to confirm the genetic imprints of Mongolians. As expected, Han-Mongolian possessed significantly more shared haplotype blocks compared with others (P << 0.05). The result that Maya shares more haplotypes with East Asian groups (Mongolian and Han) than the groups of other regions (Europe, South Asia, and Middle East) supports the close relationship between Native Americans and the Asians. Populations of Europe and South Asia possess more shared haplotypes with Mongolian than that with Maya, supporting our inference that the gene flows happened after ancestors of Native Americans moved to the New World, again likely during the period of the Mongol Empire expansion (table 4 and supplementary table S21, Supplementary Material online).
Table 4.
Genetic Imprints of Mongolian Genome on Global Populations Based on Shared Haplotype Blocks
| Group 1 |
Group 2 |
P Value | ||
|---|---|---|---|---|
| Populations | Shared Blocksa | Populations | Shared Blocks | |
| Han-Mongo. | 801 | French-Mongo. | 258 | <2.2E-16 |
| Han-Mongo. | 801 | Mongo.-Russian | 292 | <2.2E-16 |
| Han-Mongo. | 801 | Maya-Mongo. | 284 | <2.2E-16 |
| Han-Mongo. | 801 | Adygei-Mongo. | 273 | <2.2E-16 |
| Han-Mongo. | 801 | Mongo.-Brahui | 240 | <2.2E-16 |
| Han-Mongo. | 801 | Mongo.-Burusho | 162 | <2.2E-16 |
| Han-Mongo. | 801 | Mongo.-Palestinian | 222 | <2.2E-16 |
| Maya-Mongo. | 284 | French-Maya | 218 | 3.2E-3 |
| Maya-Mongo. | 284 | Maya-Russian | 232 | 2.2E-2 |
| Maya-Han | 289 | Maya-French | 289 | 1.6E-3 |
| Maya-Mongo. | 284 | Adygei-Maya | 240 | 5.5E-2 |
| Maya-Mongo. | 284 | Maya-Palestinian | 179 | 1.1E-6 |
| Maya-Mongo. | 284 | Maya-Burusho | 110 | <2.2E-16 |
| Maya-Mongo. | 284 | Maya-Brahui | 179 | 1.1E-6 |
| Brahui-Mongol. | 240 | Brahui-Maya | 179 | 2.9E-3 |
| Burusho-Mongo. | 162 | Burusho-Maya | 110 | 1.6E-3 |
| French-Mongo. | 258 | French-Maya | 218 | 6.7E-2 |
| Mongo.-Russian | 292 | Maya-Russian | 232 | 8.8E-3 |
Note.—Mongo., Mongolian. P value is the statistical significance based on the chi-square test (χ2 test, P = 0.05).
aThe shared haplotype blocks had been filtered by using the haplotype blocks of Africans.
Discussion
In this study, we selected a Mongolian male individual for assembly of the Mongolian reference genome, genetic variation map, and subsequent genetic analyses. There are approximately 10 million Mongolians currently living in the world, mostly in Asia. They have their own distinct language, life style, tradition, geographical territories, and physiological traits. Because of the expansion of the Mongolian Empire in the 13th century, the population contributed significant genetic imprints on global ethnic groups during a relatively short period of time. A high-quality Mongolian reference genome will undoubtedly lay the first step to better understand population structure and history of Mongolians and related populations.
De nova assembly of short reads and comparative analysis demonstrated a high-quality Mongolian genome draft assembly, which is comparable to the YH genome and the human reference genome. The evaluation of the assembled genome shows that the distribution of GC content is comparable to the other two genomes. A small notch at the peak of the distribution can be seen in the Mongolian genome (supplementary fig. S3, Supplementary Material online) and may have resulted from 1) randomness of genomic DNA fragmentation in libraries; 2) sequencing bias of some regions that are difficult to sequence, including high (>50%) or low GC regions (<30%), and so on; and 3) genome assembly errors.
The genome assembly is one of the few reference genomes sequenced to high depth representing a specific ethnic group to our knowledge. We also constructed a detailed personal genetic variation map. From the comparative analysis with publically available variant databases, we find that the Mongolian genome possesses a certain number of novel variants (fig. 1D and E and supplementary fig. S4, Supplementary Material online). These will be a major source of variants that characterize the population. Significantly, large proportion of 3n-bp indels in CDS (supplementary fig. S5, Supplementary Material online) as reported in nonhuman genome (Zheng et al. 2011) might have resulted from high conservation of coding regions. Although the length distribution of SVs is similar to that of YH genome, the Mongolian genome has more large SVs (>1 kb) and fewer small ones (<100 bp) (supplementary fig. S7, Supplementary Material online), compared with the YH and the reference human genome. This difference could be to individual specificity and the genome assembly method used.
We did not find homozygous disease-related SNPs in the genome, especially ones that fit a recessive Mendelian disease model. This indicates that the studied individual is representative of the normal, healthy population.
Clade D in Y haplogroup is widely found in Tibeto-Burman populations in Asia, and it has been proposed as the most ancient lineage in East Asia (Shi et al. 2008). We thus infer that the Mongolian has a common patrilineal ancestor with Tibeto-Burman populations. This observation is consistent with results from a recent study that Mongolians and Tibetans share some genetic alleles (Xing et al. 2013). The fact that D clade is frequent in Africa, relatively common in the people of Andaman island, the Tibetan Plateau, and less frequent in East and Central Asia supports the hypothesis of northward migration of modern humans into East Asia (Su et al. 1999). Our study indicates that the patrilineal ancestor of the Mongolian might be an early settler in East and Central Asia. Investigation of gene flows indicates that the genetic imprints of Mongolians approximately match the route of the Empire expansion, including the impacts of diluted Mongolian lineage on the Indian subcontinent. Mongolians share significantly more ancestral alleles with the populations in the regions of the Empire expansion than geographically adjacent populations outside the expansion route, such as Bantu in North Africa, Bedouin, and Palestinian (table 1), indicating that Mongolians played an evident role in shaping of modern Eurasian populations. This observation is consistent with inferences from a previous study (Zerjal et al. 2003).
Using different outgroups, all D tests demonstrate that different Indian populations possess different amount of shared ancestral allele with Mongolians. However, Indians possess fewer shared ancestral alleles than non-Indians, indicating that the diluted Mongolian lineage in Indian populations might have happened after the expansion of the Mongolian empire in the 13th century, possibly a result of Mughal Empire expansion in the 16th century.
The Mongolian genome provides invaluable resources for further studies of modern human origin and evolution, causal variants for Mongolian characteristic disease/traits and Mongolian personal medicine. However, the recent large scale historical activities of the ethnic group present many challenges. The genomic data of a larger number of Mongolian individuals will help to answer these questions in the population context.
Supplementary Material
Supplementary figures S1–S12 and tables S1–S21 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors sincerely thank our sample donor for generously contributing his blood. This project was supported by the National Basic Research Program of China (973 program no. 2011CB809201, 2011CB809202, and 2011CB809203), the Chinese 863 Program (2012AA02A201), the National Natural Science Foundation of China (30890032, 31161130357, 81060098, and 81160101), Foundation of the Inner Mongolia Department of Education (NJZY13169), and the Shenzhen Municipal Government of China (grants ZYC200903240080A and ZYC201105170397A). It was also supported by the intramural program of NHGRI (1ZIAHG000024), the National Institute of Health (NIH) to N.N. The authors are indebted to the Mongolian historians and anthropologists who contributed to this work, but are not included in the author list, including Dr Yuqing Bao, Dr Ming Chen, Dr Yongchun Fu, and others. They sincerely thank Dr David Reich for generously providing the genotype data of India populations. They thank Dr Francis S. Collins, Dr Steve C.J. Parker, Dr Lawrence Brody, and Dr Kevin Stuart for helpful advice, comments, and editing. They also thank two anonymous reviewers for critical comments. The authors declare that they have no competing interest.
Literature Cited
- 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers CA, et al. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21:961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Andrews RM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Attwood TK, Beck ME, Bleasby AJ, Parry-Smith DJ. PRINTS—a database of protein motif fingerprints. Genome Res. 1994;22:3590–3596. [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Genome Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J, Fry B, Maller J, Daly M. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Behar DM, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- Behar DM, et al. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am J Hum Genet. 2012;90:675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bru C, et al. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 2005;33:D212–D215. doi: 10.1093/nar/gki034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Deng W, et al. Evolution and migration history of the Chinese population inferred from Chinese Y-chromosome evidence. J Hum Genet. 2004;49:339–348. doi: 10.1007/s10038-004-0154-3. [DOI] [PubMed] [Google Scholar]
- DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenko M, et al. Phylogeographic analysis of mitochondrial DNA in northern Asian populations. Am J Hum Genet. 2007;81:1025–1041. doi: 10.1086/522933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. the Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of Dutch population. Nat Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS. Improved pairwise alignment of genomic DNA [PhD thesis] 2007. The Pennsylvania State University. [Google Scholar]
- Hulo N, et al. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karafet TM, et al. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, et al. Genetic features of Mongolian ethnic groups revealed by Y-chromosomal analysis. Gene. 2005;346:63–70. doi: 10.1016/j.gene.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi A, Nozaki J, Ohura T, Kayo T, Wada Y, et al. Genetic epidemiology of the carnitine transporter OCTN2 gene in a Japanese population and phenotypic characterization in Japanese pedigrees with primary systemic carnitine deficiency. Hum Mol Genet. 1999;8:2247–2254. doi: 10.1093/hmg/8.12.2247. [DOI] [PubMed] [Google Scholar]
- Kolman CJ, Sambuughin N, Bermingham E. Mitochondrial DNA analysis of Mongolian populations and implications for the origin of New World founders. Genetics. 1996;142:1321–1334. doi: 10.1093/genetics/142.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu F, et al. Investigation of oxidative stress and dietary habits in Mongolian people, compared to Japanese people. Nutr Metab (Lond). 2006;3:21. doi: 10.1186/1743-7075-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu F, et al. Dietary habits of Mongolian people, and their influence on lifestyle-related diseases and early aging. Curr Aging Sci. 2008;1:84–100. doi: 10.2174/1874609810801020084. [DOI] [PubMed] [Google Scholar]
- Komatsu F, Kagawa Y, Kawabata T, Kaneko Y, Ishiguro K. Relationship of dietary habits and obesity to oxidative stress in Palauan people: compared with Japanese and Mongolian people. Curr Aging Sci. 2009;2:214–222. doi: 10.2174/1874609810902030214. [DOI] [PubMed] [Google Scholar]
- Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Li R, Fan W, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, et al. Building the sequence map of the human pan-genome. Nat Biotechnol. 2010;28:57–63. doi: 10.1038/nbt.1596. [DOI] [PubMed] [Google Scholar]
- Li Y, et al. Structural variation in two human genomes mapped at single-nucleotide resolution by whole genome de novo assembly. Nat Biotechnol. 2011;29:723–730. doi: 10.1038/nbt.1904. [DOI] [PubMed] [Google Scholar]
- Longo N, Amat di San Filippo C, Pasquali M. Disorders of carnitine transport and the carnitine cycle. Am J Med Genet C Semin Med Genet. 2006;142C:77–85. doi: 10.1002/ajmg.c.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merriwether DA, Hall WW, Vahlne A, Ferrell RE. mtDNA variation indicates Mongolia may have been the source for the founding population for the New World. Am J Hum Genet. 1996;59:204–212. [PMC free article] [PubMed] [Google Scholar]
- Ogata H, et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, et al. Reconstructing Native American population history. Nature. 2012;488:370–374. doi: 10.1038/nature11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JF. The New Cambridge History of India. Cambridge: Cambridge University Press; 1993. The Mughal Empire. [Google Scholar]
- Shi H, et al. Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations. BMC Biol. 2008;6:45. doi: 10.1186/1741-7007-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Tzvetkova A, Morgenstern B. AUGUSTUS at EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol. 2006;7:S11–S11. doi: 10.1186/gb-2006-7-s1-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starikovskaya EB, et al. Mitochondrial DNA diversity in indigenous populations of the southern extent of Siberia, and the origins of Native American haplogroups. Ann Hum Genet. 2005;69:67–89. doi: 10.1046/j.1529-8817.2003.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, et al. Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am J Hum Genet. 1999;65:1718–1724. doi: 10.1086/302680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svobodova H, et al. Apolipoprotein E gene polymorphism in the Mongolian population. Folia Biol (Praha). 2007;53:138–142. doi: 10.14712/fb2007053040138. [DOI] [PubMed] [Google Scholar]
- Tanaka M, et al. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda K, Harihara S, Tanabe Y, Dashnyam B. Polymorphism of the apolipoprotein B gene and association with plasma lipid and lipoprotein levels in the Mongolian Buryat. Biochem Genet. 2012;50:249–268. doi: 10.1007/s10528-011-9468-y. [DOI] [PubMed] [Google Scholar]
- Twitchett D, Fairbank JK. Alien regimes and borders states. In: Twitchett DC, Franke H, editors. The Cambridge History of China. Vol. 6. Cambridge: Cambridge University Press; 1994. pp. 321–489. [Google Scholar]
- Tylor NP, et al. MLH3 mutation in endometrial cancer. Cancer Res. 2006;66:7502–7508. doi: 10.1158/0008-5472.CAN-06-0248. [DOI] [PubMed] [Google Scholar]
- Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherford J. Genghis Khan and the making of the modern world. New York: Crown Publisher; 2005. The Mongol world war: 1121-1261; pp. 84–207. [Google Scholar]
- Xing J, et al. Genomic analysis of natural selection and phenotypic variation in high-altitude Mongolians. PLoS Genet. 2013;9:e1003634. doi: 10.1371/journal.pgen.1003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YA, et al. SLC22A5 mutations in a patient with systemic primary canitine deficiency: the first Korean case confirmed by biological and molecular investigation. Ann Clin Lab Sci. 2012;42:424–428. [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Zerjal T, et al. The genetic legacy of the Mongols. Am J Hum Genet. 2003;72:717–721. doi: 10.1086/367774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Han Z, Lu S, Li Y, Shuyuan L. Morphological traits in peoples of Mongolian nationality of the Hulunbuir league, Inner Mongolia, China. Anthropol Anz. 2002;60:175–185. [PubMed] [Google Scholar]
- Zheng LY, et al. Genome-wide patterns of genetic variation in sweet ang grain sorghum (Sorghum bicolor) Genome Biol. 2011;12:R114. doi: 10.1186/gb-2011-12-11-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, et al. Global distribution of Y-chromosome haplogroup C reveals the prehistoric migration routes of African exodus and early settlement in East Asia. J Hum Genet. 2010;55:428–435. doi: 10.1038/jhg.2010.40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.