Abstract
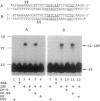
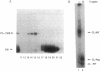
Azide moieties have been specifically placed in the backbone of DNA by chemical coupling between azidophenacyl bromide and uniquely positioned phosphorothioate residues. The derivatized DNA forms specific complexes with a DNA-binding protein and, following irradiation with 302-nm light, makes specific crosslinks to the protein. Isolation of this covalent complex, followed by tryptic digestion and Edman degradation of the resulting crosslinked peptide, identifies the portion of the protein that is near the derivatized segment of the target DNA. We use this method to probe the interaction between a specific DNA sequence and integration host factor (IHF) protein. A single IHF heterodimer is known to contact > 25 bp of DNA and thereby introduce a sharp bend. Two segments of a typical IHF site were derivatized with aryl azide. Although the segments were separated by only 5 bp, they crosslinked to different subunits of IHF. The locations of the crosslinks support our current view for the way IHF protein binds to and bends its specific targets.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker T. A., Mizuuchi K. DNA-promoted assembly of the active tetramer of the Mu transposase. Genes Dev. 1992 Nov;6(11):2221–2232. doi: 10.1101/gad.6.11.2221. [DOI] [PubMed] [Google Scholar]
- Bartholomew B., Kassavetis G. A., Braun B. R., Geiduschek E. P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990 Jul;9(7):2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin A. B., Jr, Nash H. A. Symmetry in the mechanism of bacteriophage lambda integrative recombination. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9642–9646. doi: 10.1073/pnas.89.20.9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin A. B., Pace N. R. Mapping the active site of ribonuclease P RNA using a substrate containing a photoaffinity agent. EMBO J. 1990 Dec;9(12):4111–4118. doi: 10.1002/j.1460-2075.1990.tb07633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capson T. L., Benkovic S. J., Nossal N. G. Protein-DNA cross-linking demonstrates stepwise ATP-dependent assembly of T4 DNA polymerase and its accessory proteins on the primer-template. Cell. 1991 Apr 19;65(2):249–258. doi: 10.1016/0092-8674(91)90159-v. [DOI] [PubMed] [Google Scholar]
- Edman P., Begg G. A protein sequenator. Eur J Biochem. 1967 Mar;1(1):80–91. doi: 10.1007/978-3-662-25813-2_14. [DOI] [PubMed] [Google Scholar]
- Friedman D. I. Integration host factor: a protein for all reasons. Cell. 1988 Nov 18;55(4):545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., Schwartz M. L., McClure W. R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF). Nucleic Acids Res. 1990 Sep 11;18(17):4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granston A. E., Nash H. A. Characterization of a set of integration host factor mutants deficient for DNA binding. J Mol Biol. 1993 Nov 5;234(1):45–59. doi: 10.1006/jmbi.1993.1562. [DOI] [PubMed] [Google Scholar]
- Hanna M. M., Meares C. F. Synthesis of a cleavable dinucleotide photoaffinity probe of ribonucleic acid polymerase: application to trinucleotide labeling of an Escherichia coli transcription complex. Biochemistry. 1983 Jul 19;22(15):3546–3551. doi: 10.1021/bi00284a002. [DOI] [PubMed] [Google Scholar]
- Hixson S. H., Hixson S. S. P-Azidophenacyl bromide, a versatile photolabile bifunctional reagent. Reaction with glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 1975 Sep 23;14(19):4251–4254. doi: 10.1021/bi00690a016. [DOI] [PubMed] [Google Scholar]
- Kim J. L., Nikolov D. B., Burley S. K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993 Oct 7;365(6446):520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger J. H., Hahn S., Sigler P. B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993 Oct 7;365(6446):512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Lee E. C., Hales L. M., Gumport R. I., Gardner J. F. The isolation and characterization of mutants of the integration host factor (IHF) of Escherichia coli with altered, expanded DNA-binding specificities. EMBO J. 1992 Jan;11(1):305–313. doi: 10.1002/j.1460-2075.1992.tb05053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengeritsky G., Goldenberg D., Mendelson I., Giladi H., Oppenheim A. B. Genetic and biochemical analysis of the integration host factor of Escherichia coli. J Mol Biol. 1993 Jun 5;231(3):646–657. doi: 10.1006/jmbi.1993.1316. [DOI] [PubMed] [Google Scholar]
- Nash H. A. Bending and supercoiling of DNA at the attachment site of bacteriophage lambda. Trends Biochem Sci. 1990 Jun;15(6):222–227. doi: 10.1016/0968-0004(90)90034-9. [DOI] [PubMed] [Google Scholar]
- Pendergrast P. S., Chen Y., Ebright Y. W., Ebright R. H. Determination of the orientation of a DNA binding motif in a protein-DNA complex by photocrosslinking. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10287–10291. doi: 10.1073/pnas.89.21.10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz I., Ofengand J. Photo-affinity labeling of tRNA binding sites in macromolecules. I. Linking of the phenacyl-p-azide of 4-thiouridine in (Escherichia coli) valyl-tRNA to 16S RNA at the ribosomal P site. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3951–3955. doi: 10.1073/pnas.71.10.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sylvers L. A., Wower J. Nucleic acid-incorporated azidonucleotides: probes for studying the interaction of RNA or DNA with proteins and other nucleic acids. Bioconjug Chem. 1993 Nov-Dec;4(6):411–418. doi: 10.1021/bc00024a001. [DOI] [PubMed] [Google Scholar]
- Werner M. H., Clore G. M., Gronenborn A. M., Nash H. A. Symmetry and asymmetry in the function of Escherichia coli integration host factor: implications for target identification by DNA-binding proteins. Curr Biol. 1994 Jun 1;4(6):477–487. doi: 10.1016/s0960-9822(00)00108-1. [DOI] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]
- Zulianello L., de la Gorgue de Rosny E., van Ulsen P., van de Putte P., Goosen N. The HimA and HimD subunits of integration host factor can specifically bind to DNA as homodimers. EMBO J. 1994 Apr 1;13(7):1534–1540. doi: 10.1002/j.1460-2075.1994.tb06415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]