Abstract
Aims
Krüppel-like factors (KLFs) are a family of transcription factors which play important roles in the heart under pathological and developmental conditions. We previously identified and cloned Klf6 whose homozygous mutation in mice results in embryonic lethality suggesting a role in cardiovascular development. Effects of KLF6 on pathological regulation of the heart were investigated in the present study.
Methods and results
Mice heterozygous for Klf6 resulted in significantly diminished levels of cardiac fibrosis in response to angiotensin II infusion. Intriguingly, a similar phenotype was seen in cardiomyocyte-specific Klf6 knockout mice, but not in cardiac fibroblast-specific knockout mice. Microarray analysis revealed increased levels of the extracellular matrix factor, thrombospondin 4 (TSP4), in the Klf6-ablated heart. Mechanistically, KLF6 directly suppressed Tsp4 expression levels, and cardiac TSP4 regulated the activation of cardiac fibroblasts to regulate cardiac fibrosis.
Conclusion
Our present studies on the cardiac function of KLF6 show a new mechanism whereby cardiomyocytes regulate cardiac fibrosis through transcriptional control of the extracellular matrix factor, TSP4, which, in turn, modulates activation of cardiac fibroblasts.
Keywords: Fibrosis, Angiotensin II, Krüppel-like factor, Thrombospondin, Cardiomyocyte
1. Introduction
Fibrosis is a hallmark pathological feature of end-stage organ damage.1 Persistent pathological stimulation promotes a plethora of responses including inflammatory cell infiltration, destruction of parenchymal cell structure, excess extracellular matrix (ECM) deposition by myofibroblasts, and lack of tissue reconstruction. These responses, in turn, provide for a vicious cycle, which subsequently leads to fibrosis and organ failure.2 Cardiac fibrosis is initiated by direct injury (e.g. ischaemia and viral infection), haemodynamic insult (e.g. hypertension and pressure-overload), in addition to neurohormonal stimulation (e.g. angiotensin II, Ang II).3 A milieu of cells such as cardiomyocytes, cardiac fibroblasts, inflammatory cells, and endothelial cells, in addition to bioactive molecules such as chemokines/cytokines and extracellular matrix factors, are involved in this response.4 Deciphering the underlying mechanisms of this process has proven difficult in part due to the complicated cell-to-cell hierarchy and interactions that are involved.5 Recent progress has been made to understand the origin of myofibroblasts by fate mapping; however, mechanisms and regulation of cell-to-cell interaction and communication within the heart and its constituent cells in cardiac fibrosis remain unclear.6,7 A better understanding of regulatory and mechanistic processes is important as they would provide direct therapeutic targets for modulation by drugs and/or antibody therapy in the prevention and treatment of end-organ damage.
Krüppel-like factors (KLFs) are transcription factors that play diverse roles in tissue remodelling in response to pathological stimuli such as pressure-overload and neurohormonal factors (e.g. Ang II).8–11 KLF6 has been reported to regulate the hepatic stellate cell that plays a central role in liver fibrosis,12 and hepatic cell-specific deletion of this factor has been reported to show reduced severity of liver lipofibrosis.13 In the cardiovasculature, homozygous mutation of Klf6 results in embryonic lethality with diminished yolk sac vasculature, suggesting involvement in cardiovascular development.14 We therefore hypothesized that KLF6 might regulate fibrosis of the heart and investigated roles of KLF6 in regulating pathological cardiac fibrosis in the present study, and find that KLF6 in cardiomyocytes regulates cardiac fibrosis through transcriptional control of the extracellular matrix (ECM) factor, thrombospondin 4 (TSP4), in response to Ang II stimulation. Through our studies, we identify a new intercellular regulatory mechanism whereby cardiomyocytes modulate cardiac fibroblast functions and the fibrotic/remodelling response.
2. Methods
2.1. Animals
Mice were housed in temperature-controlled rooms with a 12-h light/12-h dark cycle. All care and experimental procedures of animals were in accordance with the guidelines for the Care and Use of Laboratory Animals published by the National Institute of Health (NIH Publication, eighth edition, 2011) and subjected to prior approval by the local animal protection authority in the University of Tokyo Ethics Committee for Animal Experiments (reference no. P10-040).
Heterozygous deletion of KLF6 (Klf6+/−) in C57BL/6J mice was generated as previously described.15 To generate cardiomyocyte- and cardiac fibroblast-specific Klf6 knockout mice, Klf6flox/− and Klf6flox/flox mice were, respectively, cross-bred with αMHC-Cre and Postn-Cre mice. Tsp4 knockout mice were obtained from The Jackson Laboratory and further back-crossed with C57BL/6J mice.
2.2. Ang II induced cardiac fibrosis
To induce cardiac fibrosis, mice were subjected to continuous Ang II infusion as described previously.9 Briefly, mice (8- to 10-week-old males) were anaesthetized with intraperitoneal administration of xylazine (5 mg/kg) and ketamine (100 mg/kg). Ang II (Wako, Osaka, Japan) dissolved in 0.15 mol/L of NaCl and 1 mmol/L of acetic acid solution was subcutaneously administered by an osmotic pump (Alzet model 2002, Alza Corp., Mountain View, CA, USA) at a rate of 3.2 mg/kg/day for 3–14 days to 8- to 10-week-old mice. After 14 days of Ang II treatment, mice were sacrificed via cervical dislocation under isoflurane (Baxter, Deerfield, IL, USA) anaesthesia (1.5%), and hearts were immediately harvested for further experiments.
2.3. Pressure-overload induced by a transverse aortic constriction model
Transverse aortic constriction (TAC) was performed as described previously.10 Briefly, mice (8- to 10-week-old males) were anaesthetized with intraperitoneal administration of xylazine (5 mg/kg) and ketamine (100 mg/kg). The mice were then intubated and ventilated with a tidal volume of 0.4 mL room air at 100 breaths/min. After mid-sternal thoracotomy, the transverse portion of the aortic arch (between the innominate and left common carotid arteries) was ligated by a 10.0-silk suture tied firmly three times against a 26-gauge blunted needle. The thoracic cavity was closed by sternum and skin sutures. Mice were sacrificed via cervical dislocation under isoflurane anaesthesia (1.5%), 14 days after the operation and hearts were immediately harvested for further experiments.
2.4. Echocardiographic analysis
Mice were lightly anaesthetized with 3% inhaled isoflurane and set in a supine position. Two-dimensional (2D) M-mode and Doppler echocardiography was performed with a Vevo2100 Imaging System (Visual Sonics, Inc., Toronto, Canada) equipped with an 18–38 MHz MicroScan™ transducer. The left ventricle (LV) at the papillary muscle level was imaged in 2D mode in the parasternal short-axis view. LV diastolic posterior wall thickness (PWd), interventricular septal thickness at end-diastole (IVSd), LV diastolic dimension (LVDd), and LV end-systolic dimension (LVDs) were measured. LV fractional shortening and ejection fraction were calculated with equipped software.
2.5. Histological analysis and immunohistochemistry
Hearts from mice were embedded in paraffin, and 5 μm thick sections were prepared for Masson's trichrome (MA) staining and immunostaining. Digital images of MA-stained heart were used for measurement of fibrotic area with both Photoshop software (7.0, Adobe, Mountain View, CA, USA) and Image J software (NIH, http://rsbweb.nih.gov/ij/). For immunohistochemistry, after deparaffinization and blocking, mouse heart sections were incubated with KLF6 antibody (Santa Cruz Biotechnology or Biolegend). Secondary antibodies conjugated with horseradish peroxidase (Dako) and 3,3′-diamiobenzidine (DAB; Dako) were used to confirm labelling and counterstained with haematoxylin.
2.6. Immunofluorescent staining
For visualizing KLF6 localization in primary cultured rat neonatal cardiomyocytes and cardiac fibroblasts, anti-rabbit KLF6 antibody (Biolegend) and anti-mouse Troponin T antibody (Thermo Fischer) were used as primary antibodies, and then followed by corresponding secondary antibodies: Alex Fluor 488 or 635 (Life Technologies) was used for fluorescent labelling. Nuclear staining was performed with Hoechst33258 after the final series of washes. Finally, the specimen was mounted with Fluorescent Mounting Medium (Dako) and then visualized with a laser confocal microscope (LSM 510, Zeiss).
2.7. Microarray analysis
Mouse genome-wide gene expression analysis was performed with the Affymetrix Mouse Gene 1.0 ST Array. Whole heart RNA was extracted from wild-type and Klf6+/− mice. Total RNA was extracted by using the RNeasy fibrous tissue kit (Qiagen) and quantified. Microarray analyses were performed in duplicate from independent mice according to the standard Affymetrix Genechip protocol. Genes exhibiting significantly different expression levels in wild-type and Klf6+/− mice hearts were selected (>2.0-fold change).
2.8. Isolation of neonatal cardiomyocytes and non-myocytes
Neonatal ventricles from 1-day-old Sprague–Dawley rats euthanized by decapitation were separated and minced in ice-cold balanced salt solution as described in the Neonatal Cardiomyocyte Isolation System according to the manufacturer's instructions with minor modifications (Worthington Biochemical). To isolate cardiac fibroblasts and cardiomyocytes, minced heart was incubated in a balanced salt solution containing 300 U/mL of collagenase type 2 for 30–45 min at 37°C with agitation. The digestion steps were repeated 10 times, and the collected cell suspension was mixed with 10% volume of chilled FCS and then pelleted by centrifugation. The pellet was dissolved in chilled FCS and kept at 4°C for 12 h. Differentiation of myocytes from non-myocytes was performed by the discontinuous Percoll gradient method.16 Balanced salt solutions containing 40.5 or 58.5% of Percoll (Sigma-Aldrich) were prepared. The cardiac cell suspension was placed onto the layer of 58.5% of Percoll solution. After centrifugation (1200 g at room temperature for 30 min), the cardiomyocytes gathered in the interface layer, and the cardiac fibroblasts migrated to the layer of 40.5% of Percoll solution. The purified cardiomyoctes were then passed onto gelatin-coated 12-well culture plates at a density of 2 × 105cells/well in Medium 199 (Gibco and Life Technologies) supplemented with 10% FCS. Preserved angiotensin receptor expression in isolated cardiomyocytes was confirmed by RNA and protein expression levels (see Supplementary material online, Figure S6A and B).
2.9. RNA extraction and real-time PCR
For RNA extraction and purification, we used the RNeasy mini Kit for primary cultured cells, the RNeasy Fibrous Tissue Kit for heart samples, and the RNeasy Plus Micro Kit (all Qiagen) for FACS-sorted cells according to the manufacturer's protocols. First-strand cDNA was synthesized from 1 μg (tissue and cultured cells) or 30 ng (FACS-sorted cells) of total RNA, random primers, and SuperScript III Reverse Transcriptase (Invitrogen). Real-time PCR was performed with a LightCycler 480 SYBR Green I Master (Roche Applied Science) in a LightCycler 480 instrument according to the manufacturer's instructions (Roche Applied Science). The expression level of each gene was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (Gapdh), which served as an endogenous internal control. The sequences of the PCR primers were as follows: Gapdh, forward 5′-AGGTCGGTGTGAACGGATTTG-3′ and reverse 5′-TGTAGACCATGTAGTTGAGGTCA-3′; Klf6, forward 5′-TATCTTCAGGATGAGCCCTGCTAC-3′ and reverse 5′-AGACTTCACCAATGGGATCAGAGG-3′; Tsp4, forward 5′-TGGAAGGACTCCAGGAATGT-3′ and reverse 5′-TCATAAAAGCGCACCCTGA-3′; transforming growth factor-β1 (Tgfb1), forward 5′-CGTTACAGTGTTTCTGCCACCT-3′ and reverse 5′-AGACGAAGCACACTGGTCCAGC-3′; connective tissue growth factor (Ctgf), forward 5′-CTAAGACCTGTGGGATGGGC-3′ and reverse 5′-CTCAAAGATGTCATTGTCCCC-3′; collagen-1 (Col1a), forward 5′-TGGAGACAGGTCAGACCTG-3′ and reverse 5′-TATTCGATGACTGTCTTGCC-3′; matrix metalloproteinase 9 (Mmp9), forward 5′-ATCTCTTCTAGAGACTGGGAAGGAG-3′ and reverse 5′-AATAAAAGGTCAGAATCCACCCTAC-3′; smooth muscle α-actin (Acta2), forward 5′-AGCTGTTTTCCCATCCATTG-3′ and reverse 5′-GCGCTTCATCACCCACGTAG-3′.
2.10. Western blot analysis
Mouse heart specimens were homogenized with T-PER protein extraction buffer (Thermo Scientific) containing a protease inhibitor cocktail (Roche) and phosphatase inhibitors (Roche). Cultured cells were homogenized with M-PER protein extraction buffer (Thermo Scientific) containing a protease inhibitor cocktail (Roche) and phosphatase inhibitors (Roche). Protein concentration was assayed by the BCA protein assay kit (Pierce), and 5 or 10 µg of the protein was resolved by 10 or 12% of NuPAGE gels (Invitrogen) and then transferred to the polyvinylidene difluoride membrane (Invitrogen). The blot was probed with primary antibodies against KLF6 (Santa Cruz Biotechnology or Biolegend) and TSP4 (R&D). Membranes were washed and incubated with the corresponding horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology). The blot for GAPDH served as an internal control for protein loading. Labelling bands were detected by ECLplus (Thermo Scientific).
2.11. Preparation of plasmid constructs and recombinant adenoviruses
The KLF6 expression vector pCAG-KLF6 was obtained by inserting the KLF6-coding cDNA into pCAGMS vector.9 The adenoviral KLF6 construct was constructed as previously described.17 Primary cultured cardiomyocytes from neonatal rats were infected with adenoviral KLF6 or backbone adenovirus at 100 m.o.i. for 12 h with serum-free Medium 199. The cells were then stimulated with Ang II (10 μM) in fresh medium for 12 h, and the cells were subjected to western blot.
2.12. Co-transfection reporter assay
Co-transfection reporter assay was performed as previously described. Primary cultured rat neonatal cardiomyocytes (1.25 × 105 cells/well in 24-well plates) were transfected with 200 ng of pGL3 luciferase reporter vector containing the Tsp4 promoter region construct and 200 ng of pCAG-KLF6 and Lipofectamine 2000 (Invitrogen).10 Luciferase activity was measured with a luciferase assay system (Promega) and then normalized to the protein concentration in each cell lysate. Assays were done in duplicate, and expression levels of KLF6 were confirmed by immunoblotting with anti-KLF6 antibody (Santa Cruz Biotechnology).
2.13. Chromatin immunoprecipitation assays
Chromatin immunoprecipitation assays were performed as previously described with rat neonatal cardiomyocytes stimulated with Ang II (10 μM) for 24 h prior to crosslinking for 10 min with 1% formaldehyde.10 Chromatin was sheared by sonication to an average size of 200–1000 bp (Covaris). Immunoprecipitation was performed with anti-KLF6 antibody (Santa Cruz Biotechnology) and rabbit IgG antibody (Santa Cruz Biotechnology). PCR amplification of the Tsp4 promoter region spanning KLF-binding elements was performed using the following primers: forward: 5′- TCCGTTGTGGTCCTCTCTGCCAT-3′ and reverse 5′- CGCTTTATGGTCCAGCCACCCG-3′.
2.14. Cardiac fibroblast modulation by TSP4
Modulation of cardiac fibroblast activity by matricellular TSP4 protein was evaluated as previously described with minor modification.18 Briefly, six-well culture plates were coated with 0.1% gelatin (Wako) or 50 μg/mL of mouse recombinant TSP4 (R&D systems) for 20 h at 4°C and then blocked with 0.5% polyvinylpyrrolidone (Sigma) for 1 h at room temperature. Primary cultured cardiac fibroblasts from neonatal rats (1.5 × 105/well) were seeded with serum-free Medium 199 (Gibco). Twelve hours after passage, the cells were stimulated with Ang II (10 μM) for 24 h and were harvested for detection of mRNA expression levels.
2.15. Integrin inhibition assay
Integrin-blocking effect was assayed according to a previously described method.18 Cardiac fibroblasts from wild-type mice were pretreated for 20 min at 37°C with blocking antibodies to integrins and plated on recombinant TSP4 -coated plates then stimulated with Ang II (10 μM) for 24 h. Integrin-blocking antibodies: rabbit anti-integrin β3 (Biolegend), rat anti-α5β1, anti-α4 (Chemicon), and anti-αM (clone M1/70, Biolegend).
2.16. Statistical analysis
All data are presented as mean ± SD. Difference between two groups was analysed by the Welch's t test. Comparisons between multiple groups were done using one- or two-way ANOVA followed by a post hoc Bonferroni test. A P-value of <0.05 was considered significant.
3. Results
3.1. Klf6 haploinsufficiency results in reduced cardiac fibrosis and preserved cardiac function
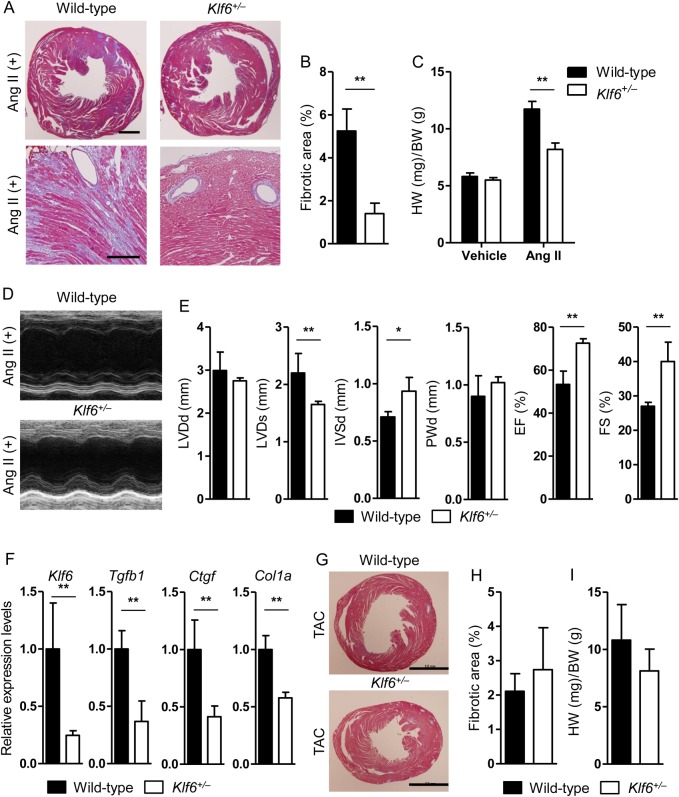
Heterozygous deletion of Klf6 (Klf6+/−) showed, in the heart, markedly decreased fibrotic deposition in response to Ang II overload (Figure 1A, left panels), and decreased interstitial fibrosis compared with that of wild-type mice (quantitative analysis, Figure 1B), whereas perivascular fibrosis showed no apparent difference (Figure 1A, right panels). These results were also confirmed by biochemical analysis (see Supplementary material online, Figure S6D). Additionally, haemodynamic analysis showed blood pressure-independent fibrotic effects by Ang II stimulation (see Supplementary material online, Figure S1A–C). Furthermore, heart weight after Ang II stimulation was increased in the wild-type group compared with that of Klf6+/− mice (Figure 1C). Functionally, the heart of Klf6+/− mice exhibited lower end-diastolic pressure (see Supplementary material online, Figure S1D and E and Supplementary material online, Table S1), preserved left ventricular geometries, and systolic and diastolic function by echocardiographic examination after Ang II treatment (Figure 1D and E, and see Supplementary material online, Figure S2A and E). Reduced cardiac fibrosis of Klf6 knockout mice was accompanied by decreased expression levels of fibrotic regulatory genes such as Tgfb1, Ctgf, and Col1a (Figure 1F and see Supplementary material online, Figure S6C). Moreover, pressure-overload by TAC to wild-type and Klf6+/− mice did not show difference in cardiac fibrosis (Figure 1G and H), relative heart weight (Figure 1I), haemodynamic analysis (see Supplementary material online, Figure S1F–H and Supplementary material online, Table S1), or echocardiographic function (see Supplementary material online, Figure S2D). To note, Ang II treatment or TAC for 14 days induced significant cardiac fibrosis in wild-type mice. However, attenuated cardiac fibrosis was detected only in Ang II treated, but not in TAC-treated, Klf6+/− mice.
Figure 1.
Klf6 haploinsufficiency resulted in decreased cardiac fibrosis and preserved cardiac function. (A and B) Klf6+/− mice showed markedly decreased interstitial cardiac fibrosis compared with wild-type mice after continuous Ang II stimulation for 14 days, and heart samples were stained with Masson's trichrome. Representative figures are shown. Scale bar: 2.0 mm (upper panels), 500 μm (lower panels). (C–E) Klf6+/− mouse hearts showed preserved left ventricular function and size, whereas wild-type mouse hearts showed decreased function and dilated left ventricular size by continuous Ang II infusion. (F) Whole heart mRNA was subjected to quantitative RT-PCR. Klf6+/− mouse hearts showed decreased expression levels of Tgfb1, Ctgf, and Col1a. (G) Representative figures of wild-type and Klf6+/− mouse hearts subjected to pressure-overload (TAC) and stained with Masson's trichrome. Scale bar: 2.0 mm. (H and I) There was no significant difference between wild-type and Klf6+/− mouse hearts in cardiac fibrosis and cardiac hypertrophy by TAC. HW, heart weight; BW, body weight. Data are presented as mean ± SD. n = 3–6 per group. *P < 0.05, **P < 0.01.
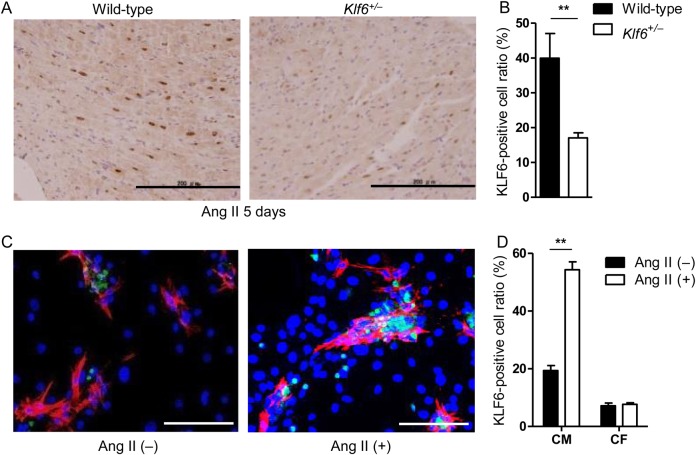
At the cellular level, immunohistochemical studies showed that KLF6 was specifically up-regulated in the nuclei of cardiomyocytes but not in those of cardiac fibroblasts under conditions of cardiac fibrosis (Figure 2A and B). KLF6 was markedly induced in cardiomyocytes but not in cardiac fibroblasts as early as 1 day after Ang II stimulation, and this induction continued during stimulation, thus suggesting that KLF6 may regulate the cardiac fibrotic process not through the cardiac fibroblast but by modulation of cardiomyocytes. Specific expression of KLF6 was further confirmed using cultured cardiomyocytes and fibroblasts in which KLF6 induction was seen after treatment with Ang II in cardiomyocytes but not in fibroblasts (Figure 2C and D). Collectively, these results show that cardiomyocyte KLF6 participates in cardiac fibrosis induced by Ang II but not by TAC.
Figure 2.
KLF6 expression increased in cardiomyocytes, but not in cardiac fibroblasts, after Ang II stimulation. (A and B) Immunohistochemistry showed KLF6 up-regulation (brown stained) in cardiomyocytes of wild-type, but not in Klf6+/− mice, after Ang II stimulation. Scale bar: 200 μm. Note that KLF6 was up-regulated in stimulated myocardium, specifically in the nuclei of cardiomyocytes. (C and D) Primary cultured rat neonatal cardiomyocytes and cardiac fibroblasts were stimulated by Ang II and subjected to immunofluorescent staining. After stimulation, KLF6 (green) was selectively expressed in cardiomyocytes. Green fluorescence, KLF6; red fluorescence, Troponin T. Scale bar: 50 μm. Data are presented as mean ± SD. n = 3–4 per group. **P < 0.01.
3.2. Cardiomyocyte KLF6 expression is essential for cardiac fibrosis
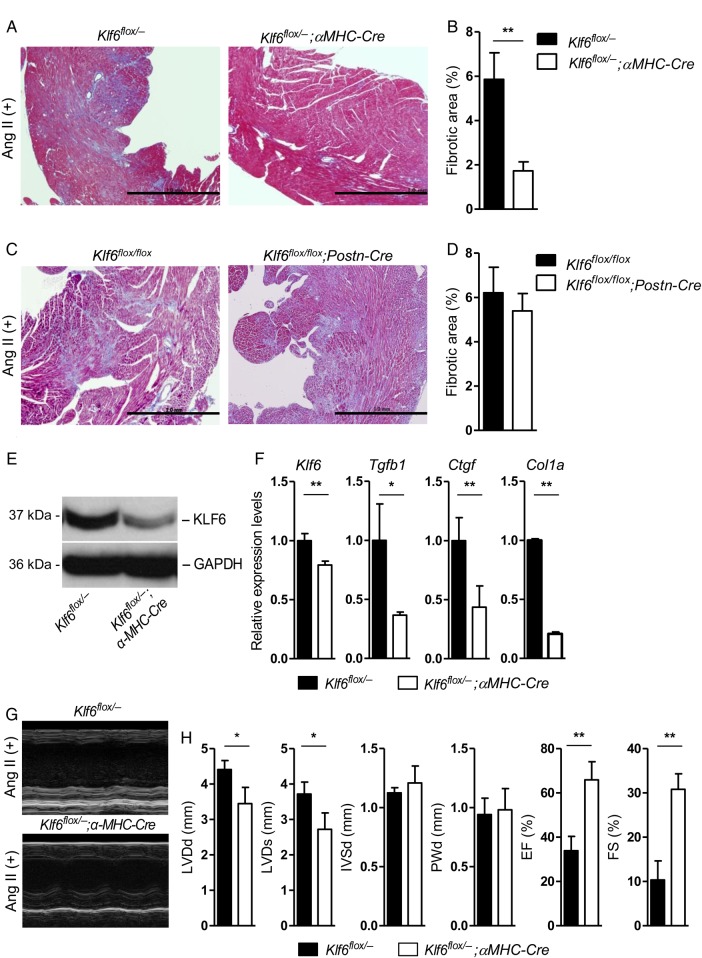
To further clarify the role and importance of cardiomyocyte-specific expression of KLF6, cell type-specific Klf6-deleted mice were generated including cardiomyocyte-specific (Klf6flox/−;αMHC-Cre) and cardiac fibroblast-specific (Klf6flox/flox;Postn-Cre)-deleted mice. Deletion of KLF6 in these conditional knockout mice was confirmed by western blot and RT-PCR analysis after cell separation by flow cytometry (Figure 3E and F, and see Supplementary material online, Figure S3A–C). At baseline, both Klf6flox/−;αMHC-Cre and Klf6flox/flox;Postn-Cre mice showed comparable cardiac function (see Supplementary material online, Figure S2B and C). Only cardiomyocyte-specific but not the fibroblast-specific Klf6-deleted mice showed decreased cardiac fibrotic deposition in response to Ang II stimulation (Figure 3A–D) which was reminiscent if not identical to the phenotype observed in the heart of Klf6+/− mice. Biochemical analysis also confirmed significantly decreased fibrotic content in Klf6flox/−;αMHC-Cre mouse hearts (see Supplementary material online, Figure S6D). Expression levels of fibrotic marker genes (Tgfb1, Ctgf, and Col1a) were also decreased in the hearts of these mice, but neither in those of control nor fibroblast-specific Klf6-deleted mice (Figure 3F and see Supplementary material online, Figure S3D). Functional analysis by echocardiography further confirmed that Klf6flox/−;αMHC-Cre mice showed lack of functional impairment with preserved left ventricular geometries and systolic function (Figure 3G and H). Klf6flox/−;αMHC-Cre mice did not show difference in systolic blood pressure under Ang II treatment or pressure-overload compared with Klf6+/− mice (see Supplementary material online, Figure S1I and J).
Figure 3.
Expression of KLF6 in cardiomyocytes is essential for cardiac fibrosis. (A–D) Conditional deletion of Klf6 in the cardiomyocyte (Klf6flox/−;αMHC-Cre) and cardiac fibroblast (Klf6flox/flox;Postn-Cre). Both mice were subjected to Ang II stimulation for 14 days, and cardiac fibrosis was analysed. Klf6 deletion in cardiomyocytes showed significantly less fibrosis than that in the cardiac fibroblast-specific Klf6-deleted mice in response to Ang II stimulation. Cardiac fibrosis area was calculated by fibrotic area/heart area, and then compared with control mice. Scale bar: 1.0 mm. (E and F) Klf6 protein and mRNA expression levels were significantly decreased in cardiomyocyte-specific Klf6-deleted mice heart. (F) Whole heart mRNA was subjected to quantitative PCR. Cardiomyocyte-specific Klf6-deleted mouse hearts also showed decreased expression levels of Tgfb1, Ctgf, and Col1a. (G and H) Klf6+/− mouse hearts showed preserved left ventricular function and size, whereas wild-type mouse hearts showed decreased function and dilated left ventricular size by continuous Ang II infusion. Data are presented as mean ± SD. n = 3–5 per group. *P < 0.05, **P < 0.01.
These results showed indispensability of KLF6 expression by cardiomyocytes for cardiac fibrotic progression, and a possible signalling pathway from cardiomyocytes to cardiac fibroblasts in this process.
3.3. Tsp4 is a direct target of KLF6 and has cardioprotective properties
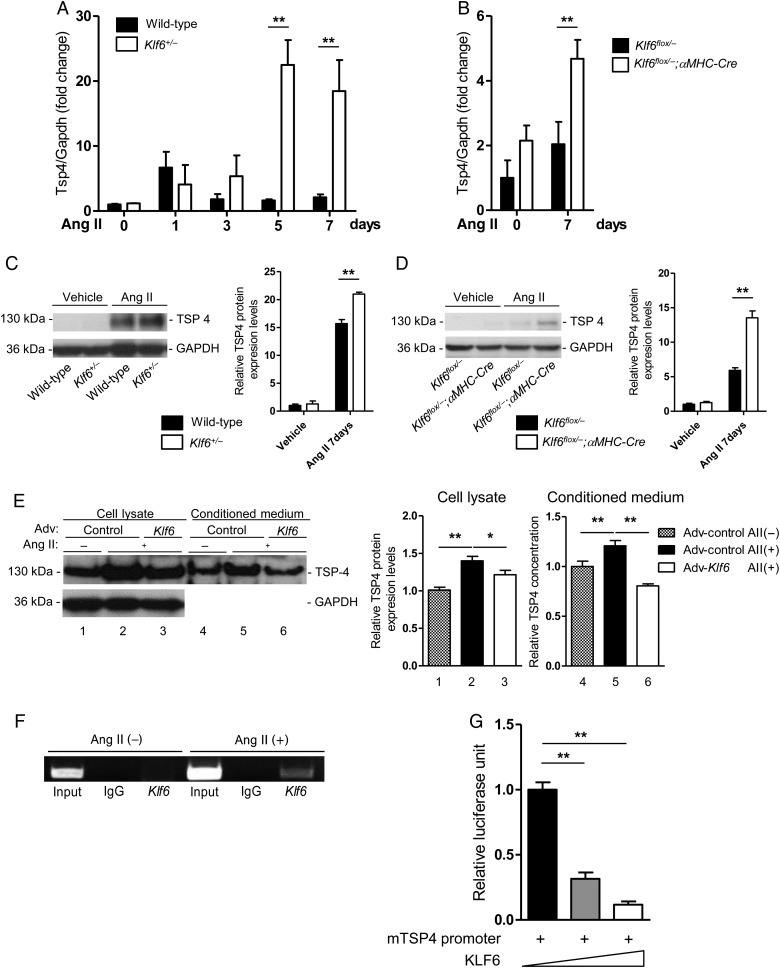
To identify the factor(s) that originate in cardiomyocytes to mediate cardiac fibrosis, we analyzed genome-wide gene expression profiles for the whole heart between wild-type mice and Klf6+/− mice under Ang II stimulation (Table 1). Expression levels of candidate factors were screened for genes encoding secreted proteins whose expression levels were differently changed between the wild-type and knockout mice by RT-PCR analysis. As TSP4 has been reported to prevent excess cardiac fibrosis and dysfunction by pressure-overload,19,20 we hypothesized that increased Tsp4 expression (3.613 up-fold change) might be involved in attenuated cardiac fibrosis in Klf6+/− mice under Ang II stimulation (Table 1). We therefore proceeded to test whether Tsp4 is a target gene of KLF6. mRNA and protein levels of TSP4 were markedly up-regulated in the hearts of both Klf6+/− (Figure 4A and C) and Klf6flox/−;αMHC-Cre mice (Figure 4B and D) after stimulation. Overexpression of KLF6 in cardiomyocytes resulted in decreased TSP4 expression levels in cell lysate and also attenuated secretion into medium (Figure 4E). Immunostaining for TSP4 showed this protein to be localized in the interstitial space of the wild-type heart with Ang II stimulation; however, increased expression levels in cardiomyocytes were recognized in the Klf6flox/−;αMHC-Cre mice (see Supplementary material online, Figure S4C and D). Primary cultured cardiomyocytes also expressed TSP4 after Ang II treatment (see Supplementary material online, Figure S4E). At the molecular level, the Tsp4 promoter contains four putative KLF-binding motifs (Sp1/Egr-1) at −290 bp from the transcriptional start site (see Supplementary material online, Figure S4A). Chromatin immunoprecipitation experiments showed direct recruitment of KLF6 to this region under Ang II stimulation (Figure 4F). Furthermore, reporter analysis of the Tsp4 promoter showed dose-dependent repression by KLF6 overexpression (Figure 4G), and mutational analysis of the Tsp4 promoter region revealed that this effect was attributed to the Sp1 site most proximal to the transcription start site (see Supplementary material online, Figure S4B). KLF6, therefore, appears to be a negative regulator of Tsp4 transcription.
Table 1.
Microarray data analysis
| Genbank no. | Gene description | Gene symbol | Fold change | Regulation |
|---|---|---|---|---|
| BC080666 | Cartilage intermediate layer protein, nucleotide pyrophosphohydrolase | Cilp | 4.961 | Up |
| D13664 | Periostin, osteoblast-specific factor | Postn | 4.923 | Up |
| BC015260 | FK506-binding protein 5 | Fkbp5 | 4.408 | Up |
| BC023373 | Angiopoietin-like 7 | Angptl7 | 3.866 | Up |
| AF102887 | Thrombospondin 4 | Tsp4 | 3.613 | Up |
| AF180805 | Microfibrillar-associated protein 5 | Mfap5 | 2.727 | Up |
| M64086 | Serine (or cysteine) peptidase inhibitor, clade A, member 3N | Serpina3n | 2.704 | Up |
| BC014722 | Secreted frizzled-related protein 2 | Sfrp2 | 2.689 | Up |
| BC051649 | Elastin | Eln | 2.653 | Up |
| BC002043 | Cyclin-dependent kinase inhibitor 1A (P21) | Cdkn1a | 2.648 | Up |
| BC116846 | Synaptotagmin XII | Syt12 | 2.616 | Up |
| BC042422 | Thrombospondin 1 | Tsp1 | 2.581 | Up |
| BC034888 | Asporin | Aspn | 2.522 | Up |
| M65142 | Lysyl oxidase | Lox | 2.383 | Up |
| BC031758 | Metallothionein 2 | Mt2 | 2.381 | Up |
| BC131907 | Collagen triple helix repeat containing 1 | Cthrc1 | 2.366 | Up |
| BC022666 | Microfibrillar-associated protein 4 | Mfap4 | 2.354 | Up |
| L38990 | Glucokinase | Gck | 2.262 | Up |
| AF033530 | Cartilage oligomeric matrix protein | Comp | 2.177 | Up |
| BC055077 | Collagen, type V, alpha 2 | Col5a2 | 2.171 | Up |
| X04684 | Tissue inhibitor of metalloproteinase 1 | Timp1 | 2.158 | Up |
| BC058275 | Platelet-derived growth factor receptor-like | Pdgfrl | 2.129 | Up |
| AB021861 | Mitogen-activated protein kinase kinase kinase 6 | Map3k6 | 2.128 | Up |
| BC064779 | Fibromodulin | Fmod | 2.116 | Up |
| BC019946 | Activating transcription factor 3 | Atf3 | 2.116 | Up |
| BC061695 | Pleiotrophin | Ptn | 2.115 | Up |
| BC025860 | Tumour necrosis factor receptor superfamily, member 12a | Tnfrsf12a | 2.099 | Up |
| BC119198 | Actin-binding Rho-activating protein | Abra | 2.098 | Up |
| BC020152 | Integrin, beta-like 1 | Itgbl1 | 2.096 | Up |
| BC054782 | Heat shock protein 1A | Hspa1a | 2.058 | Up |
| Z47205 | Zinc finger and BTB domain containing 16 | Zbtb16 | 2.044 | Up |
| BC061100 | Myosin, light polypeptide 7, regulatory | Myl7 | 2.041 | Down |
| BC002076 | Mesenchyme homeobox 2 | Meox2 | 2.028 | Down |
| U88568 | Frizzled-related protein | Frzb | 2.023 | Up |
| BC068150 | Nuclear receptor subfamily 4, group A, member 3 | Nr4a3 | 2.004 | Up |
Whole heart RNA was obtained from wild-type and Klf6+/− mice after Ang II stimulation and then subjected to microarray analysis (fold change: Klf6+/− vs. wild-type, Ang II (+), n = 2).
Figure 4.
KLF6 suppressed TSP4 expression levels in response to Ang II stimulation. (A and B) Tsp4 mRNA expression levels after Ang II stimulation by qRT-PCR. Whole heart samples were normalized by Gapdh and then by basal expression levels of wild-type mice. Klf6+/− and cardiomyocyte-specific Klf6-deleted mice heart showed remarkably increased TSP4 expression levels after Ang II stimulation. (C) TSP4 protein expression levels were assayed from wild-type or Klf6+/− mouse whole heart samples after Ang II stimulation. Right bar graph shows quantification of TSP4 protein levels normalized by GAPDH. n = 3. (D) TSP4 protein expression levels were assayed from Klf6flox/− or Klf6flox/−;αMHC-Cre mouse whole heart samples after Ang II stimulation. Right bar graph shows quantification of TSP4 protein levels normalized by GAPDH. n = 3. (E) Primary cultured neonatal rat cardiomyocytes were infected with adenoviral KLF6 and backbone adenovirus at 100 m.o.i. for 12 h. Cells were then stimulated with Ang II (10 μM) for 12 h. Cells and medium were subjected to western blot. GAPDH was used as protein-loading control for cell lysate. Right bar graphs indicate densitometric quantification of TSP4 levels (normalized by GAPDH in the cell lysate). Assays were done in triplicate. (F) KLF6 was directly recruited to the Tsp4 promoter region under Ang II stimulation. Chromatin immunoprecipitation was done with KLF6 antibody, and refined DNA samples were evaluated by PCR using primer pairs for the Tsp4 promoter region. Input was 5% of the chromatin sample. (G) The KLF6 expression vector (pCAG-KLF6) and the Tsp4 promoter reporter constructs were co-transfected and subjected to evaluation of luciferase reporter activity. Luciferase activity was normalized by protein concentration, and relative activities were calculated. Assays were done in triplicate. Data are presented as mean ± SD. *P < 0.05, **P < 0.01.
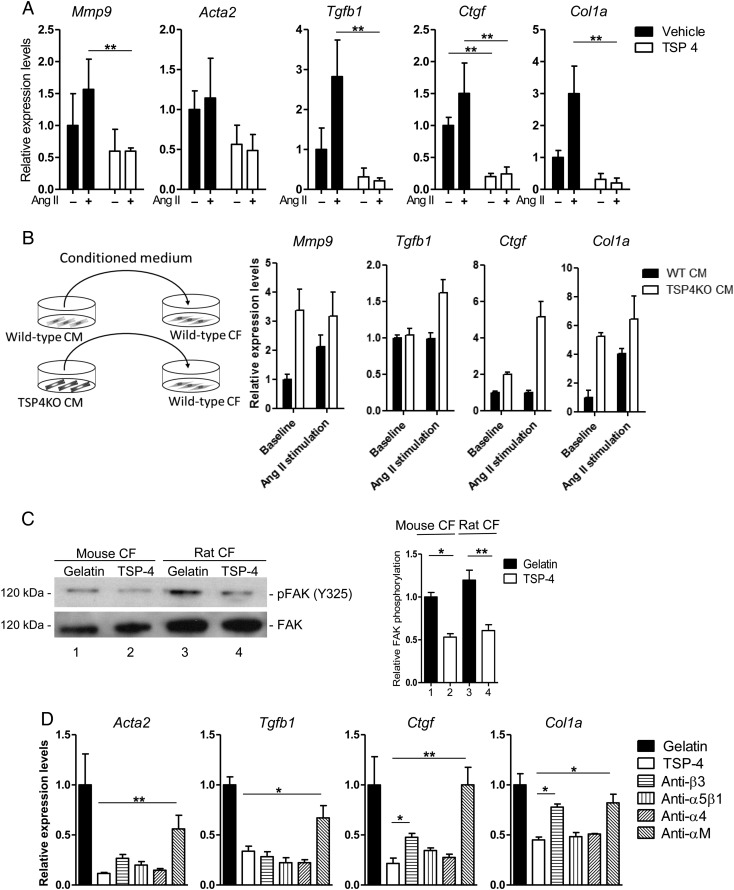
To clarify the effect of secreted TSP4 from cardiomyocytes on cardiac fibroblasts under Ang II -administered conditions, murine cardiac fibroblasts were treated with recombinant murine TSP4 and further cultured with Ang II stimulation for 24 h. Expression levels of fibroblast activation markers (Mmp9, Tgfb1, Ctgf, and Col1a) were markedly decreased in the TSP4 group (Figure 5A). We further carried out experiments using transfer of conditioned medium. Wild-type cardiac fibroblasts were cultured with medium enriched by wild-type or Tsp4KO cardiomyocytes. Incubation with medium from Tsp4KO cardiomyocytes showed higher activation, and this effect was also confirmed by co-culture of cardiac fibroblasts with Tsp4KO cardiomyocytes (Figure 5B and see Supplementary material online, Figure S4F). Suppression of cardiac fibroblast activation by TSP4 was thought to be due partly to negative regulation of FAK phosphorylation through the integrin pathway (Figure 5C and D). KLF6 deletion in the myocardium did not affect apoptosis, while proliferation of non-cardiomyocyte cells was reduced. This was thought to be due to reduced cardiac fibroblast activity (see Supplementary material online, Figure S5A–C).
Figure 5.
TSP4 negatively regulates cardiac fibroblast activity. (A) Murine cardiac fibroblasts (CF) were treated with recombinant murine TSP4 and further cultured with Ang II stimulation for 24 h. Expression levels of CF activation markers (mmp9, acta2, tgfb1, ctgf, and col1a) were compared with the vehicle group. (B) Schematic illustration of conditioned medium transfer assay. Purified cardiomyocytes (CM) from wild-type and Tsp4KO mice were incubated with Ang II for 24 h. The conditioned medium was added to culture medium of purified CF from wild-type mice, and the CF was incubated for another 24 h. Bar graphs indicate expression levels of CF activation makers as analysed by qRT-PCR. Two-way ANOVA showed a significant difference in marker gene expression levels between wild-type and Tsp4KO CM (mmp9 P = 0.0353, tgfb1 P = 0.0148, ctgf P = 0.0005, and col1a P = 0.0128). (C) Modulation of CF activity was evaluated using TSP4-coated plates. Primary cultured mouse and rat neonatal CFs were stimulated with Ang II (10 μM) for 24 h, and were harvested for evaluation of protein focal activated kinase (FAK) phosphorylation levels. FAK protein levels were used as a loading control. Right bar graph indicates quantification of FAK phosphorylation levels normalized by FAK and compared with the gelatin-treated group. (D) TSP4 exerts intracellular effects via integrin pathways. CF from wild-type mice was pretreated with blocking antibodies to integrins (β3, α5β1, α4, and αM), then plated on recombinant TSP4-coated plates and stimulated with Ang II (10 μM) for 24 h. Expression levels of CF activation markers (acta2, tgfb1, ctgf, and col1a) were analysed by qRT-PCR and compared with the group with no antibodies (TSP4). Data are presented as mean ± SD. n = 3–4 per group. *P < 0.05, **P < 0.01.
Taken together, our results showed that KLF6 in cardiomyocytes modulates cardiac fibrosis induced by Ang II stimulation through negative transcriptional control of TSP4 expression.
4. Discussion
The present study suggests that KLF6 in cardiomyocytes plays a pivotal role in progression of cardiac fibrosis through modulation of the ECM protein, thrombospondin 4 (TSP4), to regulate fibroblast activation under Ang II stimulation. Our findings may provide new physiological/pathological aspects in cell-to-cell communication between cardiomyocytes and fibroblasts in the heart.
Both whole body and cardiomyocyte-specific deletion of KLF6 resulted in decreased cardiac fibrosis accompanied by preserved cardiac function and increased expression levels of TSP4. Of interest, a phenotype of aggravated fibrosis has been reported in Tsp4 knockout mouse heart under pressure-overload.19,20 Although their results are based on TAC treatment and ours are on Ang II treatment, cardioprotective effects of TSP4 against cardiac fibrosis are consistent. Several humoral factors such as TGFβ1,21–24 FGFs,25,26 and gp130/IL6 family factors27–30 have been reported to mediate cardiac intracellular regulation. In the present study, we showed that the ECM protein, TSP4, is centrally involved in intercellular regulation of cardiac fibrosis through the cardiomyocyte to fibroblast, at least through actions of KLF6.
KLF transcription factors are known to be involved in a wide spectrum of cardiac pathologies ranging from heart development31 to remodelling32 and in response to internal and/or external stimuli.8 Disruption of KLF13 results in perinatal lethal abnormalities of heart development,33 and KLF15 deletion exhibits eccentric cardiac hypertrophy in response to pressure-overload.34 KLF5 has also been shown to be expressed specifically in the cardiac fibroblast, and to regulate cardiomyocyte hypertrophy through IGF-1 secreted from cardiac fibroblasts.10 In the present study, we showed that KLF6 in cardiomyocytes regulates activation of cardiac fibroblasts through ECM protein secretion. Others have shown that cardiomyocyte-specific KLF4 promotes enhanced cardiomyocyte hypertrophy in response to β-adrenergic stimulation.35 These accumulating data indicate the importance of understanding the role of KLFs in a single-cell manner to decipher cell-to-cell regulation in cardiac pathologies. Our findings add to our understanding of the collective and cooperative roles that the KLF-related network plays in the cardiac stress response.
TSP4 has been reported to exert a cardioprotective role under pressure-overload,36,37 and Tsp4 knockout mice show accelerated cardiac fibrosis under such conditions.19 Additionally, intracellular TSP4 located in the endoplasmic reticulum (ER) has been reported to alleviate cardiac damage by enhancing adaptation and protection to ER stress,38 suggesting cell protective effects in both intracellular and extracellular conditions. The role of TSP4 in regulating pathological remodelling responses in the heart has therefore been the attention of recent interest, and our study identifies a new functional role to this protein.39 To note, increased expression levels of secreted factors other than Tsp4 (Table 1) were also seen in the heart of Klf6-deleted mice, including periostin which is also known to be a cardioprotective matricellular protein necessary for maintaining fibroblast integrity.40,41 As KLF6 is also known as an activator of the TGF-β signalling pathway,8 we confirmed modulated TGF-β secretion and signalling (see Supplementary material online, Figure S6E–G). KLF6 therefore likely modulates cardiac fibrosis through a multitude of mechanisms and factors with further combinatorial regulation. Intriguingly, in liver fibrosis, down-regulation of KLF6 in stellate cells results in activation of fibrogenic genes, and hepatocyte-specific depletion did not alter fibrogenic gene expression or the extent of fibrosis.42 Accumulating data therefore collectively suggests that KLF6 regulates tissue fibrosis in a context-dependent and cell type-specific manner in each tissue/organ with possible cross-talk regulation which will be addressed in future investigations.43,44
In conclusion, we demonstrated that KLF6-mediated signalling from cardiomyocytes to fibroblasts regulated cardiac fibrosis. In this signalling pathway, KLF6 regulated expression levels of the matricellular protein, thrombospondin 4 (TSP4), which further deactivated cardiac fibroblasts. As cardiac fibrosis is a central regulatory process in heart failure as well as in autoimmune disease and malignancies,1 a better understanding of the underlying mechanisms of the fibrotic process has been anxiously awaited with expectations that this would lead to the development of specific therapies for fibrotic organ damage which is presently considered irreversible. KLF6 is likely a pivotal regulator of the fibrotic response, and further investigation of signalling through this molecule in the future will further aid in a better understanding not only of cardiac fibrosis but also of other pathological tissue/organ fibrosis.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This research was funded in partly by the Ministry of Health, Labour and Welfare of Japan; grants-in aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and the Japan Society for the Promotion of Science through its funding program for World-Leading Innovative R&D on Science and Technology (FIRST Program). This study was also supported by the National Institutes of Health National Cancer Institute (DK37340 and DK56621).
Acknowledgements
The murine Tsp4 promoter constructs were a generous gift from Olga Stenina Adognravi. Klf6 floxed mice were provided by Genentech.
Conflict of interest: none declared.
References
- 1.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med 2012;18:1028–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schelbert EB, Fonarow GC, Bonow RO, Butler J, Gheorghiade M. Therapeutic targets in heart failure: refocusing on the myocardial interstitium. J Am Coll Cardiol 2014;63:2188–2198. [DOI] [PubMed] [Google Scholar]
- 3.Kakkar R, Lee RT. Intramyocardial fibroblast myocyte communication. Circ Res 2010;106:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeda N, Manabe I. Cellular interplay between cardiomyocytes and nonmyocytes in cardiac remodeling. Int J Inflam 2011;2011:535241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian Y, Morrisey EE. Importance of myocyte-nonmyocyte interactions in cardiac development and disease. Circ Res 2012;110:1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeBleu VS, Taduri G, O'Connell J, Teng Y, Cooke VG, Woda C, Sugimoto H, Kalluri R. Origin and function of myofibroblasts in kidney fibrosis. Nat Med 2013;19:1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lajiness JD, Conway SJ. Origin, development, and differentiation of cardiac fibroblasts. J Mol Cell Cardiol 2014;70:2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki T, Aizawa K, Matsumura T, Nagai R. Vascular implications of the Krüppel-like family of transcription factors. Arterioscler Thromb Vasc Biol 2005;25:1135–1141. [DOI] [PubMed] [Google Scholar]
- 9.Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H, Nishimatsu H, Ishikawa T, Suzuki T, Morita H, Maemura K, Sata M, Hirata Y, Komukai M, Kagechika H, Kadowaki T, Kurabayashi M, Nagai R. Krüppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med 2002;8:856–863. [DOI] [PubMed] [Google Scholar]
- 10.Takeda N, Manabe I, Uchino Y, Eguchi K, Matsumoto S, Nishimura S, Shindo T, Sano M, Otsu K, Snider P, Conway SJ, Nagai R. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J Clin Invest 2010;120:254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiu K, Manabe I, Nagai R. Renal collecting duct epithelial cells regulate inflammation in tubulointerstitial damage in mice. J Clin Invest 2011;121:3425–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratziu V, Lalazar A, Wong L, Dang Q, Collins C, Shaulian E, Jensen S, Friedman SL. Zf9, a Krüppel-like transcription factor up-regulated in vivo during early hepatic fibrosis. Proc Natl Acad Sci USA 1998;95:9500–9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bechmann LP, Vetter D, Ishida J, Hannivoort RA, Lang UE, Kocabayoglu P, Fiel MI, Munoz U, Patman GL, Ge F, Yakar S, Li X, Agius L, Lee YM, Zhang W, Hui KY, Televantou D, Schwartz GJ, LeRoith D, Berk PD, Nagai R, Suzuki T, Reeves HL, Friedman SL. Post-transcriptional activation of PPAR alpha by KLF6 in hepatic steatosis. J Hepatol 2013;58:1000–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsumoto N, Kubo A, Liu H, Akita K, Laub F, Ramirez F, Keller G, Friedman SL. Developmental regulation of yolk sac hematopoiesis by Krüppel-like factor 6. Blood 2006;107:1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarocchi M, Hannivoort R, Hoshida Y, Lee UE, Vetter D, Narla G, Villanueva A, Oren M, Llovet JM, Friedman SL. Carcinogen-induced hepatic tumors in KLF6+/− mice recapitulate aggressive human hepatocellular carcinoma associated with p53 pathway deregulation. Hepatology 2011;54:522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada M, Itoh H, Nakagawa O, Ogawa Y, Miyamoto Y, Kuwahara K, Ogawa E, Igaki T, Yamashita J, Masuda I, Yoshimasa T, Tanaka I, Saito Y, Nakao K. Significance of ventricular myocytes and nonmyocytes interaction during cardiocyte hypertrophy: evidence for endothelin-1 as a paracrine hypertrophic factor from cardiac nonmyocytes. Circulation 1997;96:3737–3744. [DOI] [PubMed] [Google Scholar]
- 17.Miyamoto S, Suzuki T, Muto S, Aizawa K, Kimura A, Mizuno Y, Nagino T, Imai Y, Adachi N, Horikoshi M, Nagai R. Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol Cell Biol 2003;23:8528–8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolova EG, Pluskota E, Krukovets I, Burke T, Drumm C, Smith JD, Blech L, Febbraio M, Bornstein P, Plow EF, Stenina OI. Thrombospondin-4 regulates vascular inflammation and atherogenesis. Circ Res 2010;107:1313–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frolova EG, Sopko N, Blech L, Popovic ZB, Li J, Vasanji A, Drumm C, Krukovets I, Jain MK, Penn MS, Plow EF, Stenina OI. Thrombospondin-4 regulates fibrosis and remodeling of the myocardium in response to pressure overload. FASEB J 2012;26:2363–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mustonen E, Aro J, Puhakka J, Ilves M, Soini Y, Leskinen H, Ruskoaho H, Rysa J. Thrombospondin-4 expression is rapidly upregulated by cardiac overload. Biochem Biophys Res Commun 2008;373:186–191. [DOI] [PubMed] [Google Scholar]
- 21.Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. FASEB J 2004;18:816–827. [DOI] [PubMed] [Google Scholar]
- 22.Bujak M, Ren G, Kweon HJ, Dobaczewski M, Reddy A, Taffet G, Wang XF, Frangogiannis NG. Essential role of Smad3 in infarct healing and in the pathogenesis of cardiac remodeling. Circulation 2007;116:2127–2138. [DOI] [PubMed] [Google Scholar]
- 23.Schultz Jel J, Witt SA, Glascock BJ, Nieman ML, Reiser PJ, Nix SL, Kimball TR, Doetschman T. TGF-beta1 mediates the hypertrophic cardiomyocyte growth induced by angiotensin II. J Clin Invest 2002;109:787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuwahara F, Kai H, Tokuda K, Kai M, Takeshita A, Egashira K, Imaizumi T. Transforming growth factor-beta function blocking prevents myocardial fibrosis and diastolic dysfunction in pressure-overloaded rats. Circulation 2002;106:130–135. [DOI] [PubMed] [Google Scholar]
- 25.Liao S, Bodmer J, Pietras D, Azhar M, Doetschman T, Schultz Jel J. Biological functions of the low and high molecular weight protein isoforms of fibroblast growth factor-2 in cardiovascular development and disease. Dev Dyn 2009;238:249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellieux C, Foletti A, Peduto G, Aubert JF, Nussberger J, Beermann F, Brunner HR, Pedrazzini T. Dilated cardiomyopathy and impaired cardiac hypertrophic response to angiotensin II in mice lacking FGF-2. J Clin Invest 2001;108:1843–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aoyama T, Takimoto Y, Pennica D, Inoue R, Shinoda E, Hattori R, Yui Y, Sasayama S. Augmented expression of cardiotrophin-1 and its receptor component, gp130, in both left and right ventricles after myocardial infarction in the rat. J Mol Cell Cardiol 2000;32:1821–1830. [DOI] [PubMed] [Google Scholar]
- 28.Freed DH, Moon MC, Borowiec AM, Jones SC, Zahradka P, Dixon IM. Cardiotrophin-1: expression in experimental myocardial infarction and potential role in post-MI wound healing. Mol Cell Biochem 2003;254:247–256. [DOI] [PubMed] [Google Scholar]
- 29.Kuwahara K, Saito Y, Harada M, Ishikawa M, Ogawa E, Miyamoto Y, Hamanaka I, Kamitani S, Kajiyama N, Takahashi N, Nakagawa O, Masuda I, Nakao K. Involvement of cardiotrophin-1 in cardiac myocyte-nonmyocyte interactions during hypertrophy of rat cardiac myocytes in vitro. Circulation 1999;100:1116–1124. [DOI] [PubMed] [Google Scholar]
- 30.Wang F, Trial J, Diwan A, Gao F, Birdsall H, Entman M, Hornsby P, Sivasubramaniam N, Mann D. Regulation of cardiac fibroblast cellular function by leukemia inhibitory factor. J Mol Cell Cardiol 2002;34:1309–1316. [DOI] [PubMed] [Google Scholar]
- 31.Brown JL, Kassis JA. Spps, a Drosophila Sp1/KLF family member, binds to PREs and is required for PRE activity late in development. Development 2010;137:2597–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Wang Y, Liu Y, Wang N, Qi Y, Du J. Kruppel-like factor 4 transcriptionally regulates TGF-beta1 and contributes to cardiac myofibroblast differentiation. PLoS ONE 2013;8:e63424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavallee G, Andelfinger G, Nadeau M, Lefebvre C, Nemer G, Horb ME, Nemer M. The Krüppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J 2006;25:5201–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, Cui L, Kumar A, Lin Z, Sen-Banerjee S, Das H, Petersen CA, Mende U, Burleigh BA, Zhu Y, Pinto YM, Liao R, Jain MK. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc Natl Acad Sci USA 2007;104:7074–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida T, Yamashita M, Horimai C, Hayashi M. Krüppel-like factor 4 protein regulates isoproterenol-induced cardiac hypertrophy by modulating myocardin expression and activity. J Biol Chem 2014;289:26107–26118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cingolani OH, Kirk JA, Seo K, Koitabashi N, Lee DI, Ramirez-Correa G, Bedja D, Barth AS, Moens AL, Kass DA. Thrombospondin-4 is required for stretch-mediated contractility augmentation in cardiac muscle. Circ Res 2011;109:1410–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rainer PP, Hao S, Vanhoutte D, Lee DI, Koitabashi N, Molkentin JD, Kass DA. Cardiomyocyte-specific transforming growth factor beta suppression blocks neutrophil infiltration, augments multiple cytoprotective cascades, and reduces early mortality after myocardial infarction. Circ Res 2014;114:1246–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch JM, Maillet M, Vanhoutte D, Schloemer A, Sargent MA, Blair NS, Lynch KA, Okada T, Aronow BJ, Osinska H, Prywes R, Lorenz JN, Mori K, Lawler J, Robbins J, Molkentin JD. A thrombospondin-dependent pathway for a protective ER stress response. Cell 2012;149:1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doroudgar S, Glembotski CC. ATF6 [corrected] and thrombospondin 4: the dynamic duo of the adaptive endoplasmic reticulum stress response. Circ Res 2013;112:9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimazaki M, Nakamura K, Kii I, Kashima T, Amizuka N, Li M, Saito M, Fukuda K, Nishiyama T, Kitajima S, Saga Y, Fukayama M, Sata M, Kudo A. Periostin is essential for cardiac healing after acute myocardial infarction. J Exp Med 2008;205:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhn B, del Monte F, Hajjar RJ, Chang YS, Lebeche D, Arab S, Keating MT. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med 2007;13:962–969. [DOI] [PubMed] [Google Scholar]
- 42.Ghiassi-Nejad Z, Hernandez-Gea V, Woodrell C, Lang UE, Dumic K, Kwong A, Friedman SL. Reduced hepatic stellate cell expression of Krüppel-like factor 6 tumor suppressor isoforms amplifies fibrosis during acute and chronic rodent liver injury. Hepatology 2013;57:786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol 2006;7:589–600. [DOI] [PubMed] [Google Scholar]
- 44.Dobaczewski M, de Haan JJ, Frangogiannis NG. The extracellular matrix modulates fibroblast phenotype and function in the infarcted myocardium. J Cardiovasc Transl Res 2012;5:837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]