Abstract
Aims
Progress in tissue preservation (high-pressure freezing), data acquisition (tomographic electron microscopy, TEM), and analysis (image segmentation and quantification) have greatly improved the level of information extracted from ultrastructural images. Here, we combined these methods and developed analytical tools to provide an in-depth morphometric description of the intercalated disc (ID) in adult murine ventricle. As a point of comparison, we characterized the ultrastructure of the ID in mice heterozygous-null for the desmosomal gene plakophilin-2 (PKP2; mice dubbed PKP2-Hz).
Methods and results
Tomographic EM images of thin sections of adult mouse ventricular tissue were processed by image segmentation analysis. Novel morphometric routines allowed us to generate the first quantitative description of the ID intercellular space based on three-dimensional data. We show that complex invaginations of the cell membrane significantly increased the total ID surface area. In addition, PKP2-Hz samples showed increased average intercellular spacing, ID surface area, and membrane tortuosity, as well as reduced number and length of mechanical junctions compared with control. Finally, we observed membranous structures reminiscent of junctional sarcoplasmic reticulum at the ID, which were significantly more abundant in PKP2-Hz hearts.
Conclusion
We have developed a systematic method to characterize the ultrastructure of the intercellular space in the adult murine ventricle and have provided a quantitative description of the structure of the intercellular membranes and of the intercellular space. We further show that PKP2 deficiency associates with ultrastructural defects. The possible importance of the intercellular space in cardiac behaviour is discussed.
Keywords: Plakophilin-2, Intercalated disc, Ultrastructure, Tomographic EM
1. Introduction
Adult cardiac myocytes are highly organized into specific subcellular domains (e.g. sarcomeres, T-tubules, costameres, etc). Each of these subdomains is, in turn, populated by molecules that cluster together within confined spaces. In the cardiac myocytes as in other specialized cells, precise spatial organization is critical to overall cell homeostasis. Indeed, the functional output of one molecule often depends on its proper association with molecular partners that in turn localize, either primarily or exclusively, in a particular subdomain (e.g. the sodium channel in the cell end vs. the costamere1,2). Here we focus on a particular subcellular domain that is highly relevant to cell electrophysiology, namely, the intercalated disc (ID). This structure hosts complexes involved in cell–cell adhesion, intercellular electrical coupling, and excitability, integrated into a complex protein interacting network that we have dubbed ‘the connexome’.3,4
Ultrastructural observations of the ID have relied mostly on conventional 2D micrographs. As such, a description of the anatomy of a continuous ID volume at nanometric resolution is lacking. Here, we combined advanced methods of tissue preservation (high-pressure freezing), data acquisition (tomographic electron microscopy, TEM) visualization (segmentation), and quantification (image analysis) to characterize the three-dimensional ultrastructure of the ID. We place particular emphasis on the contour of the intercellular membranes, their contact with other intracellular membranous structures, and the physical characteristics of the volume contained within the intercellular space. Importantly, we develop analytical tools of potential wider application so that morphological features can be quantified and compared both within groups and between groups. As a point of comparison, we study hearts deficient in a desmosomal protein (plakophilin-2, PKP2). The possible importance of the intercellular space in the electrical behaviour of the heart in health and disease is discussed.
2. Methods
Experiments were conducted in mice 3–6 months old, of both genders. Mice were euthanized with an overdose of anaesthetic (isoflurane 20%) and confirmed death by cervical dislocation. All procedures were carried out in accordance with New York University guidelines for animal use and care and conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication 58-23, revised 2011).
Adult mouse ventricular tissue was prepared by fixing the hearts in situ with 4% paraformaldehyde in 0.1 M PBS (pH 7.4). Small pieces were cut, rapidly frozen with a high-pressure freezer, and stored under liquid nitrogen with 2% osmium tetroxide in acetone. Samples were slowly warmed by using an automated freeze substitution machine, infiltrated with acetone:Embed 812 resin 1:1, then incubated in twice pure Embed 812, and polymerized at 60°C. One hundred-nanometre-thick sections were collected on slotted copper grids and imaged with a 200 kV electron microscope (FEI TF20). Images were recorded at a set magnification of 9.6k and 11.5k on a 4k × 4k CCD camera set to 2× binning, giving an effective pixel size of 1.76 nm and 1.47 nm, respectively. Dual-axis tilt series (1° steps, ±70° per axis) were acquired using SerialEM.5 Protomo6 software was used for aligning the projection images and reconstructing the tomograms. IDs contained in the tomograms were segmented in Amira (Visage Imaging, San Diego, CA, USA) to generate their 3D models. ImageJ7 and Matlab (Mathworks, Natick, MS, USA) equipped with the Image Processing toolbox were used for quantitative analysis.
2.1. Visualization
A data set composed of 10 WT and 8 PKP2-Hz mice tomograms (3.2 × 3.2 × 0.1 μm3 in size) was processed for volume segmentation and analysis.8 For quantitative comparison, all the tomograms were selected from tissue sections where the actin filaments run parallel to the plane of section. Different structures of interest were manually traced on all the virtual slices to get their 3D rendered models. This way we obtained representative 3D pictures where the spatial interrelations of the different structures could be seen in high detail.
2.2. Image processing and analysis
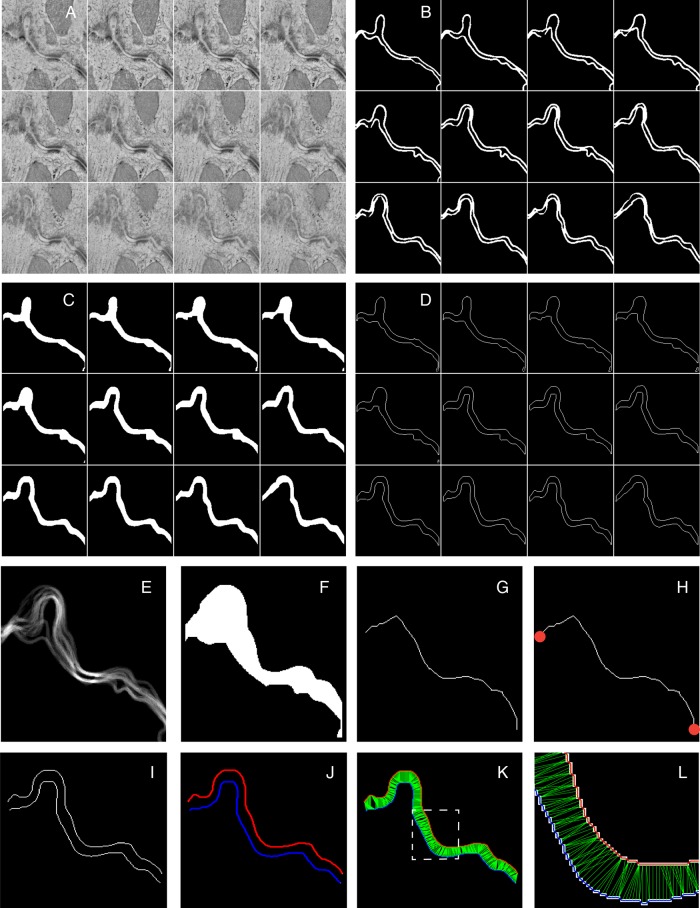
Using the three-dimensional data sets obtained by TEM, the contour of the continuous membranes forming the ID was segmented, thus generating a 3D binary mask for each sample (Figure 1A and B) where voxels corresponding to the ID were assigned a value of 1 (white) and the rest of the image received a value of 0 (black). These binary images were used to measure intercellular space volume and surface area, contour length and tortuosity, and intercellular distance.
Figure 1.
Operational workflow applied to tomographic EM images of the adult ID. (A) Gallery view of virtual XY slices along the Z-axis of a small region cropped from a tomogram (down-sampled here for simplicity). (B) The membranes of each apposing cell were manually traced on each slice to create a binary 3D mask. (C) Intercellular volume binary masks were generated by virtually filling the space located between the two segmented membranes. (D) Mask of the contours of the membranes generated by applying an edge detection algorithm. (E) 2D projection along the Z-axis of mask in B. (F) Virtually filled space located between the two most external lines. (G) Skeletonized 2D representation of the ID obtained by mathematically extracting the central line of the white region shown in F. (H) A representation of the skeletonized ID whose ends (red dots) have been automatically detected. (I) A virtual slice of the 3D binary mask of the contours generated in D. (J) Boundaries automatically detected in I. Each colour represents the membrane of a different cell. (K) For each boundary point, the closest point in the opposite boundary was found and the corresponding distance was calculated (in green). (L) Close-up of the region inside the dashed square in K. Original contours (white), boundaries (blue and red), and distances (green) are visible.
2.2.1. Intercellular space volume and surface area
For each 3D binary mask, we calculated the intercellular volume by virtually filling the space located in-between the two segmented membranes and counting all voxels in that region that acquired a value of 1 (‘imfill’; Matlab; Figure 1C). An edge detection algorithm using the Prewitt method (Matlab) was applied to get the contours of the binary image (Figure 1D). The number of exposed white voxel faces gave us the total surface area (SA). For normalization, each SA was measured relative to the SA of the smallest rectangular box containing all the white pixels (the ‘bounding box’, calculated by Matlab function ‘regionprops’; only the two large faces of the bounding box were counted). Intercellular volumes were normalized per unit of end-to-end Euclidean distance, the latter calculated as described below.
2.2.2. Tortuosity
To estimate tortuosity, we first obtained the z-projection of all 3D masks (Figure 1E); the space between the two most external white lines was filled (using Matlab function ‘imfill’; Figure 1F) and the central line of the generated white region was mathematically extracted by applying a skeletonization operation (Matlab function ‘bwmorph’; Figure 1G). ‘Contour length’ was estimated as one half of the perimeter of the central line (Matlab function ‘regionprops’); separately, the end points of the skeletonized 2D representative curve were detected (Matlab function ‘bwmorph’) and their distance defined as the end-to-end Euclidean distance (Figure 1H); tortuosity was calculated as the relation between contour length and end-to-end Euclidean distance.
2.2.3. Intercellular distance
For each virtual slice of the 3D binary mask, the membranes of the two apposing cells were defined as two boundaries (b1 and b2; Matlab function ‘bwboundaries’; Figure 1I and J), and for each point in one boundary, we identified the shortest distance to the opposing one (Figure 1K and L). Mean and maximum values were obtained and used for comparison.
2.2.4. Mechanical junctions: length and intercellular distance
The methods described above were applied to masks that contained only the regions identified as either desmosomes or mixed junctions (also called ‘area composita’). These masks were used to determine the length of the mechanical junctions along the x–y plane of the images, as well as the distance between the two opposing junctional plaques across the intercellular space. Of note, given that PKP2 is found in both desmosomes and mixed junctions, both structures were analysed within the same group.
2.3. Statistics
All values are presented as mean ± S.E.M. Student's t-test was used for statistical assessment. A probability value of <0.05 was considered to indicate a significant difference between means.
3. Results
3.1. Structural complexity of the ID
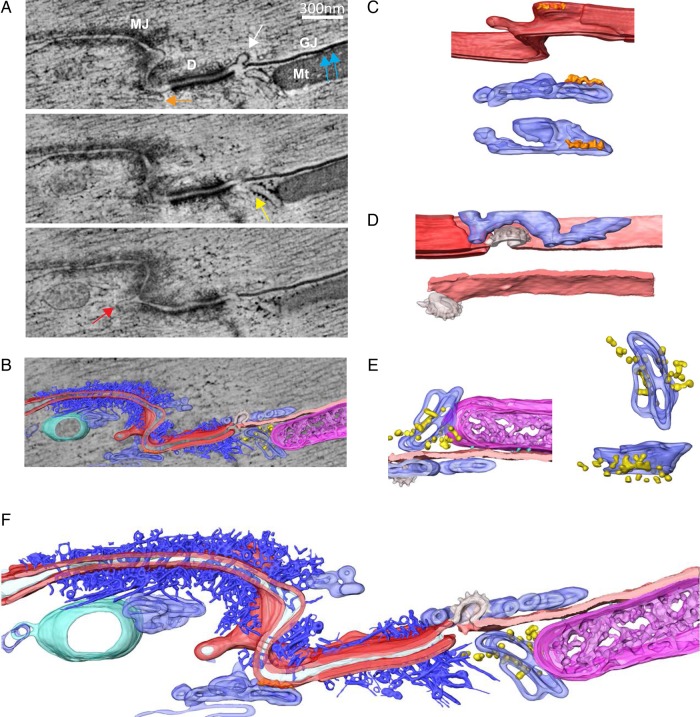
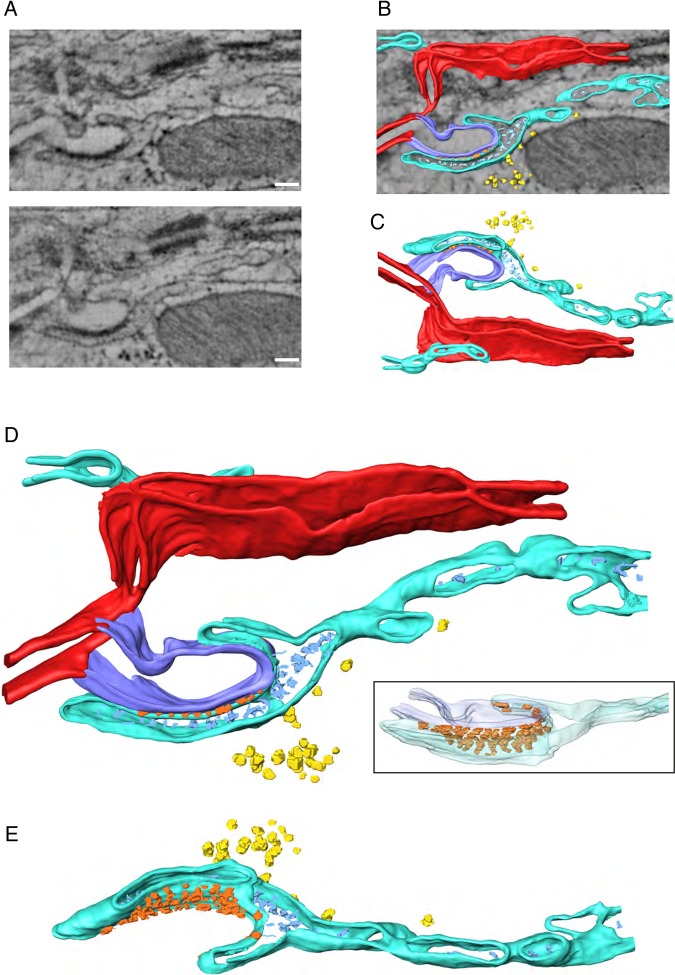
Our process of analysis included both implementation of visualization techniques aimed at enhancing the clarity of the images in 3D renderings, as well as methods for quantitative image analysis to obtain parameters that can be compared between experiments. For enhanced visualization, 3D tomographic EM images of the adult ID region were processed by volume segmentation9 (Figure 2 and Video OS). Different colours were used to highlight the various structures. Figure 2A shows three different planes from the same sample, separated by 25 nm from each other in the z-axis. Notice that fine structures were absent—or almost absent—in one plane but apparent in others. Arrows of different colours point to structural features that were followed through the planes by volume segmentation and then reconstructed (Figure 2B–E; the colour of the arrow matches the colour assigned to the structure in the subsequent panels). The sarcolemma is depicted in red. The orange arrow highlights the presence of a hollow, ellipsoidal structure with small electron-dense speckles decorating its surface and the red arrow in Figure 2A, bottom, points to a long membrane invagination projecting into the interior of the cell, forming a vesicle. Both elements are depicted in two different orientations in Figure 2C (the ‘speckles’ are depicted in orange). The white arrow in Figure 2A, top, points to a clathrin-coated vesicle emerging between a desmosome and a gap junction plaque. The rendered volume of this structure is shown in Figure 2D (two different perspectives after rotation); the image includes (in blue) a complex tubular structure surrounding the vesicle. The yellow arrow in Figure 2A, middle, points to a different hollow structure decorated with round, very dense small structures, which could correspond to ribosomes, though their identity was not fully verified in this study (see the various projections in Figure 2E; mitochondria in purple). Overall, as shown in Figure 2F, volume segmentation demonstrates that the vicinity of a gap junction plaque and a desmosome is populated by organized macrostructures that reside within distances compatible with direct protein interactions.
Figure 2.
TEM sections and 3D rendered representation of ID. (A) 3 XY sections of a tomogram, 25 nm apart from each other along the Z-axis. Different structures can be seen: GJ is gap junction; Mt is mitochondria; D is desmosome; MJ is mixed junction. White arrow points to a budding vesicle in between the gap junction and the desmosome; blue arrows point to electron-dense particles at the interface between the mitochondria and the gap junction; orange arrow points to a membranous structure in contact with a kink of the ID; yellow arrow points to some electron-dense particles attached to the membrane of a tubular structure in the vicinity of the gap junction; red arrow points to an ID membrane budding. (B) Overlay of one tomographic slice and 3D rendered models resulting from segmentation of different structures of interest: cellular membranes forming the ID (red), a gap junction (light pink), a budding vesicle with a rough surface (possibly clatherin coating) between desmosome and gap junction (white), a complex network of filaments adjacent to desmosome and mix junction (dark blue), tubular and cisternae structures forming an intricate connected network in close proximity to ID (light blue), often decorated with electron-dense particles, of dimensions compatible with ribosomes (yellow), a multivesicular body (green) and mitochondria (magenta) in close contact with gap junction. (C) 3D model of region indicated with orange and red arrows in A. A kink of the ID can be seen (red), in physical contact with a tubular network (light blue), with some electron-dense particles at the interface (orange). Middle and bottom panels correspond to rotated views of the tubular structure with particles attached. (D) Rendered model of gap junction and budding, possibly clatherin coated, vesicle. Another tubular network is visible in close proximity to these structures. Note the sneaking path, surrounding the vesicle. (E) Rendered model of area corresponding to interface between mitochondria (magenta), gap junction (light pink), and some portions of a tubular network (light blue) with attached particles (yellow), size compatible with ribosomes. Some electron-dense particles between mitochondria and gap junction are also visible (cyan). Two right panels correspond to rotated views of tubular network with decorating particles. (F) 3D rendered model of all structures of interest segmented in the tomogram, where all spatial interrelations are observed (see Supplementary material online, Video S1).
3.2. Quantitative analysis of intercellular space
A data set composed of 10 tomograms from the ventricles of adult wild-type mice, each containing a cube of 3.2 × 3.2 × 0.1 μm3 in size, was processed for analysis, following the methods described under Figure 1. The average end-to-end linear distance of ID included in the analysis and the average contour length of intercellular membrane studied in all 10 tomograms are shown in Table 1. Projections in three-dimensions of the intercellular membranes showed them not forming two perfectly parallel sheets; rather, we observed extensive vesicular activity and places where the membranes bended away from each other, resulting in heterogeneity in the separation of the two cells (Figure 2F; membranes in red). Intercellular distances varied extensively, between 0 nm in the domain of a gap junction plaque (pink in Figure 2F), to >700 nm in areas where vesicular activity was detected. The average intercellular distance, measured from all planes of acquisition and from all control samples studied, was 90.25 ± 1.83 nm (Table 1). Of note, vesicles were often detected in the immediate vicinity of gap junctions (Figure 2), creating large and abrupt expansions of intercellular space confined on one side by a region where membranes were in close apposition. An average of the maximum intercellular distances recorded in all 10 tomograms from wild-type mice is presented in Table 1. Areas occupied by mechanical junctions revealed an average intercellular distance of 63.02 ± 2.64 nm, and the average maximum distance remained below the mean overall intercellular distance, indicating that mechanical junctions represent points of relative proximity between the two cells (Table 1).
Table 1.
Summary of computed values of ID structural properties
| WT | PKP2-Het | P-value | |
|---|---|---|---|
| End to end dist (µm) | 2.30 ± 0.26 | 1.78 ± 0.23 | – |
| Contour length (µm) | 5.43 ± 0.51 | 7.77 ± 0.94 | – |
| Tortuosity index | 2.65 ± 0.37 | 4.71 ± 0.67 | 1.22 × 10−2 |
| Mean intercellular distance All ID (nm) | 90.25 ± 1.83 | 95.35 ± 3.28 | 1.81 × 10−1 |
| Max intercellular distance All ID (nm) | 316.23 ± 14.02 | 404.20 ± 13.21 | 7.56 × 10−6 |
| Mean intercellular distance MJ (nm) | 63.02 ± 2.64 | 33.50 ± 5.92 | 5.50 × 10−5 |
| Max intercellular distance MJ (nm) | 87.95 ± 2.36 | 75.96 ± 11.51 | 1.76 × 10−1 |
| Mean length of MJ (nm) | 365.54 ± 40.13 | 241.90 ± 43.75 | 6.40 × 10−2 |
| Max length of MJ (nm) | 429.95 ± 44.04 | 299.79 ± 51.41 | 7.97 × 10−2 |
Ten control and eight PKP2-Hz mice tomograms from different animals were analysed. Total number of MJ was 26 and 13 for control and PKP2-Hz groups, respectively.
All ID, data measured for the entire sample; MJ, data measured at mechanical junctions.
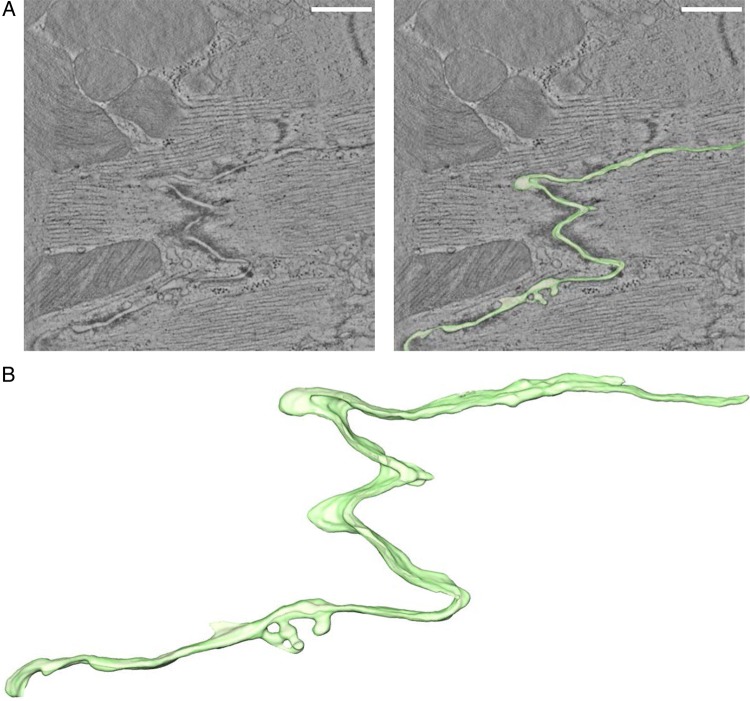
Consistent with the variations in intercellular distances, renderings of intercellular volume (Figure 3A and B) showed the intercellular space as a crevice of various widths, with larger expansions interrupted by sites of close membrane apposition, such as in the areas occupied by gap junctions. Measurements of total and normalized (per μm of end-to-end distance analysed) volume contained between two cells are reported in Table 2. The contour of the intercellular volume was taken as a replica of the intercellular membrane surface and used to measure intercellular surface area (Table 2; see also ‘Methods’). The ratio between the total surface area and the area of a rectangular bounding box indicated that the membrane circumvolutions amplify the surface area of the wild-type ID by a factor of ∼6 times (see ‘SA normalized per bounding box’ in Table 2).
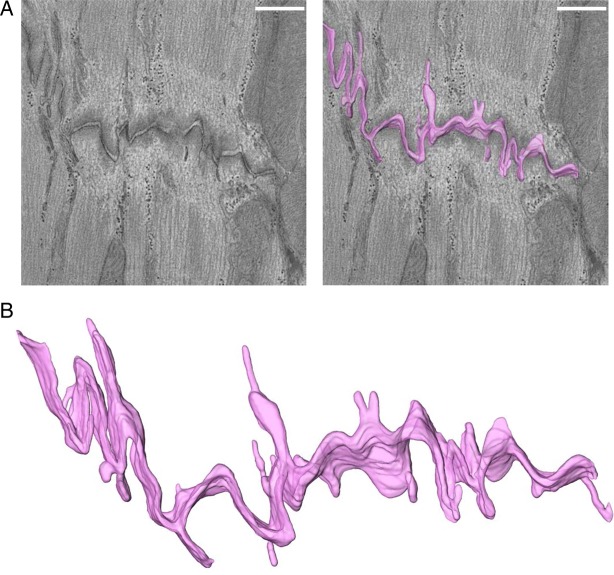
Figure 3.
Intercellular space volume segmentation and quantification of the ID in wild-type adult ventricular murine tissue. (A) 4 nm thick tomographic slice containing a region of ID (left) and overlay with 3D-rendered model of intercellular volume (right). (B) Zoomed-in view of 3D rendering. Scale bar: 500 nm.
Table 2.
Surface area (SA) and volume (Vol) measured from 3D-TEM images
| WT | PKP2-Het | P-value | |
|---|---|---|---|
| Vol (µm3 × 10−2) | 4.54 ± 0.78 | 3.52 ± 0.56 | – |
| SA (µm2) | 2.98 ± 0.35 | 3.13 ± 0.45 | – |
| Vol/unit of end-to-end dist (µm3/µm × 10−2) | 2.18 ± 0.38 | 2.20 ± 0.50 | 9.81 × 10−1 |
| SA normalized per bounding box (µm2/µm2) | 5.86 ± 0.58 | 7.92 ± 0.70 | 3.56 × 10−2 |
Ten control and eight PKP2-Hz mice tomograms from different animals were analysed.
3.3. Junctional sarcoplasmic reticulum at the ID
We investigated not only the characteristics of the intercellular space but also the structural features of membranes that came into apposition with the ID. The presence of structures reminiscent of junctional sarcoplasmic reticulum (jSR) in the area of the ID has been anecdotally reported before.10 Given the dynamic nature of the jSR,11 its dependence on microtubular stability,11 and its importance in arrhythmogenesis,12 we decided to further characterize the jSR at the ID.
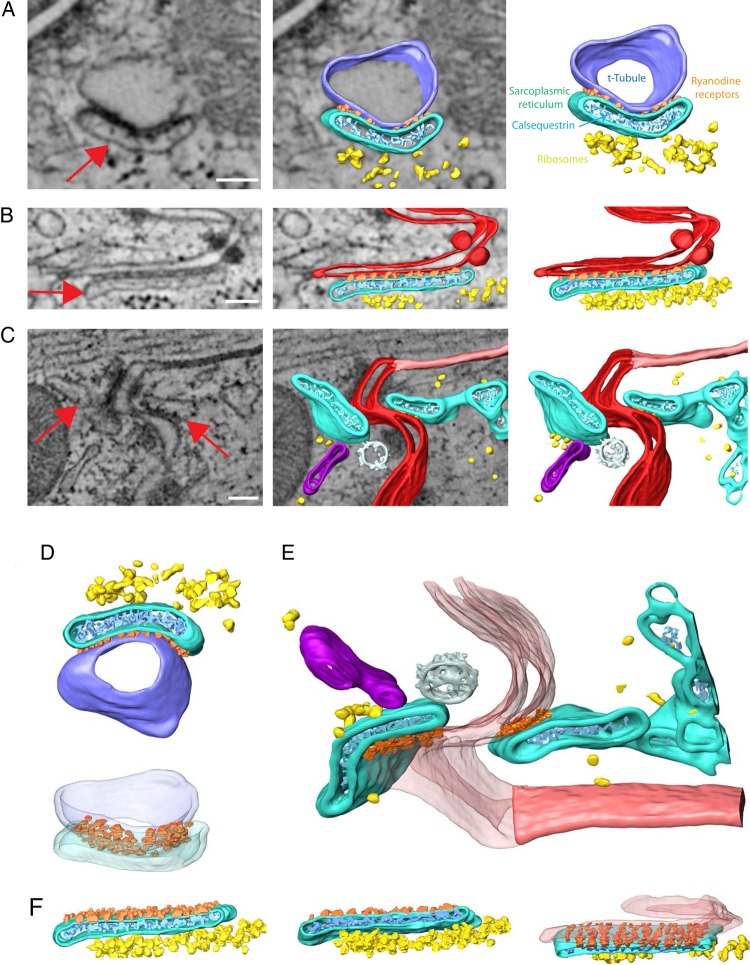
Figure 4A shows a classic dyadic structure with a T-tubule (blue) in close opposition to jSR (turquoise); electron-dense particles (orange) were apparent between structures, suggestive of RyR2 receptors (though their definite identity was not established). Particles depicted in light blue and yellow show structural features similar to those previously ascribed to calsequestrin and ribosomes, respectively. Figure 4B shows a comparable structure at the ID (intercellular membranes in red). The red arrow points to a structure with a nano-architecture similar to that of the jSR shown in A. Notice the close proximity between this ‘dyadic’ membrane complex and the neighbouring desmosome. Interestingly, we found examples where the complex would be found in both sides of the intercellular space (Figure 4C; in this case, adjacent to a gap junction, marked in light red). Figure 4D and F show alternative projections of the structures described in Figure 4A–C. Finally, Figure 5 shows an example where a dyadic structure is formed between the SR (turquoise) and an invagination of the sarcolemma that originated in the membrane facing the intercellular space and penetrated the myocyte in a direction parallel to the long axis of the cell. In other words, this dyad was formed against not a ‘transverse tubule’ but against a ‘longitudinal tubule’ that originated at the ID.
Figure 4.
Dyad structures. Fragments of jSR were found in close contact with t-tubules and intercellular membranes at ID. (A) Example of typical dyad: t-tubule (blue) and portion of SR (turquois) shown in close contact. Electron-dense particles (orange), likely ryanodine receptors, were located at the interface between structures. Calsequestrin-compatible densities (cyan) were found within the jSR; ribosomes (yellow) were consistently observed in the vicinity. (B) Example of interaction between SR (turquois) and intercellular membranes (red). Colour codes as in A. (C) example of ID fragment with jSR converging on both sides of the intercellular space. A gap junction (light pink), a coated vesicle (white), and another cisternae-shaped structure (magenta) were found in the area. (D–F) show same structures as A–C but from a different perspective. Scale bar: 100 nm.
Figure 5.
Example of dyad where sarcolemmal tubule originates in the membrane facing the intercellular space. (A) Two virtual XY sections of a tomogram, 21 nm apart from each other along Z-axis. (B) Overlay of one tomographic slice and 3D-rendered model resulting from segmentation of cellular membranes forming the intercalated disc (red), and the membrane of a tubular projection into the intracellular space (blue), the SR surrounding the tubular invagination (turquois), the electron-dense particles at the interface between t-tubule and SR (orange) attributable to ryanodine receptors, and electron-dense particles inside the SR (light blue) attributable to calsequestrin; some speckled structures, likely ribosomes, shown in yellow. (C) 3D-rendered model rotated 180° around X-axis with respect to the orientation shown in B. (D) Enlarged view of segmented structures. A different view highlighting the partially ordered arrangement of densities attributed to ryanodine receptors at the interface between the t-tubule and the SR are seen in inset. (E) Different view of 3D model where t-tubule and ID membranes have been omitted. Of notice, the SR expands towards the cell interior as a network of connected cisternae and tubules. Scale bar: 100 nm.
3.4. Ultrastructure of the ID in PKP2-deficient cells
It is generally accepted that expression of proteins classically defined as belonging to the desmosome is necessary for proper preservation of ID ultrastructure. Here, we investigated the changes that result from haploinsufficiency of a desmosomal molecule, PKP2 (these animals are referred to as PKP2-Hz). Our results showed a significant increase in the tortuosity of the intercellular membranes (Figure 6A and B; Table 1), and the presence of larger expansions of intercellular volume interposed by areas of limited width, corresponding with a significant increase in the average maximal intercellular distance, measured from all planes of acquisition and from all samples studied (Table 1). Furthermore, our data indicate that in the case of the PKP2-Hz, the membrane foldings of the ID increased the surface area by a factor of 7.92 ± 0.70, a value significantly larger than the one observed for the wild-type samples (Table 2). Interestingly, the number of identifiable mechanical junction complexes was decreased (13 in PKP2-Hz vs. 26 in control mice), but the intercellular distances in those junctions that were identified were significantly reduced compared with control (Table 1).
Figure 6.
Intercellular space volume segmentation of the ID in PKP2-Hz adult ventricular murine tissue. (A) 4 nm thick tomographic slice containing a region of ID and overlay with 3D-rendered model of intercellular volume. (B) Zoomed-in view of 3D rendering. Note the more convoluted path in the PKP2-Hz compared with the wild-type heart shown in Figure 3. Scale bar: 500 nm.
Dyadic structures such as those in Figure 4 were also observed in samples obtained from PKP2-Hz mice (Figure 7A–D). Furthermore, as shown by the bar graph in Figure 7E, the number of ID ‘dyads’ per unit of ID length was significantly more in PKP2-Hz mice than in control. These results show that loss of a desmosomal molecule can influence not only the degree of separation between cells, but also the interactions that occur between subspecialized membranes in the intracellular space.
Figure 7.
Sarcoplasmic reticulum at the intercalated disc in PKP2-Hz mice. Red arrows in A and B point to structures compatible with jSR. Structures were observed either in one (A) or in both sides (B) of intercellular membranes. Colour coding same as in Figure 5. Different perspectives are presented in C and D. (E) Bar plot with frequency of interactions between ID and SR per Angstrom of ID contour length found for control and PKP2-Hz mice. P = 1.22E−2. Number of dyads found: nWT = 14 and nPKP2-Hz = 25. Scale bar corresponds to 100 nm.
4. Discussion
We have characterized the ultrastructure of the intercellular space in the adult murine ventricle. Our data provide, to our knowledge, the first observations on the three-dimensional anatomy of the intercellular region at nanometric resolution, obtained from a 100 nm thick tissue section. Our results show that intercellular membranes represent a large and complex surface bounding a confined intercellular volume, which can be modified by reduced expression of a desmosomal/area composita protein (PKP2). We further show that the intercellular space includes structures compatible with those previously defined as dyads, and that these structures are also dependent on PKP2 expression. Finally, we set in place methods for quantitative comparisons between samples under our experimental conditions. Recently, our group has focused on (i) implementing novel visualization tools and (ii) developing corresponding methods for quantitative analysis.3,4,13 The present study is a continuation of this effort, where we follow a visual proteomics14,15 approach to the study of the cardiac ID.
4.1. Limitations
We should emphasize though that our results are limited to a small sample of the entire area of intercellular contact. Indeed, the total thickness of our sample (100 nm) is ∼1% of the entire height of a ventricular myocyte. Moreover, for all the tomograms chosen for analysis, the sections were aligned in a direction parallel to the actin fibres, but it is not possible to determine the exact position of our 100 nm thick section in relation to the top, middle, or bottom of the myocyte. On the other hand, our results are consistent between different preparations, suggesting that the patterns observed in the samples might be consistent across at least most of the intercellular space.
4.2. Free diffusion in the intercellular space?
Our results are not consistent with the idea of the intercellular cleft as a space of free diffusion, directly connected to the bulk volume surrounding the cell. Rather, the emerging picture is that of a complex environment where diffusion of ions is challenged by the presence of patches of high resistance (gap junctions), high protein density (mechanical junctions), and lengthy, convoluted paths towards the outside (intricate membrane foldings). The ability of ions to diffuse in and out of the intercellular space can be critical for function. Sodium channels, as well as the ATP-sensitive potassium channel (K-ATP), preferentially localize to the ID.16 A large potassium current flowing out of the cell and into the intercellular cleft could cause a temporary accumulation of potassium ions in that space, causing local depolarization and in doing so, inactivating sodium channels critical for electrical conduction.
4.3. jSR at the ID?
We have observed membrane invaginations, reminiscent of tubules, entering the cell from the intercellular space. The potential for ion accumulation in these ID tubules is particularly high, considering the lack of a direct connection to the bulk volume that surrounds the cell. Furthermore, our data also show the presence of structures reminiscent of jSR. Whether the structures reported here are functional as calcium-induced calcium release units, as they are in the vicinity of the T-tubules, remains to be defined. If they are, the restricted intercellular space could also represent a challenge to the maintenance of proper calcium concentration and as such, proper calcium homeostasis for the intracellular space as a whole.
Recently we used super-resolution fluorescence microscopy to demonstrate that the gap junction protein connexin43 (Cx43) and PKP2 share a common subdomain.4 We also showed that changes either in Cx43 or in PKP2 abundance17,18 decrease sodium current amplitude, and we proposed that rather than independent, molecules classically defined as belonging to desmosomes, gap junctions or the voltage-gated sodium channel actually form a single protein interacting network, which we called the connexome.4,19 The finding of jSR structures at the ID adds an interesting level of complexity. In fact, one may speculate that changes in the structure and/or molecular composition of the ID may have direct consequences on calcium homeostasis and, in turn, changes in the molecular composition of the jSR may affect ID function. A case in point is that of phospholamban, a molecule classically defined by its function in control of intracellular calcium concentration and yet, recently identified as a potential ARVC-causing molecule when mutated.20
4.4. ID structure and electric field-mediated transmission
The concept of electric field-mediated propagation in the heart21,22 (so-called ephaptic transmission23) has been recently revisited, particularly given the fact that experimental data following genetic manipulation of connexin abundance does not conform with the assumption that gap junctions are the only conduit for cell–cell transfer of charge. Mathematical modelling suggests that for this type of transmission to occur, intercellular distance is critical, and it would be most relevant if gap junction-mediated coupling is drastically impaired.22,23 Our data show that the spacing between membranes at mechanical junctions was actually shorter in the PKP2-Hz hearts. In other words, reduced abundance of a ‘mechanical junction’ molecule actually associated with the cell membranes being positioned at a closer distance from each other. The increased proximity between the membranes may have implications for electrical propagation. We have previously shown that PKP2 associates with Nav1.5, the pore forming subunit of the sodium channel complex. Additional studies from our laboratory show that Nav1.5 is in a tight complex with mechanical junction proteins (Ref. 3 and also unpublished studies). We therefore postulate that Nav1.5 may be within the electron-dense structure defined as the ‘area composita’. Close examination of Figure 3 of the original paper of Kucera et al.24 shows that narrower clefts can cause a slowing of conduction, even if gap junctional coupling is high. Furthermore, at low levels of coupling the relation is no longer monotonic, and in fact, it breaks for clefts in the range of 30–80 nm, which is the range of cleft separations we observe in the PKP2-Hz hearts. Of note, we have previously observed that PKP2-Hz hearts have a higher propensity toward arrhythmogenesis.25 In summary, the structural observation allows us to postulate that reduced PKP2 abundance leads to shortening of the intercellular gap within the mechanical junction, which can drastically impair electric field-mediated propagation and create areas of microscopic block.
The mechanism that leads to shortening of the intercellular gap is not known. We speculate that the force of cell–cell attraction resulting from the binding of the cadherin molecules in the intercellular space normally works against the force of the intermediate filaments, which tries to pull the cells apart from each other. A weaker anchoring of the intermediate filament (due to reduced PKP2 abundance) may then tilt the balance of these forces in favour of the attraction exerted by the bound cadherins. Of note, reduced intercellular space at the mixed junction following loss of a desmosomal protein can be seen in some anecdotal examples in the literature (see Figure 4 of Notari et al.;26 not the desmosome indicated by their yellow arrow but the one running at the bottom right of the figure).
4.5. ID structure and PKP2 deficiency
We have observed that the ultrastructure of PKP2-Hz hearts is altered compared with that of wild-type animals. Whether these structural changes may contribute to the increased arrhythmogenesis observed in this animal model is unclear.25 Affected intercellular membrane distances and shorter mechanical junctions could impair electric field-mediated transmission.22 The disrupted structure of the cell membrane may play a role in the sodium channel dysfunction that we have previously reported,25 particularly considering that sodium channels show mechano-sensitivity.27,28 Altered dyadic coupling can also be arrhythmogenic, particularly if its relation to sodium current is disturbed (see Refs. 29,30). It is important to emphasize that the PKP2-Hz animals do not develop a cardiomyopathy. On the other hand, in the human population, not every PKP2 mutation carrier develops a disease.31,32 The molecular mechanisms that facilitate the initiation of a clinically manifest cardiomyopathy in a subset of the gene-positive population remain unclear.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
Supported by grants R01-HL106632 and R01-GM57691 from the National Institutes of Health (M.D.), by a Leducq Foundation Transatlantic network (M.D.), and by a grant from The Children Cardiomyopathy Foundation (M.D.).
Acknowledgements
The authors thank the NYUMC Electron Microscopy Core and the New York Structural Biology Center (NYSBC) for help with sample preparation and data collection and the New York University HPC resources for computational support.
Conflict of interest: none declared.
References
- 1.Lin X, Liu N, Lu J, Zhang J, Anumonwo JM, Isom LL, Fishman GI, Delmar M. Subcellular heterogeneity of sodium current properties in adult cardiac ventricular myocytes. Heart Rhythm 2011;8:1923–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petitprez S, Zmoos AF, Ogrodnik J, Balse E, Raad N, El-Haou S, Albesa M, Bittihn P, Luther S, Lehnart SE, Hatem SN, Coulombe A, Abriel H. SAP97 and dystrophin macromolecular complexes determine two pools of cardiac sodium channels Nav1.5 in cardiomyocytes. Circ Res 2011;108:294–304. [DOI] [PubMed] [Google Scholar]
- 3.Agullo-Pascual E, Lin X, Leo-Macias A, Zhang M, Liang FX, Li Z, Pfenniger A, Lubkemeier I, Keegan S, Fenyo D, Willecke K, Rothenberg E, Delmar M. Super-resolution imaging reveals that loss of the C-terminus of connexin43 limits microtubule plus-end capture and NaV1.5 localization at the intercalated disc. Cardiovasc Res 2014;104:371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agullo-Pascual E, Reid DA, Keegan S, Sidhu M, Fenyo D, Rothenberg E, Delmar M. Super-resolution fluorescence microscopy of the cardiac connexome reveals plakophilin-2 inside the connexin43 plaque. Cardiovasc Res 2013;100:231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 2005;152:36–51. [DOI] [PubMed] [Google Scholar]
- 6.Winkler H, Taylor KA. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy 2006;106:240–254. [DOI] [PubMed] [Google Scholar]
- 7.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volkmann N. Methods for segmentation and interpretation of electron tomographic reconstructions. Methods Enzymol 2010;483:31–46. [DOI] [PubMed] [Google Scholar]
- 9.Lucic V, Rigort A, Baumeister W. Cryo-electron tomography: the challenge of doing structural biology in situ. J Cell Biol 2013;202:407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayettey AS, Navaratnam V. The T-tubule system in the specialized and general myocardium of the rat. J Anat 1978;127:125–140. [PMC free article] [PubMed] [Google Scholar]
- 11.Vega AL, Yuan C, Votaw VS, Santana LF. Dynamic changes in sarcoplasmic reticulum structure in ventricular myocytes. J Biomed Biotechnol 2011;2011:382586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Despa S, Shui B, Bossuyt J, Lang D, Kotlikoff MI, Bers DM. Junctional cleft [Ca2+] measurements using novel cleft-targeted Ca2+ sensors. Circ Res 2014;115:339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhargava A, Lin X, Novak P, Mehta K, Korchev Y, Delmar M, Gorelik J. Super-resolution scanning patch clamp reveals clustering of functional ion channels in adult ventricular myocyte. Circ Res 2013;112:1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forster F, Han BG, Beck M. Visual proteomics. Methods Enzymol 2010;483:215–243. [DOI] [PubMed] [Google Scholar]
- 15.Nickell S, Kofler C, Leis AP, Baumeister W. A visual approach to proteomics. Nat Rev Mol Cell Biol 2006;7:225–230. [DOI] [PubMed] [Google Scholar]
- 16.Hong M, Bao L, Kefaloyianni E, Agullo-Pascual E, Chkourko H, Foster M, Taskin E, Zhandre M, Reid DA, Rothenberg E, Delmar M, Coetzee WA. Heterogeneity of ATP-sensitive K+ channels in cardiac myocytes: enrichment at the intercalated disk. J Biol Chem 2012;287:41258–41267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen JA, Noorman M, Musa H, Stein M, de Jong S, van der Nagel R, Hund TJ, Mohler PJ, Vos MA, van Veen TA, de Bakker JM, Delmar M, van Rijen HV. Reduced heterogeneous expression of Cx43 results in decreased Nav1.5 expression and reduced sodium current that accounts for arrhythmia vulnerability in conditional Cx43 knockout mice. Heart Rhythm 2012;9:600–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato PY, Musa H, Coombs W, Guerrero-Serna G, Patino GA, Taffet SM, Isom LL, Delmar M. Loss of plakophilin-2 expression leads to decreased sodium current and slower conduction velocity in cultured cardiac myocytes. Circ Res 2009;105:523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agullo-Pascual E, Cerrone M, Delmar M. Arrhythmogenic cardiomyopathy and Brugada Syndrome: diseases of the connexome. FEBS Lett 2014;588:1322–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Zwaag PA, van Rijsingen IA, Asimaki A, Jongbloed JD, van Veldhuisen DJ, Wiesfeld AC, Cox MG, van Lochem LT, de Boer RA, Hofstra RM, Christiaans I, van Spaendonck-Zwarts KY, Lekanne dit Deprez RH, Judge DP, Calkins H, Suurmeijer AJ, Hauer RN, Saffitz JE, Wilde AA, van den Berg MP, van Tintelen JP. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail 2012;14:1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veeraraghavan R, Poelzing S, Gourdie RG. Intercellular electrical communication in the heart: a new, active role for the intercalated disk. Cell Commun Adhes 2014;21:161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veeraraghavan R, Lin J, Hoeker GS, Keener JP, Gourdie RG, Poelzing S. Sodium channels in the Cx43 gap junction perinexus may constitute a cardiac ephapse: an experimental and modeling study. Pflugers Arch 2015; doi:10.1007/s00424-014-1675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori Y, Fishman GI, Peskin CS. Ephaptic conduction in a cardiac strand model with 3D electrodiffusion. Proc Natl Acad Sci USA 2008;105:6463–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kucera JP, Rohr S, Rudy Y. Localization of sodium channels in intercalated disks modulates cardiac conduction. Circ Res 2002;91:1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerrone M, Noorman M, Lin X, Chkourko H, Liang FX, van der Nagel R, Hund T, Birchmeier W, Mohler P, van Veen TA, van Rijen HV, Delmar M. Sodium current deficit and arrhythmogenesis in a murine model of plakophilin-2 haploinsufficiency. Cardiovasc Res 2012;95:460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notari M, Hu Y, Sutendra G, Dedeic Z, Lu M, Dupays L, Yavari A, Carr CA, Zhong S, Opel A, Tinker A, Clarke K, Watkins H, Ferguson DJ, Kelsell DP, de Noronha S, Sheppard MN, Hollinshead M, Mohun TJ, Lu X. iASPP, a previously unidentified regulator of desmosomes, prevents arrhythmogenic right ventricular cardiomyopathy (ARVC)-induced sudden death. Proc Natl Acad Sci USA 2015;112:E973–E981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beyder A, Rae JL, Bernard C, Strege PR, Sachs F, Farrugia G. Mechanosensitivity of Nav1.5, a voltage-sensitive sodium channel. J Physiol 2010;588:4969–4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beyder A, Strege PR, Reyes S, Bernard CE, Terzic A, Makielski J, Ackerman MJ, Farrugia G. Ranolazine decreases mechanosensitivity of the voltage-gated sodium ion channel Na(v)1.5: a novel mechanism of drug action. Circulation 2012;125:2698–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin X, O'Malley H, Chen C, Auerbach D, Foster M, Shekhar A, Zhang M, Coetzee W, Jalife J, Fishman GI, Isom L, Delmar M. Scn1b deletion leads to increased tetrodotoxin-sensitive sodium current, altered intracellular calcium homeostasis, and arrhythmias in murine hearts. J Physiol 2015;593:1389–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radwanski PB, Brunello L, Veeraraghavan R, Ho HT, Lou Q, Makara MA, Belevych AE, Anghelescu M, Priori SG, Volpe P, Hund TJ, Janssen PM, Mohler PJ, Bridge JH, Poelzing S, Gyorke S. Neuronal Na+ channel blockade suppresses arrhythmogenic diastolic Ca2+ release. Cardiovasc Res 2015;106:143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delmar M, McKenna WJ. The cardiac desmosome and arrhythmogenic cardiomyopathies: from gene to disease. Circ Res 2010;107:700–714. [DOI] [PubMed] [Google Scholar]
- 32.Cerrone M, Lin X, Zhang M, Agullo-Pascual E, Pfenniger A, Chkourko Gusky H, Novelli V, Kim C, Tirasawadichai T, Judge DP, Rothenberg E, Chen HS, Napolitano C, Priori SG, Delmar M. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a brugada syndrome phenotype. Circulation 2014;129:1092–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]