Abstract
Aims
Excitation–contraction coupling in cardiomyocytes requires the proper targeting and retention of membrane proteins to unique domains by adaptor proteins like ankyrin-B. While ankyrin-B has been shown to interact with a variety of membrane and structural proteins located at different subcellular domains in cardiomyocytes, what regulates the specificity of ankyrin-B for particular interacting proteins remains elusive.
Methods and results
Here, we report the identification of two novel ankyrin-B isoforms AnkB-188 and AnkB-212 in human, rat, and mouse hearts. Novel cDNAs for both isoforms were isolated by long-range PCR of reverse-transcribed mRNA isolated from human ventricular tissue. The isoforms can be discriminated based on their function and subcellular distribution in cardiomyocytes. Heterologous overexpression of AnkB-188 increases sodium–calcium exchanger (NCX) membrane expression and current, while selective knockdown of AnkB-188 in cardiomyocytes reduces NCX expression and localization in addition to causing irregular contraction rhythms. Using an isoform-specific antibody, we demonstrate that the expression of AnkB-212 is restricted to striated muscles and is localized to the M-line of cardiomyocytes by interacting with obscurin. Selective knockdown of AnkB-212 significantly attenuates the expression of endogenous ankyrin-B at the M-line but does not disrupt NCX expression at transverse tubules in cardiomyocytes.
Conclusion
The identification and characterization of two functionally distinct ankyrin-B isoforms in heart provide compelling evidence that alternative splicing of the ANK2 gene regulates the fidelity of ankyrin-B interactions with proteins.
Keywords: Ankyrin-B, Alternative splicing, Sodium–calcium exchanger, Obscurin, Cardiomyocytes
1. Introduction
Excitation–contraction coupling requires correct subcellular distribution of ion channels and transporters in cardiomyocytes. Ankyrin-B is an adaptor protein that facilitates recruitment and retention of membrane proteins to subcellular domains in cardiomyocytes such as the transverse tubule and M-line of the sarcomere. Mutations that disrupt interaction with ankyrin-B have been linked to a variety of cardiac arrhythmias including type 4 long QT syndrome, sick sinus syndrome, and atrial fibrillation.1–3
In 1991, the cDNA for ankyrin-B 220 kD was constructed by combining overlapping but partial cDNAs identified through an expression library screen using an antibody to ankyrin-R.4 Since then, it has been assumed that this coding sequence represents the dominant ankyrin-B isoform in most tissues including the heart. Unfortunately, this assumption is not consistent with the common observation that tissues frequently express numerous alternative isoforms of ankyrin genes.4–13
Alternative splicing of ankyrin genes is an important process, because it likely confers specific functions to particular isoforms. In fact, the current paradigm that the heart only expresses ankyrin-B 220 kD is confounded by the observation that ankyrin-B interacts with a variety of ion channels and transporters that lack similarity in function and subcellular distribution. We hypothesize that the heart expresses a diverse population of ankyrin-B isoforms that are tailored through alternative splicing to impart functional specificity and distinct subcellular distribution. We previously demonstrated that the heart expresses over 25 unique iterations of ANK2 transcripts, but a limitation of that study was that we only evaluated partial mRNA transcripts.7
Here, we describe the identification and characterization of two ankyrin-B isoforms AnkB-188 and AnkB-212 that were isolated by long-range reverse transcriptase-PCR from human ventricular mRNA. Atrial and ventricular tissues from mouse, rat, and human hearts express ANK2 transcripts corresponding to these isoforms as determined by quantitative real-time PCR. We demonstrate that both isoforms bind to the sodium–calcium exchanger (NCX), but only AnkB-188 increases NCX membrane expression and current. Furthermore, siRNA knockdown of AnkB-188 in neonatal cardiomyocytes decreases NCX expression and localization at transverse tubules. We generated an isoform-specific antibody to AnkB-212 and demonstrate that this isoform is selectively expressed in heart and skeletal muscle. Moreover, endogenous AnkB-212 localizes to the M-line in adult and neonatal cardiomyocytes. Only AnkB-212 binds to obscurin, a large scaffolding protein implicated in M-line formation and maintenance, and this interaction directs AnkB-212 targeting to the M-line. Moreover, siRNA treatment to AnkB-212 significantly reduces the expression of endogenous ankyrin-B at the M-line, but has no effect on NCX localization at the T-tubules. In summary, our data provide the first description of two ankyrin-B isoforms that display unique functions and subcellular distributions in cardiomyocytes.
2. Methods
2.1. Animals
Mice used in these studies include adult and neonatal wild-type (C57BL/6) and ankyrin-B (C57BL/6) heterozygous and homozygous null animals. Neonatal Sprague-Dawley rats used in these studies were ordered from Texas Animal Specialties (Humble, TX, USA). Studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals published by the National Institute of Health following protocols that were reviewed and approved by the University of Texas Health Science Center at Houston Animal Welfare Committee.
2.2. Human tissue samples
Ventricular and atrial tissues from healthy donor hearts not suitable for transplantation were obtained through the Iowa Donors Network and the National Disease Research Interchange. Age and sex were the only identifying information acquired from tissue providers and the Iowa Human Subjects Committee determined that informed consent was not required. This investigation conforms to the principles outlined in the Declaration of Helsinki.
2.3. Isolation and cloning of full-length ANK2 transcripts from human ventricular mRNAs
RNA was isolated from ventricular tissue using a Qiagen RNeasy mini-kit and 500 ng of RNA was reverse-transcribed with a poly dT primer using SuperScript III reverse transcriptase (LifeTechnologies). cDNAs corresponding to AnkB-188 and AnkB-212 were PCR-amplified using the primer sets (atgaccaccatgttgcaaaag, cttttaattattgatccatcctc) and (atgaccaccatgttgcaaaag, cttttcaaaagctgcatcttc). PCR products were subcloned into pcDH1-MCS lenti-viral expression plasmid (System Biosciences).
2.4. Quantitative real-time-PCR analysis of ANK2 transcripts
Exon–exon boundary spanning primers were designed to the unique splice junctions encoding the C-terminal domains of AnkB-188 (exon junction 45 to 51) and AnkB-212 (exon junction 50 to 51). cDNAs were generated from ventricular and atrial mRNA that was isolated from three human and three mouse hearts. Relative expression of ANK2 transcripts was measured in triplicate by quantitative real-time (qt)-PCR using SYBR Green dye (Bio-Rad) and experiments were replicated three times. Additional details are provided in the Supplementary materials online.
2.5. AnkB-212 antibody generation
Ninety-three amino acids encoded by ANK2 exons 50 and 51 and unique to AnkB-212 were subcloned in triplicate into pET-15b (Novagen), overexpressed in BL21(DE3)pLysS competent cells (Promega), and purified over a nickel column. Polyclonal antibody production was contracted to Pocono Rabbit Farm and Laboratory (Canadensis, PA, USA).
2.6. Immunoblot assay
Heart, skeletal muscle, brain, lung, and liver lysates were isolated from neonatal wild-type and ankyrin-B null mice. Human atrial and ventricular heart lysates were obtained from donor hearts. Protein lysates were separated by SDS–PAGE, transferred to nitrocellulose membrane (Whatman), incubated with primary antibody at 4°C overnight, and visualized using SuperSignal West Pico Chemiluminescent Substrate (Pierce). Primary antibodies used were: pan-ankyrin-B (1:10001) and AnkB-212-specific antibody (1:3000), and pan-actin-5 (1:10 000; Thermo Scientific). Additional details regarding the cellular fractionation assay are provided in the Supplementary materials online.
2.7. Isolation of neonatal mouse hearts and cultures of neonatal rat ventricular myocytes
Neonatal rat ventricular myocytes (NRVM) were isolated from 1- to 2-day-old Sprague-Dawley rat hearts as previously described with a minor modification.14 In accordance with the animal protocol approved by the Animal Welfare Committee at University of Texas Health Science Center at Houston, neonatal mouse and rat pups were euthanized by decapitation. Additional details are provided in the Supplementary materials online.
2.8. Isolation of individual adult mouse cardiomyocytes
Adult mouse cardiomyocytes were isolated as described previously.14 Additional details are provided in the Supplementary materials online.
2.9. siRNA constructs and NRVM transfections
NRVMs were transfected with siRNAs at a final concentration of 50 nM with DharmaFECT (Dharmacon GE) targeting rat AnkB-188, AnkB-212, and non-targeting siRNA control (scramble). The NRVMs were transfected for 6 h, then media were changed to serum-free DMEM. Experiments were performed 48 h later for quantitative real-time PCR and 72 h later for immunoblot analysis and cell contraction recordings. Supplementary material online, Table S1 contains siRNA sequences (see Supplementary materials online, Table S1).
2.10. Fluorescent immunocytochemistry and image quantification
Primary antibodies used were: ankyrin-B (1:5001), ankyrinB-212 (1:900), α-actinin (1:1000, Sigma), myomesin (1:500, Developmental Studies Hybridoma Bank), MyBPC3 (1:250, Santa Cruz), GFP (1:250, UC Davis/NIH NeuroMab Facility). Images were obtained with a Nikon A1 confocal microscope (Nikon, Melville, NY, USA) equipped with ×100 oil, numerical aperture 1.4 objective. Additional details are provided in the Supplementary materials online.
2.11. Binding studies
AnkB-188-GFP and AnkB-212-GFP were expressed in HeLa-T7 cells and purified using an affinity-purified GFP antibody coupled to protein A-agarose beads as previously described15 and incubated with in vitro translated 35S-radiolabeled fragments of the sodium–calcium exchanger (Asp-253 to Lys-615, NM_021097) or the C-terminal domain of human obscurin (Leu-6148 to Asn-6460, NM_052843) (TnT T7-Coupled Reticulate Lysate System, Promega). Additional details are provided in the Supplementary materials online.
2.12. Patch-clamp recording
Whole-cell, patch-clamp recordings were performed using an Axopatch 200B amplifier (Molecular Devices, CA, USA) at room temperature on an inverted phase-contrast microscope. Additional details are provided in the Supplementary materials online.
2.13. Recording and analysis of NRVM contraction rates
Contracting NRVM syncytium was located on light microscope (Micromaster, Fisher) with ×10 magnification eyepiece and ×20 objective lens. A camera with 10 mp resolution (HTC One M8) was mounted on the microscope using an adapter (Snapzoom). Contractions were recorded for 2–2.5 min. The same recording method was used following stimulation with 1 µM epinephrine. Recordings were analysed using the Video Spot Tracker program according to the method described by Fassina et al.16 Additional details are provided in the Supplementary materials online.
2.14. Statistics
P-values were determined for single comparisons using unpaired Student's t-test (two-tailed). The NRVM contraction data were assessed for normality and processed to calculate the mean and the S.D. separately for each syncytial recording. To standardize the dispersion of the time difference between each contraction and the average contraction time for each respective syncytium, the coefficient of variation for each recording was calculated. The coefficients of variation across the groups were compared using the Kruskal–Wallis test with post hoc Wilcoxon rank-sum test. A P-value of <0.05 was considered statistically significant. Values are expressed as means ± SE except for the box-and-whisker plot for contractile rhythm variability where the box contains inter-quartile range covering 25–75% of the readings and the whiskers contain 90% of the readings.
3. Results
3.1. Identification and cardiac expression of ankyrin-B isoforms
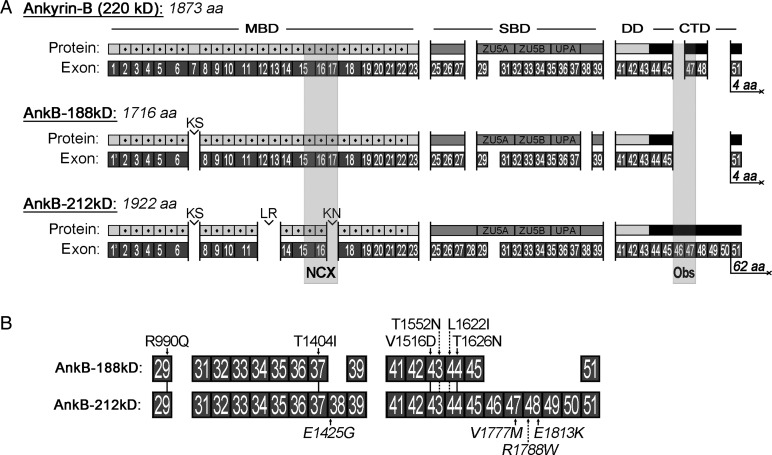
The biggest challenge to isolating tissue-specific ankyrin cDNAs is that ankyrin genes are long and complex containing over 50 exons that are alternatively spliced resulting in mRNA transcripts over 5000 bases in length. To identify full-length ANK2 cDNAs encoding ankyrin-B isoforms expressed in human heart, we performed long-range PCR on reverse-transcribed mRNA isolated from human hearts. We identified two novel ankyrin-B isoforms AnkB-188 and AnkB-212, which are labelled based on their predicted mol. wts (Figure 1A). In contrast to ankyrin-B 220 kD, which starts with exon 1, both isoforms start with exon 1′.7 Both isoforms display similar alternative splicing patterns including excision of exons 7, 24, 30, and 40. Unique alternative splicing to AnkB-188 includes excision of exons 28, 38, and 46–50. Remarkably, exons 46–50 encode the majority of the C-terminal regulatory domain including the binding site for the large scaffolding protein obscurin.17 Unique alternative splicing to AnkB-212 includes excision of exons 12–13 and 17. Previously, we have mapped the NCX-binding site to the three ANK repeats encoded by exons 15–17.15 In addition, the cDNA for AnkB-212 includes exon 50 that shifts the open reading frame by plus one such that exon 51 encodes a unique stretch of 62 amino acids terminating with the residues QEAK. In contrast, the amino acid sequences for both ankyrin-B 220 kD and AnkB-188 terminate with residues DNNE. The complete amino acid sequences for each isoform are presented in (see Supplementary material online, Figures S1 and S2). Missense mutations associated with ankyrin-B dysfunction and cardiac arrhythmias have been mapped to the spectrin-binding and C-terminal regulatory domains of both isoforms (Figure 1B). Interestingly, the mutation E1425G resides in exon 38 and this exon is not present in the AnkB-188 isoform. E1425G has been well studied and linked to a variety of arrhythmias including type 4 long QT syndrome, sick sinus syndrome, and atrial fibrillation.2,3,18–20
Figure 1.
Exon composition and protein domains of ankyrin-B isoforms AnkB-188 and AnkB-212. (A) Exon organization and functional domains of ankyrin-B 220 kD, AnkB-188, and AnkB-212. MBD, membrane-binding domain; SBD, spectrin-binding domain; DD, death domain; CTD, C-terminal regulatory domain. Individual ANK repeats are indicated by dotted grey squares and dipeptides linking ANK repeats are labelled. The ZU5A, ZU5B, and UPA domains are identified in the SBD. Binding sites for sodium–calcium exchanger (NCX) and obscurin (Obs) are shaded in grey. (B) Ten missense mutations associated with cardiac arrhythmias are mapped to both isoforms.3,7,18–20,33
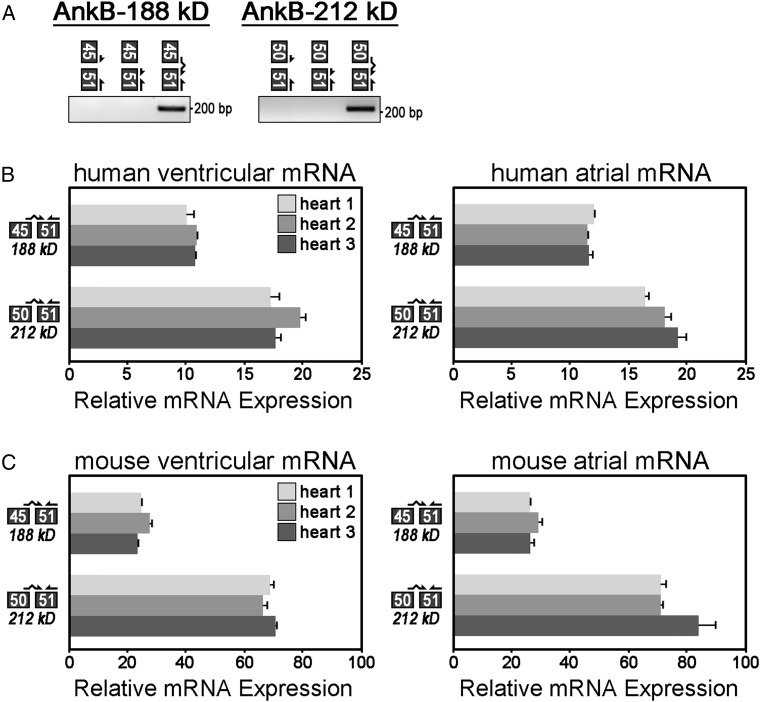
To validate and measure the expression of these isoforms in heart mRNA, we designed exon–exon boundary spanning primers that specifically amplify partial transcripts based on unique exon junctions in each isoform. For example, to detect partial mRNAs encoding AnkB-188, we designed a primer set to the unique exon junction of exons 45 to 51. Specifically, this exon–exon boundary spanning primer anneals to the terminal 14 nucleotides of exon 45 (CAGCTTTTGAAAAG) and the initial 12 nucleotides of exon 51 (GACAACAATGAG). We validated that a PCR product is only amplified using a full-length exon–exon boundary spanning primer (Figure 2A, lane 3). In contrast, no PCR products are amplified using half the exon–exon boundary spanning primer (Figure 2A, lanes 1 and 2). To amplify partial transcripts of AnkB-212, we designed a primer set to amplify the junction of exons 50 to 51. All primer sets were optimized based on nucleotide sequences and annealing temperatures such that primer efficiencies are within 90–110% (see Supplementary material online, Table S2).
Figure 2.
Relative mRNA expression of AnkB-188 and AnkB-212 in human and mouse cardiac tissues. (A) Exon–exon boundary spanning primers were used to PCR amplify alternative splice junctions unique to each isoform (AnkB-188: junction 45/51, AnkB-212: junction 50/51). (B) Relative mRNA expression of alternative splice junctions in AnkB-188 and AnkB-212 was measured in cardiac tissues from three human hearts. Expression of each splice junction was normalized to the expression of exon junction 31/32 (presumably expressed in all ANK2 transcripts and set to 100%). (C) Relative mRNA expression of alternative splice junctions in AnkB-188 and AnkB-212 was measured in cardiac tissues from three mouse hearts. For (B and C), samples were repeated in triplicate (n = 3) and experiments were repeated three times.
Using these exon–exon boundary spanning primers, we performed qt-PCR to measure the relative mRNA expression of AnkB-188 and AnkB-212 in ventricular and atrial tissues isolated from both human and mouse hearts (Figure 2B and C). Relative expression of each isoform was normalized to expression of exon junction 31/32. These exons encode the minimal spectrin-binding domain and presumably this domain is expressed in the majority of ankyrin-B isoforms. Both isoforms are expressed in atrial and ventricular tissues. Interestingly, each primer set demonstrated a similar expression pattern between human and mouse. Both human and mouse hearts express less mRNA for AnkB-188 (45/51) than AnkB-212 (50/51). While the trends are the same in human and mouse hearts, the magnitude of difference between the paired junctions (45/51 vs. 50/51) is not the same. For example, junction 50/51 is ∼3-fold greater than junction 45/51 in mouse, but only ∼1.5-fold greater in human. We speculate that variations in tissue extraction and preservation most likely account for these differences. Taken together, these findings demonstrate that mouse and human hearts express ANK2 transcripts with unique exon junctions present in AnkB-188 and AnkB-212.
3.2. AnkB-188 increases NCX membrane expression and current
In the heart, many different functions have been ascribed to ankyrin-B including the recruitment and retention of various membrane proteins including dystrophin,21 sodium potassium ATPase (NKA),3,18 IP3 receptor,3,18,22 and NCX.3,15,18 We previously demonstrated that ANK repeats 16–18 in the membrane-binding domain of ankyrin-B encodes the binding site for NCX.15 Considering AnkB-212 lacks exon 17 that encodes ANK repeat 18, we anticipated that NCX would not bind to this isoform (Figure 3A).
Figure 3.
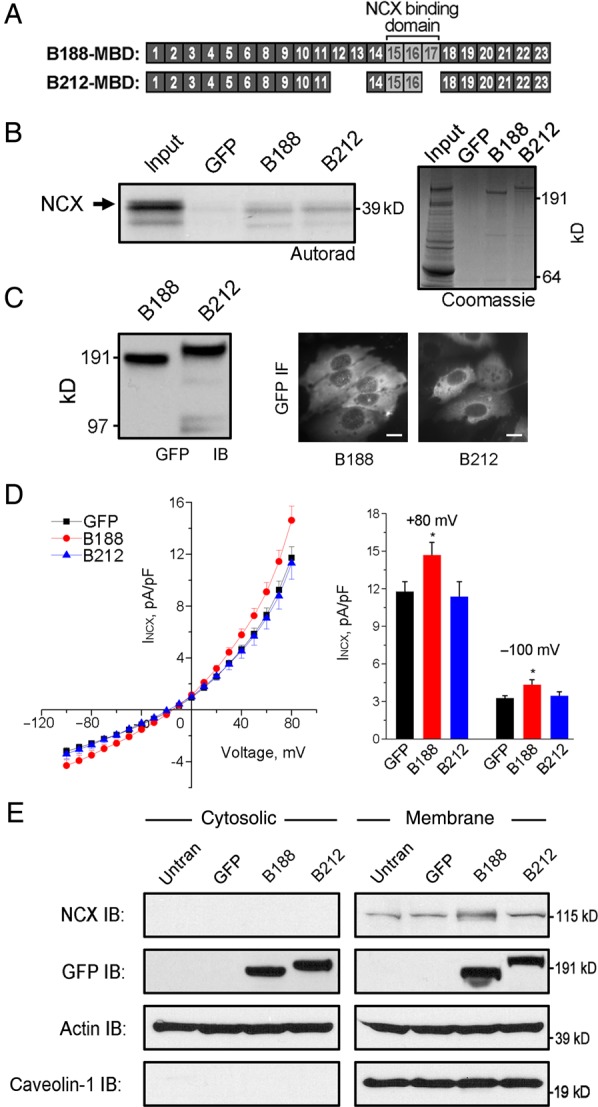
AnkB-188 increases NCX membrane expression and current. (A) Exons encoding the membrane-binding domains of AnkB-188 and AnkB-212. The NCX-binding site maps to ANK2 exons 15–17. (B) 35S-labelled, in vitro translated NCX fragment binds to both AnkB isoforms. Experiments were repeated three times. Coomassie Blue Stain demonstrates equal expression of both isoforms. (C) Detection of the GFP-tag on isoforms by immunoblot and IF in HeLa-T7 cells stably expressing NCX. Scale bar is 5 μm. (D) NCX voltage–current curves of HeLa-T7 cells transiently expressing GFP, AnkB-188, or AnkB-212 (n = 29 for GFP and AnkB-188, n = 27 for AnkB-212, *P < 0.05). (E) NCX expression is increased in the membrane fraction of HeLa-T7 cells expressing AnkB-188. Equal protein loading was demonstrated by immunoblot to pan-actin, while membrane enrichment was demonstrated by immunoblot to caveolin-1. Experiments were reproduced three times.
To assess whether the AnkB isoforms display different NCX binding, we performed an in vitro binding assay with radiolabelled NCX fragment and full-length AnkB-188 or AnkB-212 (Figure 3B). GFP-tagged AnkB isoforms were transiently expressed in HeLa-T7 cells and immunoprecipitated from lysate using a GFP antibody. Interestingly, both isoforms equally bound NCX, suggesting that the absence of ANK repeat 18 had no effect on NCX binding in this in vitro paradigm.
To examine the effect of each isoform on NCX function, we measured the NCX current in HeLa-T7 cells stably expressing NCX and transiently expressing either GFP-tagged AnkB isoform. Similar expression of each isoform was demonstrated by immunoblot analysis of the GFP-tag, and individual GFP-positive cells were measured for NCX current (Figure 3C). While a basal NCX current was detected in the stable cell lines, only the transient expression of AnkB-188 enhanced this current compared with cells expressing GFP alone or AnkB-212 (Figure 3D). HeLa-T7 cells express an endogenous isoform of ankyrin-B as detected by immunoblot (see Supplementary material online, Figure S3), which most likely accounts for the basal NCX current in the stable cell lines. qt-PCR analysis demonstrated that NCX mRNA expression was equivalent in all transfection conditions, indicating that neither ankyrin isoform increases NCX mRNA expression (see Supplementary material online, Figure S3).
To investigate the molecular basis for increased NCX current mediated by AnkB-188, we measured the expression of NCX in cytosolic and membrane fractions of cells transiently expressing GFP alone, AnkB-188, or AnkB-212 by immunoblot analysis (Figure 3E). We found that membrane expression of NCX was two-fold greater in cells expressing AnkB-188 compared with cells expressing GFP alone or AnkB-212 as determined by densitometric analysis of immunoblots from three independent experiments (see Supplementary material online, Figure S3). Taken together, these data suggest that cells expressing AnkB-188 display enhanced NCX current, because this isoform increases NCX expression in the plasma membrane.
3.3. AnkB-212 is selectively expressed in heart and skeletal muscle
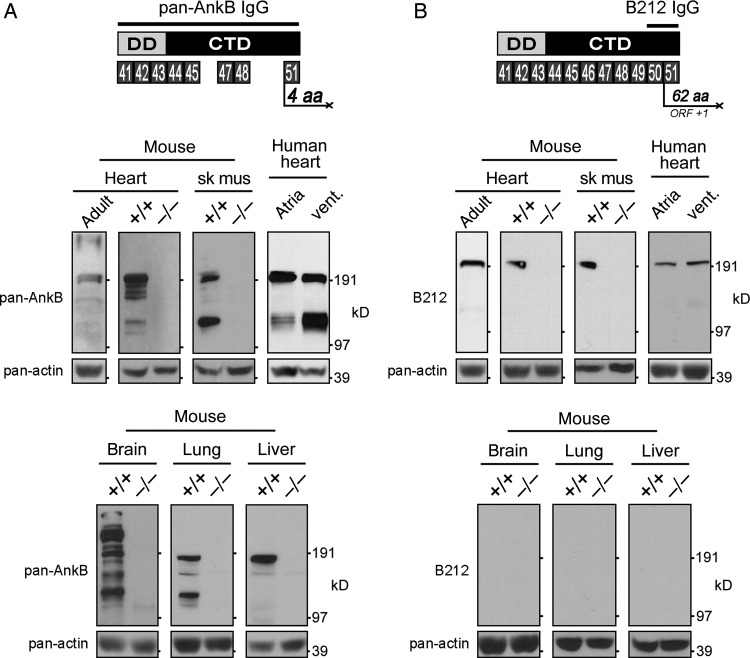
In the initial characterization of ankyrin-B, a polyclonal antibody was generated against the death and C-terminal domains (Figure 4A).4,23 We have previously demonstrated that the C-terminal regulatory domain is subject to complex alternative splicing7 presumably resulting in numerous alternative ankyrin-B isoforms. Using this pan-AnkB antibody, we demonstrate that multiple ankyrin-B isoforms are expressed in adult mouse heart and in mouse neonatal tissue including heart, skeletal muscle, brain, lung, and liver (Figure 4A). In contrast, no isoforms are detected in the same tissues isolated from neonatal AnkB null (−/−) mice. Using the same antibody, multiple ankyrin-B isoforms are also detected in atrial and ventricular tissues isolated from human heart.
Figure 4.
Expression of pan-AnkB and AnkB-212 in various tissues. (A) The antibody to pan-AnkB was generated against the death and C-terminal domains encoded by exons 41–45, 47–48, and 51.23 Expression of multiple AnkB isoforms was detected in heart, skeletal muscle, brain, lung, and liver of wild-type (+/+) mouse neonates and adult mouse heart. (B) The antibody to AnkB-212 was generated against amino acids encoded by exons 50 and 51. Only one isoform of AnkB-212 was detected in heart and skeletal muscle of wild-type neonates and adult mouse heart. Pan-actin immunoblot demonstrates equal protein loading (lower panels). For (A and B), experiments were reproduced three times.
One of the challenges in characterizing alternative isoforms is identifying a unique stretch of amino acids that could be used to generate an isoform-specific antibody. Fortunately, the coding sequence for AnkB-212 includes exon 50, which shifts the open reading frame by plus one such that exon 51 encodes a unique stretch of 62 amino acids (Figure 4B). Numerous expressed sequence tags (>10) from human cDNAs contain this unique coding sequence including DB499390.1, DA768553.1, and DA364599.1. To generate an isoform-specific antibody to AnkB-212, we made a bacterially expressed and purified His-tag fusion protein of the 93 amino acids encoded by exons 50 and 51. Rabbit anti-serum was used to detect AnkB-212 expression in AnkB wild-type (+/+) and null (−/−) tissues including heart, skeletal muscle, brain, lung, and liver (Figure 4B). Interestingly, AnkB-212 is expressed in wild-type heart (adult and neonate) and skeletal muscle, but it is not expressed in brain, lung, or liver from wild-type neonate. AnkB-212 was also detected in human atrial and ventricular tissues. These findings suggest that expression of AnkB-212 is restricted to striated muscle.
3.4. Endogenous AnkB-212 is expressed at the cardiomyocyte M-line
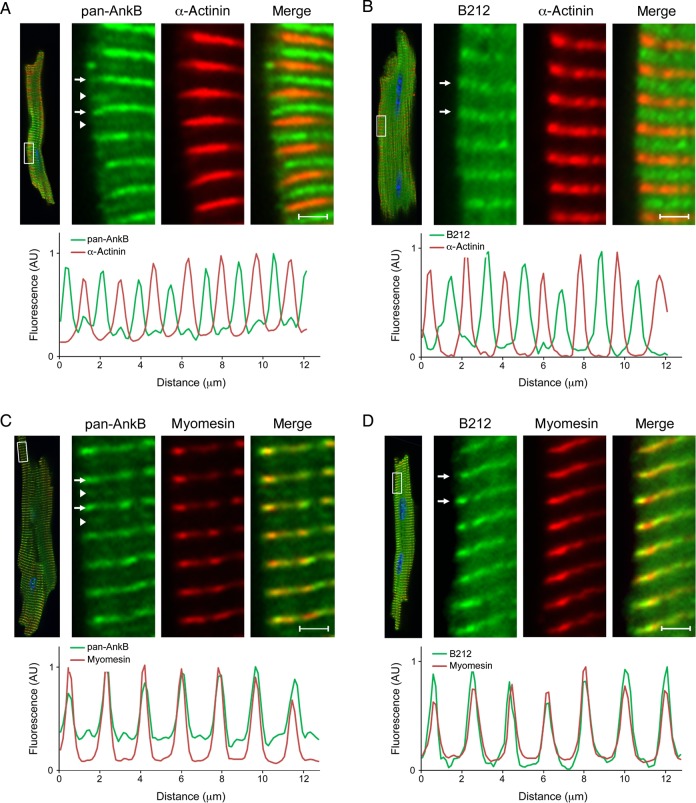
Within cardiomyocytes, ankyrin-B is expressed at the sarcomeric M-line and the sarcoplasmic reticulum/transverse (T)-tubule junctions, which generally co-localize with components of the sarcomeric Z-line. Using the pan-AnkB antibody, we demonstrate the expression of these two ankyrin-B subpopulations at the Z- and M-lines in isolated adult cardiomyocytes (Figure 5A and C). We measured the relative fluorescence intensity of this signal and found that the less abundant ankyrin-B population co-localizes with the resident Z-line marker protein α-actinin, while the more abundant population co-localizes with myomesin, a M-line resident protein. Interestingly, AnkB-212 is only detected at the M-line of adult cardiomyocytes (Figure 5B and D). Specifically, AnkB-212 co-localizes with myomesin, but not with α-actinin. Similar results were found in experiments using cardiomyocytes isolated from ankyrin-B heterozygous hearts (see Supplementary material online, Figure S4). These findings suggest that the M-line population of ankyrin-B is partially composed of the AnkB-212 isoform.
Figure 5.
Endogenous AnkB-212 localizes to the cardiomyocyte M-line. (A and C) In adult cardiomyocytes, two subpopulations of ankyrin-B are detected using the pan-AnkB antibody. The less abundant population (white arrow heads) co-localizes with the Z-line marker α-actinin (A), while the more abundant population (white arrows) co-localizes with the M-line marker myomesin (C). Relative fluorescence intensity of the insets was measured and graphed. (B and D) Only one population of AnkB-212 (white arrows) is detected in isolated cardiomyocytes and co-localizes with the M-line marker myomesin (D) and not the Z-line marker α-actinin (B). For all antibody conditions, 6–8 myocytes per condition were imaged. Scale bar is 2 μm.
3.5. Exogenous AnkB-212 is targeted to the M-line via an interaction with obscurin
To investigate the molecular basis of AnkB-212 targeting and retention at the sarcomeric M-line, we evaluated the isoforms for binding to obscurin, an 800 kD structural protein that has been implicated in M-line formation and alignment.24–26 Two ankyrin-binding sites reside in the C-terminal domain (CTD) of obscurin.27 We previously demonstrated that the ankyrin-B CTD contains two obscurin-binding sites encoded by ANK2 exons 46 and 47.17 Interestingly, the CTD of AnkB-188 lacks exons 46 through 50, while the same domain in AnkB-212 contains these exons (Figure 6A). We performed an in vitro binding assay in which the obscurin CTD was in vitro translated and radiolabelled with 35S methionine, then incubated with GFP-tagged AnkB-188 or AnkB-212, which was overexpressed and purified from HeLa-T7 cells. Obscurin only bound to AnkB-212, and not to AnkB-188 (Figure 6B). Removing exon 46 from the CTD of AnkB-212, which encodes the first obscurin-binding site, disrupts the obscurin/AnkB-212 interaction (see Supplementary material online, Figure S5).
Figure 6.
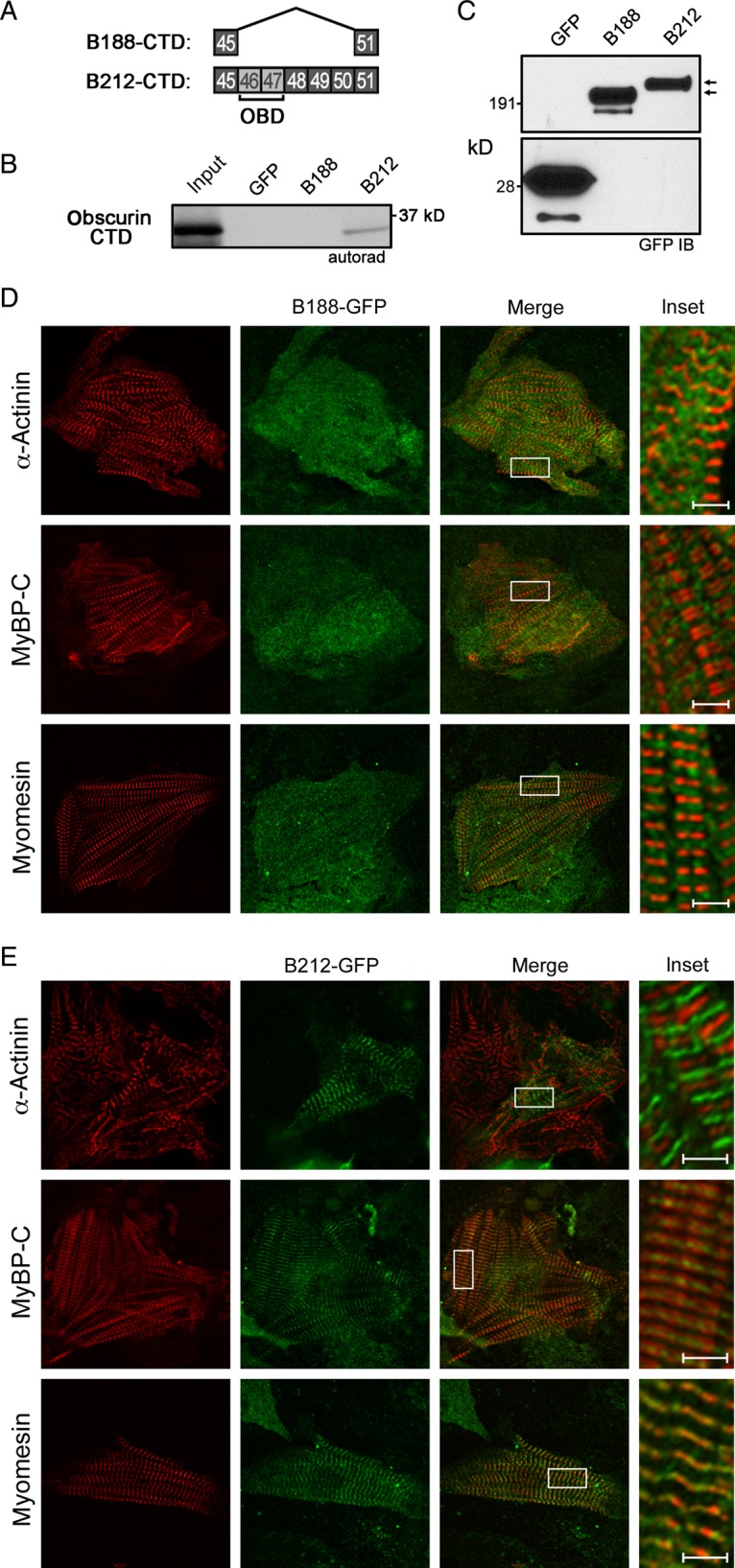
AnkB-212 binds to obscurin and is targeted to the M-line of cardiomyocytes. (A) Exons coding the C-terminal regulatory domains of AnkB-188 and AnkB-212. ANK2 exons 46 and 47 encode two obscurin-binding domains (OBD). (B) In vitro translated obscurin CTD binds GFP-tagged AnkB-212. (C) GFP immunoblot demonstrates similar expression of ankyrin isoforms. Experiments were reproduced twice. Co-localization of virally expressed GFP-tagged AnkB-188 (D) or AnkB-212 (E) with resident proteins of the Z-line (α-actinin), A-band (myosin-binding protein C—MyBP-C), and M-line (myomesin) in rat neonatal cardiomyocytes. For all antibody conditions, 6–8 myocytes per condition were imaged. Scale bar is 2 μm.
To assess whether AnkB-188 and AnkB-212 display differential subcellular localization in cardiomyocytes, we evaluated the co-localization of each isoform to three sarcomeric proteins: α-actinin (Z-line), myosin-binding protein C (A-band), and myomesin (M-line). GFP-tagged ankyrin isoforms were expressed in rat neonatal cardiomyocytes by lenti-virus and images were acquired by confocal microscopy. AnkB-188 expression was diffusely cytosolic and lacked sarcomeric localization (Figure 6D). In contrast, AnkB-212 distinctly co-localizes with myomesin and is flanked by myosin-binding protein C, suggesting that its localization does not extend beyond the M-line (Figure 6E). Removing exon 46 from the CTD of AnkB-212 disrupts the M-line targeting of this construct (see Supplementary material online, Figure S5). Taken together, these data suggest that AnkB-212 is targeted to the M-line via its interaction with obscurin, which is consistent with a report that M-line targeting of endogenous ankyrin-B is lost in skeletal muscle fibres of the obscurin knockout mouse.21
3.6. AnkB-188 knockdown decreases the expression and proper subcellular localization of NCX
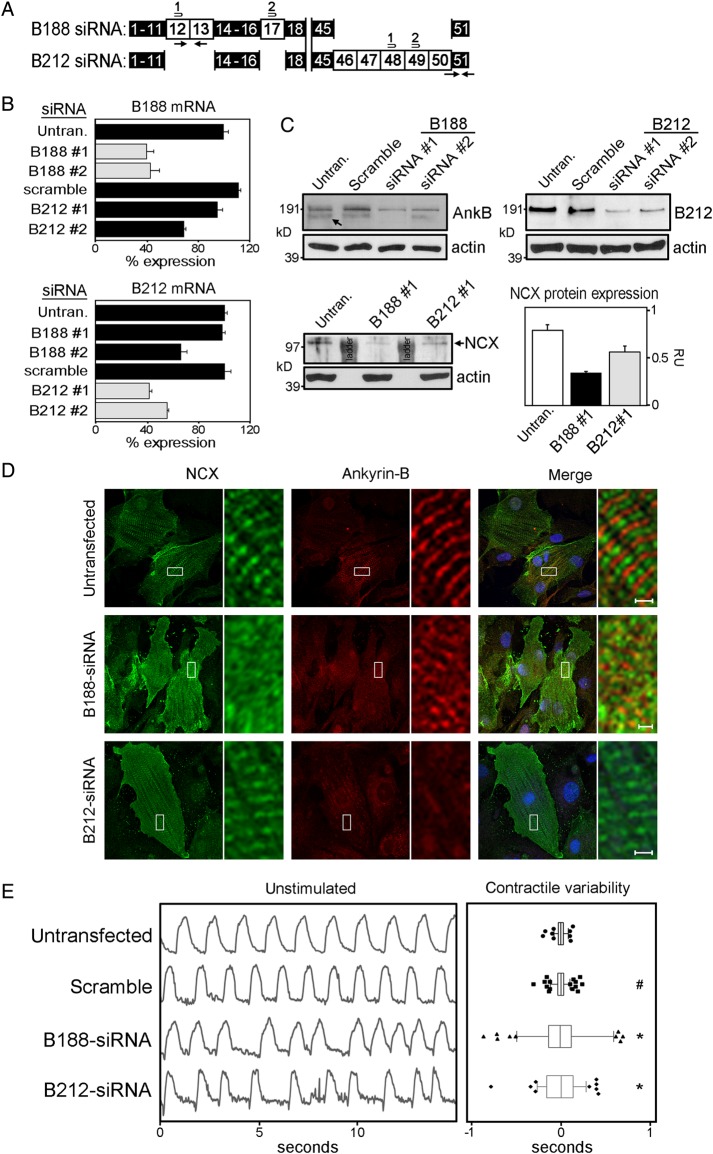
To identify potential functions associated with either ankyrin-B isoform in rat neonatal cardiomyocytes, we used siRNAs to preferentially knock-down each isoform. siRNAs were designed to unique exons in each isoform (Figure 7A). We used qt-PCR transcript analysis to evaluate the efficacy and specificity of the siRNAs against its intended isoform (Figure 7B, grey bars) vs. the other isoform (Figure 7B, black bars). siRNAs that displayed preferential knockdown of each isoform were used in subsequent assays. Immunoblot analysis was performed to confirm siRNA-mediated knockdown of each isoform. Using the pan-AnkB antibody, two high mol. wt isoforms are clearly resolved and siRNAs to AnkB-188 reduce the expression of the smaller isoform, although expression of the larger isoform also decreases but to a lesser extent (Figure 7C). The isoform-specific antibody to AnkB-212 demonstrates a marked reduction in the expression of this isoform following siRNA treatment (Figure 7C).
Figure 7.
AnkB-188 knockdown decreases the expression and subcellular localization of NCX and elicits arrhythmic contractions. (A) Isoform-specific siRNAs were designed to exons unique to AnkB-188 and AnkB-212. (B) Knockdown of isoform mRNA was assessed by qt-PCR using primer sets unique to each isoform (black arrows). qt-PCR values were normalized to GAPDH mRNA expression and expressed as a percentage of isoform expression in untransfected cardiomyocytes, which was set to 100%. Grey bars represent isoform mRNA expression targeted by the siRNAs. Black bars represent mRNA expression of controls and the other isoform. (C) Immunoblot analysis of AnkB-188, AnkB-212, and NCX in control and siRNA-treated cardiomyocytes. Pan-actin immunoblot demonstrates equal protein loading. (D) Immunofluorescent localization of NCX and ankyrin-B in siRNA-treated cardiomyocytes. Regions boxed in white are magnified in the side panel. (E) AnkB-188 or AnkB-212 siRNA treatment precipitates irregular cardiomyocyte contractions. The left panel presents representative contraction rhythms of untransfected and transfected cardiomyocytes. The right panel presents the summary data. Box-and-whisker plots represent the difference in time between individual contractions and the average contraction time over a 30 s interval. Sample size is untransfected: 6, scramble: 7, B188-siRNA: 7, and B212-siRNA: 5. #P-value » 0.05 and *P-value < 0.05 compared with the untransfected.
Previous studies have demonstrated that ankyrin-B is important for the expression and proper subcellular targeting of NCX.3,15,18 Considering AnkB-188 increases NCX membrane expression and current in HeLa-T7 cells (Figure 3D and E), we evaluated siRNA-mediated knockdown of each isoform on the expression and localization of NCX in rat neonatal cardiomyocytes. While NCX protein expression is slightly reduced following AnkB-212 knockdown, AnkB-188 knockdown results in a more significant reduction in NCX protein expression (Figure 7C). In untransfected cardiomyocytes, NCX predominantly co-localizes with α-actinin at the Z-line (see Supplementary material online, Figure S6), while two populations of ankyrin-B are detected at the M- and Z-lines (Figure 7D). AnkB-212 knockdown dramatically reduces ankyrin-B expression at the M-line in cardiomyocytes, but interestingly NCX maintains its striated patterning over the Z-lines. In contrast, the Z-line striated patterning of NCX is markedly reduced in cardiomyocytes following AnkB-188 knockdown (Figure 7D and see Supplementary material online, FigureS6) suggesting that this isoform is important for NCX subcellular localization in cardiomyocytes.
Ankyrin-B haploinsufficiency results in decreased NCX membrane expression, altered calcium dynamics, and decreased contractions in ventricular cardiomyocytes.3,15,19 Considering the numerous cardiac arrhythmias associated with ankyrin-B dysfunction, we evaluated the effect of isoform-specific knockdown on contraction rhythms of cardiomyocytes. Both untransfected and scramble siRNA-treated cardiomyocytes display uniformly timed contractions (Figure 7E). In contrast, cardiomyocytes with knockdown of AnkB-188 display patterns of arrhythmic contractions. Surprisingly, cardiomyocytes with knockdown of AnkB-212, which has no effect on NCX membrane expression or localization, also exhibit patterns of arrhythmic contractions. The same patterns are observed following epinephrine treatment (see Supplementary material online, Figure S7).
4. Discussion
Normal heart function requires the proper functioning of proteins at unique membrane and sarcomeric domains in cardiomyocytes such as the costamere, intercalated disc, and T-tubules. As an adaptor protein, ankyrin targets and maintains distinctive networks of membrane and signalling proteins at these domains by tethering them to the underlying cytoskeletal network. The alternative splicing of an ankyrin gene regulates the modular nature of ankyrin isoforms such that they can serve as a common interface between β-spectrin and unique cohorts of proteins that define functionally distinct subcellular domains.
In earlier studies that initially described ankyrin genes, there were many references to alternative isoforms. Some of these isoforms displayed tissue-specific expression, while other isoforms demonstrated unique subcellular distribution. For example, while ankyrin-G expression is relatively ubiquitous throughout tissues, a brain-specific isoform is targeted to the neuronal axon initial segment by an ∼6000 bp exon.12,28 Likewise, an alternative start site in the ankyrin-R gene gives rise to a truncated muscle-specific isoform with a novel transmembrane domain that sequesters it to the membrane of the sarcoplasmic reticulum.5,10,29
In the initial characterization of the ankyrin-B gene, two isoforms (440 and 220 kD) were detected in the brain.4,23 The cDNA for the 220 kD isoform was synthesized by combining seven partial, but overlapping cDNA clones that were identified in an expression library screen.4 The only difference between these cDNAs is that an ∼6000 bp exon is included in the cDNA of the brain-specific 440 kD isoform. With time, it became tacitly assumed that the 220 kD isoform represented the predominant isoform expressed in other tissues.
This study is the first to describe two novel ankyrin-B isoforms identified by long-range PCR of reverse-transcribed mRNA isolated from human ventricular tissue. We demonstrate the expression of mRNA and protein of these isoforms in human, rat, and mouse hearts. In addition, we can discriminate between the two isoforms based on their function and subcellular distribution in cardiomyocytes. Specifically, overexpression of AnkB-188 in HeLa-T7 cells increases NCX membrane expression and current, while siRNA-mediated knockdown of this isoform decreases NCX expression and subcellular localization at Z-lines in neonatal cardiomyocytes. Using an isoform-specific antibody, we demonstrate that expression of AnkB-212 is restricted to striated muscle and distinctly expressed at the M-line in cardiomyocytes. Moreover, the M-line targeting of AnkB-212 is regulated by its interaction with the large scaffolding protein obscurin. In contrast to AnkB-188, the preferential knockdown of AnkB-212 in neonatal cardiomyocytes does not alter the Z-line striated patterning of NCX. Interestingly, knockdown of either isoform results in arrhythmic contractions. The function of AnkB-212 at the M-line and whether these isoforms share areas of functional overlap remain unanswered questions but are important lines of inquiry for future experiments.
One unresolved question in the ankyrin field is what regulates ankyrin specificity for particular membrane proteins. Ankyrin-B 220 kD has been shown to interact with a diverse array of proteins including the sodium–calcium exchanger (NCX), sodium potassium ATPase (NKA), inositol 1,4,5-triphosphate receptor (IP3R), ATP-sensitive inward-rectifying potassium channel subunit (Kir6.2), dystrophin, and obscurin.15,17,18,21,22,30–32 Our data support the hypothesis that alternative splicing tailors ankyrin proteins such that they are specific for particular membrane or scaffolding proteins. We anticipate that the heart expresses additional ankyrin-B isoforms that have yet to be identified.
The expression of different function-specific ankyrin-B isoforms in heart would provide a fitting explanation for the diversity of arrhythmias associated with ankyrin-B dysfunction. In fact, the designation of ankyrin-B syndrome has been adopted to incorporate the various arrhythmias associated with ankyrin-B dysfunction, which partially include bradycardia, idiopathic ventricular fibrillation, catecholaminergic polymorphic ventricular tachycardia, and atrial fibrillation.1–3,19,20 Past efforts to understand the molecular basis of dysfunction stemming from these mutations have been performed in the context of ankyrin-B 220 kD. While these studies have provided insight into ankyrin-B function, it is important to re-evaluate ankyrin-B syndrome in the context of the newly identified ankyrin-B isoforms. For example, previous studies have demonstrated that the arrhythmia-associated AnkB missense mutation E1425G decreases NCX membrane expression in ventricular cardiomyocytes. Our data demonstrate that NCX membrane expression is regulated by AnkB-188, which lacks exon 38 and would not express the E1425G missense mutation. We anticipate that future experiments characterizing these isoforms will provide a more complete picture of ankyrin-B function in normal hearts and how missense mutations in different ankyrin-B isoforms result in various atrial and ventricular arrhythmias.
4.1. Limitations
While this study is an important first step in providing evidence for how ankyrin attains specificity for particular proteins, many issues have yet to be resolved. The generation of an isoform-specific antibody to AnkB-188 would have allowed for its identification by immunoblot and immunofluorescent analyses. In addition, the data generated from siRNA-mediated knockdown of AnkB-188 are consistent with the notion that this isoform regulates NCX membrane expression and localization, but siRNA technology has its shortcomings. In addition to the possibility of non-specific effects, siRNA-mediated silencing of other potentially uncharacterized isoforms will confound our understanding of ankyrin-B function in cardiomyocytes. Future experiments will address these and other unanswered questions regarding the role of ankyrin-B isoforms in cardiomyocyte cellular physiology and pathology. For example, are there other ankyrin-B isoforms expressed in the heart? If so, do they have distinct interacting protein partners? Are different cohorts of ankyrin-B isoforms involved in the trafficking and scaffolding of membrane proteins? Finally, how do arrhythmia-associated mutations alter the function of specific isoforms and does this account for the diversity of cardiac arrhythmias manifested in ankyrin-B syndrome?
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported by the National Institute of Health (grant numbers HL092232 to S.R.C.; GM101218 to H.H.).
Acknowledgements
We acknowledge CISMM at UNC-CH for the generation and dissemination of the Video Spot Tracker (VST) program (NIH NIBIB 5-P41-RR02170).
Conflict of interest: none declared.
References
- 1.Cunha SR, Hund TJ, Hashemi S, Voigt N, Li N, Wright P, Koval O, Li J, Gudmundsson H, Gumina RJ, Karck M, Schott JJ, Probst V, Le Marec H, Anderson ME, Dobrev D, Wehrens XH, Mohler PJ. Defects in ankyrin-based membrane protein targeting pathways underlie atrial fibrillation. Circulation 2011;124:1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Scouarnec S, Bhasin N, Vieyres C, Hund TJ, Cunha SR, Koval O, Marionneau C, Chen B, Wu Y, Demolombe S, Song LS, Le Marec H, Probst V, Schott JJ, Anderson ME, Mohler PJ. Dysfunction in ankyrin-B-dependent ion channel and transporter targeting causes human sinus node disease. Proc Natl Acad Sci USA 2008;105:15617–15622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohler PJ, Schott JJ, Gramolini AO, Dilly KW, Guatimosim S, duBell WH, Song LS, Haurogne K, Kyndt F, Ali ME, Rogers TB, Lederer WJ, Escande D, Le Marec H, Bennett V. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature 2003;421:634–639. [DOI] [PubMed] [Google Scholar]
- 4.Otto E, Kunimoto M, McLaughlin T, Bennett V. Isolation and characterization of cDNAs encoding human brain ankyrins reveal a family of alternatively spliced genes. J Cell Biol 1991;114:241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birkenmeier CS, Sharp JJ, Gifford EJ, Deveau SA, Barker JE. An alternative first exon in the distal end of the erythroid ankyrin gene leads to production of a small isoform containing an NH2-terminal membrane anchor. Genomics 1998;50:79–88. [DOI] [PubMed] [Google Scholar]
- 6.Birkenmeier CS, White RA, Peters LL, Hall EJ, Lux SE, Barker JE. Complex patterns of sequence variation and multiple 5′ and 3′ ends are found among transcripts of the erythroid ankyrin gene. J Biol Chem 1993;268:9533–9540. [PubMed] [Google Scholar]
- 7.Cunha SR, Le Scouarnec S, Schott JJ, Mohler PJ. Exon organization and novel alternative splicing of the human ANK2 gene: implications for cardiac function and human cardiac disease. J Mol Cell Cardiol 2008;45:724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devarajan P, Stabach PR, Mann AS, Ardito T, Kashgarian M, Morrow JS. Identification of a small cytoplasmic ankyrin (AnkG119) in the kidney and muscle that binds beta I sigma spectrin and associates with the Golgi apparatus. J Cell Biol 1996;133:819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagelin C, Constantin B, Deprette C, Ludosky MA, Recouvreur M, Cartaud J, Cognard C, Raymond G, Kordeli E. Identification of Ank(G107), a muscle-specific ankyrin-G isoform. J Biol Chem 2002;277:12978–12987. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher PG, Forget BG. An alternate promoter directs expression of a truncated, muscle-specific isoform of the human ankyrin 1 gene. J Biol Chem 1998;273:1339–1348. [DOI] [PubMed] [Google Scholar]
- 11.Hopitzan AA, Baines AJ, Ludosky MA, Recouvreur M, Kordeli E. Ankyrin-G in skeletal muscle: tissue-specific alternative splicing contributes to the complexity of the sarcolemmal cytoskeleton. Exp Cell Res 2005;309:86–98. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Bennett V. Restriction of 480/270-kD ankyrin G to axon proximal segments requires multiple ankyrin G-specific domains. J Cell Biol 1998;142:1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamankurt G, Wu HC, McCarthy M, Cunha SR. Exon organization and novel alternative splicing of ank3 in mouse heart. PLoS One 2015;10:e0128177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baskin KK, Taegtmeyer H. AMP-activated protein kinase regulates E3 ligases in rodent heart. Circ Res 2011;109:1153–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunha SR, Bhasin N, Mohler PJ. Targeting and stability of Na/Ca exchanger 1 in cardiomyocytes requires direct interaction with the membrane adaptor ankyrin-B. J Biol Chem 2007;282:4875–4883. [DOI] [PubMed] [Google Scholar]
- 16.Fassina L, Di Grazia A, Naro F, Monaco L, De Angelis MG, Magenes G. Video evaluation of the kinematics and dynamics of the beating cardiac syncytium: an alternative to the Langendorff method. Int J Artif Organs 2011;34:546–558. [DOI] [PubMed] [Google Scholar]
- 17.Cunha SR, Mohler PJ. Obscurin targets ankyrin-B and protein phosphatase 2A to the cardiac M-line. J Biol Chem 2008;283:31968–31980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohler PJ, Davis JQ, Bennett V. Ankyrin-B coordinates the Na/K ATPase, Na/Ca exchanger, and InsP3 receptor in a cardiac T-tubule/SR microdomain. PLoS Biol 2005;3:e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohler PJ, Le Scouarnec S, Denjoy I, Lowe JS, Guicheney P, Caron L, Driskell IM, Schott JJ, Norris K, Leenhardt A, Kim RB, Escande D, Roden DM. Defining the cellular phenotype of ‘ankyrin-B syndrome’ variants: human ANK2 variants associated with clinical phenotypes display a spectrum of activities in cardiomyocytes. Circulation 2007;115:432–441. [DOI] [PubMed] [Google Scholar]
- 20.Mohler PJ, Splawski I, Napolitano C, Bottelli G, Sharpe L, Timothy K, Priori SG, Keating MT, Bennett V. A cardiac arrhythmia syndrome caused by loss of ankyrin-B function. Proc Natl Acad Sci USA 2004;101:9137–9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Randazzo D, Giacomello E, Lorenzini S, Rossi D, Pierantozzi E, Blaauw B, Reggiani C, Lange S, Peter AK, Chen J, Sorrentino V. Obscurin is required for ankyrinB-dependent dystrophin localization and sarcolemma integrity. J Cell Biol 2013;200:523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohler PJ, Davis JQ, Davis LH, Hoffman JA, Michaely P, Bennett V. Inositol 1,4,5-trisphosphate receptor localization and stability in neonatal cardiomyocytes requires interaction with ankyrin-B. J Biol Chem 2004;279:12980–12987. [DOI] [PubMed] [Google Scholar]
- 23.Kunimoto M, Otto E, Bennett V. A new 440-kD isoform is the major ankyrin in neonatal rat brain. J Cell Biol 1991;115:1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ackermann MA, Hu LY, Bowman AL, Bloch RJ, Kontrogianni-Konstantopoulos A. Obscurin interacts with a novel isoform of MyBP-C slow at the periphery of the sarcomeric M-band and regulates thick filament assembly. Mol Biol Cell 2009;20:2963–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Randall WR, Bloch RJ. Obscurin regulates the organization of myosin into A bands. Am J Physiol Cell Physiol 2004;287:C209–C217. [DOI] [PubMed] [Google Scholar]
- 26.Raeker MO, Su F, Geisler SB, Borisov AB, Kontrogianni-Konstantopoulos A, Lyons SE, Russell MW. Obscurin is required for the lateral alignment of striated myofibrils in zebrafish. Dev Dyn 2006;235:2018–2029. [DOI] [PubMed] [Google Scholar]
- 27.Busby B, Willis CD, Ackermann MA, Kontrogianni-Konstantopoulos A, Bloch RJ. Characterization and comparison of two binding sites on obscurin for small ankyrin 1. Biochemistry 2010;49:9948–9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kordeli E, Lambert S, Bennett V, Ankyrin G. A new ankyrin gene with neural-specific isoforms localized at the axonal initial segment and node of Ranvier. J Biol Chem 1995;270:2352–2359. [DOI] [PubMed] [Google Scholar]
- 29.Zhou D, Birkenmeier CS, Williams MW, Sharp JJ, Barker JE, Bloch RJ. Small, membrane-bound, alternatively spliced forms of ankyrin 1 associated with the sarcoplasmic reticulum of mammalian skeletal muscle. J Cell Biol 1997;136:621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayalon G, Davis JQ, Scotland PB, Bennett V. An ankyrin-based mechanism for functional organization of dystrophin and dystroglycan. Cell 2008;135:1189–1200. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Kline CF, Hund TJ, Anderson ME, Mohler PJ. Ankyrin-B regulates Kir6.2 membrane expression and function in heart. J Biol Chem 2010;285:28723–28730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kline CF, Kurata HT, Hund TJ, Cunha SR, Koval OM, Wright PJ, Christensen M, Anderson ME, Nichols CG, Mohler PJ. Dual role of K ATP channel C-terminal motif in membrane targeting and metabolic regulation. Proc Natl Acad Sci USA 2009;106:16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith SA, Sturm AC, Curran J, Kline CF, Little SC, Bonilla IM, Long VP, Makara M, Polina I, Hughes LD, Webb TR, Wei Z, Wright P, Voigt N, Bhakta D, Spoonamore KG, Zhang C, Weiss R, Binkley PF, Janssen PM, Kilic A, Higgins RS, Sun M, Ma J, Dobrev D, Zhang M, Carnes CA, Vatta M, Rasband MN, Hund TJ, Mohler PJ. Dysfunction in the βII spectrin-dependent cytoskeleton underlies human arrhythmia. Circulation 2015;131:695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]