Abstract
Aims
After injury, the adult zebrafish can regenerate the heart. This requires the activation of the endocardium and epicardium as well as the proliferation of pre-existing cardiomyocytes to replace the lost tissue. However, the molecular mechanisms involved in this process are not completely resolved. In this work, we aim to identify the proteins involved in zebrafish heart regeneration and to explore their function.
Methods and results
Using a proteomic approach, we identified Hyaluronan-mediated motility receptor (Hmmr), a hyaluronic acid (HA) receptor, to be expressed following ventricular resection in zebrafish. Moreover, enzymes that produce HA, hyaluronic acid synthases (has), were also expressed following injury, suggesting that this pathway may serve important functions in the regenerating heart. Indeed, suppression of HA production, as well as depletion of Hmmr, blocked cardiac regeneration. Mechanistically, HA and Hmmr are required for epicardial cell epithelial–mesenchymal transition (EMT) and their subsequent migration into the regenerating ventricle. Furthermore, chemical inhibition of Focal Adhesion Kinase (FAK) or inhibition of Src kinases, downstream effectors of Hmmr, also prevented epicardial cell migration, implicating a HA/Hmmr/FAK/Src pathway in this process. In a rat model of myocardial infarction, both HA and HMMR were up-regulated and localized in the infarct area within the first few days following damage, suggesting that this pathway may also play an important role in cardiac repair in mammals.
Conclusion
HA and Hmmr are required for activated epicardial cell EMT and migration involving the FAK/Src pathway for proper heart regeneration.
Keywords: Zebrafish heart regeneration, Hyaluronic acid, Hmmr, Epicardial cell migration, pFAK
1. Introduction
Ischemic heart disease is one of the most common causes of mortality in developed countries. After myocardial infarction (MI), billions of cardiomyocytes undergo apoptosis, pyroptosis, and necrosis, and a non-contractile collagen scar that limits the cardiac function is formed.1 The ability of the mammalian heart to replace lost cardiomyocytes is limited.2 Neonatal mice are able to regenerate after amputation of the ventricular apex,3 and after MI,4 but regeneration is restricted to post-natal day 7 (P7), after which the majority of cardiomyocytes become post-mitotic. More recently, cardiac ischaemic injury during the phase of cardiac growth in adolescent mice (P15) also exhibited regenerative capacity, suggesting that under certain conditions regeneration can occur in mammals.5 In contrast, adult zebrafish (Danio rerio) can efficiently regenerate the heart after amputation of the ventricle apex throughout its lifespan.6,7 A key factor of the regenerative process in zebrafish is the ability of pre-existing cardiomyocytes to undergo proliferation following organ damage.8–11 Another feature is the activation and proliferation of epicardial cells to undergo epithelial–mesenchymal transition (EMT), followed by their migration into the injury site to promote angiogenesis.9,12–14 In addition, within 3 h of ventricular injury, the endocardium undergoes morphological changes and induces the expression of retinoic acid (RA)-synthesizing enzyme aldh1a2, indicating that the endocardium is also a dynamic player in zebrafish heart regeneration.13 The molecular pathways that direct these processes are beginning to be elucidated with evidence that fibroblast growth factors (FGF),11,14 transforming growth factor-β (TGF-β),15 platelet derived growth factor β (PDGFβ),16 insulin-like growth factor 2 (IGF2),17 RA,13 Jak1/Stat3,18 neuregulin 1 (Nrg1),19 Hedgehog,20 and Notch signaling21 playing important roles to promote cardiomyocyte proliferation, endocardium activation, and epicardial EMT.
In this pilot study, we analysed the proteomic changes following cardiac resection of the ventricular apex in adult zebrafish. We identified increased expression of an hyaluronic acid (HA) receptor (Hmmr) and hypothesized that the HA pathway could play a crucial role in cardiac regeneration. HA is a large, linear, non-sulfated GAG component of extracellular matrix (ECM). Following injury, HA is produced in the inner side of the plasma membrane by HA Synthases (HAS) and is extruded onto the cell surface where it accumulates in the wound to promote cellular proliferation and migration to support tissue remodelling and healing.22,23 Chemical suppression of HA synthesis as well as knockdown of Hmmr blocked cardiac regeneration. Mechanistically, we observed decreased migration of activated epicardial cells into the regenerating heart and reduced coronary vasculature, suggesting that HA is important for epicardial EMT. Our studies document the importance of HA and its receptor in epicardial cell migration into the clot tissue for remodelling of the nascent coronary vasculature.
2. Methods
2.1. Zebrafish maintenance, ventricular amputation, and retro-orbital injections
The zebrafish experiments were performed according to protocol approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pittsburgh that conforms to the NIH guidelines. Adult (6–18 months) wild type AB* and transgenic Tg(myl7:EGFP)f1,24 Tg(fli1a:EGFP)y1,25 Tg(wt1b:EGFP)li1 (a kind gift from Christoph Englert)26 zebrafish were maintained at 28°C. Zebrafish were anaesthetized by immersing in 0.168 g/L ethyl 3-aminobenzoate methanesulfonate salt (MS-222; Sigma) for 3–5 min. Zebrafish were placed onto a wet sponge with the abdomen facing up. Approximately 20% of the ventricle apex was resected as described by Poss et al.,6 and zebrafish returned to the system for recovery before retro-orbital injections of drugs or with Vivo Morpholinos (VMO) at one day post-surgery as described by Pugach et al.27 Retro-orbital injections were performed daily for 2, 4, 6, or 10 days at which point hearts were extracted. To suppress HA production, zebrafish were injected with 3 µL of 500 µM 7-hydroxy-4-methylcoumarin (HMC) (Acros Organics) (also known as 4-methylumbelliferone, 4-Mu), or with PBS as vehicle control. To inhibit Focal Adhesion Kinase (FAK), 3 µL of 100 µM of PF-573228 (Sigma) dissolved in DMSO (Sigma) was injected. To suppress Src Kinase, 3 µL of 200 µM Src Inhibitor 1 (SKI-1) (Sigma) or vehicle DMSO was injected. To knockdown Hmmr, 1 µL of 3 mg/kg Hmmr E414 spliced VMO (Gene Tools, LLC) (TGTGCAAACAGATGTACCTCTTTCT) was injected. For controls, 5 bp mismatch VMO (Mut-MO) (TGAGGAAACACATCTACGTCTTTCT) was retro-orbitally injected.
2.2. Difference gel electrophoresis and MS/MS analysis
A detailed description of difference gel electrophoresis (DiGE) and MS/MS analysis is provided in the Supplementary material online.
2.3. In situ hybridization, immunostaining, clot area measurement, and cell counting
For histological examination, zebrafish were euthanized using 0.168 g/L ethyl 3-aminobenzoate methanesulfonate salt for 15 min, and hearts were collected in cold PBS and fixed in 4% paraformaldehyde (PFA) overnight at 4°C. Hearts were cryopreserved with sucrose before immersion in embedding media (Instrumedics). Fourteen micrometre cryosections were collected, and consecutive sections were used for in situ hybridization (ISH), immunostaining, and Acid Fuchsin Orange G (AFOG). ISH was performed in cryosections, using digoxygenin-labelled cRNA probes as described.14 BM purple (Roche) was used as AP substrate. AFOG staining was performed as described by Poss et al.6 Images were captured with Leica MZ 16 microscope and Q Imaging Retiga 1300 camera. A detailed description of clot area measurement and cell counting is provided in Supplementary material online.
2.4. RNA extraction, cDNA synthesis, PCR, and quantitative PCR
Total RNA was isolated from uninjured hearts and hearts at 1, 3, and 7 days post-amputation (dpa) using TRIzol reagent (Invitrogen), and RNeasy Micro kit (Qiagen), according to manufacturer's instructions. Eight hearts were pooled together for each condition. One microgram of total RNA was reverse transcribed to cDNA with SuperScript (Invitrogen) using random hexamers. PCR was performed to test the efficacy of hmmr Spliced V-MO knockdown. Eukaryotic translation elongation factor 1 alpha 1, like 1 (eef1a1l1) was used as reference gene. The primers sequences for PCR are the following: eef1a1l1-F ATCTACAAATGCGGTGGAAT; eef1a1l1-R ATACCAGCCTCAAACTCACC; hmmr-Ex3-F GGACCATGTCTGTTGATGGTTTGGCTG; hmmr-Ex-7-R GACCTTTACCTTTCCTTCTGAGC. RT–PCR products were electrophoresed on agarose gels and stained with ethidium bromide. The primer sequences used for quantitative PCR (Q-PCR) were designed using Beacon designer and are listed in Supplementary material online, Table S1. Two step real-time PCR was performed using SYBR Green (Bio-rad) in a PCR system iQ5 thermal cycler (Bio-rad). Primers set efficiency was calculated and adjusted using LinRegPCR. β-Actin and RNA polymerase were used to normalize gene expression in the Q-PCR experiments. Experiments were done in triplicate.
2.5. Western blot
Proteins were extracted from a pool of five ventricles apex of each condition, using 200 µL of Laemmli Buffer (Bio-rad), containing β-mercaptoethanol (Fisher). Samples were heated at 95°C for 5 min, and 0.1 M dithiothreitol (Calbiochem) was added before loading. After SDS–PAGE, proteins were transferred in nitrocellulose membrane (Li-cor). Membrane was blocked in Odyssey Blocking Buffer (Li-cor) for 1 h. Antibodies were diluted in Odyssey Blocking Buffer, containing 0.2% Tween 20 (National diagnostics). Blots were scanned using Li-cor Odyssey CLx Infrared imaging system, and band intensity was quantified and normalized using Image Studio software. Primary antibody used were anti-Twist1 (Sigma) (1:1000), anti-ERK-2 (Sigma) (1:500), anti-actin (Sigma) (1:5000), and anti-FAK [pY397] (Invitrogen) (1:1000). Secondary antibody used for western blot was IRDye 800 donkey anti-rabbit IgG (H + L) (Li-cor) (1:15000), and IRDye 680 goat anti mouse (H + L) (Li-cor).
2.6. Ex vivo cell migration assay
Epicardial cell migration was measured in ex vivo assay as described by Kim et al.,28 using Tg(wt1b:EGFP)li1 fish. Briefly, 3 dpa hearts were extracted from zebrafish retro-orbital injected for 2 days with HMC, hmmr VMO, PF-573228, SKI-1, or vehicles PBS and DMSO. Hearts were cultured in 24-well plate pre-coated with fibrin and incubated at 28°C for 3 days. Cell migration was measured using ImageJ from the edge of the heart to the edge of the cell monolayer at 1, 2, and 3 days post-extraction.
2.7. Rat maintenance and myocardial infarct induction
A detailed description of rat maintenance and MI induction is described in Supplementary material online.
2.8. Statistical analysis
Statistical significance was analysed by the Student's t-test, one-way ANOVA, and two-way ANOVA and shown as mean ± S.D. For one-way ANOVA, Tukey post hoc tests were performed. P-values were considered significant when <0.05.
3. Results
3.1. Atp5a1, desmuslin, and Hmmr are up-regulated after heart injury
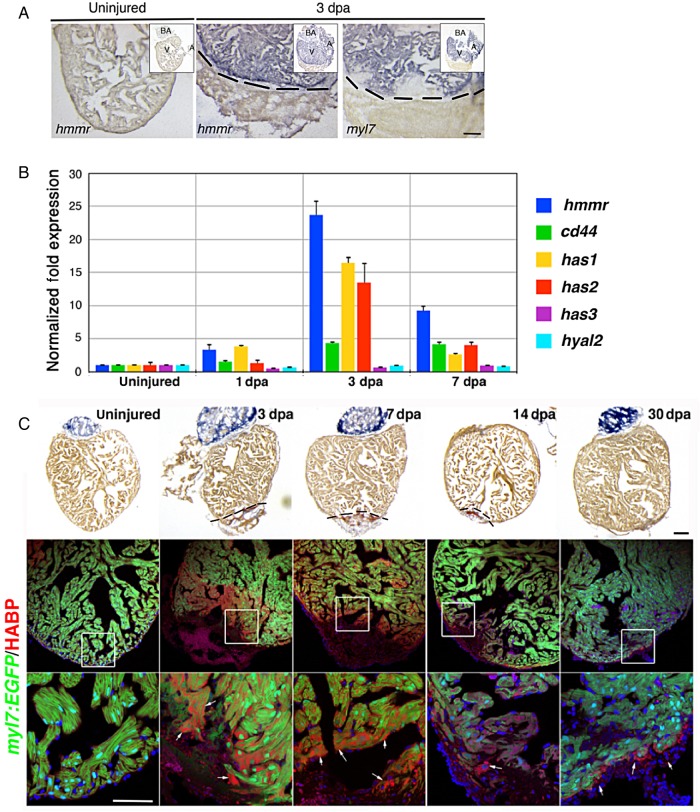
To identify the proteins that are differentially expressed during zebrafish heart regeneration, we performed a pilot DiGE experiment on 3 dpa and control uninjured hearts (see Supplementary material online, Figure S1A and B). We picked the most prominent protein spots that were within the detection range of MALDI-TOF/TOF-MS for protein identification. We identified ATP Synthase 5a1 (Atp5a1), desmuslin, and hyaluronan-mediated motility receptor (Hmmr), to be increased in heart samples at 3 dpa (see Supplementary material online, Figure S1A–C). Previous work revealed the importance of HA in cardiac development and tail regeneration.29,30 However, the role for HA receptor in regeneration is not known. Therefore, we sought to understand the function of Hmmr in zebrafish cardiac regeneration. We verified that hmmr gene transcription was also increased following injury by in situ hybridization (ISH) and Q-PCR. hmmr expression was absent in uninjured hearts but was markedly increased throughout the whole heart after injury (Figure 1A) and confirmed by Q-PCR (Figure 1B). A rapid increase in hmmr expression was observed at 3 dpa, but expression declined at 7 dpa (Figure 1B). Similarly, expression of cluster of differentiation-44 (cd44), a co-receptor for HA, was induced at 3 and 7 dpa. Moreover, the enzymes that synthesize HA and HA synthases (has1 and has2) were also up-regulated at 3 dpa (Figure 1B). In contrast, expression of hyaluronidases2 (hyal2), an enzyme responsible for HA degradation, did not change, suggesting that upon injury HA accumulates in the heart.
Figure 1.
Increased expression of hmmr, cd44, has-1 and −2, and HA after ventricular resection. (A) ISH in hearts showing that hmmr is highly expressed at 3 dpa (n = 5), compared with the uninjured hearts (n = 5). myl7 labels myocardium. A, Atrium; V, Ventricle; BA, Bulbus Arteriosus. Black dashed lines delimitate the injured area. (B) Q-PCR analyses of hmmr, cd44, hyaluronan synthase 1 (has 1), 2, and 3, and hyaluronidases 2 (hyal2) expression at 1, 3, and 7 dpa. Values were normalized to α-actin and RNAP expression. (C) AFOG staining of uninjured hearts (n = 6) and hearts at 3 (n = 13), 7 (n = 11), 14 (n = 5), and 30 dpa (n = 5). AFOG stains intact cardiac muscle in yellow-orange, fibrin in red, and collagen in blue. Confocal images of Tg(myl7:EGFP) hearts showing intact cardiac muscle in green and accumulation of HA after injury in red. HA, detected using biotinylated HABP (bHABP), was found in proximity of the clot (arrows). White boxes show the area where the higher magnification picture was taken. Sections were counterstained with DAPI (blue). Scale bars, 100 μm.
To confirm the presence of HA in the heart, we used biotinylated HA binding protein (bHABP) to detect HA accumulation following cardiac injury.31 In uninjured hearts, HA was not detectable, but at 3 and 7 dpa substantial HA was localized in cells adjacent to the wound and also within the clot (Figure 1C). By 14 and 30 dpa, the HA was still present, but it was restricted to the clot tissue (Figure 1C). Furthermore, immunostaining for Has proteins revealed that expression was restricted in the clot area and co-localized with HA, confirming that upon injury, HA accumulates within the damaged tissue (see Supplementary material online, Figure S2A).32 Thus, Has proteins and HA receptors are rapidly induced following cardiac damage, suggesting that these factors are important in zebrafish heart regeneration.
3.2. HA is required for proper heart regeneration
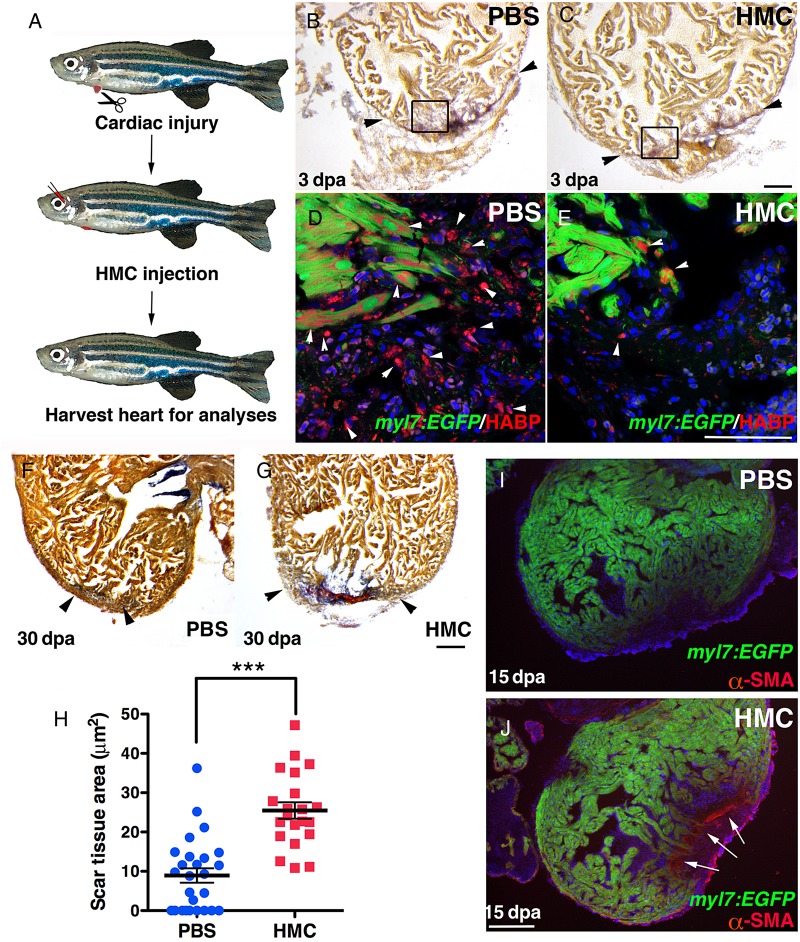
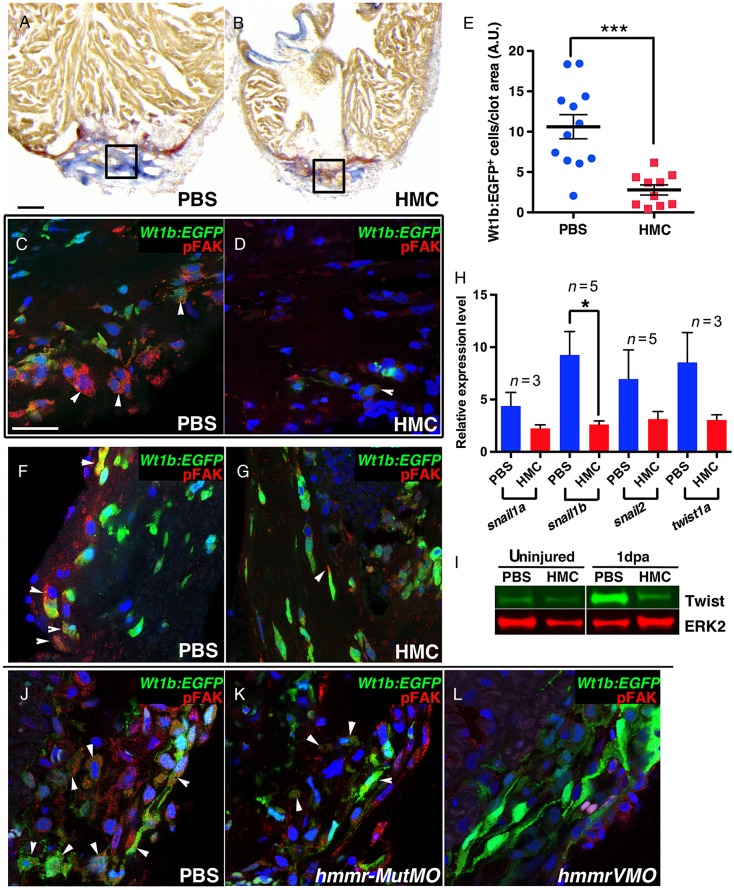
To determine the importance of HA during cardiac regeneration, we suppressed HA synthesis after ventricular resection using HMC, an inhibitor of Has enzymes by depleting its cellular substrate, UDP-glucoronic acid.33,34 HMC was delivered by retro-orbital injections following cardiac injury (Figure 2A). At 3 dpa, HA in the clot area was greatly diminished in HMC-injected fish (Figure 2B–E and see Supplementary material online, Figure S2B and C). By 30 dpa, when regeneration is complete6 (Figure 2F and H), HMC-injected zebrafish still contained significant scar tissue (Figure 2G and H). In concordance with these observations, HMC-injected zebrafish contained activated myofibroblasts as indicated by the presence of α-smooth muscle actin (α-SMA) within the clot tissue at 15 dpa (Figure 2I and J). Activated myofibroblasts were even detected at 60 dpa, suggesting that blocking HA production can completely block the regenerative process resulting in permanent scar tissue deposited in the heart (see Supplementary material online, Figure S3).
Figure 2.
Suppressing HA production blocked cardiac regeneration. (A) Experimental outline. Amputation of ventricle apex was performed at Day 0. At 1 dpa, zebrafish were retro-orbital injected daily to deliver HMC or PBS. (B–E) HMC injections decreased HA levels in the wound. AFOG staining of Tg(myl7:EGFP) hearts at 3 dpa injected with PBS (n = 7) (B) or with HMC (n = 7) (C). Black boxes outline area of higher magnification images. Black arrowheads mark scar tissue. (D) HA (red) (white arrowheads) was detected within the injury site in control PBS injected, but was markedly diminished after HMC injection (E). (F and G) Hearts at 30 dpa had minimal or no scar tissue in PBS controls (n = 26) (F), but after HMC injections (n = 21) significant scar tissue was observed (G). Black arrowheads mark scar tissue. (H) Graph of measured scar tissue at 30 dpa in PBS and HMC-injected zebrafish. Black bars indicate the mean and error bars the SEM. (J and I) αSMA staining for myofibroblasts in Tg(myl7:EGFP) hearts at 15 dpa, injected for 6 days with PBS (I; n = 4) or HMC (J; n = 5). Sections were counterstained with DAPI (blue). White arrows indicate α-SMA staining. Scale bars, 100 μm. ***P < 0.001. Student's t-test.
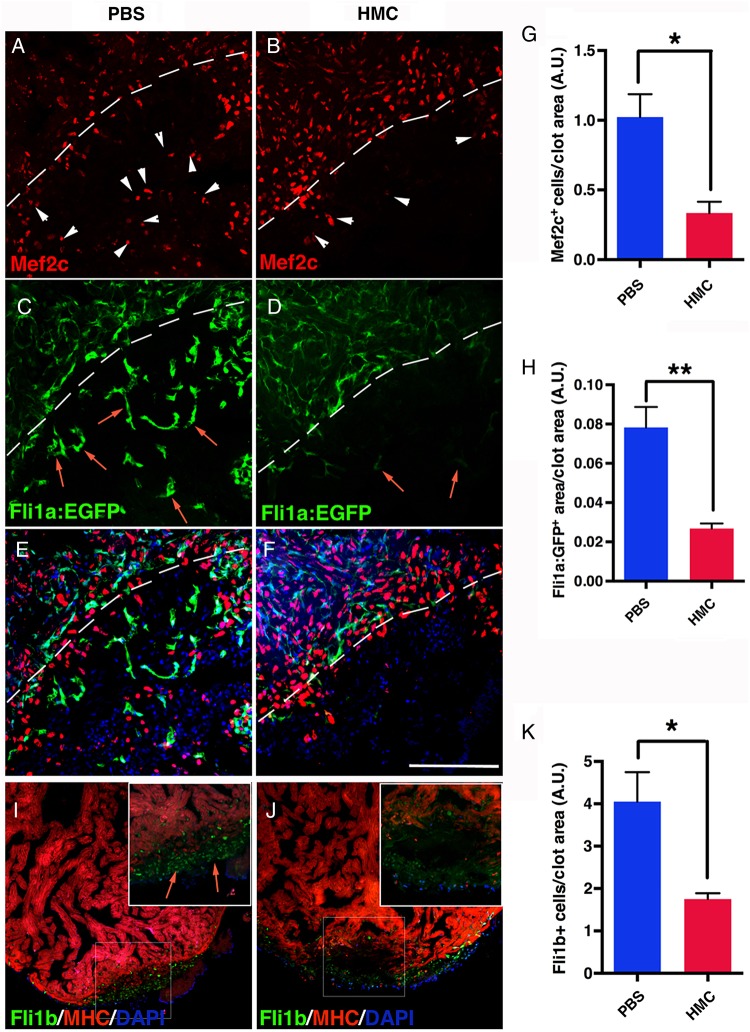
To understand mechanistically how HA may function in cardiac regeneration, we measured cardiomyocyte proliferation in HMC-injected adults. At 7 dpa, the number of cardiomyocytes (Mef2c) in S phase (PCNA) was not statistically different from control PBS-injected zebrafish (see Supplementary material online, Figure S4). However at 10 dpa, significant reduction in cardiomyocytes inside the clot was observed compared with controls, implicating a deficiency in cardiomyocyte repopulating the clot tissue (Figure 3A, B, and G) that persisted at 60 dpa (see Supplementary material online, Figure S5).10,35 The lack of cardiomyocytes in the clot tissue could reflect a migratory defect and their survival in HMC-injected hearts after the proliferative phase. Another important aspect of cardiac regeneration is the establishment of the coronary vasculature to support the regenerating cardiac tissue. Therefore, we next determined whether the formation of the coronary vasculature was also disrupted. HMC injections into a transgenic line that labels blood vessels with green fluorescent protein (Tg(fli1a:EGFP)) showed a significant reduction in coronary vasculature in the clot area (Figure 3C–F and H, and see Supplementary material online, Figure S6). Moreover, the endothelial cells that were detected in HMC-injected zebrafish were sparse and failed to organize into vessel-like structures. The lack of coronary vasculature was confirmed by immunostaining for the presence of Fli1b, another endothelial-expressed Ets-transcription factor important for vessel formation.36 In HMC-injected zebrafish, there was a pronounced decrease in Fli1b+ cells within the clot (Figure 3I, J, and K). Taken together, these results show that HA is produced following cardiac injury and plays a critical role in the formation of the coronary vasculature and in the repopulation of cardiomyocytes in the clot tissue.
Figure 3.
Has inhibition suppressed angiogenesis in regenerating hearts. (A) Tg(fli1a:EGFP) hearts at 10 dpa, injected for 6 days with PBS (n = 9) and (B) HMC (n = 5). Mef2c immunostaining revealed the presence of cardiomyocytes (white arrowheads) that have populated into the clot area in PBS controls, (A), but was clearly decreased after HMC treatment (B). White dashed lines delineate the resection plane. The formation of coronary vasculature at 10 dpa was observed through endothelial EGFP expression (red arrows) in Tg(fli1a:EGFP) hearts (C). In HMC-treated zebrafish (D), the presence of EGFP+ cells was reduced. (E and F) Merged images of A and C, B and D. DAPI is shown in blue. (G) Graph showing cardiomyocyte number in the regenerating area. (H) Quantification of angiogenesis after injection of HMC and PBS, expressed as new vessel area inside the clot. (I and J) Hearts at 10 dpa, injected for 6 days with PBS (I; n = 5) or HMC (J; n = 5) stained for Fli1b expression and MHC. (K) Quantification of Fli1b+ cells per unit clot area. Black bars in graphs indicate the mean and error bars the SEM. Scale bars, 100 μm. *P < 0.05; **P < 0.01. Student's t-test.
3.3. Hmmr is necessary for heart regeneration
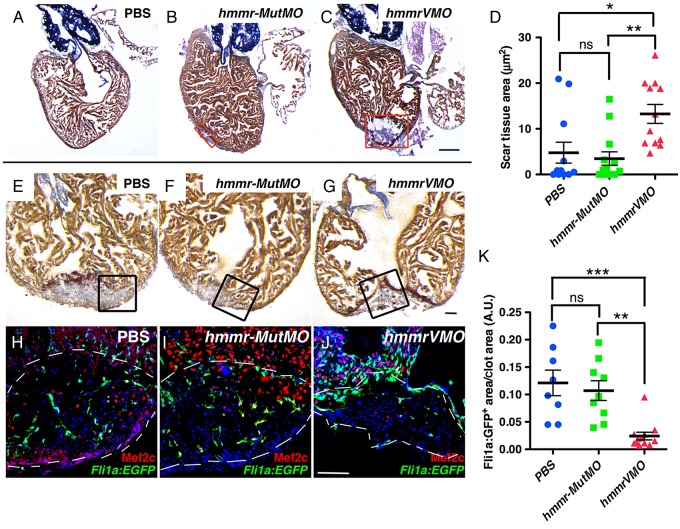
Given the importance of HA in heart regeneration, we next determined whether the Hmmr is also required in this process. hmmr was knocked down using VMO. VMOs have been shown to be effective in targeting the depletion of protein expression in several tissues in mice and in adult zebrafish.37 The hmmrVMO was designed to suppress intron splicing between exons 4 and 5 (see Supplementary material online, Figure S7A). RT–PCR experiments confirmed that hmmr transcripts in pooled injured hearts were reduced in zebrafish injected with hmmrVMO, but not in control hmmr-MutMO (5-bp mismatch VMO) (see Supplementary material online, Figure S7B). Following ventricular resection, hmmrVMO, hmmr-MutMO, or PBS were retro-orbitally injected from 1 dpa until 10 dpa, and fish were allowed to recover until time of analyses. At 30 dpa, scar tissue area was significantly larger in hmmrVMO-injected zebrafish compared with PBS and to the hmmr-MutMO controls (Figure 4A–D). As noted with suppressing HA production, knockdown of hmmr also decreased angiogenesis at 10 dpa (Figure 4E–K and see Supplementary material online, Figure S8), without affecting cardiomyocyte proliferation (see Supplementary material online, Figure S9). Thus, depletion of hmmr resulted in decreased regenerative capacity of the zebrafish heart.
Figure 4.
Cardiac regeneration is inhibited after Hmmr knockdown. (A–C) AFOG staining of hearts at 30 dpa injected for 10 days with PBS (n = 12) (A), 5 bp mismatch VMO (hmmr-MutMO) (n = 13) (B), and hmmrVMO (n = 12) (C). Red boxes indicate the scar tissue area. Zebrafish injected with hmmrVMO contained extensive scar tissue at 30 dpa. (D) Graph depicting clot area at 30 dpa. (E–J) Hearts at 10 dpa showing clot by AFOG staining (E–G), and cardiomyocytes (Mef2c) and coronary vasculature (Fli1a:EGFP+) (H–J). Injection of hmmrVMO resulted in decreased Fli1a:EGFP+ cells into the injury site (J; n = 8), compared with controls (H; n = 16 and I; n = 6). (K) Quantification of Fli1a:EGFP+ cells in the clot area at 10 dpa after Hmmr knockdown. Black boxes indicate the area where the confocal picture was taken. White dashed lines delimitate the injured area. DAPI is shown in blue. Black bars in graphs indicate the mean and error bars the SEM. Scale bars, 100 μm. *P < 0.05; **P < 0.01; ***P < 0.001 ns, not significant. One-way ANOVA.
3.4. Inhibition of Has and Hmmr blocks epicardial EMT and their migration
Previous studies have demonstrated the importance of epicardial cell activation and migration into the clot to support angiogenesis.14 We observed a marked reduction in vessels structures in the regenerating ventricle after HMC or hmmrVMO injections, suggesting a defective epicardial response after injury. The lack of coronary vasculature could have arisen from a failure of either activation of epicardial cells or their subsequent EMT and migration into the clot tissue. We determined that HA was localized within the epicardium implicating a role for HA in this tissue (see Supplementary material online, Figure S10A). Previous work has shown that HA can stabilize Integrin signalling through phosphorylation of FAK, thus facilitating proper cell migration.38 FAK is a non-receptor cytosolic tyrosine kinase that is present at the sites of contact between cells and the ECM.39 In epicardial cells that have migrated into the clot, pFAK (Y-397) was detected in a punctate pattern (see Supplementary material online, Figure S10B). To test whether HA is required for epicardial cell activation and migration, HMC was injected into Tg(wt1b:EGFP) zebrafish after ventricular resection. Although epicardial Wt1b:EGFP+ cells activation was similar between PBS and HMC-injected fish (see Supplementary material online, Figure S10C and D), we observed diminished number of EGFP+ cells inside the clot (Figure 5A–E) in HMC-injected hearts. HMC treatment also reduced pFAK levels in epicardial cells (see Supplementary material online, Figure S10E). Moreover, in PBS-injected hearts, Wt1b:EGFP+ cells appeared more round in shape rather than elongated, suggesting by morphology that epicardial cells are undergoing EMT (Compare Figure 5F–G and see Supplementary material online, Figure S10F). To confirm that epicardial EMT was suppressed after HMC injections, we performed Q-PCR to measure expression of EMT genes.16 At 3 dpa, HMC injections reduced the expression levels of snail1a, snail1b, snail2, and twist1a (Figure 5H and I), supporting the notion that HA is required for epicardial EMT. These findings were confirmed with knockdown of Hmmr as the presence of epicardial pFAK was reduced (Figure 5J–L). Our studies suggest a role where HA and its receptor Hmmr are required for proper epicardial cell EMT and migration into the wound.
Figure 5.
HA is required for epicardial cell EMT. (A–D) Tg(wt1b:EGFP) zebrafish hearts at 5 dpa injected with PBS (n = 8) (A–C) or HMC (n = 6) (B–D) had similar clot area, as shown by AFOG staining (A and B), but the number of Wt1b:EGFP+ cells in the clot was greatly reduced after HMC injections (D), compared with PBS (C). Black boxes demarcate area of higher magnification. (E) Graph showing the number of Wt1b:EGFP+ cells in the clot at 7 dpa. (F and G) Confocal images of activated wt1b+ cells at the epicardial region at 3 dpa after injection of control PBS (F; n = 4) or HMC (G; n = 5). In controls (F), some Wt1b:EGFP+ cells were pFAK positive (white arrowhead) and appeared round in shape. In contrast, Wt1b:EGFP+ cells after HMC injections (G) remained elongated. (H) Q-PCR analysis of snail1a, −1b, −2, and twist1a expression in ventricular tissue at 3 dpa after injections of HMC. HMC reduced the expression of EMT genes. n indicates the number of independent replicates performed for each gene. (I) Western blot analysis of Twist1 expression in uninjured hearts and hearts at 1 dpa. Increased Twist1 expression was detected in resected hearts, but was suppressed after HMC injections. Total Erk2 was used as loading control. (J–L) Tg(wt1b:EGFP) heart at 3 dpa immunostained for pFAK after injection of PBS (J; n = 4), control mutant MO (K; n = 4), or hmmrVMO (L; n = 3). hmmrVMO decreased the number of round Wt1b:EGFP+ cells and the number of cells expressing pFAK (L). Black bars in graphs indicate the mean and error bars the SEM. Scale bars, 100 μm (A and B), 25 μm (C, D, F, G, J, K, and L). *P < 0.05; ***P < 0.001. Student's t-test.
3.5. HA signals through FAK and Src for epicardial EMT
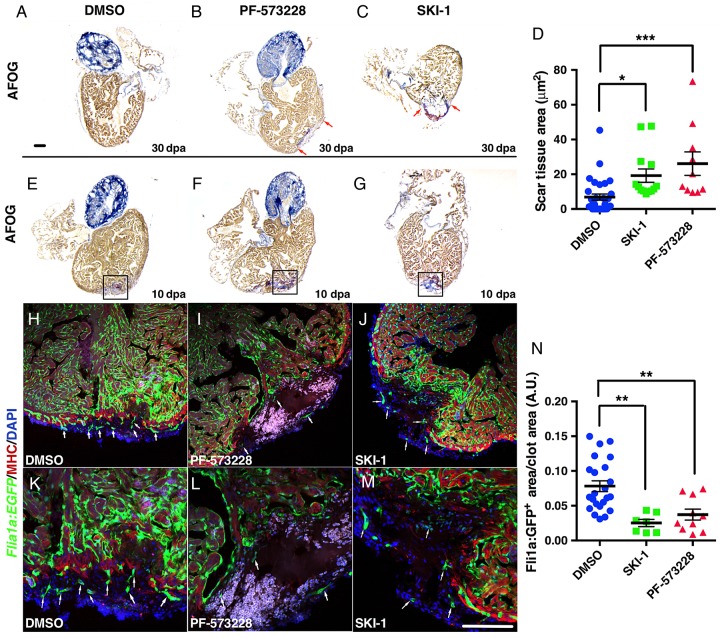
We next addressed the potential role for FAK and Src in heart regeneration as downstream effectors of HA/Hmmr. FAK is autoactivated through tyrosine autophosphorylation at the Y397 site by Integrin/Hmmr interaction and is known to be a critical regulator of cardiac growth and remodelling.40 This pY397 creates a high-affinity binding site for the SH2 domain of Src family kinases that leads to further phosphorylation of FAK by Src.41 The activation of FAK/Src complex then regulates downstream signalling pathways that control cell proliferation, spreading, motility, and EMT.42 In this study, we used chemical inhibition of FAK with PF-57322843 and the Src kinase inhibitor (SKI-1) to suppress Src,44 to dissect the role of these proteins in heart regeneration. Inhibiting either FAK or Src suppressed heart regeneration (Figure 6A–D and see Supplementary material online, Figure S11A and B). As observed with blocking HA production, the formation of new vessels was also significantly reduced after injection of PF-573228 or SKI-1 (Figure 6E–N and see Supplementary material online, Figure S11C–F), and the migration of epicardial cells inside the clot was reduced after SKI-1 injection (see Supplementary material online, Figure S11G and H), suggesting that Hmmr activation of pFAK and Src is important for epicardial cell function.
Figure 6.
FAK and Src inhibition suppressed angiogenesis in regenerating hearts. (A–C) At 30 dpa, hearts injected for 10 days with PF-573228, a FAK inhibitor (B) (n = 10), or with SKI-1 (C) (n = 13), a Src inhibitor, failed to properly regenerate the heart compared with DMSO controls (A) (n = 31). Red arrows delineate scar. (D) Graph showing clot area at 30 dpa after injections of PF-573228 or SKI-1. (E–M) PF-573228 and SKI-1 injections inhibited angiogenesis. (E–G) AFOG staining of hearts at 10 dpa, injected with DMSO (E) (n = 15), PF-573228 (F) (n = 10), or SKI-1 (G) (n = 10). (H–M) Tg(fli1a:EGFP) hearts injected with DMSO (H and K), PF-573228 (I and L), or with SKI-1 (J and M), immunostained for MHC to detect resection plane. White arrows indicate new vessels formed inside the clot. (N) Quantification fli1a+:EGFP area in the clot at 10 dpa. FAK and Src inhibition significantly reduced new vessel formation. Black boxes delimitate the areas of confocal imaging. Black bars in graphs indicate the mean and error bars the SEM. Scale bars, 100 μm. *P < 0.05; **P < 0.01; ***P < 0.001 one-way ANOVA.
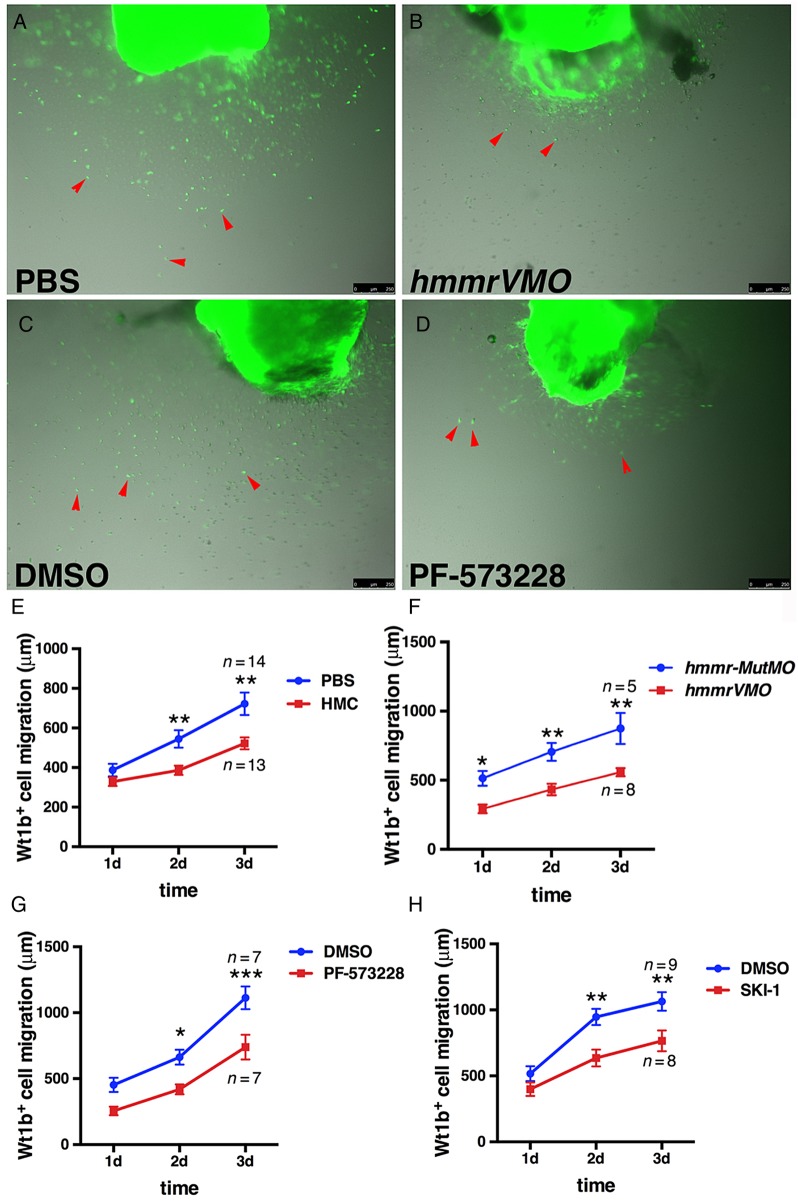
In support of these findings, we employed an ex vivo assay to monitor epicardial cell migration from intact hearts grown in culture.28 Decreased epicardial cell spreading from the heart on fibrin-coated wells after hmmrVMO injections was observed (Figure 7A, B, and F). Moreover, suppressing FAK (Figure 7C, D, and G), HA production (Figure 7E), or Src activity (Figure 7H) also reduced epicardial cell spreading. Taken together, our results implicate a function for HA and Hmmr in directing activated epicardial migration through FAK and Src kinases.
Figure 7.
HA and its receptor Hmmr are necessary for epicardial cell migration in ex vivo culture. (A–D) Ex vivo epicardial migration assay showing decreased Wt1b:EGFP+ migratory behaviour after hmmrVMO (B) (n = 8) and PF-573228 (D) (n = 7) treatments compared with controls (A) (n = 14) and (C) (n = 7). Images were captured at 3 days post-extraction of the heart. The migration of Wt1b:EGFP+ epicardial cells (red arrows) was measured from Day 1 to Day 3. In control PBS, DMSO, or hmmr-MutMO treatments, epicardial cells migrated on the fibrin-coated plates. In contrast, treatment with hmmrVMO, PF-573228 or SKI-1 significantly suppressed epicardial cell migration. (E–H) Graphs showing migration of Wt1b:EGFP+ cells under various treatments, including suppressing HA production in the hearts with HMC (E) (n = 13), hmmrVMO (F) (n = 8), PF-573228 (G) (n = 7), and with SKI-1, the Src kinase inhibitor (H) (n = 8). Black bars in graphs indicate the mean and error bars the SEM. Scale bars, 250 μm. *P < 0.05; **P < 0.01; ***P < 0.001. Two-way ANOVA.
3.6. Accumulation of HA and HMMR is conserved in a mammalian model of MI
In contrast to zebrafish, after MI, adult mammals fail to regenerate injured myocardial tissue. One key difference between these animals is that in mammals cardiomyocytes are post-mitotic, often existing as binuclear or polyploid cells.45 However, there are some similarities with respect to how the epicardium responds to injury.46 HA has been reported to accumulate after injury in many tissues and in different species,30,47 but the presence and function of HMMR remain understudied in injured mammalian hearts. In uninjured rat hearts, neither HMMR, nor HA was detected (see Supplementary material online, Figure S12A, D and G). However, after MI, HMMR and HA were detected in the scar tissue at 7 days post-MI (dpMI) (see Supplementary material online, Figure S12B, E and H) and at 8 weeks post-MI (wpMI), but not in the myocardium (see Supplementary material online, Figure S12C, F and I). These data support the notion that there is an evolutionary conserved response to cardiac damage with the production of HA and induction of its receptor in scar tissue.
4. Discussion
Using a proteomic approach, we identified increased expression of Atp5a1, desmuslin, and Hmmr in the zebrafish regenerating heart at 3 dpa. Atp5a1 is a critical enzyme in the energetic pathways of cells, and its induction can be interpreted as increasing of energy demand of the damaged heart. This is consistent with results in rat ischaemic myocardium where ATP synthase expression was increased.48 Desmuslin is an intermediate filament, important for the structure of the cell. During embryogenesis, mutation of Desmuslin correlates with the development of cardiomyopathies.49 Desmuslin could play a role in the activation and replacement of contractile apparatus, maintaining the cell integrity and restoring the damaged cells in the injured area.
Hmmr is a cell surface receptor for HA, and its main function is to promote cell motility in wound healing.50 HMMR is particularly interesting because it is detected on the plasma membrane as well as in the cytoplasm and in the nucleus with distinct extracellular and intracellular functions.51 On the cell surface, HMMR promotes PDGF receptor aggregation to generate sufficient signalling for tissue remodelling.52 In Xenopus laevis tail regeneration studies, HA, has2, and cd44 are important for mesenchymal cell proliferation.30 While studies in heart development point to an essential role for Has2 in endocardial cushion, cardiac valve, and epicardium formation,53–55 as Has2 knockdown mice die during mid-gestion (E9.5–E10) due to severe cardiac and vascular deformations.53 In zebrafish, has2 is strongly expressed in cardiac progenitor cells and is necessary for cardiomyocyte migration and cardiac cone rotation.29,56,57 However, the exact role for HA and Hmmr in cardiac regeneration has not been explored. Consistent with studies in newts,58 we report that HA accumulates in adult zebrafish and rat hearts following injury. In this study, we focused on the potential role of HA and Hmmr in regulating cellular migration and proliferation. We observed that decreased production of HA reduced epicardial cell EMT and their migration into the injured area, which is an important step to support coronary vasculature formation. These observations are consistent with studies showing a role for HA to induce EMT through either TGF-β1 or EGF in lung and breast cancer cells.59 It is also noteworthy that HA can modulate the crosstalk between TGF-β1, PDGF-BB, and CD44 signalling, suggesting that HA is a key player in regulating signalling through these pathways.60 In zebrafish heart regeneration, PDGF-BB signalling is required for cardiomyocyte proliferation.11 More recent studies highlight the importance of pdgfrβ in epicardial proliferation and EMT.16 Our studies could point to a potential role for HA and Hmmr in regulating PDGF signalling in epicardial cell EMT. This is further supported with observations that chemical inhibition of FAK activity also resulted in diminished epicardial cell migration. PDGF has been shown to directly activate FAK phosphorylation in lipids rafts.61 Given that PDGF induced specific FAK tyrosine phosphorylation at position 397 is known to activate Src,38,61 we also observed a similar lack of epicardial cell migration in zebrafish treated with a Src Kinase Inhibitor. Taken together, these studies suggest that HA and Hmmr play a critical role in epicardial cell EMT after injury through FAK and Src signalling. Our data imply that HA is not only a component of the ECM, but could function as an instructive molecule with PDGF to promote epicardial EMT. Future studies using tissue and cell specific gene knockout of components in this pathway will determine their exact function in epicardial cell EMT and zebrafish heart regeneration.
The relevance of these findings to mammals is supported by the observation that expression of HMMR and HA was induced in rat hearts following ischaemic injury. Other studies have shown that application of HA-based hydrogels into rat hearts after MI can induce neovascularization, reduce infarct area, and more important, improve cardiac function.62,63 These findings open the possibility of applying HA in mammals to improve cardiac repair after ischaemic injury. Since HA can be present in different sizes and given that the functions of HA can vary according to its molecular weight (MW),64 it would be of interest to determine whether low or high MW-HA is deposited in the zebrafish ventricle after injury. In conclusion, our study highlights the importance of HA and hmmr in epicardial EMT in the regenerating zebrafish heart.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Conflict of interest: none declared.
Funding
This work was supported by funding from the American Heart Association (14GRNT20480183), the National Institute of Health (NHLBI/NIH R01HL088016), and the American Recovery and Reinvestment Act (ARRA) supplemental funding.
References
- 1.Schaper J. Ultrastructural changes of the myocardium in regional ischaemia and infarction. Eur Heart J 1986;7(Suppl. B):3–9. [DOI] [PubMed] [Google Scholar]
- 2.Andersen DC, Ganesalingam S, Jensen CH, Sheikh SP. Do neonatal mouse hearts regenerate following heart apex resection? Stem Cell Rep 2014;2:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science 2011;331:1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, Mammen PP, Rothermel BA, Olson EN, Sadek HA. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci USA 2013;110:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naqvi N, Li M, Calvert JW, Tejada T, Lambert JP, Wu J, Kesteven SH, Holman SR, Matsuda T, Lovelock JD, Howard WW, Iismaa SE, Chan AY, Crawford BH, Wagner MB, Martin DI, Lefer DJ, Graham RM, Husain A. A proliferative burst during preadolescence establishes the final cardiomyocyte number. Cell 2014;157:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science 2002;298:2188–2190. [DOI] [PubMed] [Google Scholar]
- 7.Itou J, Kawakami H, Burgoyne T, Kawakami Y. Life-long preservation of the regenerative capacity in the fin and heart in zebrafish. Biol Open 2012;1:739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010;464:606–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, Poss KD. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 2011;138:2895–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 2010;464:601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLoS Biol 2006;4:e260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Rosa JM, Peralta M, Mercader N. Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev Biol 2012;370:173–186. [DOI] [PubMed] [Google Scholar]
- 13.Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, Poss KD. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell 2011;20:397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 2006;127:607–619. [DOI] [PubMed] [Google Scholar]
- 15.Chablais F, Jazwinska A. The regenerative capacity of the zebrafish heart is dependent on TGFbeta signaling. Development 2012;139:1921–1930. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Wu Q, Zhang Y, Wiens KM, Huang Y, Rubin N, Shimada H, Handin RI, Chao MY, Tuan TL, Starnes VA, Lien CL. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc Natl Acad Sci USA 2010;107:17206–17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Harrison MR, Osorio A, Kim J, Baugh A, Duan C, Sucov HM, Lien CL. Igf signaling is required for cardiomyocyte proliferation during zebrafish heart development and regeneration. PLoS One 2013;8:e67266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang Y, Gupta V, Karra R, Holdway JE, Kikuchi K, Poss KD. Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc Natl Acad Sci USA 2013;110:13416–13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gemberling M, Karra R, Dickson AL, Poss KD. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife 2015;4; doi:10.7554/eLife.05871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Cao J, Dickson AL, Poss KD. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 2015;522:226–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao L, Borikova AL, Ben-Yair R, Guner-Ataman B, MacRae CA, Lee RT, Burns CG, Burns CE. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc Natl Acad Sci USA 2014;111:1403–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slevin M, Krupinski J, Gaffney J, Matou S, West D, Delisser H, Savani RC, Kumar S. Hyaluronan-mediated angiogenesis in vascular disease: uncovering RHAMM and CD44 receptor signaling pathways. Matrix Biol 2007;26:58–68. [DOI] [PubMed] [Google Scholar]
- 23.Philipson LH, Schwartz NB. Subcellular localization of hyaluronate synthetase in oligodendroglioma cells. J Biol Chem 1984;259:5017–5023. [PubMed] [Google Scholar]
- 24.Burns CG, Milan DJ, Grande EJ, Rottbauer W, MacRae CA, Fishman MC. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat Chem Biol 2005;1:263–264. [DOI] [PubMed] [Google Scholar]
- 25.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 2002;248:307–318. [DOI] [PubMed] [Google Scholar]
- 26.Perner B, Englert C, Bollig F. The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev Biol 2007;309:87–96. [DOI] [PubMed] [Google Scholar]
- 27.Pugach EK, Li P, White R, Zon L. Retro-orbital injection in adult zebrafish. J Vis Exp 2009; doi:10.3791/1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Rubin N, Huang Y, Tuan TL, Lien CL. In vitro culture of epicardial cells from adult zebrafish heart on a fibrin matrix. Nat Protoc 2012;7:247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klewer SE, Yatskievych T, Pogreba K, Stevens MV, Antin PB, Camenisch TD. Has2 expression in heart forming regions is independent of BMP signaling. Gene Expr Patterns 2006;6:462–470. [DOI] [PubMed] [Google Scholar]
- 30.Contreras EG, Gaete M, Sanchez N, Carrasco H, Larrain J. Early requirement of Hyaluronan for tail regeneration in Xenopus tadpoles. Development 2009;136:2987–2996. [DOI] [PubMed] [Google Scholar]
- 31.Toole BP, Yu Q, Underhill CB. Hyaluronan and hyaluronan-binding proteins. Probes for specific detection. Methods Mol Biol 2001;171:479–485. [DOI] [PubMed] [Google Scholar]
- 32.Rosa F, Sargent TD, Rebbert ML, Michaels GS, Jamrich M, Grunz H, Jonas E, Winkles JA, Dawid IB. Accumulation and decay of DG42 gene products follow a gradient pattern during Xenopus embryogenesis. Dev Biol 1988;129:114–123. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura T, Funahashi M, Takagaki K, Munakata H, Tanaka K, Saito Y, Endo M. Effect of 4-methylumbelliferone on cell-free synthesis of hyaluronic acid. Biochem Mol Biol Int 1997;43:263–268. [DOI] [PubMed] [Google Scholar]
- 34.Kakizaki I, Kojima K, Takagaki K, Endo M, Kannagi R, Ito M, Maruo Y, Sato H, Yasuda T, Mita S, Kimata K, Itano N. A novel mechanism for the inhibition of hyaluronan biosynthesis by 4-methylumbelliferone. J Biol Chem 2004;279:33281–33289. [DOI] [PubMed] [Google Scholar]
- 35.Itou J, Oishi I, Kawakami H, Glass TJ, Richter J, Johnson A, Lund TC, Kawakami Y. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development 2012;139:4133–4142. [DOI] [PubMed] [Google Scholar]
- 36.Moore JC, Sheppard-Tindell S, Shestopalov IA, Yamazoe S, Chen JK, Lawson ND. Post-transcriptional mechanisms contribute to Etv2 repression during vascular development. Dev Biol 2013;384:128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thummel R, Bai S, Sarras MP Jr, Song P, McDermott J, Brewer J, Perry M, Zhang X, Hyde DR, Godwin AR. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn 2006;235:336–346. [DOI] [PubMed] [Google Scholar]
- 38.Sieg DJ, Hauck CR, Ilic D, Klingbeil CK, Schaefer E, Damsky CH, Schlaepfer DD. FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol 2000;2:249–256. [DOI] [PubMed] [Google Scholar]
- 39.Heffler M, Golubovskaya VM, Conroy J, Liu S, Wang D, Cance WG, Dunn KB. FAK and HAS inhibition synergistically decrease colon cancer cell viability and affect expression of critical genes. Anticancer Agents Med Chem 2013;13:584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hakim ZS, DiMichele LA, Rojas M, Meredith D, Mack CP, Taylor JM. FAK regulates cardiomyocyte survival following ischemia/reperfusion. J Mol Cell Cardiol 2009;46:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolos V, Gasent JM, Lopez-Tarruella S, Grande E. The dual kinase complex FAK-Src as a promising therapeutic target in cancer. Onco Targets Ther 2010;3:83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey KM, Liu J. Caveolin-1 up-regulation during epithelial to mesenchymal transition is mediated by focal adhesion kinase. J Biol Chem 2008;283:13714–13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slack-Davis JK, Martin KH, Tilghman RW, Iwanicki M, Ung EJ, Autry C, Luzzio MJ, Cooper B, Kath JC, Roberts WG, Parsons JT. Cellular characterization of a novel focal adhesion kinase inhibitor. J Biol Chem 2007;282:14845–14852. [DOI] [PubMed] [Google Scholar]
- 44.Calalb MB, Polte TR, Hanks SK. Tyrosine phosphorylation of focal adhesion kinase at sites in the catalytic domain regulates kinase activity: a role for Src family kinases. Mol Cell Biol 1995;15:954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brodsky WY, Arefyeva AM, Uryvaeva IV. Mitotic polyploidization of mouse heart myocytes during the first postnatal week. Cell Tissue Res 1980;210:133–144. [DOI] [PubMed] [Google Scholar]
- 46.Smart N, Risebro CA, Melville AA, Moses K, Schwartz RJ, Chien KR, Riley PR. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature 2007;445:177–182. [DOI] [PubMed] [Google Scholar]
- 47.Calve S, Isaac J, Gumucio JP, Mendias CL. Hyaluronic acid, HAS1, and HAS2 are significantly upregulated during muscle hypertrophy. Am J Physiol Cell Physiol 2012;303:C577–C588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grover GJ, Atwal KS, Sleph PG, Wang FL, Monshizadegan H, Monticello T, Green DW. Excessive ATP hydrolysis in ischemic myocardium by mitochondrial F1F0-ATPase: effect of selective pharmacological inhibition of mitochondrial ATPase hydrolase activity. Am J Physiol Heart Circ Physiol 2004;287:H1747–H1755. [DOI] [PubMed] [Google Scholar]
- 49.Olive M, Goldfarb L, Moreno D, Laforet E, Dagvadorj A, Sambuughin N, Martinez-Matos JA, Martinez F, Alio J, Farrero E, Vicart P, Ferrer I. Desmin-related myopathy: clinical, electrophysiological, radiological, neuropathological and genetic studies. J Neurol Sci 2004;219:125–137. [DOI] [PubMed] [Google Scholar]
- 50.Savani RC, Wang C, Yang B, Zhang S, Kinsella MG, Wight TN, Stern R, Nance DM, Turley EA. Migration of bovine aortic smooth muscle cells after wounding injury. The role of hyaluronan and RHAMM. J Clin Invest 1995;95:1158–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tolg C, Hamilton SR, Nakrieko KA, Kooshesh F, Walton P, McCarthy JB, Bissell MJ, Turley EA. Rhamm-/- fibroblasts are defective in CD44-mediated ERK1,2 motogenic signaling, leading to defective skin wound repair. J Cell Biol 2006;175:1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheung WF, Cruz TF, Turley EA. Receptor for hyaluronan-mediated motility (RHAMM), a hyaladherin that regulates cell responses to growth factors. Biochem Soc Trans 1999;27:135–142. [DOI] [PubMed] [Google Scholar]
- 53.Camenisch TD, Spicer AP, Brehm-Gibson T, Biesterfeldt J, Augustine ML, Calabro A Jr, Kubalak S, Klewer SE, McDonald JA. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest 2000;106:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lagendijk AK, Szabo A, Merks RM, Bakkers J. Hyaluronan: a critical regulator of endothelial-to-mesenchymal transition during cardiac valve formation. Trends Cardiovasc Med 2013;23:135–142. [DOI] [PubMed] [Google Scholar]
- 55.Craig EA, Austin AF, Vaillancourt RR, Barnett JV, Camenisch TD. TGFbeta2-mediated production of hyaluronan is important for the induction of epicardial cell differentiation and invasion. Exp Cell Res 2010;316:3397–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakkers J, Kramer C, Pothof J, Quaedvlieg NE, Spaink HP, Hammerschmidt M. Has2 is required upstream of Rac1 to govern dorsal migration of lateral cells during zebrafish gastrulation. Development 2004;131:525–537. [DOI] [PubMed] [Google Scholar]
- 57.Smith KA, Chocron S, von der Hardt S, de Pater E, Soufan A, Bussmann J, Schulte-Merker S, Hammerschmidt M, Bakkers J. Rotation and asymmetric development of the zebrafish heart requires directed migration of cardiac progenitor cells. Dev Cell 2008;14:287–297. [DOI] [PubMed] [Google Scholar]
- 58.Mercer SE, Odelberg SJ, Simon HG. A dynamic spatiotemporal extracellular matrix facilitates epicardial-mediated vertebrate heart regeneration. Dev Biol 2013;382:457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L, Qi L, Liang Z, Song W, Liu Y, Wang Y, Sun B, Zhang B, Cao W. Transforming growth factor-beta1 induces EMT by the transactivation of epidermal growth factor signaling through HA/CD44 in lung and breast cancer cells. Int J Mol Med 2015;36:113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Porsch H, Mehic M, Olofsson B, Heldin P, Heldin CH. Platelet-derived growth factor beta-receptor, transforming growth factor beta type I receptor, and CD44 protein modulate each other's signaling and stability. J Biol Chem 2014;289:19747–19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seong J, Ouyang M, Kim T, Sun J, Wen PC, Lu S, Zhuo Y, Llewellyn NM, Schlaepfer DD, Guan JL, Chien S, Wang Y. Detection of focal adhesion kinase activation at membrane microdomains by fluorescence resonance energy transfer. Nat Commun 2011;2:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abdalla S, Makhoul G, Duong M, Chiu RC, Cecere R. Hyaluronic acid-based hydrogel induces neovascularization and improves cardiac function in a rat model of myocardial infarction. Interact Cardiovasc Thorac Surg 2013;17:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonafe F, Govoni M, Giordano E, Caldarera CM, Guarnieri C, Muscari C. Hyaluronan and cardiac regeneration. J Biomed Sci 2014;21:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tolg C, Telmer P, Turley E. Specific sizes of hyaluronan oligosaccharides stimulate fibroblast migration and excisional wound repair. PLoS One 2014;9:e88479. [DOI] [PMC free article] [PubMed] [Google Scholar]