Abstract
Although CRISPR-Cas9 nucleases are widely used for genome editing1, 2, the range of sequences that Cas9 can recognize is constrained by the need for a specific protospacer adjacent motif (PAM)3–6. As a result, it can often be difficult to target double-stranded breaks (DSBs) with the precision that is necessary for various genome editing applications. The ability to engineer Cas9 derivatives with purposefully altered PAM specificities would address this limitation. Here we show that the commonly used Streptococcus pyogenes Cas9 (SpCas9) can be modified to recognize alternative PAM sequences using structural information, bacterial selection-based directed evolution, and combinatorial design. These altered PAM specificity variants enable robust editing of endogenous gene sites in zebrafish and human cells not currently targetable by wild-type SpCas9, and their genome-wide specificities are comparable to wild-type SpCas9 as judged by GUIDE-Seq analysis7. In addition, we identified and characterized another SpCas9 variant that exhibits improved specificity in human cells, possessing better discrimination against off-target sites with non-canonical NAG and NGA PAMs and/or mismatched spacers. We also found that two smaller-size Cas9 orthologues, Streptococcus thermophilus Cas9 (St1Cas9) and Staphylococcus aureus Cas9 (SaCas9), function efficiently in the bacterial selection systems and in human cells, suggesting that our engineering strategies could be extended to Cas9s from other species. Our findings provide broadly useful SpCas9 variants and, more importantly, establish the feasibility of engineering a wide range of Cas9s with altered and improved PAM specificities.
CRISPR-Cas9 nucleases enable efficient genome editing in a wide variety of organisms and cell types1, 2. Target site recognition by Cas9 is programmed by a chimeric single guide RNA (sgRNA) that encodes a sequence complementary to a target protospacer5, but also requires recognition of a short neighboring PAM3–6. SpCas9, the most robust and widely used Cas9 to date, primarily recognizes NGG PAMs and is consequently restricted to sites that contain this motif5, 8. It can therefore be challenging to implement genome editing applications that require precision, such as: homology-directed repair (HDR), which is most efficient when DSBs are placed within 10–20 bps of a desired alteration9–11; the introduction of variable-length insertion or deletion (indel) mutations into small size genetic elements such as microRNAs, splice sites, short open reading frames, or transcription factor binding sites by non-homologous end-joining (NHEJ); and allele-specific editing, where PAM recognition might be exploited to differentiate alleles.
One potential solution to address targeting range limitations would be to engineer Cas9 variants with novel PAM specificities. A previous attempt to alter SpCas9 PAM specificity mutated R1333 and R1335 residues that contact the guanine nucleotides at the second and third PAM positions; however, the R1333Q/R1335Q variant failed to cleave a site harboring the expected NAA PAM in vitro12. Using a human cell-based U2OS EGFP reporter gene disruption assay in which nuclease-induced indels lead to loss of fluorescence13, 14, we confirmed that an R1333Q/R1335Q SpCas9 variant failed to efficiently cleave target sites with NAA PAMs (Fig. 1a). Additionally, we found that single R1333Q and R1335Q variants each failed to efficiently cleave target sites containing the expected NAG and NGA PAMs, respectively (Fig. 1a), suggesting that re-engineering PAM specificity might require additional mutations.
Figure 1. Evolution and characterization of SpCas9 variants with altered PAM specificities.
a, Activity of wild-type and mutant SpCas9s assessed via U2OS human cell-based EGFP disruption. Frequencies were quantified by flow cytometry; error bars represent s.e.m., n = 3; mean level of background EGFP loss represented by the dashed red line for this and subsequent panels (c, g, h, and j). b, Schematic of the positive selection assay (see also Extended Data Fig. 1). c, Combinatorial assembly and human cell testing of mutations obtained from the positive selection for SpCas9 variants that can cleave a target site containing an NGA PAM, using the EGFP disruption assay. d, Schematic of the negative selection assay, adapted to profile Cas9 PAM specificity by generating a library of plasmids that contain a randomized sequence adjacent to the 3’ end of the protospacer (see also Extended Data Fig. 3b). e, Scatterplot of the post-selection PAM depletion values (PPDVs) of wild-type SpCas9 with two randomized PAM libraries (each with a different protospacer). PAMs are plotted by their 2nd/3rd/4th positions. The red dashed line indicates statistically significant depletion (obtained from a dCas9 control experiment, see Extended Data Fig. 3c), and the gray dashed line represents five-fold depletion (PPDV of 0.2). f, PPDV scatterplots for the VQR and EQR variants. g, EGFP disruption frequencies for wild-type, VQR, and EQR SpCas9 on sites with NGAN and NGNG PAMs. h, Combinatorial assembly and human cell testing of mutations obtained from the positive selection for SpCas9 variants that can cleave a target site containing an NGC PAM, using the EGFP disruption assay. i, PPDV scatterplot for the VRER variant. j, EGFP disruption frequencies for wild-type and VRER SpCas9 on sites with NGCN and NGNG PAMs.
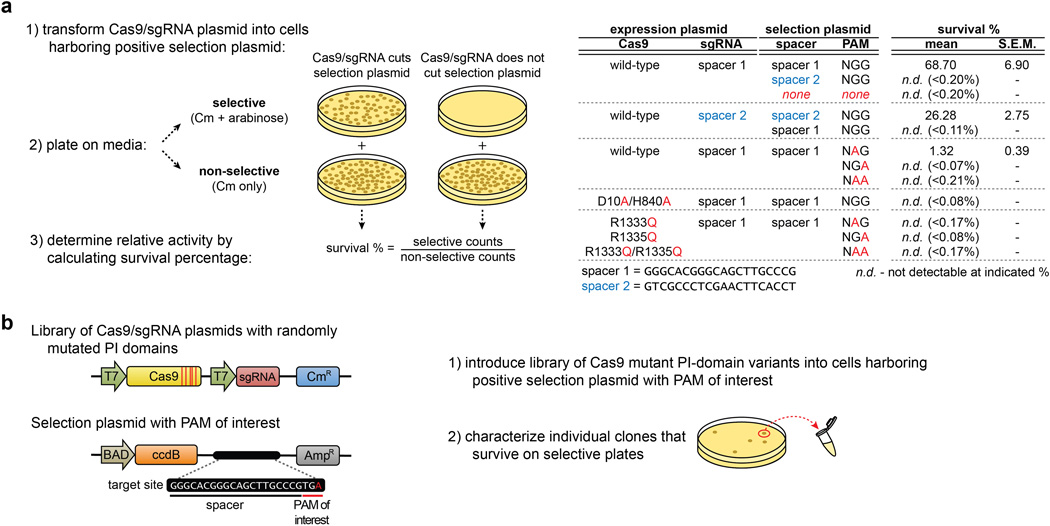
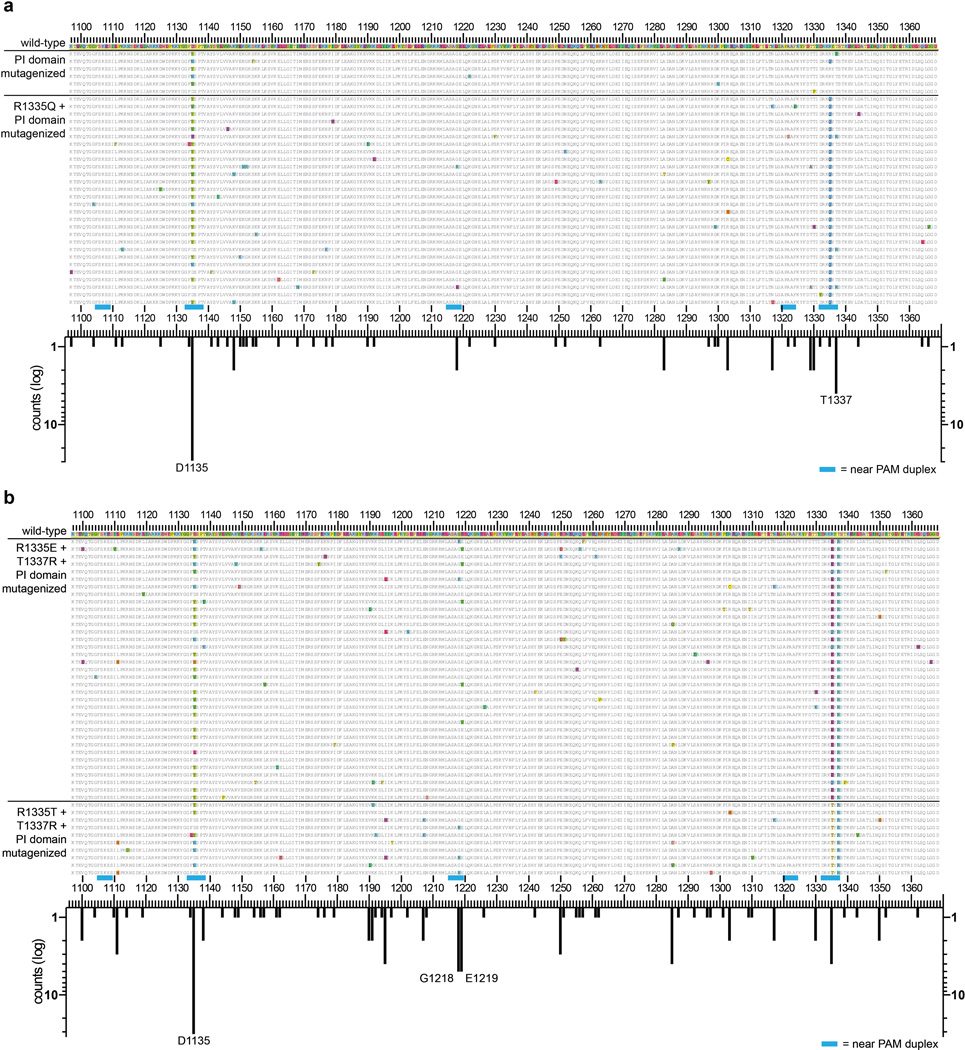
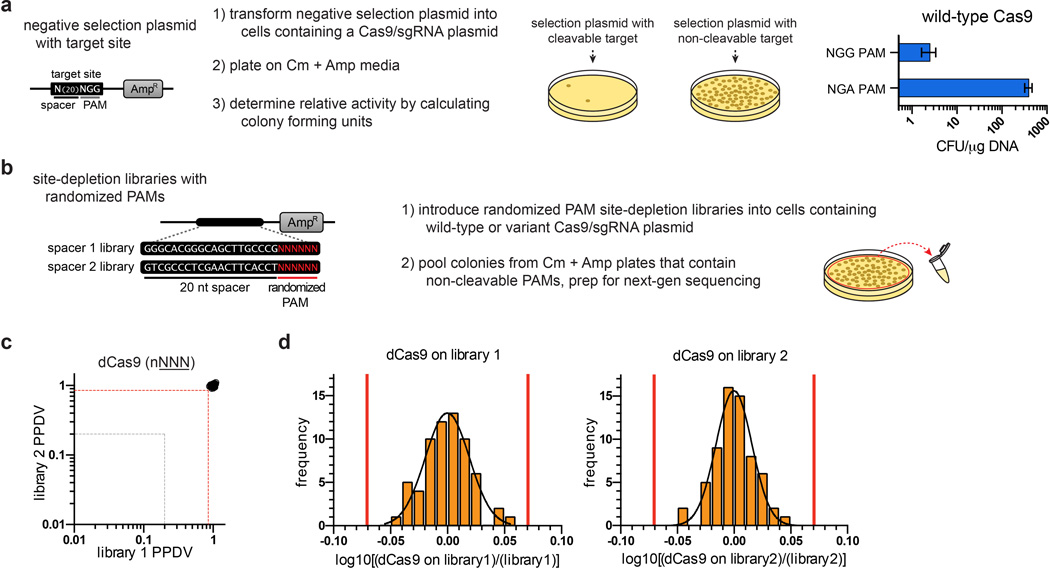
To identify such mutations, we adapted a bacterial selection system (hereafter referred to as the positive selection) previously used to study properties of homing endonucleases15, 16. In our adaptation of this system, survival is enabled by Cas9-mediated cleavage of a selection plasmid encoding an inducible toxic gene (Fig. 1b, Extended Data Fig. 1a). We mutagenized the PAM-interacting (PI) domains of wild-type and R1335Q SpCas9 and performed selections against an NGA PAM target site (Extended Data Fig. 1b, Online Methods). Sequences of surviving clones from both libraries revealed the most frequent substitutions were D1135V/Y/N/E, R1335Q, and T1337R (Extended Data Fig. 2a). After testing all combinations of these mutations using the human cell-based EGFP disruption assay, two variants were chosen for further characterization because they possessed the greatest discrimination between NGA and NGG PAMs: D1135V/R1335Q/T1337R and D1135E/R1335Q/T1337R (hereafter referred to as the VQR and EQR variants, respectively) (Fig. 1c).
To define the global PAM specificity profiles of these SpCas9 variants, we used a bacterial-based negative selection system (Fig. 1d, Extended Data Fig. 3a) similar to other methods previously used to identify PAM preferences of Cas98, 17. In this site-depletion assay, a library of plasmids bearing 6 randomized base pairs adjacent to a protospacer is tested for cleavage by Cas9 in E. coli (Extended Data Fig. 3b). Plasmids with PAM sequences refractory to Cas9 enable cell survival due to the presence of an antibiotic resistance gene, whereas plasmids bearing targetable PAMs are depleted from the library (Fig. 1d, Extended Data Fig. 3b). Sequencing the uncleaved population of plasmids enables the calculation of a post-selection PAM depletion value (PPDV), an estimate of Cas9 activity against those PAMs (post-selection frequency relative to the pre-selection frequency). Site-depletion data obtained with catalytically inactive Cas9 (dCas9) on two randomized PAM libraries (each with a different protospacer) enabled us to define what represents a statistically significant change in PPDV for any given PAM or group of PAMs (Extended Data Fig. 3c), and PPDVs observed for wild-type SpCas9 recapitulated its previously described profile of targetable PAMs8 (Fig. 1e).
Using the site-depletion assay, we obtained PAM specificity profiles for the VQR and EQR variants. The VQR variant strongly depleted sites bearing NGAN and NGCG PAMs, while the EQR variant appeared more specific for an NGAG PAM (Fig. 1f). The human cell EGFP disruption assay paralleled these results, with the VQR variant robustly cleaving sites bearing NGAN PAMs (with relative efficiencies NGAG>NGAT=NGAA>NGAC), and also sites bearing NGNG PAMs with generally lower efficiencies (Fig. 1g). Similarly, the EQR variant preferred NGAG to the other NGAN and NGNG PAMs in human cells, again at lower activities than with the VQR variant (Fig. 1g). The activities of the VQR and EQR variants in human cells therefore recapitulated what was observed with the bacterial site-depletion assay and suggested that PPDVs of 0.2 (five-fold depletion) provide a reasonable predictive threshold for activity in human cells (Extended Data Fig. 4).
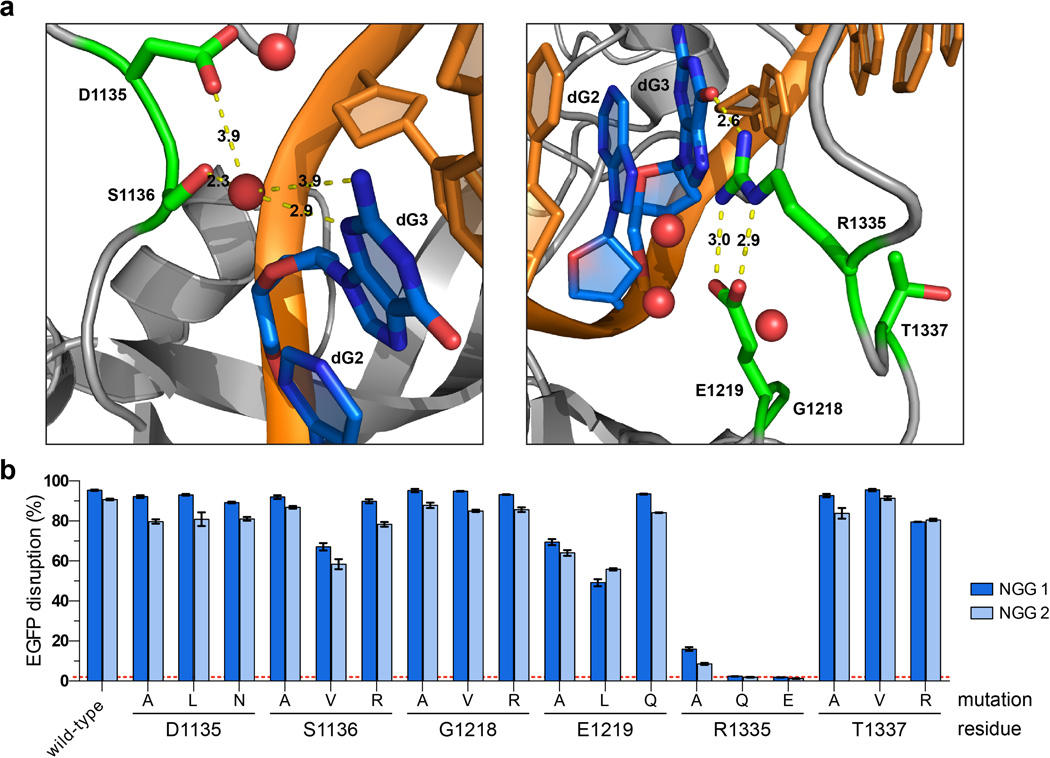
We next sought to extend the generalizability of our engineering strategy by identifying SpCas9 variants capable of recognizing an NGC PAM. Selections using libraries bearing pre-existing R1335E/T1337R and R1335T/T1337R substitutions (Online Methods) yielded surviving colonies harboring a variety of additional mutations (Extended Data Fig. 2b). Testing all possible combinations of the most common mutations using the EGFP disruption assay established that the quadruple mutant VRER variant (D1135V/G1218R/R1335E/T1337R) displayed the highest activity on an NGC PAM and minimal activity on an NGG PAM (Fig. 1h). Analysis of the VRER variant using the site-depletion assay revealed it to be highly specific for NGCG PAMs (Fig. 1i). Consistent with this result, EGFP disruption assays revealed efficient cleavage of sites with NGCG PAMs, and inconsistent or low activity against NGCH and NGNG PAMs (Fig. 1j). Notably, the mutations critical for altering the specificity of SpCas9 are spatially oriented near the PAM (Extended Data Fig. 5a), and the nature and effect of the mutations imply that they are most likely gain of function (Extended Data Fig. 5b). For example, the T1337R mutation appears to confer a preference for a fourth PAM base, especially in the case of the VRER variant.
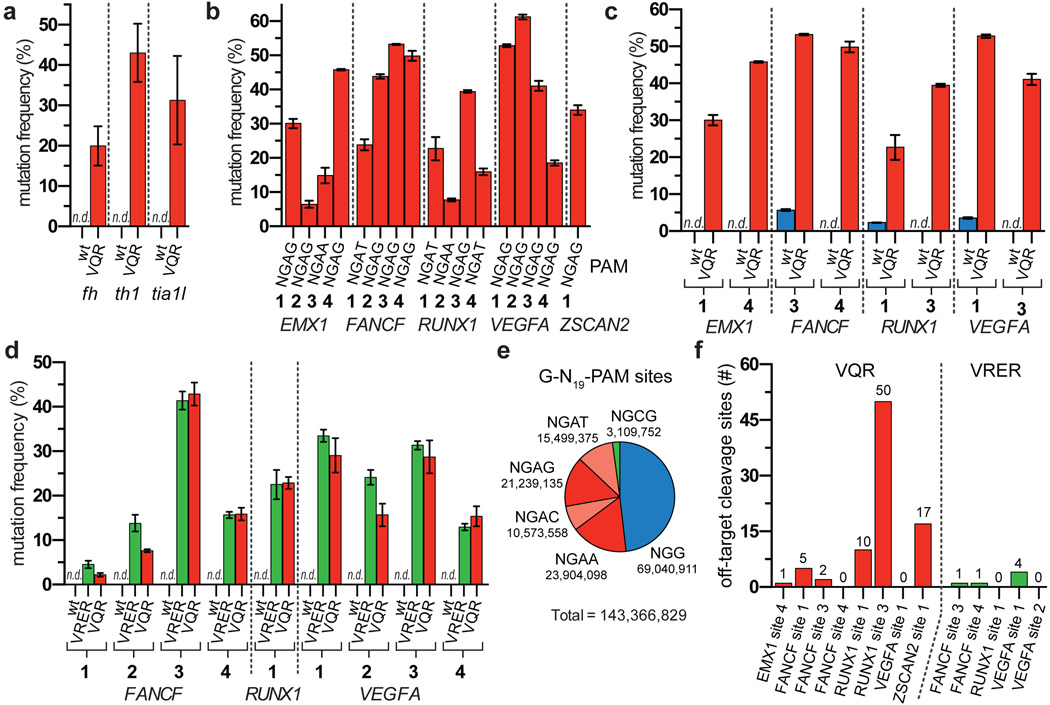
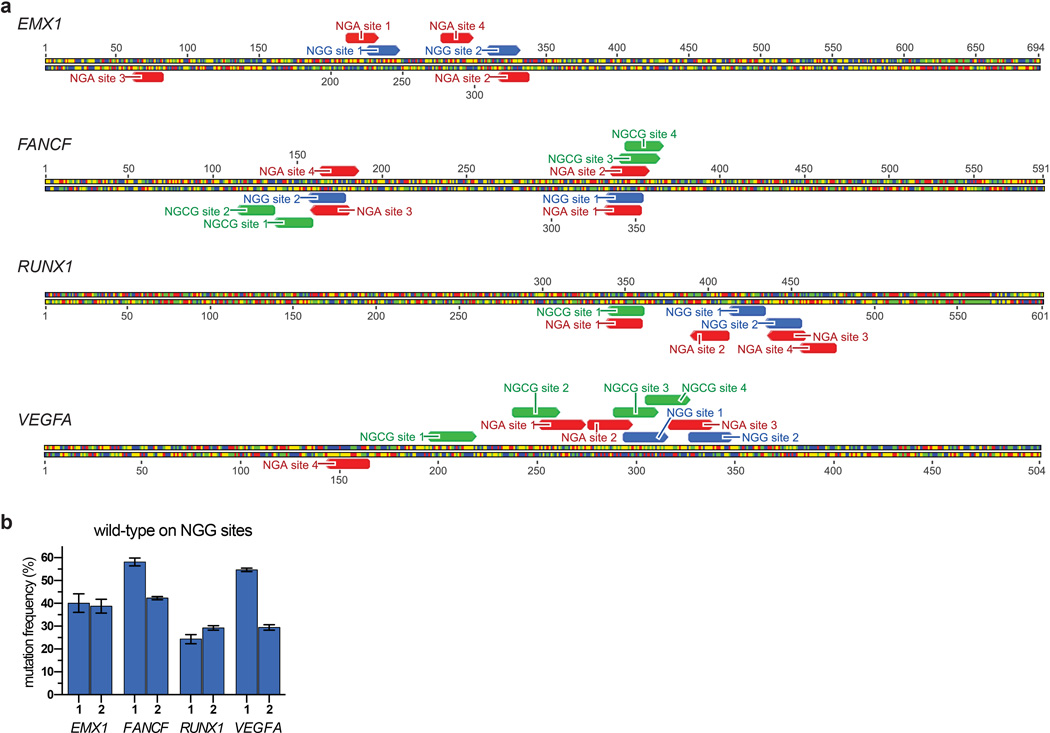
To demonstrate directly that the SpCas9 variants broaden the targeting range of SpCas9, we tested their activities against endogenous genes in zebrafish embryos and human cells. In zebrafish embryos, the VQR variant efficiently modified sites bearing NGAG PAMs (range of 20 to 43%, Fig. 2a) with the indels originating at the predicted cleavage sites (Extended Data Fig. 6). In human cells, the VQR variant robustly modified endogenous sites that harbored NGA PAMs (again, with a preference for NGAG>NGAT=NGAA, range of 6 to 53%) (Fig. 2b, Extended Data Fig. 7a). Importantly, wild-type SpCas9 was unable to robustly alter NGA PAM sites in zebrafish and human cells (Figs. 2a, 2c), yet able to efficiently modify neighboring sites bearing NGG PAMs (Extended Data Fig. 7b). Similarly, when examining VRER variant activity at endogenous human sites with NGCG PAMs, we also observed robust disruption frequencies (range of 5 to 36%) (Fig. 2d). Consistent with the site-depletion data (Figs. 1e, 1f), the VQR variant also altered NGCG PAM sites while wild-type SpCas9 was unable to do so (Fig. 2d). Taken together, these results demonstrate that the VQR and VRER variants enable modification of previously inaccessible sites in zebrafish embryos and human cells, and computational analysis of the reference human genome reveals that they double the targeting potential of SpCas9 (Fig. 2e). To identify target sites for the engineered variants, we have developed a web-based tool called CasBLASTR (http://www.CasBLASTR.org).
Figure 2. SpCas9 PAM variants robustly modify endogenous sites in zebrafish embryos and human cells.
a, Mutagenesis frequencies in zebrafish embryos induced by wild-type or VQR SpCas9 at endogenous gene sites bearing NGAG PAMs. Mutation frequencies were determined using the T7E1 assay; n.d., not detectable by T7E1; error bars represent s.e.m., n = 5 to 9 embryos. b, Endogenous gene disruption activity of the VQR variant quantified by T7E1 assay. Error bars represent s.e.m., n = 3. c, Endogenous gene disruption activity of wild-type SpCas9 against NGA PAM sites quantified by T7E1 assay, where VQR data is re-presented from panel b for ease of comparison. Error bars represent s.e.m., n = 3. d, Mutation frequencies of wild-type, VRER, and VQR SpCas9 at endogenous human cell sites containing NGCG PAMs quantified by T7E1 assay; error bars represent s.e.m., n = 3. e, Representation of the number sites in the human genome with 20 nt spacers targetable by wild-type, VQR, and VRER SpCas9. The 5’-G is included for expression from a U6 promoter. f, Number of off-target cleavage sites identified by GUIDE-seq for the VQR and VRER variants using sgRNAs from panels b and d.
To determine the genome-wide specificity of the VQR and VRER SpCas9 nucleases, we used the recently described GUIDE-seq method7 to profile off-target cleavage events in human cells. The total number of detectable off-target DSBs induced by the SpCas9 variants in human cells (Fig. 2f) are comparable to (or, in the case of the VRER variant, perhaps better than) what has been previously observed with wild-type SpCas97. The off-target sites observed generally possess the expected PAM sequences predicted by our site-depletion experiments (compare Figs. 1f, 1i to Extended Data Fig. 8), and the mismatches observed in the off-target sites of the variants are similar to the profiles previously observed with wild-type SpCas9 for sgRNAs targeted to non-repetitive sequences7. The stringent genome-wide specificity observed with the VRER variant might result from its extension of the PAM by 1 bp, and perhaps from the relative depletion of NGCG PAMs in the human genome (Fig. 2e)18.
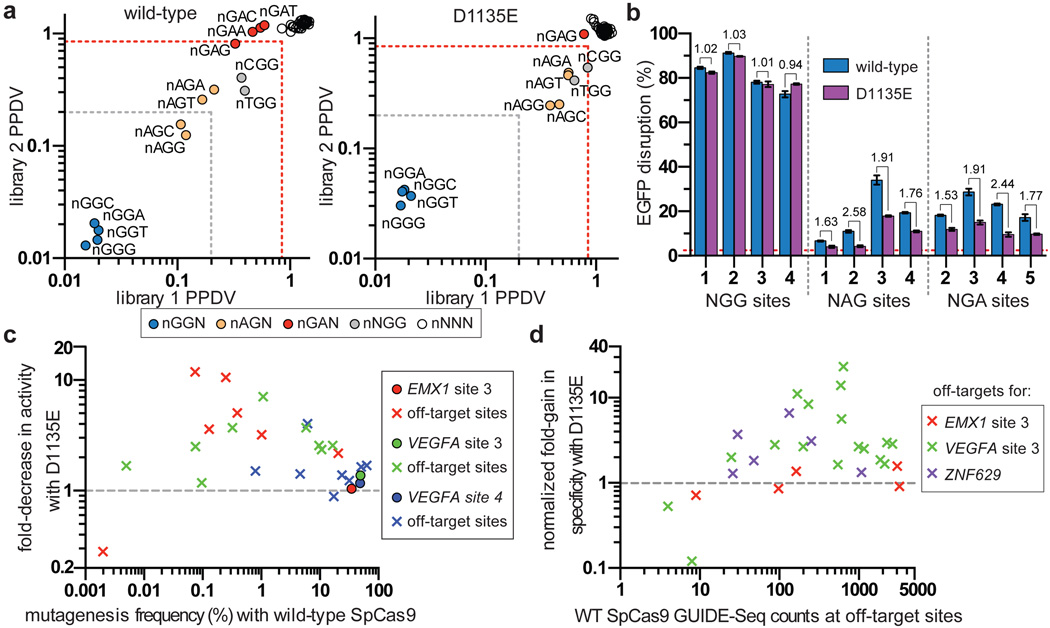
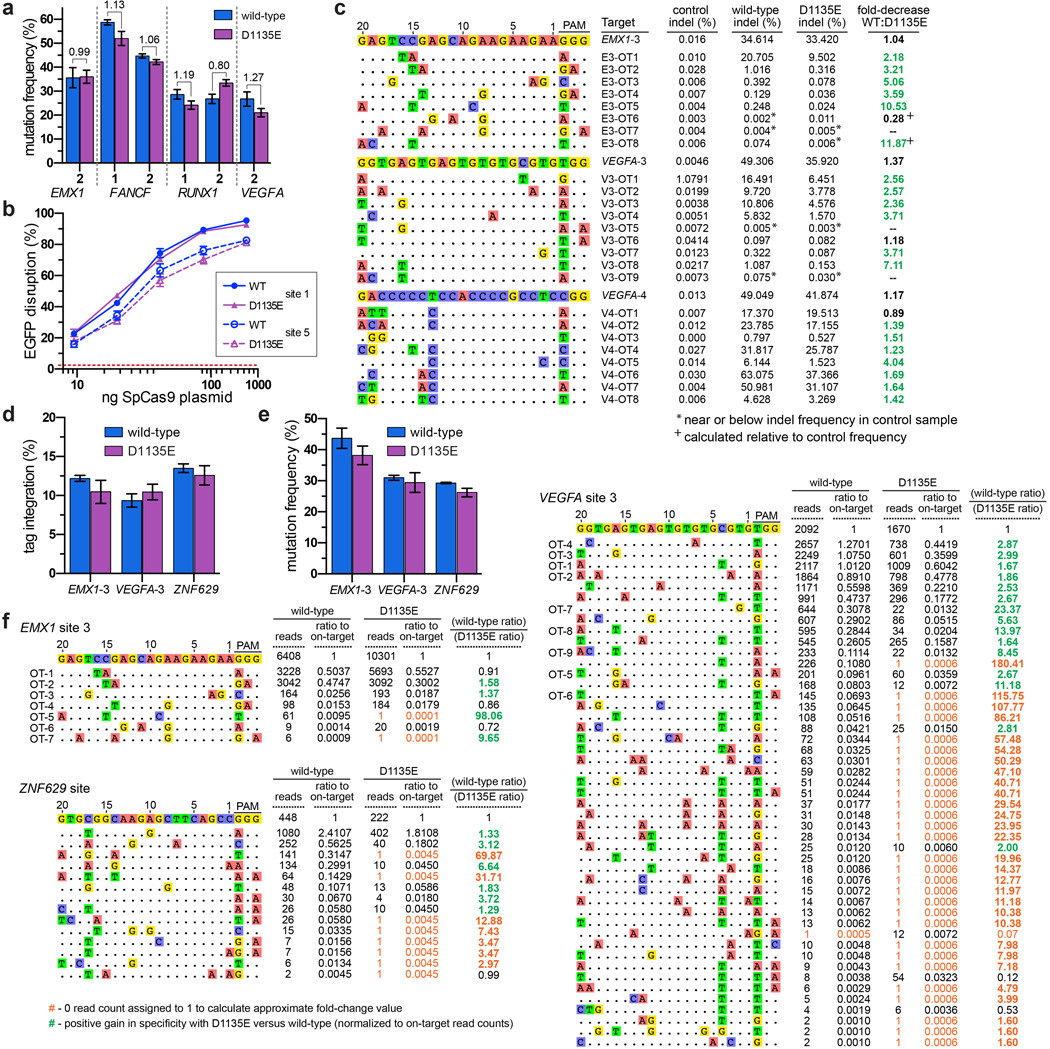
Previous studies have shown that imperfect PAM recognition by SpCas9 can lead to recognition of non-canonical PAMs7, 8, 19–21. While engineering the VQR variant, we noticed that a D1135E mutant appeared to better discriminate between NGG and NGA PAMs compared with wild-type SpCas9 (Fig. 1c). Using the site-depletion assay to assess the D1135E variant, we observed a decrease in activity against non-canonical NAG, NGA, and NNGG PAMs relative to wild-type SpCas9, with this effect being more prominent for one protospacer (Fig. 3a). Improved PAM specificity was also observed in human cell EGFP disruption assays, where NAG and NGA PAM sites were less efficiently cleaved by D1135E compared to wild-type SpCas9 (Fig. 3b, mean fold-decrease in activity of 1.94). Importantly, wild-type and D1135E SpCas9 had comparable activities against canonical NGG PAM sites when targeted to the EGFP reporter or endogenous human gene sites (mean fold-decrease in activity of 1.04) (Figs. 3b, Extended Data Fig. 9a, respectively). It is unlikely that the enhanced specificity of the D1135E variant is the result of protein destabilization, because titration experiments revealed no substantial differences in activity compared with wild-type SpCas9 (Extended Data Fig. 9b).
Figure 3. A D1135E mutation improves the PAM recognition and spacer specificity of SpCas9.
a, PPDV scatterplots for wild-type and D1135E SpCas9 for the two randomized PAM libraries. PAMs are plotted by their 2nd/3rd/4th positions, and wild-type data is the same as shown in Fig. 1d for ease of comparison. The red dashed line indicates PAMs that are statistically significantly depleted (see Extended Data Fig. 3c), and the gray dashed line indicates five-fold depletion (PPDV of 0.2). b, EGFP disruption activities of wild-type and D1135E SpCas9 on sites that contain canonical and non-canonical PAMs in human cells. Disruption frequencies were quantified by flow cytometry; mean background level of EGFP loss represented by the dashed red line; error bars represent s.e.m., n = 3; fold change in activity is shown. c, Summary of targeted deep-sequencing data demonstrating specificity gains at off-target sites when using D1135E (see also Extended Data Fig. 9c). d, Summary of GUIDE-seq detected changes in specificity between wild-type and D1135E at off-target sites (see also Extended Data Fig. 9f). Estimated fold-gain in specificity at sites without read-counts for D1135E are not plotted (see Extended Data Fig. 8c).
To more directly assess the effect of D1135E on off-target effects, we examined the mutation rates induced by wild-type and D1135E SpCas9 on 25 previously known off-target sites of three sgRNAs7, 14, 19. Deep-sequencing revealed that D1135E improved specificity for 19 of the 22 off-target sites with mutation frequencies above background indel rates, when compared to the relative mutation frequencies observed at the on-target sites (Figs. 3c, Extended Data Fig. 9c). Interestingly, the gains in specificity with D1135E are not restricted to sites with non-canonical PAMs. To more thoroughly assess the improvements in specificity associated with the D1135E variant, we performed GUIDE-seq using three different sgRNAs and observed a generalized improvement in genome-wide specificity relative to wild-type SpCas9 (Fig. 3d, Extended Data Figs. 9d–f). Collectively, these results show that the D1135E substitution increases the specificity of SpCas9.
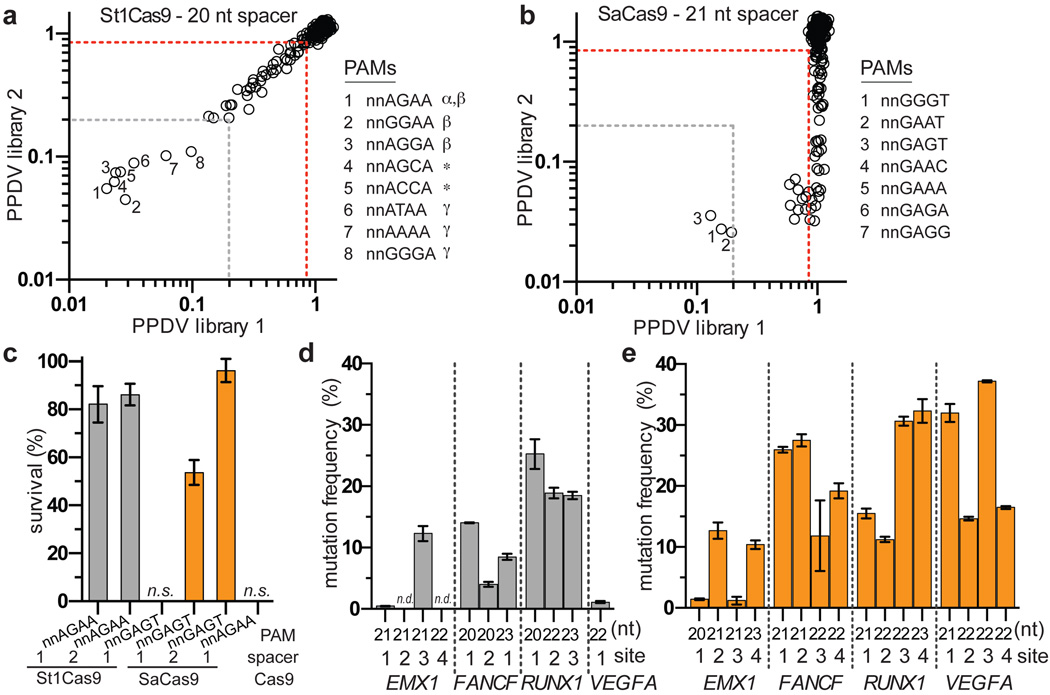
The many Cas9 orthologues from other bacteria make attractive candidates for characterizing and engineering Cas9s with novel PAM specificities22, 23. To explore this, we determined whether two smaller-size orthologues, Streptococcus thermophilus Cas9 from the CRISPR1 locus (St1Cas9)24, 25 and Staphyloccocus aureus (SaCas9)23 could function in the bacterial selection assays. Although the PAM of St1Cas9 has previously been characterized as NNAGAA17, 22, 24, 25, our attempts to bioinformatically derive the SaCas9 PAM using a previously described approach22 failed to yield a consensus sequence. Therefore, we used the site-depletion assay to determine the PAM for SaCas9 and, as a positive control, St1Cas9. For St1Cas9, we identified two novel PAMs in addition to six PAMs that had been previously described17, 22, 25 (Fig. 4a, Extended Data Figs. 10a, 10b). For SaCas9, only three PAMs were depleted greater than 5-fold in all experiments (NNGGGT, NNGAAT, NNGAGT, Fig. 4b), although additional PAMs were targetable when using the second protospacer library (Extended Data Figs. 10c, 10d). These results are consistent with a recent definition of SaCas9 PAM specificity23. We also found that St1Cas9 and SaCas9 can function efficiently in the bacterial positive selection system (Fig. 4c), suggesting that their PAM specificities could potentially be modified by mutagenesis and selection.
Figure 4. Characterization of St1Cas9 and SaCas9 in bacteria and human cells.
a, b, PPDV scatterplots for St1Cas9 (panel a) and SaCas9 (panel b), with PAMs plotted by their 3rd/4th/5th/6th positions. The red dashed line indicates PAMs that are statistically significantly depleted (Extended Data Fig. 3c), and the gray dashed line represents five-fold depletion (PPDV of 0.2); α, PAM previously predicted by a bioinformatic approach25; β, PAMs previously identified under stringent experimental conditions17; *, novel PAMs discovered in this study; γ, PAMs previously identified under moderate experimental conditions17 c, Survival percentages of St1Cas9 and SaCas9 in the bacterial positive selection when challenged with selection plasmids that harbor different target sites and PAMs. d, e, Mutation frequencies of St1Cas9 (panel d) and SaCas9 (panel e) quantified by T7E1 assay at sites in four endogenous human genes. Error bars represent s.e.m., n = 3; n.d., not detectable by T7E1.
Because not all Cas9 orthologues function efficiently outside of their native context17, 23, we tested whether St1Cas9 and SaCas9 can modify sites in human cells. St1Cas9 has been previously shown to function as a nuclease in human cells but only on four sites17, 23, 26, and a recently published manuscript assessed SaCas9 activity23. In EGFP disruption experiments, St1Cas9 displayed high activity at three of five target sites and SaCas9 efficiently targeted eight sites (Extended Data Fig. 10e). No obvious correlation between activity and length of spacer was observed (Extended Data Fig. 10e, 10f). When examining activity on endogenous loci, St1Cas9 efficiently targeted 7 out of 11 sites (1 to 25% disruption; Fig. 4d), SaCas9 displayed more robust activity at 16 sites (1% to 37%; Fig. 4e), and again no distinct spacer length requirement was observed (Extended Data Fig. 10g). Collectively, these results demonstrate that St1Cas9 and SaCas9 function in human cells, making them attractive candidates for engineering additional variants with novel PAM specificities.
The VQR and VRER variants engineered in this study enhance the opportunities to utilize the CRISPR-Cas9 platform to practice efficient HDR, to generate NHEJ-mediated indels in small genetic elements, and to exploit the requirement for a PAM to distinguish between different alleles in the same cell. Importantly, the VQR, VRER, and D1135E variants all have similar (or better) genome-wide specificities compared to wild-type SpCas9. These variants can be rapidly incorporated into existing and widely used SpCas9 vectors by simple site-directed mutagenesis, and we expect that the variants should also work with other previously described improvements to the SpCas9 platform (e.g., truncated sgRNAs7, 27, SpCas9 nickases20, 28, or dimeric FokI-dCas9 fusions29, 30). Collectively, our results establish engineering PAM recognition and characterization of additional Cas9 orthologues (as previously described)17, 22, 23 as complementary approaches to provide researchers with an expanded repertoire of genome-editing reagents, while also demonstrating the feasibility of engineering Cas9 nuclease variants with useful new properties.
Online Methods
Plasmids and oligonucleotides
DNA sequences for parent constructs used in this study can be found in Supplementary Information. Sequences of oligonucleotides used to generate the positive selection plasmids, negative selection plasmids, and site-depletion libraries are available in Supplementary Table 1. Sequences of all sgRNA targets in this study are available in Supplementary Table 2. Point mutations in Cas9 were generated by PCR. For cloning purposes, please note the low copy number origins of these plasmids. All new plasmids described in this study will be deposited with the non-profit plasmid distribution service Addgene: http://www.addgene.org/crispr-cas.
Bacterial Cas9/sgRNA expression plasmids were constructed with two T7 promoters to separately express Cas9 and the sgRNA. These plasmids encode human codon optimized versions of Cas9 for S. pyogenes (BPK764, SpCas9 sequence subcloned from JDS24614), S. thermophilus Cas9 from CRISPR locus 1 (MSP1673, St1Cas9 sequence modified from previous published description17), and S. aureus (BPK2101, SaCas9 sequence codon optimized from Uniprot J7RUA5). Previously described sgRNA sequences were utilized for SpCas931, 32 and St1Cas917, while the SaCas9 sgRNA sequence was determined by searching the European Nucleotide Archive sequence HE980450 for crRNA repeats using CRISPRfinder (http://crispr.u-psud.fr/Server/) and identifying the tracrRNA using a bioinformatic approach similar to one previously described33. Annealed oligos to complete the spacer complementarity region of the sgRNA were ligated into BsaI cut BPK764 and BPK2101, or BspMI cut MSP1673 (append 5’-ATAG to the spacer to generate the top oligo and append 5’-AAAC to the reverse compliment of the spacer sequence to generate the bottom oligo). A 5’-GG dinucleotide was included on all bacterial plasmid sgRNAs for proper expression from the T7 promoter.
Residues 1097–1368 of SpCas9 were randomly mutagenized using Mutazyme II (Agilent Technologies) at a rate of ~5.2 substitutions/kilobase to generate mutagenized PAM-interacting (PI) domain libraries. For NGA PAM selections, wild-type SpCas9 and R1335Q were utilized as templates for mutagenesis. For NGC PAM selections, we first designed Cas9 mutants bearing amino acid substitutions of R1335 that might be expected to interact with a cytosine (D, E, S, or T) and found no activity on an NGC PAM site using the positive selection system (data not shown). We then randomly mutagenized the PAM-interacting domain of each of these singly substituted variants but still failed to obtain surviving colonies in positive selections (data not shown). Because the T1337R mutation had increased the activities of our VQR and EQR variants, we combined this mutation with R1335 substitutions of A, D, E, S, T, or V, and again randomly mutagenized their PAM-interacting domains. Selections using two of these six mutagenized libraries (bearing pre-existing R1335E/T1337R and R1335T/T1337R substitutions) yielded surviving colonies harboring a variety of additional mutations (Extended Data Fig. 2b). The theoretical complexity of each PI domain library was estimated to be greater than 107 clones based on the number of transformants obtained. Positive and negative selection plasmids were generated by ligating annealed target site oligos into XbaI/SphI or EcoRI/SphI cut p11-lacY-wtx115, respectively.
Two randomized PAM libraries (each with a different protospacer sequence) were constructed using Klenow(-exo) to fill-in the bottom strand of oligos that contained six randomized nucleotides directly adjacent to the 3’ end of the protospacer (see Supplementary Table 1). The double-stranded product was cut with EcoRI to leave EcoRI/SphI ends for ligation into cut p11-lacY-wtx1. The theoretical complexity of each randomized PAM library was estimated to be greater than 106 based on the number of transformants obtained.
SpCas9 and variants were expressed in human cells from vectors derived from JDS24614. For St1Cas9 and SaCas9, the Cas9 ORFs from MSP1673 and BPK2101 were subcloned into a CAG promoter vector to generate MSP1594 and BPK2139, respectively. Plasmids for U6 expression of sgRNAs (into which desired spacer oligos can be cloned) were generated using the sgRNA sequences described above for the SpCas9 sgRNA (BPK1520), the St1Cas9 sgRNA (BPK2301), and the SaCas9 sgRNA (VVT1). Annealed oligos to complete the spacer complementarity region of the sgRNA were ligated into the BsmBI overhangs of these vectors (append 5’-CACC to the spacer to generate the top oligo and append 5’-AAAC to the reverse complement of the spacer sequence to generate the bottom oligo). A 5’-G of target spacer sequences was included when designing human cell sgRNAs, for proper expression from the U6 promoter (and thus included in the calculation in Fig. 2e).
Bacterial-based positive selection assay for evolving SpCas9 variants
Competent E.coli BW25141(λDE3)34 containing a positive selection plasmid (with embedded target site) were transformed with Cas9/sgRNA-encoding plasmids. Following a 60 minute recovery in SOB media, transformations were plated on LB plates containing either chloramphenicol (non-selective) or chloramphenicol + 10 mM arabinose (selective). Cleavage of the positive selection plasmid was estimated by calculating the survival frequency: colonies on selective plates / colonies on non-selective plates (see also Extended Data Fig. 1).
To select for SpCas9 variants that can target novel PAMs, PI-domain mutagenized Cas9/sgRNA plasmid libraries were electroporated into E.coli BW25141(λDE3) cells containing a positive selection plasmid that encodes a target site and PAM of interest. Generally ~50,000 clones were screened to obtain between 50–100 survivors. The PI domains of surviving clones were subcloned into fresh backbone plasmid and re-tested in the positive selection. Clones that had greater than 10% survival in this secondary screen for activity were sequenced. Mutations observed in the sequenced clones were chosen for further assessment based on their frequency in surviving clones, type of substitution, proximity to the PAM bases in the SpCas9/sgRNA crystal structure (PDB:4UN3)12, and (in some cases) activities in a human cell-based EGFP disruption assay.
Bacterial-based site-depletion assay for profiling Cas9 PAM specificities
Competent E.coli BW25141(λDE3) containing a Cas9/sgRNA expression plasmid were transformed with negative selection plasmids harboring cleavable or non-cleavable target sites. Following a 60 minute recovery in SOB media, transformations were plated on LB plates containing chloramphenicol + carbenicillin. Cleavage of the negative selection plasmid was estimated by calculating the colony forming units per µg of DNA transformed (see also Extended Data Fig. 3).
The negative selection was adapted to determine PAM specificity profiles of Cas9 nucleases by electroporating each randomized PAM library into E.coli BW25141(λDE3) cells harboring an appropriate Cas9/sgRNA plasmid. Between 80,000–100,000 colonies were plated at a low density spread on LB + chloramphenicol + carbenicillin plates. Surviving colonies containing negative selection plasmids refractory to cleavage by Cas9 were harvested and plasmid DNA isolated by maxi-prep (Qiagen). The resulting plasmid library was amplified by PCR using Phusion Hot-start Flex DNA Polymerase (New England BioLabs) followed by an Agencourt Ampure XP cleanup step (Beckman Coulter Genomics). Dual-indexed Tru-Seq Illumina deep-sequencing libraries were prepared using the KAPA HTP library preparation kit (KAPA BioSystems) from ~500 ng of clean PCR product for each site-depletion experiment. The Dana-Farber Cancer Institute Molecular Biology Core performed 150-bp paired-end sequencing on an Illumina MiSeq Sequencer.
The raw FASTQ files outputted for each MiSeq run were analyzed with a Python program to determine relative PAM depletion. The program (see Supplementary Information) operates as follows: First, a file dialog is presented to the user from which all FASTQ read files for a given experiment can be selected. For these files, each FASTQ entry is scanned for the fixed spacer region on both strands. If the spacer region is found, then the six variable nucleotides flanking the spacer region are captured and added to a counter. From this set of detected variable regions, the count and frequency of each window of length 2–6 nt at each possible position was tabulated (see Supplementary Table 3 for the 6 nt output). The site-depletion data for both randomized PAM libraries was analyzed by calculating the post-selection PAM depletion value (PPDV): the post-selection frequency of a PAM in the selected population divided by the pre-selection library frequency of that PAM. PPDV analyses were performed for each experiment across all possible 2–6 length windows in the 6 bp randomized region. The windows we used to visualize PAM preferences were: the 3 nt window representing the 2nd, 3rd, and 4th PAM positions for wild-type and variant SpCas9 experiments, and the 4 nt window representing the 3rd, 4th, 5th, 6th PAM positions for St1Cas9 and SaCas9.
Two significance thresholds for PPDVs were determined based on: 1) a statistical significance threshold based on the distribution of dCas9 versus pre-selection library log read count ratios (see Extended Data Fig. 3c & 3d), and 2) a biological activity threshold based on an empirical correlation between depletion values and activity in human cells. The statistical threshold was set at 3.36 standard deviations from the mean PPDV for dCas9 (equivalent to a relative PPDV of 0.85), corresponding to a normal distribution two-sided p-value of 0.05 after adjusting for multiple comparisons (i.e. p=0.05/64). The biological activity threshold was set at 5-fold depletion (equivalent to a PPDV of 0.2) because this level of depletion serves as a reasonable predictor of activity in human cells (see also Extended Data Fig. 4). The 95% confidence intervals in Extended Data Fig. 4 were calculated by dividing the standard deviation of the mean, by the square root of the sample size multiplied by 1.96.
Human cell culture and transfection
U2OS cells obtained from our collaborator Toni Cathomen (Freiburg) and U2OS.EGFP cells harboring a single integrated copy of a constitutively expressed EGFP-PEST reporter gene13 were cultured in Advanced DMEM media (Life Technologies) supplemented with 10% FBS, 2 mM GlutaMax (Life Technologies), penicillin/streptomycin, at 37 °C with 5% CO2. Additionally, U2OS.EGFP cells were cultured in 400 µg/ml of G418. The identity of U2OS and U2OS.EGFP cell lines were validated by STR profiling (ATCC) and deep sequencing, and cells were tested bi-weekly for mycoplasma contamination. Cells were co-transfected with 750 ng of Cas9 plasmid and 250 ng of sgRNA plasmid (unless otherwise noted) using the DN-100 program of a Lonza 4D–nucleofector according to the manufacturer’s protocols. Cas9 plasmid transfected together with an empty U6 promoter plasmid was used as a negative control for spontaneous background EGFP loss for all human cell EGFP disruption experiments, and all engodenous gene disruption experiments (none of which showed detectable activity by T7E1). Target sites for endogenous gene experiments were selected within 200 bp of NGG sites cleavable by wild-type SpCas9 (see Extended Data Fig. 6a and Supplementary Table 2).
Zebrafish care and injections
Zebrafish care and use was approved by the Massachusetts General Hospital Subcommittee on Research Animal Care. Cas9 mRNA was transcribed with PmeI-digested JDS246 (wild-type SpCas9) or MSP469 (VQR variant) using the mMESSAGE mMACHINE T7 ULTRA Kit (Life Technologies) as previously described32. All sgRNAs in this study were prepared according to the cloning-independent sgRNA generation method35. sgRNAs were transcribed by the MEGAscript SP6 Transcription Kit (Life Technologies), purified by RNA Clean & Concentrator-5 (Zymo Research), and eluted with RNase-free water.
sgRNA- and Cas9-encoding mRNA were co-injected into one-cell stage zebrafish embryos. Each embryo was injected with ~2–4.5 nL of solution containing 30 ng/µL sgRNA and 300 ng/µL Cas9 mRNA. The next day, injected embryos were inspected under a stereoscope for normal morphological development, and genomic DNA was extracted from 5 to 9 embryos.
Human cell EGFP disruption assay
EGFP disruption experiments were performed as previously described14. Transfected cells were analyzed for EGFP expression ~52 hours post-transfection using a Fortessa flow cytometer (BD Biosciences). Background EGFP loss was gated at approximately 2.5% for all experiments (graphically represented as a dashed red line).
T7E1 assay, targeted deep-sequencing, and GUIDE-seq to quantify nuclease-induced mutations
T7E1 assays were performed as previously described for human cells13 and zebrafish32. For U2OS.EGFP human cells, genomic DNA was extracted from transfected cells ~72 hours post-transfection using the Agencourt DNAdvance Genomic DNA Isolation Kit (Beckman Coulter Genomics). Target loci from zebrafish or human cell genomic DNA were amplified using the primers listed in Supplementary Table 1. Roughly 200 ng of purified PCR product was denatured, annealed, and digested with T7E1 (New England BioLabs). Mutagenesis frequencies were quantified using a Qiaxcel capillary electrophoresis instrument (QIagen), as previously described for human cells13 and zebrafish32.
For targeted deep-sequencing, previously characterized on- and off-target sites7, 14, 27 were amplified using Phusion Hot-start Flex with the primers listed in Supplementary Table 1. Genomic loci were amplified for a control condition (empty sgRNA), wild-type, and D1135E SpCas9. An Agencourt Ampure XP cleanup step (Beckman Coulter Genomics) was performed prior to pooling ~500 ng of DNA from each condition for library preparation. Dual-indexed Tru-Seq Illumina deep-sequencing libraries were generated using the KAPA HTP library preparation kit (KAPA BioSystems). The Dana-Farber Cancer Institute Molecular Biology Core performed 150-bp paired-end sequencing on an Illumina MiSeq Sequencer. Mutation analysis of targeted deep-sequencing data was performed as previously described30. Briefly, Illumina MiSeq paired end read data was mapped to human genome reference GRChr37 using bwa36. High-quality reads (quality score >= 30) were assessed for indel mutations that overlapped the target or off-target sites. 1-bp indel mutations were excluded from the analysis unless they occurred within 1-bp of the predicted breakpoint. Changes in activity at on- and off-target sites comparing D1135E versus wild-type SpCas9 were calculated by comparing the indel frequencies from both conditions (for rates above background control amplicon indel levels).
GUIDE-seq experiments were performed as previously described7. Briefly, phosphorylated, phosphorothioate-modified double-stranded oligodeoxynucleotides (dsODNs) were transfected into U2OS cells along with Cas9 and sgRNA expression plasmids, as described above. dsODN-specific amplification, high-throughput sequencing, and mapping were performed to identify genomic intervals containing DSB activity. For wild-type versus D1135E experiments, off-target read counts were normalized to the on-target read counts to correct for sequencing depth differences between samples. The normalized ratios for wild-type and D1135E SpCas9 were then compared to calculate the fold-change in activity at off-target sites. To determine whether wild-type and D1135E samples for GUIDE-seq had similar oligo tag integration rates at the intended target site, restriction fragment length polymorphism (RFLP) assays were performed by amplifying the intended target loci with Phusion Hot-Start Flex from 100 ng of genomic DNA (isolated as described above) using primers listed in Supplementary Table 1. Roughly 150 ng of PCR product was digested with 20 U of NdeI (New England BioLabs) for 3 hours at 37 °C prior to clean-up using the Agencourt Ampure XP kit. RFLP results were quantified using a Qiaxcel capillary electrophoresis instrument (QIagen) to approximate oligo tag integration rates. T7E1 assays were performed for a similar purpose, as described above.
Extended Data
Extended Data Figure 1. Bacterial-based positive selection used to engineer altered PAM specificity variants of SpCas9.
a, Expanded schematic of the positive selection from Fig. 1b (left panel), and validation that SpCas9 behaves as expected in the positive selection (right panel). b, Schematic of how the positive selection was adapted to select for SpCas9 variants that have altered PAM recognition specificities. A library of SpCas9 clones with randomized PAM-interacting (PI) domains (residues 1097-1368) is challenged by a selection plasmid that harbors an altered PAM. Variants that survive the selection by cleaving the positive selection plasmid are sequenced to determine the mutations that enable altered PAM specificity.
Extended Data Figure 2. Amino acid sequences of clones that cleave target sites bearing alternate PAMs in the bacterial-based positive selection system.
a, Sequences of variants that survived >10% when re-tested in the positive selection assay against an NGA PAM site (see Online Methods). Variants were selected from libraries containing randomly mutagenized PAM-interacting (PI) domains (residues 1097–1368) with or without a starting R1335Q mutation. Sequence differences compared with wild-type SpCas9 are highlighted. The histogram represents the number of changes at each position (not counting the starting R1335Q mutation). b, Sequences of variants that survived >10% when re-tested in the positive selection assay against a site containing an NGC PAM. Variants were selected from libraries containing randomly mutagenized PAM-interacting (PI) domains (residues 1097–1368) with starter mutation pairs of R1335E/T1337R or R1335T/T1337R. Sequence differences compared with wild-type SpCas9 (shown at the top) are highlighted. The histogram below illustrates the number of changes at each position (not counting starter mutations at R1335 or T1337).
Extended Data Figure 3. Bacterial cell-based site-depletion assay for profiling the global PAM specificities of Cas9 nucleases.
a, Expanded schematic illustrating the negative selection from Fig. 1d (left panel), and validation that wild-type SpCas9 behaves as expected in a screen of sites with functional (NGG) and non-functional (NGA) PAMs (right panel). b, Schematic of how the negative selection was used as a site-depletion assay to screen for functional PAMs by constructing negative selection plasmid libraries containing 6 randomized base pairs in place of the PAM. Selection plasmids that contain PAMs cleaved by a Cas9/sgRNA of interest are depleted while PAMs that are not cleaved (or poorly cleaved) are retained. The frequencies of the PAMs following selection are compared to their pre-selection frequencies in the starting libraries to calculate the post-selection PAM depletion value (PPDV). c, d, A cutoff for statistically significant PPDVs was established by plotting the PPDV of PAMs for catalytically inactive SpCas9 (dCas9) (grouped and plotted by their 2nd/3rd/4th positions) for the two randomized PAM libraries (c). A threshold of 3.36 standard deviations from the mean PPDV for the two libraries was calculated (red lines in (d)), establishing that any PPDV deviation below 0.85 is statistically significant compared to dCas9 treatment (red dashed line in (c)). The gray dashed line in (c) indicates a five-fold depletion in the assay (PPDV of 0.2).
Extended Data Figure 4. Concordance between the site-depletion assay and EGFP disruption activity.
Data points represent the average EGFP disruption of the two NGAN and NGNG PAM sites for the VQR and EQR variants (Fig. 1g) plotted against the mean PPDV observed for library 1 and 2 (Fig. 1f) for the corresponding PAM. The red dashed line indicates PAMs that are statistically significantly depleted (PPDV of 0.85, see Extended Data Fig. 3c), and the gray dashed line represents five-fold depletion (PPDV of 0.2). Mean values are plotted with the 95% confidence interval.
Extended Data Figure 5. Structural and functional roles of D1135, G1218, and T1337 in PAM recognition by SpCas9.
a, Structural representations of the six residues implicated in PAM recognition. The left panel illustrates the proximity of D1135 to S1136, a residue that makes a water-mediated, minor groove contact to the 3rd base position of the PAM12. The right panel illustrates the proximity of G1218, E1219, and T1337 to R1335, a residue that makes a direct, base-specific major groove contact to the 3rd base position of the PAM12. Angstrom distances indicated by yellow dashed lines; non-target strand guanine bases dG2 and dG3 of the PAM are shown in blue; other DNA bases shown in orange; water molecules shown in red; images generated using PyMOL from PDB:4UN3. b, Mutational analysis of six residues in SpCas9 that are implicated in PAM recognition. Clones containing one of three types of mutations at each position were tested for EGFP disruption with two sgRNAs targeted to sites harboring NGG PAMs. For each position, we created an alanine substitution and two non-conservative mutations. S1136 and R1335 were previously reported to mediate contacts to the 3rd guanine of the PAM12, and D1135, G1218, E1219, and T1337 are reported in this study. EGFP disruption activities were quantified by flow cytometry; background control represented by the dashed red line; error bars represent s.e.m., n = 3.
Extended Data Figure 6. Insertion or deletion mutations induced by the VQR SpCas9 variant at endogenous zebrafish sites containing NGAG PAMs.
For each target locus, the wild-type sequence is shown at the top with the protospacer highlighted in yellow (highlighted in green if present on the complementary strand) and the PAM is marked as red underlined text. Deletions are shown as red dashes highlighted in gray and insertions as lower case letters highlighted in blue. The net change in length caused by each indel mutation is shown on the right (+, insertion; –, deletion). Note that some alterations have both insertions and deletions of sequence and in these instances the alterations are enumerated in parentheses. The number of times each mutant allele was recovered (if more than once) is shown in brackets.
Extended Data Figure 7. Endogenous genes targeted by wild-type and evolved variants of SpCas9.
a, Sequences targeted by wild-type, VQR, and VRER SpCas9 are shown in blue, red, and green, respectively. Sequences of sgRNAs and primers used to amplify these loci for T7E1 are provided in Supplementary Tables 1 and 2. b, Mean mutagenesis frequencies detected by T7E1 for wild-type SpCas9 at eight target sites bearing NGG PAMs in the four different endogenous human genes (corresponding to the annotations in the top panel). Error bars represent s.e.m., n = 3.
Extended Data Figure 8. Specificity profiles of the VQR and VRER SpCas9 variants determined using GUIDE-seq7.
The intended on-target site is marked with a black square, and mismatched positions within off-target sites are highlighted. a, The specificity of the VQR variant was assessed in human cells by targeting endogenous sites containing NGA PAMs: EMX1 site 4, FANCF site 1, FANCF site 3, FANCF site 4, RUNX1 site 1, RUNX1 site 3, VEGFA site 1, and ZSCAN2. b, The specificity of the VRER variant was assessed in human cells by targeting endogenous sites containing NGCG PAMs: FANCF site 3, FANCF site 4, RUNX1 site 1, VEGFA site 1, and VEGFA site 2.
Extended Data Figure 9. Activity differences between D1135E and wild-type SpCas9.
a, Mutagenesis frequencies detected by T7E1 for wild-type and D1135E SpCas9 at six endogenous sites in human cells. Error bars represent s.e.m., n = 3; mean fold change in activity is shown. b, Titration of the amount of wild-type or D1135E SpCas9-encoding plasmid transfected for EGFP disruption experiments in human cells. The amount of sgRNA plasmid used for all of these experiments was fixed at 250 ng. Two sgRNAs targeting different EGFP sites were used; error bars represent s.e.m., n = 3. c, Targeted deep-sequencing of on- and off-target sites for 3 sgRNAs using wild-type and D1135E SpCas9. The on-target site is shown at the top, with off-target sites listed below highlighting mismatches to the on-target. Fold decreases in activity with D1135E relative to wild-type SpCas9 at off-target sites greater than the change in activity at the on-target site are highlighted in green; control indel levels for each amplicon are reported. d, Mean frequency of GUIDE-seq oligo tag integration at the on-target sites, estimated by restriction fragment length polymorphism analysis. Error bars represent s.e.m., n = 4. e, Mean mutagenesis frequencies at the on-target sites detected by T7E1 for GUIDE-seq experiments. Error bars represent s.e.m., n = 4. f, GUIDE-seq read-count differences between wild-type SpCas9 and D1135E at 3 endogenous human cell sites. The on-target site is shown at the top and off-target sites are listed below with mismatches highlighted. In the table, a ratio of off-target activity to on-target activity is compared between wild-type and D1135E to calculate the normalized fold-changes in specificity (with gains in specificity highlighted in green). For sites without detectable GUIDE-seq reads, a value of 1 has been assigned to calculate an estimated change in specificity (indicated in orange). Off-target sites analyzed by deep-sequencing in panel c are numbered to the left of the EMX1 site 3 and VEGFA site 3 off-target sites
Extended Data Figure 10. Additional PAMs for St1Cas9 and SaCas9 and activities based on spacer lengths in human cells.
a, PPDV scatterplots for St1Cas9 comparing the sgRNA complementarity lengths of 20 and 21 nucleotides obtained with a randomized PAM library for spacer 1 (top panel) or spacer 2 (bottom panel). PAMs were grouped and plotted by their 3rd/4th/5th/6th positions. The red dashed line indicates PAMs that are statistically significantly depleted (see Extended Data Fig. 3c) and the gray dashed line represents five-fold depletion (PPDV of 0.2). b, Table of PAMs with PPDVs of less than 0.2 for St1Cas9 under each of the four conditions tested. PAM numbering shown on the left is the same as in Fig. 4a. c, PPDV scatterplots for SaCas9 comparing the sgRNA complementarity lengths of 21 and 23 nucleotides obtained with a randomized PAM library for spacer 1 (top panel) or spacer 2 (bottom panel). PAM were grouped and plotted by their 3rd/4th/5th/6th positions. The red and gray dashed lines are the same as in (a). d, Table of PAMs with PPDVs of less than 0.2 for SaCas9 under each of the four conditions tested. PAM numbering is the same as in Fig. 4b. e, f, Human cell activity of St1Cas9 and SaCas9 across various spacer lengths via EGFP disruption (panel e, data from Figs. 4d, 4e) and endogenous gene mutagenesis detected by T7E1 (panel f, data from Figs. 4f, 4g). Activity for all replicates shown (n = 3 or 4); bars illustrate mean and 95% confidence interval; number of sites per spacer length indicated.
Supplementary Material
Acknowledgements
We thank James Angstman and Vikram Pattanayak for discussion and comments on the manuscript. This work was supported by a National Institutes of Health (NIH) Director's Pioneer Award (DP1 GM105378) and NIH R01 GM107427 to J.K.J., NIH R01 GM088040 to J.K.J. and R.T.P., The Jim and Ann Orr Research Scholar Award (to J.K.J.), and a National Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (to B.P.K.).
Footnotes
Supplementary Information is included with this submission.
Author Contributions
B.P.K., M.S.P., S.Q.T., and N.T.N. performed all bacterial and human cell-based experiments. A.P.W.G. and Z.L. performed all zebrafish experiments. S.Q.T., V.T., Z.Z., and M.J.A. analyzed the site-depletion, targeted deep-sequencing, and GUIDE-seq data. B.P.K., R.T.P., J.-R.J.Y., and J.K.J. directed the research and interpreted experiments. B.P.K. and J.K.J. wrote the manuscript with input from all the authors.
Conflict of interest statement: J.K.J. is a consultant for Horizon Discovery. J.K.J. has financial interests in Editas Medicine, Hera Testing Laboratories, Poseida Therapeutics, and Transposagen Biopharmaceuticals. J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
All new reagents described in this work will be deposited with the non-profit plasmid distribution service Addgene (http://www.addgene.org/crispr-cas). A web-tool to design sgRNA sites for the engineered variants and orthogonal Cas9 nucleases described in this study can be found at http://www.CasBLASTR.org.
References
- 1.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 3.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 4.Shah SA, Erdmann S, Mojica FJ, Garrett RA. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. doi: 10.4161/rna.23764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, Zhao H. A highly sensitive selection method for directed evolution of homing endonucleases. Nucleic Acids Res. 2005;33:e154. doi: 10.1093/nar/gni148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J Am Chem Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 17.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 19.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci Rep. 2014;4:5405. doi: 10.1038/srep05405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fonfara I, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. doi: 10.1093/nar/gkt1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015 doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horvath P, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleinstiver BP, Fernandes AD, Gloor GB, Edgell DR. A unified genetic, computational and experimental framework identifies functionally relevant residues of the homing endonuclease I-BmoI. Nucleic Acids Res. 2010;38:2411–2427. doi: 10.1093/nar/gkp1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gagnon JA, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.