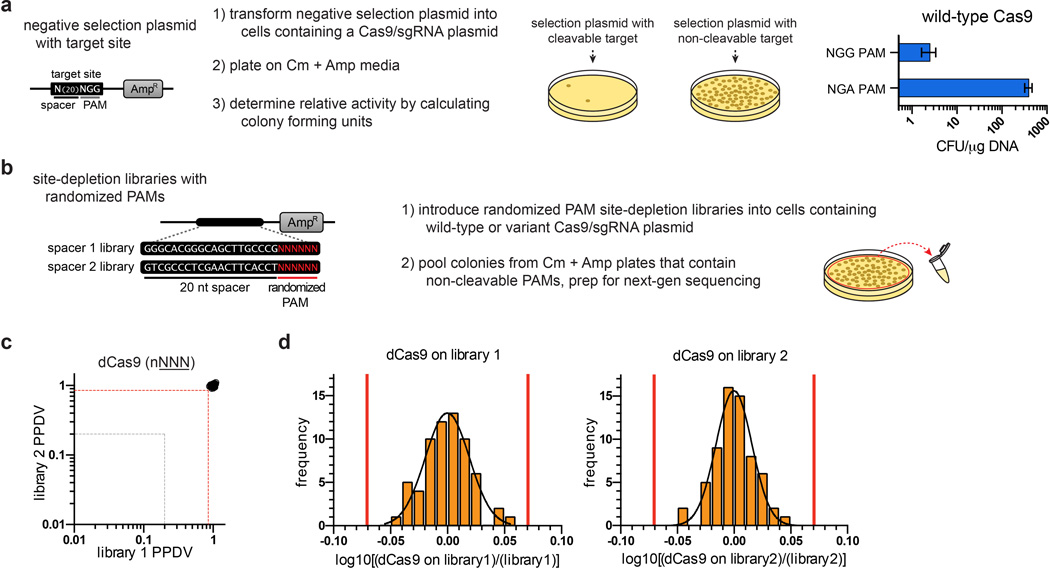
Extended Data Figure 3. Bacterial cell-based site-depletion assay for profiling the global PAM specificities of Cas9 nucleases.
a, Expanded schematic illustrating the negative selection from Fig. 1d (left panel), and validation that wild-type SpCas9 behaves as expected in a screen of sites with functional (NGG) and non-functional (NGA) PAMs (right panel). b, Schematic of how the negative selection was used as a site-depletion assay to screen for functional PAMs by constructing negative selection plasmid libraries containing 6 randomized base pairs in place of the PAM. Selection plasmids that contain PAMs cleaved by a Cas9/sgRNA of interest are depleted while PAMs that are not cleaved (or poorly cleaved) are retained. The frequencies of the PAMs following selection are compared to their pre-selection frequencies in the starting libraries to calculate the post-selection PAM depletion value (PPDV). c, d, A cutoff for statistically significant PPDVs was established by plotting the PPDV of PAMs for catalytically inactive SpCas9 (dCas9) (grouped and plotted by their 2nd/3rd/4th positions) for the two randomized PAM libraries (c). A threshold of 3.36 standard deviations from the mean PPDV for the two libraries was calculated (red lines in (d)), establishing that any PPDV deviation below 0.85 is statistically significant compared to dCas9 treatment (red dashed line in (c)). The gray dashed line in (c) indicates a five-fold depletion in the assay (PPDV of 0.2).