Abstract
The cytoplasmic events that control mammalian gene expression, primarily mRNA stability and translation, potently influence the cellular response to internal and external signals. The ubiquitous RNA-binding protein (RBP) HuR is one of the best-studied regulators of cytoplasmic mRNA fate. Through its post-transcriptional influence on specific target mRNAs, HuR can alter the cellular response to proliferative, stress, apoptotic, differentiation, senescence, inflammatory and immune stimuli. In light of its central role in important cellular functions, HuR’s role in diseases in which these responses are aberrant is increasingly appreciated. Here, we review the mechanisms that control HuR function, its influence on target mRNAs, and how impairment in HuR-governed gene expression programs impact upon different disease processes. We focus on HuR’s well-recognized implication in cancer and chronic inflammation, and discuss emerging studies linking HuR to cardiovascular, neurological, and muscular pathologies. We also discuss the progress, potential, and challenges of targeting HuR therapeutically.
Keywords: RNA-binding protein, elav, post-transcriptional gene regulation, mRNA turnover, translational regulation
2. INTRODUCTION
Mammalian gene expression is regulated at many levels, both transcriptional and post-transcriptional. Post-transcriptional gene regulation can occur at the levels of pre-mRNA splicing and maturation, as well as mRNA transport, editing, storage, stability, and translation (1, 2). Among these steps, the cytoplasmic control of mRNA turnover and translation is particularly effective in eliciting rapid adaptive changes in expressed proteins in response to environmental alterations and internal cues. The turnover and translation regulatory RNA-binding proteins (TTR-RBPs) and noncoding RNAs (particularly microRNAs) are two main classes of trans factors that associate with specific cis elements present in mRNAs whose stability and translation are subject to regulation (3–5).
Some TTR-RBPs control one specific post-transcriptional process; for example tristetraprolin (TTP), butyrate response factor-1 (BRF1), and KH domain-containing RBP (KSRP) selectively accelerate mRNA degradation (6–9). However, most TTR-RBPs, including AU-binding factor 1 (AUF1), T-cell intracellular antigen-1 (TIA-1) and TIA-1-related (TIAR) proteins, polypyrimidine tract-binding protein (PTB), nuclear factor 90 (NF90), and other TTR-RBPs (10–14; reviewed in reference 15), can influence both mRNA turnover and translation. In addition, most TTR-RBPs often function jointly, cooperating, competing, or acting sequentially on shared target mRNAs. As many disease-associated proteins (e.g., tumor suppressors, oncoproteins, cell cycle factors, and cytokines) are encoded by mRNAs which bear TTR cis elements, their aberrant post-transcriptional expression in disease processes has been the focus of intense research over the past decade (see accompanying articles in this issue).
First described in Drosophila as elav (embryonic lethal abnormal vision), the mammalian Hu/elav family of TTR-RBPs comprises the ubiquitous HuR (HuA) and the primarily neuronal proteins HuB, HuC and HuD (16). The neuronal Hu proteins have been implicated in neuronal development, neuronal plasticity, and memory (reviewed in 17, 18). Since its identification in 1996 (19), HuR has been found to interact with dozens of mRNAs, many of them encoding proteins linked to specific pathologies. Although HuR was originally described as a stabilizing TTR-RBP (20), it was later shown to modulate the translation of target mRNAs, generally promoting translation, but sometimes inhibiting it (reviewed in 15, 16). The regulation of HuR function, the fate of [HuR-mRNA] ribonucleoprotein (RNP) complexes, and the impact of HuR-mediated gene regulation in disease processes are the focus of this review.
3. HuR TARGET mRNAs
Through its three RNA recognition motifs (RRMs), HuR interacts with target mRNAs. Many HuR target mRNAs bear U- and AU-rich elements in their 3′-untranslated region (UTR), where they are termed AREs, but HuR has also been found to interact with U- and AU-rich sequences in the 5′UTR of HuR some target mRNAs (15, 16, 19–22). Although HuR is predominantly nuclear, its influence upon the expression of target mRNAs is linked to its localization in the cytoplasm, a process controlled by numerous transport mechanisms (see section 4.2).
3.1. Stabilized HuR target mRNAs
HuR stabilizes a large subset of target mRNAs, including many which encode proteins implicated in different pathologies, particularly cancer and inflammation. These mRNAs are the templates for proteins such as c-Fos, the cyclin-dependent kinase (cdk) inhibitor p21, cyclins (A2, B1, E1, D1), inducible nitric oxide synthase (iNOS), granulocyte macrophage-colony stimulating factor (GM-CSF), eukaryotic initiation factor (eIF)-4E, murine double minute (mdm)2, vascular endothelial growth factor (VEGF), transforming growth factor (TGF)-β, sirtuin 1 (SIRT1), tumor necrosis factor (TNF)-α, B-cell leukemia (Bcl)-2, myeloid leukemia cell differentiation protein (Mcl)-1, oncostatin M (OSM), cyclooxygenase (COX)-2, γ-glutamylcysteine synthetase heavy subunit (γ-GCSh), survival of motor neuron (SMN), SH2D1A, the regulator of G-protein signaling 4 (RGS4), parathyroid hormone-related protein (PTHrP), Fas ligand (FasL), Myogenin, MyoD, acetylcholinesterase (AChE), p53, ARHI [aplasia Ras homolog member I (DIRAS3)], nitric oxide/soluble guanylyl cyclase (sGC), urokinase plasminogen activator (uPA) and its receptor (uPAR), neurofibromatosis type 1 (NF1), von Hippel-Lindau protein (pVHL), toll-like receptor 4 (TLR4), Snail, matrix metalloprotease (MMP)-9, c-Fms, the mitogen-activated protein kinase (MAPK) phosphatase (MKP)-1, interferon (IFN)-γ, HuR itself, and interleukin (IL)-3, IL-4, IL-6, and IL-8 (Table 1 and discussed below). The exact mechanisms whereby HuR protects mRNAs from decay are unknown; however, binding of HuR to a target mRNA is widely believed to block the association of other TTR-RBPs or microRNAs (associated with the RNA-induced silencing complex or RISC) capable of recruiting the mRNA to sites of mRNA decay like the exosome or processing bodies (PBs) (e.g., 5, 23).
Table 1. Influence of HuR upon target mRNAs involved in disease processes.
Listed are HuR target mRNAs encoding proteins linked to disease (column 1), the influence of HuR on mRNA stability and/or translation [enhanced (↑) or reduced, (↓), column 2], the cellular processes affected by the HuR-mRNA interactions (column 3), and the cell model or disease model in which [HuR-mRNA] regulation has been studied (column 4).
| HuR target | Influence of HuR | Processes | ||
|---|---|---|---|---|
| mRNA | on mRNA | Regulated | Cell/Disease Model | References |
| c-Fos | ↑ Stability | Proliferation | Oral squamous carcinoma | (109) |
| c-Myc | ↓ Translation | Proliferation, survival | Cervical carcinoma | (31) |
| p21 | ↑ Stability | Proliferation, survival | Carcinoma (breast, colon) | (68, 69) |
| p27 | ↓ Translation | Proliferation, survival | Cervical carcinoma | (28) |
| cyclin A2 | ↑ Stability ↑ Translation |
Proliferation | Carcinoma (colon, gastric, oral) | (66, 107, 109) |
| cyclin B1 | ↑ Stability | Proliferation | Oral carcinoma | (109) |
| cyclin E1 | ↑ Stability | Proliferation | Breast carcinoma | (86) |
| cyclin D1 | ↑ Stability | Proliferation | Carcinoma (oral, colon) | (109, 24) |
| OSM | ↑ Stability | Proliferation | Lymphoma | (117) |
| eIF4E | ↑ Stability | Proliferation, survival | Pharyngeal carcinoma | (67) |
| EGF | ↑ Stability | Proliferation | Prostate carcinoma | (98) |
| VEGF | ↑ Stability ↑ Translation |
Angiogenesis, proliferation | Carcinoma (colon, non-small cell lung, kidney, pancreatic, prostate), glioma, meningioma, astrocytoma, ischemia, amyotrophic lateral sclerosis) | (59, 63, 102, 104, 105, 150, 154,155, 168) |
| HIF-1α | ↑ Stability ↑ Translation |
Angiogenesis, survival | Cervical carcinoma, ischemia | (70, 150) |
| COX-2 | ↑ Stability | Angiogenesis, survival, inflammation | Carcinoma (colon, ovarian, gastric, oral, prostate), central nervous system malignancies, rheumatoid cartilage, osteoarthritic cartilage, inflammatory, bowel disease | (63, 64, 93–95, 97, 100, 106, 109) |
| iNOS | ↑ Stability | Angiogenesis, survival, inflammation | Colon carcinoma, muscle wasting | (121, 123) |
| TSP1* | ↑ Translation | Angiogenesis | Breast carcinoma | (73) |
| TGF-β | ↑ (n.d.) | Immunity, inflammation | Tumors of the central nervous system | (63) |
| MKP-1 | ↑ Stability ↑ Translation |
Signaling, immunity | Cervical carcinoma | (74) |
| Mdm2 | ↑ Stability | Survival | Intestinal epithelium function | (170) |
| SIRT1 | ↑ Stability | Survival, stem cell development | Carcinoma (cervical, prostate) | (56, 97) |
| Bcl-2 | ↑ Stability ↑ Translation |
Survival | Carcinoma (cervical, prostate, epidermoid), leukemia, ischemia-reperfusion injury | (71, 72, 151) |
| Mcl-1 | ↑ (n.d.) | Survival | Cervical carcinoma | (71) |
| XIAP | ↑ Translation | Survival | Untransformed cells | (24) |
| Cyto c | ↑ Translation | Survival | Cervical carcinoma | (27) |
| uPA | ↑ Stability | Invasion, migration | Breast carcinoma | (80) |
| uPAR | ↑ Stability | Invasion, migration | Breast carcinoma | (80) |
| MMP-9 | ↑ Stability | Invasion | Fibrosarcoma, myeloid leukemia, fibrosis, left ventricular function and remodeling | (81, 82, 128) |
| Snail | ↑ Stability | Invasion | Breast carcinoma | (83) |
| dCK | ↑ (n.d.) | Chemotherapy | Pancreatic carcinoma | (92) |
| ARHI/DRAS3* | ↑ Stability | Tumor suppression | Ovarian carcinoma | (94) |
| p53 | ↑ Stability ↑ Translation |
Tumor suppression | Carcinoma (cervical, gastric, liver, colon), intestinal epithelium function, myocardial infarction | (25, 171) |
| pVHL | ↑ Stability | Tumor suppression | VHL syndrome, kidney carcinoma | (40, 173) |
| BRCA1 | (↓) n.d. | Tumor suppression | Breast carcinoma | (174) |
| ER | ↑ Stability | Tumorigenesis | Breast carcinoma | (175) |
| Wnt5a | ↓ Translation | Tumorigenesis | Breast carcinoma | (90) |
| c-Fms | ↑ Stability | Tumorigenesis | Breast carcinoma | (87) |
| GATA3 | ↑ Stability | Tumorigenesis | Breast carcinoma | (176) |
| GM-CSF | ↑ Stability | Inflammation, immunity | Asthma, T cell activation, atherogenesis | (115, 138) |
| TNF-α | ↑ Stability | Inflammation, immunity | Muscle wasting, malignant glioma, rheumatoid arthritis, atherosclerosis | (63, 115, 135) |
| TM | ↓ Translation | Inflammation | Sepsis | (30) |
| RGS4 | ↑ Stability | Inflammation | Smooth muscle contraction, cardiac development | (119) |
| TLR4 | ↑ Stability | Inflammation, immunity | Vascular smooth muscle hyperplasia | (139) |
| IL-6 | ↑ Stability | Inflammation, immunity | Tumors of the central nervous system, viral infection, atherosclerosis | (63, 111, 113) |
| IL-8 | ↑ Stability | Inflammation, immunity | Carcinoma (breast, colon, gastric), glioma | (63, 64, 89) |
| IL-13 | ↑ Stability | Inflammation | Allergy | (132) |
| SMN | ↑ Stability | Neuropathology | Spinal muscle atrophy | (156) |
| SH2D1A | ↑ Stability | Proliferation | X-linked lymphoproliferative disease | (163) |
| NF1 | ↑ Stability | Signaling | Neurofibromatosis | (153) |
| PROX1 | ↑ Stability | Endothelial differentiation | Kaposi’s sarcoma | (143) |
| Eotaxin | ↑ Stability | Inflammation | Asthma | (118) |
| ProTα | ↑ Translation | Tumorigenesis | Cervical carcinoma | (177) |
| IGF-1R | ↓ Translation | Proliferation | Breast carcinoma | (29) |
| HuR | ↑ Stability ↑ Translation |
(above processes) | (above cell/disease models) | (3, 34, 35) |
In column 1, * indicates mRNAs whose regulation by HuR primarily involves dissociation of HuR from the mRNA. See text for further details.
3.2. Translationally upregulated target mRNAs
HuR also promotes the translation of several target mRNAs encoding proteins which are involved in disease processes, including Cyclin A2, prothymosin α (ProTα), hypoxia-inducible factor (HIF)-1α, Bcl-2, VEGF, thrombospondin (TSP)-1, MKP-1, p53, the cationic amino acid transporter (CAT)-1, the intrinsic cellular caspase inhibitor XIAP, and cytochrome c (24, 25, Table 1 and discussed below). It is also unclear how HuR promotes the translation of each of these target mRNAs, but HuR was recently shown to associate with the internal ribosome entry site (IRES) of the XIAP 5′UTR and directly enhanced XIAP translation (24). Models of HuR-elicited exclusion of translational repressors (other TTR-RBPs or microRNA/RISC) from target mRNAs have been reported in some instances of translational upregulation by HuR [e.g., CAT-1 and cytochrome c mRNAs (25–27)].
3.3. Translationally repressed target mRNAs
HuR inhibits the translation of a small subset of target mRNAs that encode disease-associated proteins. HuR binds to the 5′UTRs of p27, IGF-1R, and thrombomodulin (TM) mRNAs and represses their translation; this inhibitory action was proposed to result from disruption of IRESs in these 5′UTRs (28–30). HuR was also found to bind to the 3′UTRs of Wnt5a and c-Myc mRNAs and repress their translation; the mechanism of Wnt5a repression is not yet known, but the reduced translation of c-Myc was linked to HuR’s recruitment of the let-7/RISC complex to the c-Myc 3′UTR (31).
4. REGULATION OF HuR FUNCTION
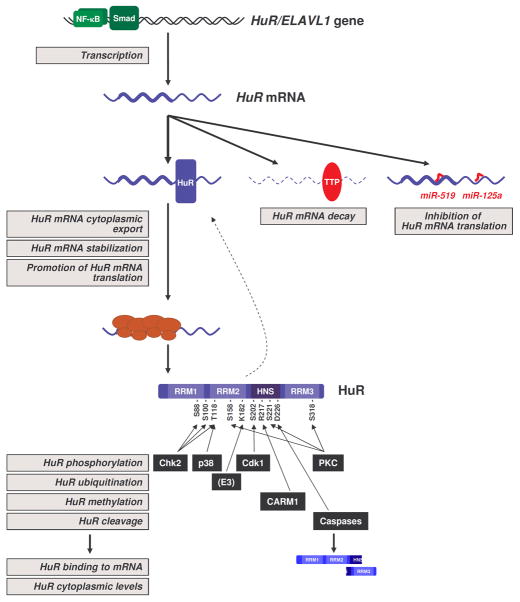
The function of HuR is controlled at multiple levels. The initial studies focused on HuR cytoplasmic export as a critical way to control expression of HuR target mRNAs, but recent work has revealed that the abundance and integrity of HuR protein, as well as post-translational modifications affecting HuR binding to mRNAs all potently influence HuR function (Figure 1).
Figure 1.
Regulation of HuR expression and function. The schematic depicts the current understanding of HuR regulation. Transcription of the HuR/ELAVL1 gene is controlled by the transcription factor NF-κB. The HuR mRNA is positively regulated by enhanced export to the cytoplasm, stabilization, and enhanced translation but HuR itself; the HuR mRNA is negatively regulated by TTR-RBP tristetraprolin (TTP), which promotes HuR mRNA decay, and by microRNAs miR-125a and miR-519, which repress HuR translation. HuR protein is subject to phosphorylation by Chk2, which affects [HuR-mRNA] interactions, by Cdk1, which affects HuR levels in the cytoplasm, and by p38 and PKC, which affect both [HuR-mRNA] interactions and cytoplasmic HuR levels. Methylation by CARM1 can also affect HuR subcellular distribution and binding to mRNAs. Ubiquitination of HuR by an as-yet unknown E3 ligase controls HuR protein stability, and caspases can cleave HuR into two fragments with different cellular properties. Gray squares indicate steps in which HuR expression or function are regulated. See text for further details.
4.1. Regulation of HuR abundance
The steady-state levels of HuR protein are regulated in a number of ways. The transcriptional control of HuR expression is poorly understood, but HuR transcription is positively regulated by the nuclear factor (NF)-κB (32) and by Smads (33). By contrast, the abundance of HuR mRNA and HuR protein are subject to multiple regulatory mechanisms (Figure 1).
4.1.1. HuR auto-regulation
HuR binds the HuR mRNA, in keeping with the ability of many TTR-RBPs to associate with the very mRNAs that encode them (3). Among different polyadenylation variants of the HuR mRNA, HuR was found to bind to and stabilize a long HuR mRNA bearing a distal AU-rich element; this effect that was opposed by the mRNA decay-promotion actions of TTP (34). HuR binding to the HuR 3′UTR also enhanced the cytoplasmic export of the HuR mRNA (35).
4.1.2. Downregulation of HuR by microRNAs
The HuR mRNA is the target of two microRNAs. miR-519 was computationally predicted to associate with the HuR mRNA coding region (CR) and a distal segment of the HuR 3′UTR, but only the CR interaction was functional (36). Acting upon the HuR CR, miR-519 selectively repressed HuR translation; in turn, cells which overexpressed miR-519 showed reduced cell proliferation in culture, displayed features of cellular senescence, and developed into significantly smaller tumors in a xenograft model (37, 38). miR-125a associated with the HuR 3′UTR and similarly repressed HuR production by inhibiting HuR translation. In breast cancer cells, miR-125a overexpression enhanced apoptosis and suppressed cell proliferation and cell migration (39).
4.1.3. HuR ubiquitination
In response to moderate heat shock, HuR protein levels declined transiently. This reduction was linked to HuR ubiquitination at residue Lys-182 followed by proteasome-mediated proteolysis. The transient degradation of HuR, which enhanced cell survival after heat shock, was antagonized by phosphorylation of HuR by Chk2 (40).
4.1.4. Caspase-mediated HuR cleavage
Recently, cleavage of HuR at Asp-226 was identified as a key component of the apoptotic cell death program (41). In response to lethal damage by staurosporine, HuR cleavage involved the apoptotic proteins FADD, caspase-8, and caspase-3 (42). In muscle cells, the larger HuR cleavage product (a 24-kDa fragment named CP1) was shown to bind to transportin 2 and to block the nuclear import of HuR, thereby promoting myogenesis (43).
4.2. Regulation of HuR localization
Although HuR is predominantly nuclear, its nuclear function is largely unknown except for a poorly defined role in pre-mRNA splicing, as shown for the pre-RNAs encoding Fas and HuD (44, 45). The transport of HuR across the nuclear envelope requires a specific HuR domain (the HuR nucleocytoplasmic shuttling domain or HNS) and several transport machinery components, including transportins 1 and 2, the chromosome region maintenance 1 (CRM1), and importin-1α (46–49). HuR nucleocytoplasmic transport is also influenced by kinases [Cdk1, AMP-activated protein kinase (AMPK), PKC, and p38] that phosphorylate HuR and HuR transport proteins (50–55), as explained in 4.3. and shown in Figure 1.
4.3. Post-translational modification of HuR
Phosphorylation of HuR at different residues by a number of kinases as well as methylation by the methyltransferase CARM1 affect the subcellular localization of HuR and/or its interaction with target mRNAs. In general, modification of residues within the RRMs affect HuR binding to target mRNAs, while modification of residues within or near the HNS alter HuR subcellular localization (Figure 1).
4.3.1. Chk2
Phosphorylation by the checkpoint kinase Chk2 at HuR residues Ser-88, Ser-100, and Thr-118 (located between and within RRM1 and RRM2) modulates HuR binding to several target mRNAs. Oxidative damage activated Chk2, which in turn phosphorylates Ser-100, triggering the dissociation of HuR from SIRT1 mRNA and other mRNAs (56).
4.3.2. Cdk1
Also known as Cdc2, this kinase phosphorylates HuR at Ser-202, triggering the nuclear retention of HuR mediated by nuclear 14-3-3θ, which interacts with HuR. Under conditions of stress, Cdk1 is inactive, the Ser-202 residue of HuR is unphosphorylated, and the protein can be mobilized to the cytoplasm (53).
4.3.3. PKC
HuR is a substrate for protein kinase C. Phosphorylation by PKCα at HuR Ser-158 and Ser-221 in response to ATP treatment, and phosphorylation by PKCδ of Ser-221 and Ser-318 in response to angiotensin II (AngII) have been shown to promote the cytoplasmic export and the binding activity of HuR (50, 51, 57, 58). PKCβ was also recently shown to phosphorylate HuR in a model of diabetic retinopathy, although the specific residues were not identified (59). These regulatory processes were associated with an increase in the expression of HuR target mRNAs encoding angiogenic, proliferative and proinflammatory proteins like cyclins, VEGF, and COX-2.
4.3.4. p38MAPK
In cells exposed to DNA damage (γ irradiation), this MAPK phosphorylates HuR at Thr-118, triggering its translocation to the cytoplasm and increasing its association with p21 mRNA (60).
4.3.5. CARM1
HuR methylation at Asp-271 by CARM1 (coactivator-associated arginine methyltransferase 1) in response to lipopolysaccharide stimulation of macrophages results in stabilization of TNF-α mRNA (61).
5. HuR IN CANCER
With the discovery that HuD was an antigen in paraneoplastic encephalomyelitis associated in patients with small-cell lung cancer (62), Hu/ELAV proteins were among the first TTR-RBPs found to be implicated in carcinogenesis. The earliest reports of HuR being elevated cancer were from the King and Prescott laboratories; their findings of upregulated HuR in brain and colon cancers were linked to the enhanced expression of COX-2, VEGF, TGF-β, IL-8, and other cancer-associated proteins (63, 64). Subsequent studies revealed that HuR was broadly elevated in virtually all malignancies tested, including cancers of the breast, colon, stomach, kidney, pancreas, esophagus, prostate, skin, lung, and thyroid (65). HuR was proposed to play a causal role in tumor development, since cultured carcinoma cells with ectopically elevated HuR developed into larger tumors in a mouse xenograft model, while forced reduction of HuR reduced the tumor size (38, 65). The work of numerous laboratories has led to the identification of dozens of additional HuR target mRNAs encoding cancer-related proteins (as reviewed in 22).
5.1. HuR modulates cancer traits
HuR binds to many mRNAs that encode proteins responsible for implementing five major cancer-associated phenotypes: enhanced cell proliferation, increased cell survival, elevated local angiogenesis, evasion of immune recognition, and facilitated cancer cell invasion and metastasis (22).
5.1.1. Enhanced proliferation
For a tumor to grow, cells must divide actively. Many HuR target mRNAs encode proteins implicated in cell cycle progression and cell division. In this manner, HuR promotes the expression of several cyclins (D1, E1, A2, B1) and other factors that enhance cell proliferation [c-Fos, epithelial growth factor (EGF), ProTα, eIF4E]. Additionally, HuR represses expression of proteins with growth inhibitory roles, such as p27 and Wnt5a. HuR was linked to elevation in these proteins in numerous malignancies, including cervical, colorectal, breast, and ovarian cancers (66, 67; reviewed in 22).
5.1.2. Anti-apoptotic phenotype
Tumor cells must also acquire resistance to death signals. HuR associates with and promotes the expression of numerous mRNAs that encode pro-survival proteins, as shown for ProTα, Bcl-2, Mcl-1, SIRT1, p21, XIAP, and Mdm2 in various tumor types (22, 24, 68–72). Moreover, HuR binds to the c-Myc mRNA, and represses the synthesis of the encoded protein c-Myc, which can have a pro-apoptotic function (31).
5.1.3. Increased angiogenesis
The development of local vasculature delivers oxygen and nutrients needed for the tumor to thrive. HuR interacts with the mRNAs that encode the pro-angiogenic factors HIF-1α, VEGF, and COX-2 and promotes their expression (22, 70). HuR can also associate with the mRNA that encodes TSP1, an inhibitor of angiogenesis, but this interaction declines in a model of breast carcinogenesis (73).
5.1.4. Reduced immunosurveillance
As the immune system can eliminate tumor cells, escaping immune recognition is advantageous for tumor cells. By binding to the MKP-1 mRNA and potently enhancing MKP-1 expression (70), HuR could suppress the function of immune cells, a key action of MKP-1 (75, 76). Another important HuR target, TGF-β mRNA, encodes a cytokine that enables advanced-stage tumor cells to escape immune recognition (77–79).
5.1.5. Invasion and metastasis
Tumor cells will often invade adjacent tissues and colonize distant organs. HuR promotes the expression of extracellular proteases and proteins which alter the interaction of the cancer cell with its local environment and can promote epithelial-to-mesenchymal transition. Examples include HuR target mRNAs encoding Snail, MMP-9, uPA and the uPA receptor (22, 80–83).
5.2. Implication of HuR in specific cancer types
HuR interacts with and regulates many mRNAs encoding cancer-related proteins. Since HuR is upregulated in virtually all cancer types (65), it has been proposed to coordinate the expression of cancer genes and thereby impact upon phenotypic traits central to tumorigenesis (section 5.1). Over the past decade, numerous studies have examined the levels of HuR in individual cancers and cancer cell models (Table 1).
In breast carcinomas, elevated cytoplasmic HuR levels were associated with tumor grade and poor patient outcome (83, 85). In breast cancer cells, HuR increased expression of cyclin E1, IL-8, estrogen receptor, TSP1, and c-Fms (73, 86–89), while HuR repressed the translation of Wnt5a, a protein that inhibits tumor growth (90). In pancreatic cancer, high HuR levels correlated with high levels of VEGF (91) and with poor patient prognosis (92); paradoxically, however, high HuR was associated with improved survival of patients treated with the chemotherapeutic drug gemcitabine, as discussed below (section 8.). This effect was linked to HuR’s interaction with the deoxycytidine kinase (dCK) mRNA, leading to the increased expression of dCK, an enzyme that activates gemcitabine into an active drug (92).
The high abundance of HuR in colon cancer contributed to the increased expression of COX-2 and VEGF levels and was associated with advanced tumor stage (64, 93). Overexpression of HuR increased the growth of colon cancer cells in an athymic mouse xenograft model (65). COX-2 levels were also upregulated in ovarian carcinomas, where both nuclear and cytoplasmic HuR were found to be elevated. Interestingly, HuR association with the ARHI/DRAS3 mRNA, which encodes a tumor suppressor, was reduced in ovarian cells (94). As in other tumors, increased HuR levels were associated with high ovarian tumor grade and poor prognosis (95, 96). In prostate cancers, HuR abundance was linked to increased levels of COX-2, prostate-specific antigen (PSA), SIRT1, and EGF (97–99). In keeping with the view that cytoplasmic HuR promotes prostate tumor development and relapse, patients who had elevated cytoplasmic HuR expressed higher COX-2 and adverse prognosis with shorter disease-free survival times (97, 100). HuR was also upregulated in oral, lung, gastric, and pharyngeal carcinomas, as well as in cancers of the central nervous system (e.g., meningioma, glioma, astrocytoma), where COX-2, c-Fos, VEGF, eIF4E, cyclin D1, cyclin A, and other HuR target mRNAs were found to be elevated (63, 67, 101–109).
6. HuR IN INFLAMMATION
HuR has been implicated in promoting inflammation and inflammatory diseases. The pro-inflammatory influence of HuR is linked to its interaction with mRNAs encoding pro-inflammatory proteins, leading to their increased production in a variety of cell types. In addition, HuR function was notably inhibited by anti-inflammatory factors (Table 1).
6.1 HuR promotes expression of pro-inflammatory factors
HuR associates with several mRNAs encoding pro-inflammatory cytokines, most prominently TNF-α and IL-6, stabilizes them and promotes the expression of the encoded proteins in different cell types, including fibroblasts, T-cells, and macrophages (63, 64, 110–113). In macrophages, endothelial cells, intestinal epithelial cells, and in colon, gastric and cervical carcinoma cells, HuR binds to mRNAs encoding the proinflammatory cytokines IL-8, TGF-β, and IFN-γ, and enhances their expression (63, 64, 92, 112, 114–116). HuR interacts with the oncostatin M (OSM) mRNA in lymphoma cells and induces expression of the encoded pro-inflammatory cytokine OSM (117) and with the eotaxin mRNA in lung epithelial cells, where it promotes expression of the encoded chemokine (118). Other proteins that modulate inflammatory responses, such as the G-protein signaling RGS4, are also regulated by HuR (119).
HuR binds to and stabilizes the mRNAs that encode two major pro-inflammatory mediators, the enzymes COX-2 and iNOS. An inducible enzyme, COX-2 catalyzes the conversion of arachidonic acid into prostanoids (including prostaglandins, prostacyclin and thromboxane) in many different cell types. HuR controls COX-2 expression in macrophages, renal mesangial cells, and many cancers, including carcinomas of the colon, breast, ovaries, prostate, larynx, and stomach (63, 64, 120, 121). Also an inducible enzyme, iNOS catalyzes the catalyze the conversion of L-arginine into nitric oxide (NO), a major trigger of inflammation. Interestingly, NO in turn promotes the cytoplasmic localization of HuR and its association with target mRNAs (74). HuR enhances iNOS expression in muscle cells, in hepatocytes, and in carcinomas of the lung and colon (122–124). Besides sharing a common upstream regulatory (HuR), COX-2 and iNOS are functionally interconnected in different ways (125). In addition to inducing the expression of pro-inflammatory factors, HuR inhibits the production of anti-inflammatory proteins such as thrombomodulin (30); this effect was linked to the HuR-mediated disruption of the TM 5′UTR IRES (30).
6.2. HuR function inhibited by anti-inflammatory factors
Some anti-inflammatory cytokines can repress HuR function. IL-10, also known as human cytokine synthesis inhibitory factor (SCIF), is a pleiotropic cytokine that can potently repress inflammation (126, 127). As shown by the Kishore laboratory, IL-10 inhibits inflammation at least partly by repressing the HuR-mediated stabilization of mRNAs encoding proinflammatory cytokines in monocytes and inflammatory cells in the myocardium (128, 129). Similarly, the anti-inflammatory cytokine IL-19 can lower HuR abundance and repress HuR function, thereby reducing the inflammatory response of vascular smooth muscle cells (VSMCs) to injury (130). In a related regulatory paradigm, HuR binds to IL-4 mRNA and promotes IL-4 expression in T-cells (131). Since IL-4 promotes the activation of repair macrophages which secrete IL-10, an inhibitor of inflammation, perhaps IL-4 can function as a negative feedback loop to shut off the production of pro-inflammatory factors by HuR. The biological effects of IL-4 are linked to the activity of transcription factor, signal transducer and activator of transcription 6 (STAT6); like IL-4, IL-13 is encoded by another HuR target mRNA and can also suppress pro-inflammatory responses and activate STAT6 (132, 133).
6.3. Implication of HuR in specific inflammatory diseases
Given its positive influence on the expression of inflammatory proteins, HuR has been implicated in different disease states. The involvement of HuR in rheumatoid arthritis was suggested based on its promotion of TNF-α expression (134, 135), while HuR-regulated COX-2 was associated with the development of rheumatoid and osteoarthritic cartilage (136). The upregulation of COX-2 by HuR in colonic epithelium was also linked to inflammatory bowel disease (137). In patients treated with human immunodeficiency virus (HIV) protease inhibitors (PIs), the chronic inflammatory disease atherosclerosis was linked to the HuR-mediated upregulation of TNF-α and IL-6 in response to HIV PI therapy (113). Also a chronic inflammatory disease, asthma has been linked to HuR function through its upregulation of factors like TNF-α, GM-CSF (138). In response to lipopolisaccharide (LPS), HuR upregulation of toll-like receptor 4 (TLR4) mRNA was linked to hyperplasia of vascular smooth muscle cells in a model of vascular inflammation. (139). Although the vast majority of evidence supports a role for HuR in promoting inflammation, one study of HuR overexpression in mouse macrophages suggests that HuR may be anti-inflammatory (140).
7. HuR IN OTHER DISEASE PROCESSES
Many studies implicating HuR in additional diseases are rapidly emerging. In most of these, HuR elicits phenotypic traits previously described in sections 5 and 6, including the promotion of inflammation, proliferation, angiogenesis, and resistance to apoptosis.
7.1. Virus replication and infection
The alphavirus Sindbis virus expresses many U-rich RNAs and recruitment of HuR to these sequences helps to express viral proteins and is necessary to maintain a high viral yield and a productive viral infection (141). Yeast-two hybrid analysis identified HuR as a protein that interacted with the HIV enzyme reverse transcriptase (RT); this association was found to be necessary for successful HIV infection (142). The Kaposi’s sarcoma herpes virus (KSHV) protein kaposin-B increased the cytoplasmic levels of HuR; in turn, HuR associated with the cellular mRNA encoding PROX1 (prospero homeobox 1), a key protein involved in the reprogramming of lymphatic endothelial cells (143). HuR also associated with the hepatitis C virus RNA, although the consequences of this interaction were not reported (144).
7.2. Cardiovascular disease
HuR was shown to associate with the β(2)-adrenergic receptor mRNA (145). Since HuR also associates with mRNAs that encode angiotensin receptors, iNOS, COX-2, and TNF-α, its putative involvement in cardiovascular diseases such as heart failure, myocardial infarction and hypertension has been long recognized (146). HuR binds to the mRNAs encoding the soluble guanylyl cyclase (sGC)-α1 and sGC-β1 and enhances their expression. In models of hypoxia-induced and spontaneous hypertension, the reduced expression of sGC was proposed to be due to the reduced ability of HuR to form complexes with the sGCα and sGCβ mRNAs (147–149). In an animal model of hypoxic adaptation involving enhanced angiogenesis, the levels of HuR, as well as those of its targets VEGF and HIF-1α were significantly upregulated; these observations prompted the authors to suggest a role for HuR in ischemia (150). The influence of HuR on Bcl-2 expression was also implicated in ischemia-reperfusion injury in the kidney (151). Elevated HuR levels were also detected in several vascular pathologies, including intimal hyperplasia, atherosclerosis, sclerosis of venous graft, and fibromuscular dysplasia (152).
7.3. Neurological pathologies
Neurofibromatosis type 1 (NF1) is an autosomal dominant disease caused by deficiency of the NF1 gene, which encodes the tumor suppressor protein neurofibromin. HuR was reported to interact with the 3′UTR of the NF1 mRNA and was thereby proposed to regulate neurofibromin abundance post-transcriptionally (153).
Amyotrophic lateral sclerosis (ALS, also known as Lou Gehrig’s disease), a neurodegenerative disease caused by the degeneration of motor neurons, has been linked to mutations in copper-zinc superoxide dismutase 1 (SOD1). Mutant SOD1 competes with HuR for binding to the VEGF 3′UTR and leads to sequestration of HuR in RNA aggregates, effectively impeding HuR from enhancing the production of VEGF, a neuroprotective factor (154, 155).
Another neurodegenerative disease, spinal muscle atrophy, is characterized by the loss of alpha motor neurons leading to progressing muscle atrophy. The disease arises from mutation or deletion of the SMN1 gene, which encodes the protein SMN (survival of motor neuron). Recent efforts to develop therapies that compensate for the loss of SMN1, aimed at expressing SMN from the SMN2 gene, have revealed that p38 promotes HuR binding to the SMN mRNA, resulting in its stabilization (156).
Paraneoplastic encephalomyelitis is a disorder that causes inflammation of the central nervous system associated with a distant cancer, often small-cell lung carcinoma. The serologic hallmark of paraneoplastic encephalomyelitis is the presence of anti-Hu auto-antibodies which recognize the three neuronal Hu/ELAV proteins (HuB, HuC, and HuD), but also reacts against HuR (157, 158). As all Hu/ELAV proteins are highly abundant in the nervous system, the pathogenesis of this disorder is considered to be autoimmune.
7.4. Muscular disorders
A number of muscle pathologies have been linked to HuR function. Inclusion body myositis (IBM) is one of a group of muscle diseases known as inflammatory myopathies characterized by chronic, progressive muscle inflammation accompanied by muscle weakness. In muscle fibers from IBM patients, both the poly(A)-binding protein 1 and HuR were found to aggregate in RNA deposits; these observations were suggested to reflect an impairment in mRNA turnover and translation in IBM (159).
Muscle wasting (cachexia) is characterized by an excessive loss in skeletal muscle mass. It is often seen in patients with cancer, AIDS, congestive heart failure, and obstructive lung disease. The onset of cachexia is linked to the activation of transcription factors NF-κB and STAT1 by inflammatory cytokines TNF-α and IFN-γ, causing the transcriptional upregulation of iNOS mRNA. As reported by Gallouzi and colleagues, TNF-α and IFN-γ trigger the association of HuR with iNOS mRNA, enhancing iNOS mRNA stability and promoting iNOS synthesis and NO production (122). NO accelerates the loss of MyoD, likely resulting from a reduction in the transcription of the MyoD gene or the stability of MyoD mRNA mediated by decay-promoting TTR-RBPs. While these observations indicate that HuR participates in cytokine-induced muscle wasting, other studies have shown that HuR associates with MyoD mRNA and promotes MyoD expression during myogenesis (160, 161). Therefore, in response to different extracellular stimuli, HuR can either promote muscle formation (160, 161) or trigger muscle decay (122); these distinct responses may be mediated by specific posttranslational modifications of HuR (section 4) and/or by the influence of other as-yet unknown post-transcriptional factors. It is interesting to note that treatment with NO triggers the association of HuR with numerous target mRNAs (162).
7.5. Lymphoproliferative disease
The X-linked lymphoproliferative disease (XLP) is a rare immunodeficiency condition characterized by frequent childhood mononucleosis triggered by Epstein-Barr virus (EBV), followed by hypogammaglobulinemia and a markedly higher risk of lymphoma and other lymphoproliferative diseases. The SH2D1A gene, which is altered or deleted in XLP patients, encodes the small protein SAP that is expressed in T and NK cells. HuR was shown to interact with the SH2D1A 3′UTR and was proposed to contribute to its stabilization (163).
8. HuR A THERAPEUTIC TARGET? CONCLUDING REMARKS
The heightened function of HuR in virtually all cancers examined to-date suggests that HuR could be a useful marker for cancer diagnosis. Indeed, the levels of HuR, its cytoplasmic abundance, and its binding to mRNAs are all regulated by several factors (Chk2, PKC, CARM1, Cdk1, p38, caspases, microRNAs) implicated in cancer. In addition, HuR appears to be a valuable prognostic indicator, since the vast majority of studies show significant correlations between HuR abundance in cancer and poor patient outcome.
Given HuR’s capacity to promote protein expression programs advantageous to cancer cells (e.g., proliferative, pro-angiogenic, and pro-survival), HuR might also represent a useful therapeutic target. Interventions to modulate HuR kinases could be fruitful, although PKC, Cdc2, Cdk1, and p38 are broad-spectrum kinases which affect many cellular processes. Approaches to decrease HuR levels by small interfering (si)RNA or microRNAs are effective in cultured cells and could be attempted in tumors, provided that they are sufficiently specific. Mukherjee and colleagues recently reported that resveratrol triggered changes in the subcellular localization of HuR, and further implicated HuR in the changes in gene expression elicited by resveratrol (164). Small chemical inhibitors of HuR have also been reported, but their usefulness in organisms remains untested (165, 166).
Besides considering how targeting HuR might inhibit or reduce tumorigenesis, it is important to keep in mind that most studies on HuR and cancer have not examined how HuR affects anticancer therapy. In pancreatic cancer, the elevated presence of cytoplasmic HuR in pancreatic cancer cells paradoxically correlated with better prognosis for patients treated with the standard drug of choice, gemcitabine; the finding that HuR increased the expression of dCK, which metabolizes and thus activates gemcitabine, partly explained why elevated HuR correlated with positive response to therapy (92). Similarly, low levels of HuR were associated with high risk of breast cancer recurrence, although the specific mediators of this effect were not identified (167). Therefore, interventions to reduce HuR function should be designed carefully, since in some cases, the elevated presence of HuR may be advantageous for other therapies. It is possible to envision therapeutic regimens in which it is more appropriate to lower HuR after use of conventional chemotherapies whose effectiveness may rely, at least in part, on functional HuR.
Strategies to inhibit HuR function could also be beneficial for treating chronic inflammatory diseases. The ability of HuR to induce major proinflammatory factors, including TNF-α, IL-6, COX-2, suggests that lowering HuR levels or function might decrease inflammatory conditions. However, thus far, there has been little work assessing directly the usefulness of HuR as a therapeutic target in chronic inflammatory diseases. In one study, HuR was implicated in the response of rheumatoid arthritis patients to the drug infliximab, although only HuR mRNA levels were measured, it is unclear if HuR protein levels and HuR binding to mRNAs encoding inflammatory factors followed the same expression pattern (134). It is also important to remember that TNF-α and other pro-inflammatory factors are necessary for clearing infections, so like with cancer therapy, HuR-directed interventions to avoid tissue damage during chronic inflammation must be devised thoughtfully.
The studies examining the therapeutic potential of HuR in other diseases are also still very limited. Patients receiving immunosuppressive therapy show high cancer incidence. Some of these therapies can mobilize HuR to the cytoplasm causing increases in expression of VEGF and other pro-tumorigenic proteins, suggesting that targeting HuR could lower the development and aggressiveness of cancers in patients undergoing immunosuppressive therapy (168).
In closing, while the use of cultured cells has advanced greatly our understanding of HuR function and influence on proteins associated with cancer, chronic inflammation, and other diseases, more efforts must now focus on mammalian models of disease. HuR overexpression in mouse macrophages suggested a systemic anti-inflammatory role for HuR, but HuR-null thymocytes showed aberrant cell division cycle, activation, selection, and survival (169). A role for HuR in the establishment of a physiologic thymocyte pool was confirmed in another mouse model with inducible global HuR-null phenotype, which showed widespread death of progenitor cells in hematopoietic organs and in the intestinal epithelium (170). While the mouse phenotypes generally agree with HuR’s roles in proliferation and survival, more studies are needed to fully understand the influence of HuR on tumorigenesis, chronic inflammatory diseases, and other human pathologies.
Acknowledgments
M. G. and S. S. are supported by the Intramural Research Program of the National Institute on Aging, NIH.
Abbreviations
- ARE
AU-rich element
- CR
coding region
- TTR-RBP
turnover and translation regulatory RNA-binding protein
- UTR
untranslated region
References
- 1.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 2.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr Opin Genet Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 3.Pullmann R, Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M. Analysis of stability and translation regulatory RBP expression through binding to cognate mRNAs. Mol Cell Biol. 2007;27:6265–6278. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 5.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 6.Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor a mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-a production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 8.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 9.Stoecklin G, Colombi M, Raineri I, Leuenberger S, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T, Moroni C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, De Maria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein. a multifunctional RNA-binding protein. Biochem Soc Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 12.Xu YH, Grabowski GA. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid b-glucosidase and other mRNAs. Mol Genet Metab. 1999;68:441–454. doi: 10.1006/mgme.1999.2934. [DOI] [PubMed] [Google Scholar]
- 13.Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M. Translational repression by RNA-binding protein TIAR. Mol Cell Biol. 2006;26:2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deschenes-Furry J, Perrone-Bizzozero N, Jasmin BJ. The RNA-binding protein HuD: a regulator of neuronal differentiation, maintenance and plasticity. Bioessays. 2006;28:822–833. doi: 10.1002/bies.20449. [DOI] [PubMed] [Google Scholar]
- 18.Pascale A, Amadio M, Quattrone A. Defining a neuron: neuronal ELAV proteins. Cell Mol Life Sci. 2008;65:128–140. doi: 10.1007/s00018-007-7017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 20.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdelmohsen K, Gorospe M. Post-transcriptional regulation of cancer traits by HuR. WIRES RNA. 2010 doi: 10.1002/wrna.4. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durie D, Lewis SM, Liwak U, Kisilewicz M, Gorospe M, Holcik M. RNA-binding protein HuR mediates cytoprotection through stimulation of XIAP translation. Oncogene. 2010 doi: 10.1038/onc.2010.527. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazan-Mamczarz K, Galbán S, López de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 27.Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng Z, Jackson NL, Choi H, King PH, Emanuel PD, Blume SW. Alterations in RNA-binding activities of IRES-regulatory proteins as a mechanism for physiological variability and pathological dysregulation of IGF-IR translational control in human breast tumor cells. J Cell Physiol. 2008;217:172–183. doi: 10.1002/jcp.21486. [DOI] [PubMed] [Google Scholar]
- 30.Yeh CH, Hung LY, Hsu C, Le SY, Lee PT, Liao WL, Lin YT, Chang WC, Tseng JT. RNA-binding protein HuR interacts with thrombomodulin 5′ untranslated region and represses internal ribosome entry site-mediated translation under IL-1 beta treatment. Mol Biol Cell. 2008;19:3812–3822. doi: 10.1091/mbc.E07-09-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang MJ, Ryu BK, Lee MG, Han J, Lee JH, Ha TK, Byun DS, Chae KS, Lee BH, Chun HS, Lee KY, Kim HJ, Chi SG. NF-κB activates transcription of the RNA-binding factor HuR, via PI3K-AKT signaling, to promote gastric tumorigenesis. Gastroenterology. 2008;135:2030–2042. doi: 10.1053/j.gastro.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 33.Jeyaraj SC, Singh M, Ayupova DA, Govindaraju S, Lee BS. Transcriptional control of human antigen R by bone morphogenetic protein. J Biol Chem. 2010;285:4432–4440. doi: 10.1074/jbc.M109.062216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-Ahmadi W, Al-Ghamdi M, Al-Haj L, Al-Saif M, Khabar KS. Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU-rich elements and auto-Regulation. Nucleic Acids Res. 2009;37:3612–3624. doi: 10.1093/nar/gkp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi J, Chang N, Liu X, Guo G, Xue L, Tong T, Gorospe M, Wang W. Reduced nuclear export of HuR mRNA by HuR is linked to the loss of HuR in replicative senescence. Nucleic Acids Res. 2010;38:1547–1558. doi: 10.1093/nar/gkp1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci USA. 2008;105:20297–20302. doi: 10.1073/pnas.0809376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marasa BS, Srikantan S, Martindale JL, Kim MM, Lee EK, Gorospe M, Abdelmohsen K. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging. 2010;2:333–343. doi: 10.18632/aging.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdelmohsen K, Kim MM, Srikantan S, Mercken EM, Brennan SE, Wilson GM, de Cabo R, Gorospe M. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle. 2010;9 doi: 10.4161/cc.9.7.11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo X, Wu Y, Hartley RS. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 2009;6:575–583. doi: 10.4161/rna.6.5.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abdelmohsen K, Srikantan S, Yang X, Lal A, Kim HH, Kuwano Y, Galban S, Becker KG, Kamara D, de Cabo R, Gorospe M. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009;28:1271–1282. doi: 10.1038/emboj.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazroui R, Di Marco S, Clair E, von Roretz C, Tenenbaum SA, Keene JD, Saleh M, Gallouzi IE. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis. J Cell Biol. 2008;180:113–127. doi: 10.1083/jcb.200709030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Roretz C, Gallouzi IE. Protein kinase RNA/FADD/caspase-8 pathway mediates the proapoptotic activity of the RNA-binding protein human antigen R (HuR) J Biol Chem. 2010;285:16806–16813. doi: 10.1074/jbc.M109.087320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beauchamp P, Nassif C, Hillock S, van der Giessen K, von Roretz C, Jasmin BJ, Gallouzi IE. The cleavage of HuR interferes with its transportin-2-mediated nuclear import and promotes muscle fiber formation. Cell Death Differ. 2010;17:1588–1599. doi: 10.1038/cdd.2010.34. [DOI] [PubMed] [Google Scholar]
- 44.Izquierdo JM. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J Biol Chem. 2008;283:19077–19084. doi: 10.1074/jbc.M800017200. [DOI] [PubMed] [Google Scholar]
- 45.Wang H, Molfenter J, Zhu H, Lou H. Promotion of exon 6 inclusion in HuD pre-mRNA by Hu protein family members. Nucleic Acids Res. 2010;38:3760–3770. doi: 10.1093/nar/gkq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallouzi IE, Steitz JA. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science. 2001;294:1895–1901. doi: 10.1126/science.1064693. [DOI] [PubMed] [Google Scholar]
- 47.Fan XC, Steitz JA. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Güttinger S, Mühlhäusser P, Koller-Eichhorn R, Brennecke J, Kutay U. Transportin2 functions as importin and mediates nuclear import of HuR. Proc Natl Acad Sci USA. 2004;101:2918–2923. doi: 10.1073/pnas.0400342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rebane A, Aab A, Steitz JA. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA. 2004;10:590–599. doi: 10.1261/rna.5224304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doller A, Akool el-S, Huwiler A, Müller R, Radeke HH, et al. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cδ elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol Cell Biol. 2008;28:2608–2625. doi: 10.1128/MCB.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doller A, Huwiler A, Müller R, Radeke HH, Pfeilschifter J, et al. Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol Biol Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang W, Fan J, Yang X, Fürer-Galban S, López de Silanes I, et al. AMP-activated kinase regulates cytoplasmic HuR. Mol Cell Biol. 2002;22:3425–3436. doi: 10.1128/MCB.22.10.3425-3436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim HH, Abdelmohsen K, Lal A, Pullmann R, Jr, Yang X, et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008;22:1804–1815. doi: 10.1101/gad.1645808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim HH, Yang X, Kuwano Y, Gorospe M. Modification at HuR(S242) alters HuR localization and proliferative influence. Cell Cycle. 2008;7:3371–3377. doi: 10.4161/cc.7.21.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim HH, Gorospe M. Phosphorylated HuR shuttles in cycles. Cell Cycle. 2008;7:3124–3126. doi: 10.4161/cc.7.20.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doller A, Schlepckow K, Schwalbe H, Pfeilschifter J, Eberhardt W. Tandem phosphorylation of serines 221 and 318 by protein kinase Cδ coordinates mRNA binding and nucleocytoplasmic shuttling of HuR. Mol Cell Biol. 2010;30:1397–1410. doi: 10.1128/MCB.01373-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Doller A, Pfeilschifter J, Eberhardt W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell Signal. 2008;20:2165–2173. doi: 10.1016/j.cellsig.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 59.Amadio M, Bucolo C, Leggio GM, Drago F, Govoni S, Pascale A. The PKCβ/HuR/VEGF pathway in diabetic retinopathy. Biochem Pharmacol. 2010;80:1230–1237. doi: 10.1016/j.bcp.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 60.Lafarga V, Cuadrado A, López de Silanes I, Bengoechea R, Fernandez-Capetillo O, Nebreda AR. p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol Cell Biol. 2009;29:4341–4351. doi: 10.1128/MCB.00210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, Stallcup MR, Laird-Offringa IA. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J Biol Chem. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]
- 62.Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner JB, Furneaux HM. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- 63.Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2001;61:2154–2161. [PubMed] [Google Scholar]
- 64.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.López de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 66.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Topisirovic I, Siddiqui N, Orolicki S, Skrabanek LA, Tremblay M, Hoang T, Borden KL. Stability of eukaryotic translation initiation factor 4E mRNA is regulated by HuR, and this activity is dysregulated in cancer. Mol Cell Biol. 2009;29:1152–1162. doi: 10.1128/MCB.01532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giles KM, Daly JM, Beveridge DJ, Thomson AM, Voon DC, Furneaux HM, Jazayeri JA, Leedman PJ. The 3′-untranslated region of p21WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J Biol Chem. 2003;278:2937–2946. doi: 10.1074/jbc.M208439200. [DOI] [PubMed] [Google Scholar]
- 69.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Galbán S, Kuwano Y, Pullmann R, Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang Y, Liu JO, Lewis SM, Holcik M, Gorospe M. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol Cell Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. 2007;6:1288–1292. doi: 10.4161/cc.6.11.4299. [DOI] [PubMed] [Google Scholar]
- 72.Ishimaru D, Ramalingam S, Sengupta TK, Bandyopadhyay S, Dellis S, Tholanikunnel BG, Fernandes DJ, Spicer EK. Regulation of Bcl-2 expression by HuR in HL60 leukemia cells and A431 carcinoma cells. Mol Cancer Res. 2009;7:1354–1366. doi: 10.1158/1541-7786.MCR-08-0476. [DOI] [PubMed] [Google Scholar]
- 73.Mazan-Mamczarz K, Hagner PR, Corl S, Srikantan S, Wood WH, Becker KG, Gorospe M, Keene JD, Levenson AS, Gartenhaus RB. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene. 2008;27:6151–6163. doi: 10.1038/onc.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, et al. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol Cell Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008;27:253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- 76.Wang X, Liu Y. Regulation of innate immune response by MAP kinase phosphatase-1. Cell Signal. 2007;19:1372–1382. doi: 10.1016/j.cellsig.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bierie B, Moses HL. Tumour microenvironment: TGFβ: the molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 78.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 79.Beck C, Schreiber H, Rowley D. Role of TGF-β in immune-evasion of cancer. Microsc Res Tech. 2001;52:387–395. doi: 10.1002/1097-0029(20010215)52:4<387::AID-JEMT1023>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 80.Tran H, Maurer F, Nagamine Y. Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol Cell Biol. 2003;23:7177–7188. doi: 10.1128/MCB.23.20.7177-7188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Annabi B, Bouzeghrane M, Currie JC, Dulude H, Daigneault L, Garde S, Rabbani SA, Panchal C, Wu JJ, Béliveau R. Inhibition of MMP-9 secretion by the anti-metastatic PSP94-derived peptide PCK3145 requires cell surface laminin receptor signaling. Anticancer Drugs. 2006;17:429–438. doi: 10.1097/01.cad.0000203388.68034.06. [DOI] [PubMed] [Google Scholar]
- 82.Annabi B, Currie JC, Moghrabi A, Béliveau R. Inhibition of HuR and MMP-9 expression in macrophage-differentiated HL-60 myeloid leukemia cells by green tea polyphenol EGCg. Leuk Res. 2007;31:1277–1284. doi: 10.1016/j.leukres.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 83.Dong R, Lu JG, Wang Q, He XL, Chu YK, Ma QJ. Stabilization of Snail by HuR in the process of hydrogen peroxide induced cell migration. Biochem Biophys Res Commun. 2007;356:318–321. doi: 10.1016/j.bbrc.2007.02.145. [DOI] [PubMed] [Google Scholar]
- 84.Heinonen M, Fagerholm R, Aaltonen K, Kilpivaara O, Aittomäki K, et al. Prognostic role of HuR in hereditary breast cancer. Clin Cancer Res. 2007;13:6959–6963. doi: 10.1158/1078-0432.CCR-07-1432. [DOI] [PubMed] [Google Scholar]
- 85.Heinonen M, Bono P, Narko K, Chang SH, Lundin J, et al. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res. 2005;65:2157–2161. doi: 10.1158/0008-5472.CAN-04-3765. [DOI] [PubMed] [Google Scholar]
- 86.Guo X, Hartley RS. HuR contributes to cyclin E1 deregulation in MCF-7 breast cancer cells. Cancer Res. 2006;66:7948–7956. doi: 10.1158/0008-5472.CAN-05-4362. [DOI] [PubMed] [Google Scholar]
- 87.Woo HH, Zhou Y, Yi X, David CL, Zheng W, et al. Regulation of non-AU-rich element containing c-Fms proto-oncogene expression by HuR in breast cancer. Oncogene. 2009;28:1176–1186. doi: 10.1038/onc.2008.469. [DOI] [PubMed] [Google Scholar]
- 88.Heinonen M, Bono P, Narko K, Chang SH, Lundin J, et al. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res. 2005;65:2157–2161. doi: 10.1158/0008-5472.CAN-04-3765. [DOI] [PubMed] [Google Scholar]
- 89.Suswam EA, Nabors LB, Huang Y, Yang X, King PH. IL-1β induces stabilization of IL-8 mRNA in malignant breast cancer cells via the 3′-untranslated region: Involvement of divergent RNA-binding factors HuR, KSRP and TIAR. Int J Cancer. 2005;113:911–919. doi: 10.1002/ijc.20675. [DOI] [PubMed] [Google Scholar]
- 90.Leandersson K, Riesbeck K, Andersson T. Wnt-5a mRNA translation is suppressed by the Elav-like protein HuR in human breast epithelial cells. Nucleic Acids Res. 2006;34:3988–3999. doi: 10.1093/nar/gkl571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Richards NG, Rittenhouse DW, Freydin B, Cozzitorto JA, Grenda D, Rui H, Gonye G, Kennedy EP, Yeo CJ, Brody JR, Witkiewicz AK. HuR status is a powerful marker for prognosis and response to gemcitabine-based chemotherapy for resected pancreatic ductal adenocarcinoma patients. Ann Surg. 2010;252:499–505. doi: 10.1097/SLA.0b013e3181f1fd44. [DOI] [PubMed] [Google Scholar]
- 92.Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, et al. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Upregulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res. 2009;69:4567–4572. doi: 10.1158/0008-5472.CAN-09-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Young LE, Sanduja S, Bemis-Standoli K, Pena EA, Price RL, Dixon DA. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136:1669–1679. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu Z, Luo RZ, Peng H, Rosen DG, Atkinson EN, et al. Transcriptional and posttranscriptional downregulation of the imprinted tumor suppressor gene ARHI (DRAS3) in ovarian cancer. Clin Cancer Res. 2006;12:2404–2413. doi: 10.1158/1078-0432.CCR-05-1036. [DOI] [PubMed] [Google Scholar]
- 95.Erkinheimo TL, Lassus H, Sivula A, Sengupta S, Furneaux H, et al. Cytoplasmic HuR expression correlates with poor outcome and with cyclooxygenase 2 expression in serous ovarian carcinoma. Cancer Res. 2003;63:7591–7594. [PubMed] [Google Scholar]
- 96.Denkert C, Weichert W, Pest S, Koch I, Licht D, et al. Overexpression of the embryonic-lethal abnormal vision-like protein HuR in ovarian carcinoma is a prognostic factor and is associated with increased cyclooxygenase 2 expression. Cancer Res. 2004;64:189–195. doi: 10.1158/0008-5472.can-03-1987. [DOI] [PubMed] [Google Scholar]
- 97.Niesporek S, Kristiansen G, Thoma A, Weichert W, Noske A, et al. Expression of the ELAV-like protein HuR in human prostate carcinoma is an indicator of disease relapse and linked to COX-2 expression. Int J Oncol. 2008;32:341–347. [PubMed] [Google Scholar]
- 98.Sheflin LG, Zou AP, Spaulding SW. Androgens regulate the binding of endogenous HuR to the AU-rich 3′UTRs of HIF-1alpha and EGF mRNA. Biochem Biophys Res Commun. 2004;322:644–651. doi: 10.1016/j.bbrc.2004.07.173. [DOI] [PubMed] [Google Scholar]
- 99.Kojima K, Fujita Y, Nozawa Y, Deguchi T, Ito M. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate. 2010;70:1501–1512. doi: 10.1002/pros.21185. [DOI] [PubMed] [Google Scholar]
- 100.Barbisan F, Mazzucchelli R, Santinelli A, López-Beltran A, Cheng L, et al. Overexpression of ELAV-like protein HuR is associated with increased COX-2 expression in atrophy, high-grade prostatic intraepithelial neoplasia, and incidental prostate cancer in cystoprostatectomies. Eur Urol. 2008;56:105–112. doi: 10.1016/j.eururo.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 101.Cho NP, Han HS, Soh Y, Son HJ. Overexpression of cyclooxygenase-2 correlates with cytoplasmic HuR expression in salivarymucoepidermoid carcinoma but not in pleomorphic adenoma. J Oral Pathol Med. 2007;36:297–303. doi: 10.1111/j.1600-0714.2007.00526.x. [DOI] [PubMed] [Google Scholar]
- 102.Wang J, Zhao W, Guo Y, Zhang B, Xie Q, Xiang D, Gao J, Wang B, Chen Z. The expression of RNA-binding protein HuR in non-small cell lung cancer correlates with vascular endothelial growth factor-C expression and lymph node metastasis. Oncology. 2009;76:420–429. doi: 10.1159/000216837. [DOI] [PubMed] [Google Scholar]
- 103.Hasegawa H, Kakuguchi W, Kuroshima T, Kitamura T, Tanaka S, et al. HuR is exported to the cytoplasm in oral cancer cells in a different manner from that of normal cells. Br J Cancer. 2009;100:1943–1948. doi: 10.1038/sj.bjc.6605084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ido K, Nakagawa T, Sakuma T, Takeuchi H, Sato K, Kubota T. Expression of vascular endothelial growth factor-A and mRNA stability factor HuR in human astrocytic tumors. Neuropathology. 2008;28:604–611. doi: 10.1111/j.1440-1789.2008.00926.x. [DOI] [PubMed] [Google Scholar]
- 105.Sakuma T, Nakagawa T, Ido K, Takeuchi H, Sato K, Kubota T. Expression of vascular endothelial growth factor-A and mRNA stability factor HuR in human meningiomas. J Neurooncol. 2008;88:143–155. doi: 10.1007/s11060-008-9559-8. [DOI] [PubMed] [Google Scholar]
- 106.Mrena J, Wiksten JP, Thiel A, Kokkola A, Pohjola L, et al. Cyclooxygenase-2 is an independent prognostic factor in gastric cancer and its expression is regulated by the messenger RNA stability factor HuR. Clin Cancer Res. 2005;11:7362–7368. doi: 10.1158/1078-0432.CCR-05-0764. [DOI] [PubMed] [Google Scholar]
- 107.Mrena J, Wiksten JP, Kokkola A, Nordling S, Haglund C, et al. Prognostic significance of cyclin A in gastric cancer. Int J Cancer. 2006;119:1897–1901. doi: 10.1002/ijc.21944. [DOI] [PubMed] [Google Scholar]
- 108.Koljonen V, Böhling T, Haglund C, Ristimäki A. Expression of HuR in Merkel cell carcinoma and in normal skin. J Cutan Pathol. 2008;35:10–14. doi: 10.1111/j.1600-0560.2007.00762.x. [DOI] [PubMed] [Google Scholar]
- 109.Kakuguchi W, Kitamura T, Kuroshima T, Ishikawa M, Kitagawa Y, Totsuka Y, Shindoh M, Higashino F. HuR knockdown changes the oncogenic potential of oral cancer cells. Mol Cancer Res. 2010;8:520–8. doi: 10.1158/1541-7786.MCR-09-0367. [DOI] [PubMed] [Google Scholar]
- 110.Sung SC, Kim K, Lee KA, Choi KH, Kim SM, Son YH, Moon YS, Eo SK, Rhim BY. 7-Ketocholesterol upregulates interleukin-6 via mechanisms that are distinct from those of tumor necrosis factor-alpha, in vascular smooth muscle cells. J Vasc Res. 2009;46:36–44. doi: 10.1159/000135663. [DOI] [PubMed] [Google Scholar]
- 111.Gealy C, Denson M, Humphreys C, McSharry B, Wilkinson G, Caswell R. Posttranscriptional suppression of interleukin-6 production by human cytomegalovirus. J Virol. 2005;79:472–485. doi: 10.1128/JVI.79.1.472-485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang JG, Collinge M, Ramgolam V, Ayalon O, Fan XC, Pardi R, Bender JR. LFA-1-dependent HuR nuclear export and cytokine mRNA stabilization in T cell activation. J Immunol. 2006;176:2105–2113. doi: 10.4049/jimmunol.176.4.2105. [DOI] [PubMed] [Google Scholar]
- 113.Zhou H, Jarujaron S, Gurley EC, Chen L, Ding H, Studer E, Pandak WM, Jr, Hu W, Zou T, Wang JY, Hylemon PB. HIV protease inhibitors increase TNF-alpha and IL-6 expression in macrophages: involvement of the RNA-binding protein HuR. Atherosclerosis. 2007;195:e134–143. doi: 10.1016/j.atherosclerosis.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 114.Winzen R, Gowrishankar G, Bollig F, Redich N, Resch K, Holtmann H. Distinct domains of AU-rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen-activated protein kinase or HuR. Mol Cell Biol. 2004;24:4835–4847. doi: 10.1128/MCB.24.11.4835-4847.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tschernatsch MM, Mlecnik B, Trajanoski Z, Zechner R, Zimmermann R. LPL-mediated lipolysis of VLDL induces an upregulation of AU-rich mRNAs and an activation of HuR in endothelial cells. Atherosclerosis. 2006;189:310–317. doi: 10.1016/j.atherosclerosis.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 116.Subramaniam D, Ramalingam S, May R, Dieckgraefe BK, Berg DE, Pothoulakis C, Houchen CW, Wang TC, Anant S. Gastrin-mediated interleukin-8 and cyclooxygenase-2 gene expression: differential transcriptional and posttranscriptional mechanisms. Gastroenterology. 2008;134:1070–1082. doi: 10.1053/j.gastro.2008.01.040. [DOI] [PubMed] [Google Scholar]
- 117.Bandyopadhyay S, Sengupta TK, Spicer EK. PMA induces stabilization of oncostatin M mRNA in human lymphoma U937 cells. Biochem J. 2008;410:177–186. doi: 10.1042/BJ20070311. [DOI] [PubMed] [Google Scholar]
- 118.Atasoy U, Curry SL, López de Silanes I, Shyu AB, Casolaro V, Gorospe M, Stellato C. Regulation of eotaxin gene expression by TNF-alpha and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J Immunol. 171:4369–4378. doi: 10.4049/jimmunol.171.8.4369. [DOI] [PubMed] [Google Scholar]
- 119.Li F, Hu DY, Liu S, Mahavadi S, Yen W, Murthy KS, Khalili K, Hu W. The RNA-binding protein HuR regulates RGS4 mRNA stability in rabbit colonic smooth muscle cells. Am J Physiol Cell Physiol. 2010 doi: 10.1152/ajpcell.00093.2010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sengupta S, Jang BC, Wu MT, Paik JH, Furneaux H, Hla T. The RNA-binding protein HuR regulates the expression of cyclooxygenase-2. J Biol Chem. 2003;278:25227–25233. doi: 10.1074/jbc.M301813200. [DOI] [PubMed] [Google Scholar]
- 121.Cok SJ, Acton SJ, Morrison AR. The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J Biol Chem. 2003;278:36157–36162. doi: 10.1074/jbc.M302547200. [DOI] [PubMed] [Google Scholar]
- 122.Di Marco S, Mazroui R, Dallaire P, Chittur S, Tenenbaum SA, Radzioch D, Marette A, Gallouzi IE. NF-κB-mediated MyoD decay during muscle wasting requires nitric oxide synthase mRNA stabilization, HuR protein, and nitric oxide release. Mol Cell Biol. 2005;25:6533–6545. doi: 10.1128/MCB.25.15.6533-6545.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, Kleinert H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Matsui K, Nishizawa M, Ozaki T, Kimura T, Hashimoto I, Yamada M, Kaibori M, Kamiyama Y, Ito S, Okumura T. Natural antisense transcript stabilizes inducible nitric oxide synthase messenger RNA in rat hepatocytes. Hepatology. 2008;47:686–697. doi: 10.1002/hep.22036. [DOI] [PubMed] [Google Scholar]
- 125.Kim SF, Huri DA, Snyder SH. Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science. 2005;310:1966–1970. doi: 10.1126/science.1119407. [DOI] [PubMed] [Google Scholar]
- 126.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 127.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O’Garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815–3822. [PubMed] [Google Scholar]
- 128.Krishnamurthy P, Rajasingh J, Lambers E, Qin G, Losordo DW, Kishore R. IL-10 inhibits inflammation and attenuates left ventricular remodeling after myocardial infarction via activation of STAT3 and suppression of HuR. Circ Res. 2009;104:e9–18. doi: 10.1161/CIRCRESAHA.108.188243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rajasingh J, Bord E, Luedemann C, Asai J, Hamada H, Thorne T, Qin G, Goukassian D, Zhu Y, Losordo DW, Kishore R. IL-10-induced TNF-alpha mRNA destabilization is mediated via IL-10 suppression of p38 MAP kinase activation and inhibition of HuR expression. FASEB J. 2006;20:2112–2114. doi: 10.1096/fj.06-6084fje. [DOI] [PubMed] [Google Scholar]
- 130.Cuneo AA, Herrick D, Autieri MV. Il-19 reduces VSMC activation by regulation of mRNA regulatory factor HuR and reduction of mRNA stability. J Mol Cell Cardiol. 2010;49:647–654. doi: 10.1016/j.yjmcc.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yarovinsky TO, Butler NS, Monick MM, Hunninghake GW. Early exposure to IL-4 stabilizes IL-4 mRNA in CD4+ T cells via RNA-binding protein HuR. J Immunol. 2010;177:4426–4435. doi: 10.4049/jimmunol.177.7.4426. [DOI] [PubMed] [Google Scholar]
- 132.Casolaro V, Fang X, Tancowny B, Fan J, Wu F, Srikantan S, Asaki SY, De Fanis U, Huang SK, Gorospe M, Atasoy U, Stellato C. Posttranscriptional regulation of IL-13 in T cells. role of the RNA-binding protein HuR. J Allergy Clin Immunol. 2008;121:853–859. doi: 10.1016/j.jaci.2007.12.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kuperman DA, Schleimer RP. Interleukin-4, interleukin-13, signal transducer and activator of transcription factor 6, and allergic asthma. Curr Mol Med. 2008;8:384–392. doi: 10.2174/156652408785161032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sugihara M, Tsutsumi A, Suzuki E, Wakamatsu E, Suzuki T, Ogishima H, Hayashi T, Chino Y, Ishii W, Mamura M, Goto D, Matsumoto I, Ito S, Sumida T. Effects of infliximab therapy on gene expression levels of tumor necrosis factor alpha, tristetraprolin, T cell intracellular antigen 1, and Hu antigen R in patients with rheumatoid arthritis. Arthritis Rheum. 2007;56:2160–2169. doi: 10.1002/art.22724. [DOI] [PubMed] [Google Scholar]
- 135.Suzuki E, Tsutsumi A, Sugihara M, Mamura M, Goto D, Matsumoto I, Ito S, Ikeda K, Ochiai N, Sato Y, Sumida T. Expression of TNF-alpha, tristetraprolin, T-cell intracellular antigen-1 and Hu antigen R genes in synovium of patients with rheumatoid arthritis. Int J Mol Med. 2006;18:273–278. [PubMed] [Google Scholar]
- 136.Nieminen R, Vuolteenaho K, Riutta A, Kankaanranta H, van der Kraan PM, Moilanen T, Moilanen E. Aurothiomalate inhibits COX-2 expression in chondrocytes and in human cartilage possibly through its effects on COX-2 mRNA stability. Eur J Pharmacol. 2008;587:309–316. doi: 10.1016/j.ejphar.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 137.Di Mari JF, Saada JI, Mifflin RC, Valentich JD, Powell DW. HETEs enhance IL-1-mediated COX-2 expression via augmentation of message stability in human colonic myofibroblasts. Am J Physiol Gastrointest Liver Physiol. 2007;293:G719–728. doi: 10.1152/ajpgi.00117.2007. [DOI] [PubMed] [Google Scholar]
- 138.Esnault S, Malter JS. Hyaluronic acid or TNF-alpha plus fibronectin triggers granulocyte macrophage-colony-stimulating factor mRNA stabilization in eosinophils yet engages differential intracellular pathways and mRNA binding proteins. J Immunol. 2003;171:6780–6787. doi: 10.4049/jimmunol.171.12.6780. [DOI] [PubMed] [Google Scholar]
- 139.Lin FY, Chen YH, Lin YW, Tsai JS, Chen JW, Wang HJ, Chen YL, Li CY, Lin SJ. The role of human antigen R, an RNA-binding protein, in mediating the stabilization of toll-like receptor 4 mRNA induced by endotoxin: a novel mechanism involved in vascular inflammation. Arterioscler Thromb Vasc Biol. 2006;26:2622–2629. doi: 10.1161/01.ATV.0000246779.78003.cf. [DOI] [PubMed] [Google Scholar]
- 140.Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G, Kontoyiannis DL. HuR as a negative posttranscriptional modulator in inflammation. Mol Cell. 2005;19:777–789. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 141.Sokoloski KJ, Dickson AM, Chaskey EL, Garneau NL, Wilusz CJ, Wilusz J. Sindbis virus Usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microbe. 2010;8:196–207. doi: 10.1016/j.chom.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lemay J, Maidou-Peindara P, Bader T, Ennifar E, Rain JC, Benarous RL, Liu X. HuR interacts with human immunodeficiency virus type 1 reverse transcriptase, and modulates reverse transcription in infected cells. Retrovirology. 2008;5:47. doi: 10.1186/1742-4690-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yoo J, Kang J, Lee HN, Aguilar B, Kafka D, Lee S, Choi I, Lee J, Ramu S, Haas J, Koh CJ, Hong YK. Kaposin-B enhances the PROX1 mRNA stability during lymphatic reprogramming of vascular endothelial cells by Kaposi’s sarcoma herpes virus. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001046. pii: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Spångberg K, Wiklund L, Schwartz S. HuR, a protein implicated in oncogene and growth factor mRNA decay, binds to the 3′ ends of hepatitis C virus RNA of both polarities. Virology. 2000;274:378–390. doi: 10.1006/viro.2000.0461. [DOI] [PubMed] [Google Scholar]
- 145.Blaxall BC, Pellett AC, Wu SC, Pende A, Port JD. Purification and characterization of beta-adrenergic receptor mRNA-binding proteins. J Biol Chem. 2000;275:4290–4297. doi: 10.1074/jbc.275.6.4290. [DOI] [PubMed] [Google Scholar]
- 146.Misquitta CM, Iyer VR, Werstiuk ES, Grover AK. The role of 3′-untranslated region (3′-UTR) mediated mRNA stability in cardiovascular pathophysiology. Mol Cell Biochem. 2001;224:53–67. doi: 10.1023/a:1011982932645. [DOI] [PubMed] [Google Scholar]
- 147.de Frutos S, Nitta CH, Caldwell E, Friedman J, González Bosc LV. Regulation of soluble guanylyl cyclase-alpha1 expression in chronic hypoxia-induced pulmonary hypertension. role of NFATc3 and HuR. Am J Physiol Lung Cell Mol Physiol. 2009;297:L475–486. doi: 10.1152/ajplung.00060.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Priviero FB, Zemse SM, Teixeira CE, Webb RC. Oxidative stress impairs vasorelaxation induced by the soluble guanylyl cyclase activator BAY 41–2272 in spontaneously hypertensive rats. Am J Hypertens. 2009;22:493–499. doi: 10.1038/ajh.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Klöss S, Rodenbach D, Bordel R, Mülsch A. Human-antigen R (HuR) expression in hypertension: downregulation of the mRNA stabilizing protein HuR in genetic hypertension. Hypertension. 2005;45:1200–1206. doi: 10.1161/01.HYP.0000165674.58470.8f. [DOI] [PubMed] [Google Scholar]
- 150.Avivi A, Shams I, Joel A, Lache O, Levy AP, Nevo E. Increased blood vessel density provides the mole rat physiological tolerance to its hypoxic subterranean habitat. FASEB J. 2005;19:1314–1316. doi: 10.1096/fj.04-3414fje. [DOI] [PubMed] [Google Scholar]
- 151.Ayupova DA, Singh M, Leonard EC, Basile DP, Lee BS. Expression of the RNA-stabilizing protein HuR in ischemia-reperfusion injury of rat kidney. Am J Physiol Renal Physiol. 2009;297:F95–F105. doi: 10.1152/ajprenal.90632.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pullmann R, Jr, Juhaszova M, López de Silanes I, Kawai T, Mazan-Mamczarz K, Halushka MK, Gorospe M. Enhanced proliferation of cultured human vascular smooth muscle cells linked to increased function of RNA-binding protein HuR. J Biol Chem. 2005;280:22819–22826. doi: 10.1074/jbc.M501106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Haeussler J, Haeusler J, Striebel AM, Assum G, Vogel W, Furneaux H, Krone W. Tumor antigen HuR binds specifically to one of five protein-binding segments in the 3′-untranslated region of the neurofibromin messenger RNA. 34. Biochem Biophys Res Commun. 2000;267:726–732. doi: 10.1006/bbrc.1999.2019. [DOI] [PubMed] [Google Scholar]
- 154.Li X, Lu L, Bush DJ, Zhang X, Zheng L, Suswam EA, King PH. Mutant copper-zinc superoxide dismutase associated with amyotrophic lateral sclerosis binds to adenine/uridine-rich stability elements in the vascular endothelial growth factor 3′-untranslated region. J Neurochem. 2009;108:1032–1044. doi: 10.1111/j.1471-4159.2008.05856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lu L, Zheng L, Viera L, Suswam E, Li Y, Li X, Estévez AG, King PH. Mutant Cu/Zn-superoxide dismutase associated with amyotrophic lateral sclerosis destabilizes vascular endothelial growth factor mRNA and downregulates its expression. J Neurosci. 2007;27:7929–7938. doi: 10.1523/JNEUROSCI.1877-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Farooq F, Balabanian S, Liu X, Holcik M, MacKenzie A. p38 Mitogen-activated protein kinase stabilizes SMN mRNA through RNA binding protein HuR. Hum Mol Genet. 2009;18:4035–4045. doi: 10.1093/hmg/ddp352. [DOI] [PubMed] [Google Scholar]
- 157.Nabors LB, Furneaux HM, King PH. HuR, a novel target of anti-Hu antibodies, is expressed in non-neural tissues. J Neuroimmunol. 1998;92:152–159. doi: 10.1016/s0165-5728(98)00196-9. [DOI] [PubMed] [Google Scholar]
- 158.King PH, Redden D, Palmgren JS, Nabors LB, Lennon VA. Hu antigen specificities of ANNA-I autoantibodies in paraneoplastic neurological disease. J Autoimmun. 1999;13:435–443. doi: 10.1006/jaut.1999.0337. [DOI] [PubMed] [Google Scholar]
- 159.Nakano S, Shinde A, Ito H, Ito H, Kusaka H. Messenger RNA degradation may be inhibited in sporadic inclusion body myositis. Neurology. 2005;65:420–425. doi: 10.1212/01.wnl.0000171341.76482.15. [DOI] [PubMed] [Google Scholar]
- 160.van der Giessen K, Di-Marco S, Clair E, Gallouzi IE. RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J Biol Chem. 2003;278:47119–47128. doi: 10.1074/jbc.M308889200. [DOI] [PubMed] [Google Scholar]
- 161.Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Muñoz-Canoves P, Gorospe M, Muñoz A. Role of HuR in skeletal myogenesis through coordinate regulation of muscle differentiation genes. Mol Cell Biol. 2003;23:4991–5004. doi: 10.1128/MCB.23.14.4991-5004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kuwano Y, Rabinovic A, Srikantan S, Gorospe M, Demple B. Analysis of nitric oxide-stabilized mRNAs in human fibroblasts reveals HuR-dependent heme oxygenase 1 upregulation. Mol Cell Biol. 2009;29:2622–2635. doi: 10.1128/MCB.01495-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Okamoto S, Ji H, Howie D, Clarke K, Gullo C, Manning S, Coyle AJ, Terhorst C. Expression of the SH2D1A gene is regulated by a combination of transcriptional and post-transcriptional mechanisms. Eur J Immunol. 2004;34:3176–3186. doi: 10.1002/eji.200324755. [DOI] [PubMed] [Google Scholar]
- 164.Mukherjee N, Lager PJ, Friedersdorf MB, Thompson MA, Keene JD. Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol Syst Biol. 2009;5:288. doi: 10.1038/msb.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat Chem Biol. 2007;3:508–515. doi: 10.1038/nchembio.2007.14. [DOI] [PubMed] [Google Scholar]
- 166.Chae MJ, Sung HY, Kim EH, Lee M, Kwak H, Chae CH, Kim S, Park WY. Chemical inhibitors destabilize HuR binding to the AU-rich element of TNF-alpha mRNA. Exp Mol Med. 2009;41:824–831. doi: 10.3858/emm.2009.41.11.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ortega AD, Sala S, Espinosa E, González-Barón M, Cuezva JM. HuR and the bioenergetic signature of breast cancer: a low tumor expression of the RNAbinding protein predicts a higher risk of disease recurrence. Carcinogenesis. 2008;29:2053–2061. doi: 10.1093/carcin/bgn185. [DOI] [PubMed] [Google Scholar]
- 168.Basu A, Datta D, Zurakowski D, Pal S. Altered VEGF mRNA stability following treatments with immunosuppressive agents. implications for cancer development. J Biol Chem. 2010;285:25196–25202. doi: 10.1074/jbc.M110.119446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Papadaki O, Milatos S, Grammenoudi S, Mukherjee N, Keene JD, et al. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J Immunol. 2009;182:6779–6788. doi: 10.4049/jimmunol.0900377. [DOI] [PubMed] [Google Scholar]
- 170.Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, et al. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J Clin Invest. 2009;119:3530–3543. doi: 10.1172/JCI38263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zou T, Mazan-Mamczarz K, Rao JN, Liu L, Marasa BS, Zhang AH, Xiao L, Pullmann R, Gorospe M, Wang JY. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem. 2006;281:19387–19394. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- 172.Zhao J, Chen J, Lu B, Dong L, Wang H, Bi C, Wu G, Guo H, Wu M, Guo Y. TIP30 induces apoptosis under oxidative stress through stabilization of p53 messenger RNA in human hepatocellular carcinoma. Cancer Res. 2008;68:4133–4141. doi: 10.1158/0008-5472.CAN-08-0432. [DOI] [PubMed] [Google Scholar]
- 173.Danilin S, Sourbier C, Thomas L, Rothhut S, Lindner V, Helwig JJ, Jacqmin D, Lang H, Massfelder TT. von Hippel-Lindau tumor suppressor gene-dependent mRNA stabilization of the survival factor parathyroid hormone-related protein in human renal cell carcinoma by the RNA-binding protein HuR. Carcinogenesis. 2009;30:387–396. doi: 10.1093/carcin/bgn275. [DOI] [PubMed] [Google Scholar]
- 174.Saunus JM, French JD, Edwards SL, Beveridge DJ, Hatchell EC, Wagner SA, Stein SR, Davidson A, Simpson KJ, Francis GD, Leedman PJ, Brown MA. Posttranscriptional regulation of the breast cancer susceptibility gene BRCA1 by the RNA binding protein HuR. Cancer Res. 2008;68:9469–9478. doi: 10.1158/0008-5472.CAN-08-1159. [DOI] [PubMed] [Google Scholar]
- 175.Pryzbylkowski P, Obajimi O, Keen JC. Trichostatin A and 5 Aza-2′ deoxycytidine decrease estrogen receptor mRNA stability in ER positive MCF7 cells through modulation of HuR. Breast Cancer Res Treat. 2008;111:15–25. doi: 10.1007/s10549-007-9751-0. [DOI] [PubMed] [Google Scholar]
- 176.Licata LA, Hostetter CL, Crismale J, Sheth A, Keen JC. The RNA-binding protein HuR regulates GATA3 mRNA stability in human breast cancer cell lines. Breast Cancer Res Treat. 2010;122:55–63. doi: 10.1007/s10549-009-0517-8. [DOI] [PubMed] [Google Scholar]
- 177.Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 2005;24:1852–1862. doi: 10.1038/sj.emboj.7600661. [DOI] [PMC free article] [PubMed] [Google Scholar]