SUMMARY
Chlamydia trachomatis is a leading cause of genital and ocular infections for which no vaccine exists. Upon entry into host cells, C. trachomatis resides within a membrane bound compartment—the inclusion--and secretes inclusion membrane proteins (Incs) that are thought to modulate the host-bacterium interface. To expand our understanding of Inc function(s), we subjected putative C. trachomatis Incs to affinity purification-mass spectroscopy (AP-MS). We identified Inc-human interactions for 38/58 Incs with enrichment in host processes consistent with Chlamydia’s intracellular lifecycle. There is significant overlap between Inc targets and viral proteins, suggesting common pathogenic mechanisms among obligate intracellular microbes. IncE binds to sorting nexins (SNXs) 5/6, components of the retromer, resulting in SNX5/6 relocalization to the inclusion membrane and enhanced inclusion membrane tubulation. Depletion of retromer components enhances progeny production, revealing that retromer restricts Chlamydia infection. This study demonstrates the value of proteomics in unveiling host-pathogen interactions in genetically challenging microbes.
Graphical abstract

Introduction
Intracellular pathogens that replicate within a membrane-bound compartment employ secreted virulence factors to subvert the host and facilitate survival. Decoding these interactions has been especially difficult for genetically challenging organisms, such as Chlamydiae. These obligate intracellular pathogens are important causes of human disease for which no effective vaccine exists. C. trachomatis is the major cause of non-congenital blindness worldwide and a leading cause of sexually transmitted diseases and non-congenital infertility in Western countries (Mandell et al., 2010). C. pneumoniae is an important cause of respiratory infections and is linked to a number of chronic diseases (Leonard and Borel, 2014). Although treatable with antibiotics, no drug is cost-effective enough for widespread elimination of disease in developing nations.
All Chlamydiae share a common intracellular life cycle, alternating between an infectious, spore-like elementary body (EB), and a non-infectious, metabolically active reticulate body (RB) (reviewed in (Bastidas et al., 2013)). Upon entry into non-phagocytic cells, the EB resides within a membrane bound compartment—the inclusion—and quickly diverges from the canonical endo-lysosomal pathway. The EB differentiates into an RB and replication commences. After replicating within the ever-enlarging inclusion over 24–72 hrs, the RB redifferentiates to an EB and is then released, ready to infect neighboring cells.
How Chlamydia establishes its replicative niche is incompletely understood. Chlamydia manipulates the actin cytoskeleton and microtubule-based motors, obtains nutrients, interacts with numerous host cell organelles, and inhibits the innate immune system, autophagy, and programmed cell death (Bastidas et al., 2013). However, the Chlamydia factors and host cell proteins that mediate these interactions are largely unknown.
Despite their small genome size, Chlamydia are estimated to encode a disproportionate number of secreted virulence effectors, ~10–15% of their genome (Betts-Hampikian and Fields, 2010). A large subset of effectors, termed inclusion membrane proteins (Incs), are translocated by the Type III secretion system and inserted into the inclusion membrane (Moore and Ouellette, 2014). Their defining feature is the presence of one or more unique bilobed domain, typically composed of two closely spaced transmembrane regions separated by a short loop (Bannantine et al., 2000) (Figure 1A). Once inserted into the inclusion membrane, Incs are predicted to extend their termini into the host cytoplasm (Rockey et al., 2002), ideally positioning them at the host-pathogen interface. Given the only recent and limited ability to genetically modify Chlamydia, a detailed understanding of the roles of Incs during infection has been challenging. Indeed, only a few host-binding partners of Incs have been identified (Moore and Ouellette, 2014). Furthermore, Incs share little homology to each other or to known proteins, providing limited insight to their functions (Dehoux et al., 2011; Lutter et al., 2012).
Figure 1. Constructing the Chlamydia Inc-Human Interactome.
(A) Predicted topology of Inc proteins in the Chlamydia inclusion membrane.
(B) Workflow summary.
(C) Overlap of Chlamydia prey with previously published AP-MS interactomes of HIV (Jager et al., 2011), HCV (Ramage et al., 2015), and KSHV (Davis et al., 2014). p-values determined by the hyper-geometric test.
(D) Inc-Virus prey overlap. Parentheses contain the number of prey shared between Chlamydia Incs and HIV, KSHV and/or HCV.
(E) Bar graph depicting the percentage of Inc prey. For the 354 prey identified for 38 Incs (“All”), 54% of prey were affinity purified by core Incs while 46% of prey were affinity purified by noncore Incs. Of the 98 prey shared by Chlamydia Incs and one or more virus (“with viral overlap”), there was a statistically significant increase in core Incs that overlap with viral prey (68%) versus noncore Incs (32%). There was no significant change in the percentage of prey bound to core versus noncore Incs for the 294 prey not shared with viruses (“without viral overlap”). p< 0.001 determined by the hyper-geometric test. See also Figure S1 and Table S1 and S3.
We used large-scale affinity purification/mass spectrometry (AP-MS) to comprehensively identify protein-protein interactions (PPIs) between C. trachomatis Incs and the host proteome to decode mechanisms by which this pathogen establishes its privileged intracellular niche. Our analysis has uncovered a wealth of previously unidentified Chlamydia Inc-host interactions. We analyzed in detail the interaction between IncE, an early expressed Inc of unknown function, and the SNX-BAR proteins, a subset of retromer components. We found that IncE directly binds the PX-domains of SNX5/6, that IncE is sufficient to disrupt retromer trafficking, and that retromer restricts C. trachomatis infection. Our study underscores the power of using comprehensive proteomics to study host-pathogen interactions, particularly for genetically challenging organisms such as Chlamydia.
RESULTS
AP-MS Identifies High Confidence Inc-Human PPIs
We cloned nearly all (58) of the 62 predicted C. trachomatis Incs (Dehoux et al., 2011; Lutter et al., 2012) from two C. trachomatis orfeomes (Roan and Starnbach, 2006; Sisko et al., 2006), including the full length proteins and/or the predicted cytoplasmic domains. Twenty-four of these Incs are evolutionarily conserved among 5 or more Chlamydia species (“core” Incs) (Dehoux et al., 2011; Griffiths et al., 2006; Lutter et al., 2012). A total of 78 Inc constructs were fused to Strep-tags, transiently expressed in HEK293T cells, and affinity purified (AP) over Strep-Tactin beads. Entire eluates were subjected to MS (Figure 1B). All APs were performed in at least triplicate and confirmed by immunobloting with an anti-Strep antibody and silver staining (data not shown). From over 250 AP-MS runs, we identified 30,924 PPIs (Table S1), representing 2,982 unique human proteins.
A set of high confidence PPIs was established by analyzing the complete data set using two algorithms: MiST (Jager et al., 2011) and CompPASS (Sowa et al., 2009) (Table S1). Using stringent criteria (MiST score ≥ 0.7 or the top 1% of CompPASS scores), we identified 354 high confidence Inc-human PPIs, representing 335 unique host binding partners for 38/58 Incs (Table S1, Figure S1). Importantly, IncD and CT228 co-eluted their known human targets, CERT (Derre et al., 2011) and MYPT1 (Lutter et al., 2013), respectively, as the highest confidence PPI (Table S1). These findings establish the validity of our scoring system and demonstrate that our pipeline correctly identifies biologically confirmed PPIs for transmembrane proteins such as the Incs.
Constructing a Comprehensive Inc-Human PPI Network
We assembled an Inc-human PPI network using two databases of known human-human protein interactions (CORUM (Ruepp et al., 2010) and STRING (Franceschini et al., 2013)) to identify multi-protein complexes and potential connections between Incs (Figure 2). Within this subset, we found individual Incs that associate with several members of multi-protein complexes, including retromer, COP9 signalosome, condensin II, GINS, and dynactin. We also observed numerous examples of two or more individual Incs that interact with the same host complex. For example, five Incs (CT005, CT556, CT195, CT058, and CT819) interacted with members of the TIM-TOM complex, host machinery responsible for mitochondrial protein import (Bauer et al., 2000). Three Incs (CT135, CT565 and CT357) each bind to different subunits of the vacuolar ATPase, a multi-protein enzyme that controls acidification of intracellular organelles (Marshansky and Futai, 2008). We also identified several Incs that target proteins involved in ubiquitination, consistent with a recent report that C. trachomatis remodels the host proteome (Olive et al., 2014). Finally, we found a subset of host proteins (RTN4, VKORC1L1, CPVL, LRRC59, WDR6 VPRBP and RNH1) that were each targeted by two individual Incs, suggesting that modulating these targets may be critical for Chlamydia’s intracellular survival.
Figure 2. Network Representation of the Inc-Host Interactome.
The high-confidence Inc-host network contains 38 Inc proteins (core Incs, dark grey; non-core Incs, light blue) and 335 unique human prey (light grey). Inc-human interactions (blue lines) were identified by AP-MS. Interactions between human proteins (dark grey lines) were curated from CORUM and STRING databases. Inc human prey in common with HIV, KSHV, or HCV prey (Davis et al., 2014; Jager et al., 2011; Ramage et al., 2015) are outlined in red. A subset of identified host complexes or proteins with similar functions are labeled. See also Table S2.
The Inc-Human Interactome Predicts Inc Functions During Chlamydia Development
Analysis of the entire dataset with gene ontology (GO) and protein database (KEGG and PFAM) terms revealed that Inc-host protein interactions were enriched for many compartments and pathways consistent with Chlamydia’s intracellular life cycle (Figures 3, S2, S3 and Table S2). For example, we found increased representation for host proteins that localize to the ER, Golgi, mitochondria, endosomes, lysosomes, and actin cytoskeleton, all of which associate with the C. trachomatis inclusion (Bastidas et al., 2013). We also identified host targets enriched for biological processes and molecular functions known to be modulated during infection, such as endocytosis, ubiquitination, apoptosis, cell cycle/division, and DNA damage/repair (Bastidas et al., 2013).
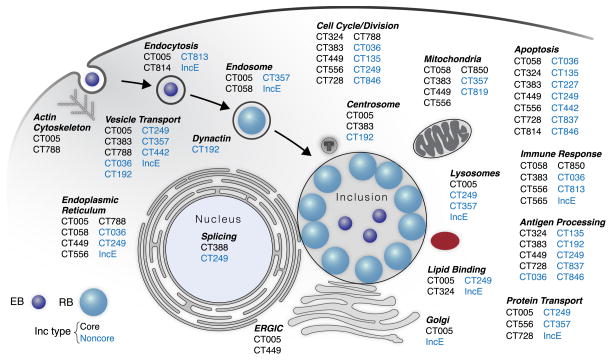
Figure 3. Predicted Functions for Specific Incs.
Schematic diagram of the Chlamydia developmental cycle, indicating predicted functions of putative Incs derived from GO, KEGG, and PFAM enrichment terms from our entire PPI dataset (see Table S2). Core Incs, black; Noncore Incs, blue. We note that some predicted Incs have not been observed on the inclusion membrane at 24 hpi(Li et al., 2008, Dehoux, 2011 #3312). See also Table S1 and Figures S2 and S3.
Our analysis uncovered cellular processes that may be modulated by Chlamydia, such as chromosome condensation (Condensin II complex), splicing (SMN complex), and retromer trafficking (SNXs) (Figures 2, S2, and Table S2), as well as with DNA/RNA sensors (Figure S3 and Table S2) involved in the cytosolic surveillance response. Our PPI dataset allows us to now associate Incs to specific host cell processes important for infection (Figure 3).
Chlamydial and Viral Effectors Target Common Host Proteins
To determine whether diverse intracellular pathogens target similar host processes, we compared our Inc-human interactome to three recently assembled virus-human interactomes: HIV (Jager et al., 2011), KSHV (Davis et al., 2014) and HCV (Ramage et al., 2015). These viral interactomes were derived using the same pipeline that we employed, providing an opportunity for cross-pathogen analyses. We found a significant overlap in high-confidence prey between Chlamydia and each of the three viruses (Figure 1C and Table S3). We identified 98 high confidence C. trachomatis prey shared with one or more virus, 10 shared with two or more viruses, and 2 shared with all 3 viruses (Figure 1D). Host complexes or pathways targeted by Chlamydia and one or more virus included lysosomal ATPases, DNA repair, the TIM-TOM complex, dynactin complex, ubiquitination, and Golgi trafficking (Figure 2 and Table S3). Strikingly, we found core Incs were more likely to overlap with viral targets compared to non-core Incs (p < 0.001; Figure 1E and Table S3), suggesting that diverse intracellular pathogens target overlapping host proteins.
IncE Interacts with Retromer-Associated Sorting Nexins (SNXs)
Our interactome revealed that IncE was enriched for retromer-associated proteins, including SNX1, SNX2, SNX5 and SNX6 (SNX1/2/5/6) (Figure S2, Figure 2, and Table S2). IncE, located in an operon together with IncD, IncF, and IncG, is expressed within the first 2 hrs of C. trachomatis infection (Scidmore-Carlson et al., 1999), but its function(s) and host binding partners were unknown. The retromer complex is composed of the vacuolar protein sorting (VPS) trimer and a heterodimer of SNX-BAR proteins, SNX1/2-SNX5/6 (Cullen and Carlton, 2012; Teasdale and Collins, 2012). SNX-BAR proteins are comprised of a phosphoinositide-binding Phox homology (PX) domain, which functions in endosomal membrane trafficking, membrane remodeling, and organelle motility (Cullen and Korswagen, 2012; Seaman, 2012), and a Bin-amphiphysin-Rvs (BAR) domain, which both senses and induces membrane curvature (Teasdale and Collins, 2012). By inducing tubulation and then recruiting additional factors that lead to scission and formation of vesicles, the retromer SNX-BARS facilitate sorting of protein receptors between endosomes and the trans-Golgi network (Cullen and Carlton, 2012; Seaman, 2012). All four retromer SNX-BARs (SNX1/2/5/6) were identified as very high confidence targets of IncE (MiST > 0.96; top 0.002% of CompPASS) (Table S1).
We confirmed that IncE interacts specifically with retromer SNX-BARs in vivo. Endogenous retromer SNX-BARs co-affinity purified with transiently expressed IncE, but not with IncD, IncF, or IncG (Figures 4A and S4A). In C. trachomatis-infected cells, IncE co-immunoprecipitated with endogenous SNX6 (Figure 4B) or with transfected FLAG-SNX5, but not transfected FLAG-SNX14 (Figure 4C), revealing that IncE specifically binds SNX5/6 in vivo.
Figure 4. IncE Interacts with Retromer SNX-BARs in vivo and in vitro.
(A) Affinity purifications of HEK293T cells transiently expressing Strep-tagged Incs and immunblotted with the indicated antibodies.
(B and C) IncE interacts with endogenous SNX6 (B) and transiently expressed SNX5-FLAG (C) in vivo. Immunoblot analysis of immunoprecipitations with anti-SNX6, -FLAG or -goat IgG in HeLa cells uninfected (−) or infected (+) with C. trachomatis (C.t.) for 24 hrs. Input represents 1% of lysates used for immunoprecipitation. Immunoblots are representative of 3 independent experiments. Untransfected, UT.
(D) Schematic of IncE deletion constructs. The predicted transmembrane (TM) domains are shaded in black, the predicted N- and C-terminal cytosolic domains are shaded in grey. The numbers refer to amino acids. Constructs that co-affinity purify with SNX-BARs are indicated.
(E) Affinity purifications of Strep-tagged IncE deletion constructs transiently expressed in HEK293T cells.
(F) IncE101-132 binds the PX domains of SNX5 and SNX6 in vitro. Purified 6xHis-MBP-Inc-Strep protein was immobilized to Strep-Tactin beads and incubated with the indicated purified 8xHis-SNXPX, subjected to SDS-PAGE and visualized by Coomassie Blue stain. Arrow, IncE or IncD. Arrowhead, SNXPX. *, Strep-Tactin. Molecular weight markers are indicated. See also Figure S4.
We tested informative deletion mutants of IncE-Strep (Figure 4D) by transient expression in HEK293T cells and found that only mutants containing the IncE C-terminal cytoplasmic domain (IncE101-132) co-affinity purified with retromer SNX-BARs (Figures 4D, 4E, and S4A). AP-MS of IncE101-132 also identified retromer SNX-BARs as the highest confidence PPIs (Table S1). Finally, IncE101-132-EGFP co-localized with endogenous SNX1, 2, 6, and transiently expressed FLAG-SNX5 in endosomes, as visualized by confocal microscopy (Figure S4C). Together, these results show that IncE101-132 is both necessary and sufficient to interact specifically with retromer SNX-BARs.
IncE101-132 Binds Directly to the PX Domains of SNX5 and SNX6
To determine whether IncE101-132 binds directly to one or more retromer SNX-BAR, we performed in vitro pulldowns with 6xHis-MBP-IncE-Strep and 8xHis- or 6xHis-MBP-SNXs individually purified from E. coli. IncE101-132 directly bound to the PX domains of SNX5 and SNX6, but failed to bind the corresponding PX domains of SNX1 or SNX2. The C-terminal cytoplasmic domain of IncD, IncD95-141, did not bind the PX domains of retromer SNX-BARs (Figure 4F). Neither IncE101-132 nor IncD95-141 bound the BAR domains of retromer SNX-BARs in vitro (Figure S4D). Thus, IncE101-132 is sufficient to bind SNX5/6 PX domains in the absence of other human proteins. PSIPRED protein sequence analysis revealed that IncE101-132 contains a putative β-hairpin structure (Figure S4E). In vitro pulldowns of IncE101-132 truncations (Figure S4E) demonstrated that the β-hairpin region was required for SNX5 binding (Figure S4F), suggesting that this secondary structure may play a role in the direct binding of IncE to SNX5/6.
Retromer SNX-BARs Colocalize with IncE on the Inclusion Membrane
Retromer SNX-BARs primarily colocalize with PtdIns(3)-rich endosomal membranes (Cullen and Carlton, 2012; Seaman, 2012). Confocal microscopy of C. trachomatis-infected cells stained with antibodies to SNXs and to IncE revealed that at 6 and 24 hrs post-infection (hpi), endogenous SNX1, 2, 6 and transfected FLAG-SNX5 colocalized with IncE on the inclusion membrane (Figures 5A, S5B, S5C, and data not shown). We also observed IncE on tubules emanating from inclusions that were positive for SNX1, SNX2, FLAG-SNX5, SNX6, and IncA (Figures 5A, S5A–C). In contrast, SNX27, a retromer-associated SNX involved in endosome-to-plasma membrane cargo recycling (Lauffer et al., 2010), was not recruited to the inclusion, but instead remained localized in punctate structures (Figures 5B). Thus, Chlamydia inclusions and tubules specifically associate with retromer SNX-BARs.
Figure 5. IncE Colocalizes with Retromer SNX-BARs on the Inclusion.
(A–C) HeLa cells were infected with C. trachomatis (C.t.) for 24 hrs and analyzed by confocal microscopy for the localization of (A) endogenous SNX1/2/6 or transfected SNX5-FLAG and IncE, (B) SNX27-EGFP (pseudocolored red) and IncE or (C) endogenous SNX1 and VPS35. Panels are (A) single Z-slices or (B and C) maximum intensity projections of 0.3 μm Z-slices. I, inclusion. N, nucleus. UI, uninfected. C.t., C. trachomatis-infected. Arrowheads point to tubules. Scale bar = 10 μm. See also Figure S5.
Retromer functions in trafficking cargo-containing vesicles along microtubules to the trans-Golgi through the interaction of SNX6 with dynactin subunit 1 (Hong et al., 2009). To determine whether Chlamydia inclusion tubulation requires microtubule polymerization, infected cells were exposed to nocodazole, and tubules were examined by confocal microscopy. Microtubules were required for the formation of SNX and IncE-positive tubules at 6 hpi (Figure S5B) and 24 hpi (Figure S5C). However, microtubule polymerization was not required for SNX recruitment to inclusions (Figures S5B and S5C), consistent with our results demonstrating that IncE directly binds to and likely recruits SNX5/6 to the inclusion.
Chlamydia Inclusions Do Not Stably Associate with the VPS Subcomplex
In addition to SNX heterodimers, retromer is also associated with the tripartite cargo recruitment complex composed of VPS26A, VPS29, and VPS35 (Seaman, 2012). Intriguingly, while AP-MS analysis demonstrated that retromer SNX-BARs co-purified with IncE, the VPS subunits and other retromer-associated SNXs (Table S1, Fig 4E). Confocal microscopy of C. trachomatis-infected cells demonstrated that, in contrast to the robust recruitment of SNX-BARs to the inclusion membrane and tubules, VPS35 (Figure 5C) and GFP-VPS29 (data not shown) remained in punctate structures. Indeed, SNX1 and VPS35 colocalization was significantly decreased in Chlamydia-infected cells (Figure S5D, p < 0.0001). Total protein levels of retromer SNX-BARS were unchanged during infection (Figure S5E), suggesting that SNX-BARs relocalize from VPS35-positive compartments to the inclusion membrane.
SNX1 is recruited to early Salmonella-containing vacuoles (SCVs) (Bujny et al., 2008), so we assessed whether other retromer components are recruited to SCVs. We confirmed SNX1 recruitment (Figure S5F) and also found that SNX2 and VPS35 clearly decorated SCVs and associated tubules (Figures S5G and S5H). Thus, in contrast to endosomal membrane tubules or SCVs, Chlamydia inclusions and tubules do not maintain a stable association with the VPS complex.
IncE is Sufficient to Recruit Retromer SNX-BARs and Induce Inclusion Tubulation
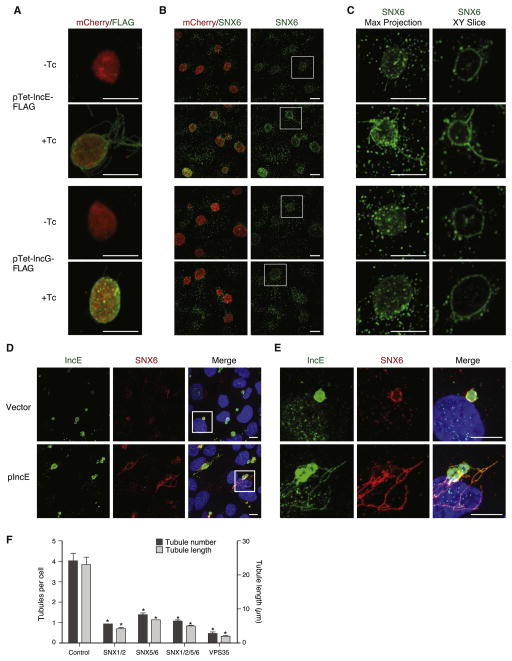
BAR domain-containing proteins form a helical coat, which impose membrane curvature and remodel membranes into tubular profiles (Frost et al., 2008). Based on our observation that retromer SNX-BARs co-localized with IncE on Chlamydia inclusion membrane tubules, we predicted that overexpression of IncE would be sufficient to enhance inclusion tubulation. Therefore, we transformed C. trachomatis serovar L2, a strain with lower baseline levels of tubulation compared to serovar D, with plasmids engineered to express C-terminally FLAG-tagged IncE (pTet-IncE-FLAG) or IncG (pTet-IncG-FLAG) under a tetracycline (Tc)-inducible promoter. As expected, upon induction with Tc, IncE-FLAG and IncG-FLAG were readily detected on the inclusion membrane and tubules (Figure 6A). Importantly, induction of IncE-FLAG was sufficient to enhance recruitment of SNX6 to the inclusion (Figure 6B and 6C) and to enhance formation of IncA-positive (Figure S6A) and SNX6-positive tubules (Figure 6B) compared to IncG-FLAG or uninduced controls. We also observed increased SNX6 recruitment and SNX6-positive tubules in C. trachomatis serovar L2 transformed with a vector expressing untagged IncE (pIncE) under control of its native promoter compared to C. trachomatis transformed with empty vector (Figure 6D and 6E). IncE expression in C. trachomatis-transformed strains was verified by immunoblot analysis (Figures S6B and S6C). We conclude that IncE expression is sufficient to enhance SNX6 inclusion recruitment and inclusion tubulation.
Figure 6. IncE is Sufficient to Enhance SNX-BAR Recruitment and Inclusion Tubulation.
(A–C) HeLa cells were infected with C. trachomatis serovar L2 transformed with pTet-IncE-FLAG or pTet-IncG-FLAG. As indicated, Tc was added at 1 hpi. At 20 hpi, cells were analyzed by confocal microscopy for (A) FLAG and (B and C), SNX6. (A and B) Maximal intensity projections of 0.2 μm Z-slices. Panel (C) shows maximal intensity projections and single XY slices from the boxed region in panel (B). Scale bar = 10 μm.
(D and E) HeLa cells were infected with C. trachomatis serovar L2 transformed with empty vector or pIncE. At 15 hpi, cells were fixed and stained with anti-SNX6, IncE, and DAPI. Panel (E) shows enlargement of boxed region of Panel (D). All images are maximum intensity projections. Scale bar = 10 μm.
(F) Quantitation of inclusion tubules per cell (dark grey bars) and length of the longest tubule per cell (light grey bars) (μm) in infected HeLa cells depleted of the indicated retromer components by siRNA. Data are mean +/− SEM from ≥3 independent experiments. *p < 0.001 compared to corresponding control, unpaired (two-tailed) t-test. See also Figure S6.
We used siRNA depletion to determine if retromer function is required for inclusion tubulation. Due to functional redundancy of retromer SNX-BARs (Seaman, 2012), we simultaneously depleted cells of SNX1 and SNX2, SNX5 and SNX6, all four SNXs, or VPS35 (Figure S6D). Depletion of any retromer component decreased both the length and number of inclusion tubules (Figures 6F and S6E), consistent with our model that recruitment of SNX-BARS enhances inclusion tubulation.
IncE Disrupts Retromer-Dependent Trafficking of the Cation-Independent Mannose-6-Phosphate Receptor (CI-MPR)
Since IncE bound SNX5/6 in vitro (Figure 4F), and C. trachomatis infection caused relocalization of SNX5/6 from endosomes to the inclusion membrane (Figures 5A), we tested whether IncE could interfere with retromer-dependent trafficking. For these experiments, we transfected cells with IncE101-132-EGFP, or, as a control, IncD95-141-EGFP, and analyzed localization of the retromer trafficked CI-MPR, a well-established assay for retromer function (Arighi et al., 2004; Wassmer et al., 2007). Compared to untransfected cells or transfection with IncD95-141, transfection with IncE101-132-EGFP changed the distribution of CI-MPR from juxtanuclear TGN46 positive compartments to a more disperse collection of large vesicles (Figure 7A). Concomitantly, there was decreased co-localization between CI-MPR and TGN46 (Figure 7B). Instead, CI-MPR largely overlapped with VPS35 and IncE101-132-EGFP (Figure 7C), indicating that IncE101-132-EGFP expression is sufficient to maintain CI-MPR in retromer-containing compartments, thereby disrupting efficient CI-MPR trafficking to the trans-Golgi.
Figure 7. IncE101-132-EGFP Disrupts Retromer Trafficking and Retromer Depletion Restricts C. trachomatis Infection.
(A) Localization of endogenous CI-MPR is perturbed in HeLa cells transiently transfected with IncE101-132-EGFP, but not IncD95-141-EGFP, compared to untransfected (UT). Cells were fixed and stained with antibodies to TGN46, CI-MPR, and costained with DAPI. Shown are single Z-slices from confocal images. Scale bar = 10 μm.
(B) Quantitation of TGN46 colocalization with CI-MPR in cells expressing IncE101-132-EGFP or IncD95-141-EGFP. Mean Pearson’s correlation coefficients from 2 two independent experiments (n~20 cells per experiment). *p < 0.05; one-way ANOVA, post hoc Tukey test.
(C) CI-MPR remains localized to VPS35-positive compartments in cells expressing IncE101-132-EGFP, but not IncD95-141-EGFP. HeLa cells were fixed and stained antibodies to VPS35, CI-MPR, and costained with DAPI. Shown are single Z-slices from confocal images. White arrows, VPS35-positive compartment; Yellow arrows, CI-MPR-positive compartment. Left panel (Merge), scale bar = 5 μm. Right panels show enlargements of boxed areas. Scale bar = 2 μm.
(D and E) Quantitation of infectious progeny (D) or primary infection (inclusion formation) (E) in HeLa cells infected with C. trachomatis serovar D following depletion of indicated retromer components. Data are mean +/− SEM from = 3 independent experiments. *p < 0.05 compared to control, unpaired (two-tailed) t-test.
Retromer SNX-BARs Restrict C. trachomatis Infection
As IncE101-132 expression disrupted retromer trafficking, we tested the hypothesis that retromer may be detrimental to C. trachomatis infection. Depletion of SNX5/6 or all four retromer SNX-BARs significantly enhanced production of infectious progeny (Figure 7D), suggesting that retromer SNX-BARs restrict C. trachomatis infection. While we observed a trend towards enhanced progeny with VPS35 depletion, it did not reach statistical significance (p = 0.0574). Enhanced progeny could not be explained by an increase in primary infection; in fact, we noted a significant decrease in primary inclusion formation upon SNX1/2 depletion (Figure 7E). Thus, SNX1/2 may participate in early stages of infection, which could explain the more modest effect of SNX1/2 depletion on infectious progeny production (Figure 7D). Altogether, our results support a model (Figure S7) whereby IncE directly binds SNX5/6, which redirects the retromer SNX-BAR subcomplex to the inclusion membrane, with two functional consequences. First, recruitment of SNX-BARS induces inclusion membrane tubulation, likely through the well-studied ability of BAR domain proteins to induce membrane curvature. Second, by sequestering SNX-BARs at the inclusion and potentially disrupting retromer function, C. trachomatis relieves the restriction that retromer imposes on infection.
DISCUSSION
Assigning functions to individual Chlamydia effectors has been difficult in the absence of robust genetic approaches. In this study, we applied an unbiased systematic AP-MS approach to comprehensively identify putative host targets of Incs, effectors that are ideally poised to orchestrate host-pathogen interactions. Our work is especially valuable, as very few Inc targets had been identified.
As with any AP-MS approach, false positives and false negatives are unavoidable. We employed two stringent scoring algorithms, MiST and CompPASS, to minimize false positives. Technical limitations of AP-MS may lead to false negatives (Verschueren et al., 2015), which could explain our failure to identify the following interactions: 14-3-3β and IncG (Scidmore and Hackstadt, 2001)), Rab4 and CT229, (Rzomp et al., 2006), or VAMPs and IncA (Delevoye et al., 2008). Our inability to identify high confidence PPIs for ~1/3 of Incs could be explained if (i) the Inc serves a structural role in the inclusion membrane (Mital et al., 2013), (ii) the host target is a lipid (Mital et al., 2013), or (iii) multiple Incs function together to form a binding interface (Gauliard et al., 2015; Mital et al., 2010). Importantly, some putative Incs have not been observed on the inclusion membrane at 24 hpi ((Dehoux et al., 2011; Li et al., 2008); summarized in Table S1), however, their localization may be stage-, cell type- or serovar-specific. Of the 20 Incs whose membrane localization has been experimentally verified (Dehoux et al., 2011; Li et al., 2008), we identified high confidence host targets for 15 Incs (75%) and validated two published Inc-host interactions (Derre et al., 2011; Lutter et al., 2013). Overall, this Inc-host interactome allows us to link host processes to specific Incs and identifies a plethora of Inc-human interactions of significant biological interest that merit further investigation.
Our standardized AP-MS pipeline provides an unprecedented opportunity to learn whether diverse intracellular pathogens use common strategies to survive. Not only did we observe a significant overlap between the interactomes of C. trachomatis and 3 human viruses, but we found that the more evolutionarily conserved core Incs are more likely to share host targets with viral effectors. Comparison of the overlap among all 4 pathogens revealed 2 common targets, MSH2 and UBE2O, suggesting that these host proteins may be critical for intracellular pathogen survival. Modulating UBE2O, one of ~35 human E2 ubiquitin ligases, is particularly intriguing, as it may be an efficient way for intracellular pathogens to regulate the stability of a distinct subset of host proteins.
Our Inc-human PPI network reveals an interaction between IncE and retromer SNX-BARs, which are involved in tubular-based endosomal trafficking (Seaman, 2012). SNX-BAR recruitment to membranes typically involves binding of SNX-PX domains to endosome-specific PtdIns(3)P (Cullen and Carlton, 2012; Seaman, 2012). Our work suggests that SNX5/6 bind directly to IncE independently of phosphoinositides and that the predicted IncE C-terminal β-hairpin is required. Our findings expand the emerging concept that PX domains function not only as lipid recognition modules, but also as PPI domains (Teasdale and Collins, 2012). While the molecular details of the IncE-SNX5/6 interaction await further investigation, we note that SNX5/6 contain a unique double PXXP motif with an adjacent ~30 amino acid insertion that forms a long helical hairpin (Koharudin et al., 2009), that distinguishes SNX5/6 from SNX1/2. This feature may contribute to the recognition of SNX5/6 by IncE.
Our data predicts that by sequestering SNX-BARs, IncE could disrupt retromer trafficking. To examine retromer trafficking in Chlamydia-infected cells, we used an assay that did not directly involve retromer cargo as a readout since full length IncE may bind cargo directly (Table S1) and the CI-MPR localizes to the inclusion membrane ((van Ooij et al., 1997) and unpublished data). We examined processing and release of cathepsin D, a secreted lysosomal protease that is missorted when retromer trafficking is disrupted (Rojas et al., 2008). We were unable to demonstrate robust intracellular accumulation of the incompletely processed forms (unpublished data), as reported upon depletion of VPS26 or Rab7 (Rojas et al., 2008), which could be explained if Chlamydia also alters secretory pathways. We do show that ectopic expression of IncE101-132, which does not appear to bind cargo (Table S1), alters CI-MPR localization in a manner similar to what is observed upon SNX5/6 depletion (Wassmer et al., 2007).
We observed a profound increase in infectious progeny production upon SNX5/6 depletion, which we postulate recapitulates IncE-mediated recruitment of the SNX-BAR complex away from retromer-containing compartments to the inclusion membrane. As primary inclusion formation was not enhanced, we predict that SNX5/6 restricts Chlamydia infection at later steps in the life cycle, such as replication, RB to EB conversion, EB escape or EB infectivity. While the mechanism by which SNX5/6 restricts bacterial development is not clear, it could be through retromer-dependent or retromer-independent pathways. For example, retromer trafficking may contribute to the recognition or clearance of Chlamydia. Alternatively, since retromer is required to maintain the integrity of the trans-Golgi (Wassmer et al., 2007), IncE-mediated sequestration of retromer SNX-BARS may promote Golgi fragmentation, a process that facilitates lipid acquisition by C. trachomatis and enhances progeny production (Heuer et al., 2009). Finally, SNX5/6 may have other roles outside of retromer trafficking (Sun et al., 2013) that contribute to Chlamydia restriction.
Our results provide mechanistic insights into the formation of Chlamydia inclusion tubulation. We demonstrate that IncE and SNX-BARs co-localize on the inclusion membrane and tubules, that ectopic expression of IncE is sufficient to induce inclusion tubulation, and that depletion of retromer components limit inclusion tubulation. The direct recruitment of BAR-domain containing proteins to the inclusion membrane provides a mechanistic link to tubulation, as these proteins are necessary and sufficient for membrane deformation and endosomal tubule formation (Frost et al., 2009). While the role of the tubules is not entirely understood, they are proposed to play a role in secondary inclusion formation (Suchland et al., 2005). We demonstrate that inclusion tubulation and infectious progeny production can be uncoupled: SNX5/6 depletion, which abrogates tubulation, leads to increased infectious progeny.
Our work highlights the strategies employed by diverse intracellular pathogens to repurpose retromer. Silencing of retromer components abrogates HIV (Groppelli et al., 2014), HPV (Lipovsky et al., 2013), Coxiella (McDonough et al., 2013) and Salmonella (Bujny et al., 2008) infections. Our finding that VPS35 is readily recruited to Salmonella-containing vacuoles reveals that Salmonella recruits the VPS complex in addition to retromer SNX-BARs. In contrast, Chlamydia IncE selectively recruits retromer SNX-BARs without stably recruiting VPS components or SNX27. Interestingly, through an entirely different mechanism, Legionella also disassociates retromer subcomplexes by sequestering VPS components and blocking SNX-PtdIns(3)P binding (Finsel et al., 2013).
In summary, we have applied AP-MS to create a global network of C. trachomatis Inc-host PPIs, providing a comprehensive bacterial effector-host interactome. Our analysis has uncovered a wealth of previously unidentified Chlamydia Inc-host interactions. By linking individual Incs to specific host processes, our work, in conjunction with the development of genetic tools to generate targeted null mutants in Chlamydia (Johnson and Fisher, 2013), sets the stage to test the functional role of Incs. This methodology has broad applicability to the study of pathogenesis and is equally effective for identifying the targets of both membrane-bound and soluble effectors. We have uncovered overlapping and potentially druggable pathways targeted by viral and bacterial pathogens. Finally, investigation of the IncE-SNX5/6 interaction may provide insight into retromer biology.
EXPERIMENTAL PROCEDURES
Cell Culture, Transformations, and Bacterial Propagation
HeLa and HEK293T cells were maintained under standard conditions. C. trachomatis serovar D and L2 were propagated as previously described (Elwell et al., 2011). Chlamydia transformations were performed as previously described (Agaisse and Derre, 2013; Wang et al., 2013).
Affinity Purification and Mass Spectrometry
Affinity purifications were performed as previously described (Jager et al., 2011). Eluates were processed, trypsin digested, and concentrated for LC-MS/MS. Digested peptide mixtures were analyzed on a Thermo Scientific Velos Pro ion trap MS system equipped with a Proxeon Easy nLC II high pressure liquid chromatography and autosampler system.
Scoring and Analyzing the Inc-Host Interactome
AP-MS samples were scored with CompPASS (Sowa et al., 2009) and MIST algorithms, using MIST weights optimized for the KSHV-host interactome (Davis et al., 2014). The scored dataset included a subset of Chlamydia secreted effectors that will be published independently. All bait-prey pairs with a MIST score ≥ 0.70 or the top 1% of the CompPASS WD scores were combined with human protein interactions from CORUM and STRING databases that connect prey. The resulting network diagram was plotted using Cytoscape, v.3.1.2 (Smoot et al., 2011). All scored preys were queried against GO, KEGG and PFAM ontologies for functional and domain annotations. Terms were manually curated and baits were analyzed for significantly enriched terms with a resampling procedure using MIST scores. Viral overlap was preformed as previously described (Davis et al., 2014), and statistical analysis was preformed using the hyper-geometric test.
Infections and RNAi
Cells were infected with Chlamydia for 1 hr and then incubated for 15–24 hrs (primary infection, tubulation or immunoprecipitation) or 72 hrs (progeny production). For some experiments, nocodazole or tetracycline were added 1 hpi. HeLa cells were transfected with siRNAs (Dharmacon) according to manufacturer’s protocols and infected with Chlamydia at 48 hrs. Protein depletion was assayed by immunoblot.
Transfections and Immunoprecipitations
Lysates from HeLa cells collected 24 hpi were immunoprecipitated with control IgG or anti-SNX6 antibody using Protein G Dynabeads (Invitrogen). Alternatively, HeLa cells were transfected (Effectene, Qiagen) with the indicated FLAG-SNX construct, infected 24 hrs later with C. trachomatis, collected at 24 hpi, immunoprecipitated with anti-FLAG beads (Sigma) and eluted with FLAG peptide.
Microscopy
HeLa cells were grown on glass coverslips, infected with Chlamydia, fixed, stained with the indicated primary and fluorophore-conjugated secondary antibodies, and mounted with Vectashield containing DAPI (Vector Laboratories). Images were acquired using Yokogawa CSU-X1 spinning disk confocal mounted on a Nikon Eclipse Ti inverted microscope equipped with an Andora Clara digital camera. Images were acquired and processed using NIS-Elements software 4.10 (Nikon). Quantitations of tubule number/length and colocalization were performed using Nikon Elements. Inclusions were quantified using the Spot function in Imaris. Statistics were performed using Instat software; p-values less than 0.05 were considered statistically significant.
In Vitro Pulldowns
Purified 6xHis-MBP-Inc-Strep constructs were immobilized to Strep-Tactin Sepharose beads according to manufacturer’s instructions (IBA), incubated with purified 8xHis-SNXPX or 6xHis-MBP-SNXBAR domains (1:2 molar ratio), the beads were washed extensively, and complexes boiled and analyzed by Coomassie Blue staining.
Supplementary Material
Acknowledgments
We thank Drs. Deborah Dean, Jai-Jai Liu, Ted Hackstadt, Dan Rockey, Denise Monack, Mark von Zastrow, Sourav Bandyopadhyay, and John Jascur for reagents; the UCSF Biological Imaging Development Center for use of software; and members of the Engel and Sil labs for advice. We acknowledge grant support from NIH (J.E.: R01 AI073770, AI105561; N.J.K: P50 GM082250; PO1 AI090935, PO1 090935,P50 GM081879, PO1 091575, U19 AI106754, U54AI081680, DARPA-10-93-Prophecy-PA-008; R.V.: AI081694 and AI100759; I.D.: R01AI101441; O.R.: K08AI091656; B.P.: P0065678 A121844 (CA-0063244)), UCSF Microbial Pathogenesis and Host Defense T32 (K.M.M.), American Heart Association (K.M.M.) and the UCSF Program for Breakthrough in Biomedical Research (J.E. and N.K.).
Footnotes
AUTHOR CONTRIBUTIONS
K.M.M., C.A.E., N.J.K, J.E. conceived, designed, and analyzed experiments. K.M.M., C.A.E., A.G, I.D. and J.E. wrote the manuscript. E.V., J.V.D., J.R.J., N.G., S.J., and T.J. performed MS or bioinformatics analysis. K.M.M., C.A.E., A.F., G.J., T.J., A.M.G., I.D., J.S. conducted experiments. O.R., R.V., M.N.S., J.D.D., A.O, and B.P. provided reagents or advice. M.S. assisted in figure design and graphics.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agaisse H, Derre I. A C. trachomatis cloning vector and the generation of C. trachomatis strains expressing fluorescent proteins under the control of a C. trachomatis promoter. PLoS One. 2013;8:e57090. doi: 10.1371/journal.pone.0057090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165:123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Griffiths RS, Viratyosin W, Brown WJ, Rockey DD. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2000;2:35–47. doi: 10.1046/j.1462-5822.2000.00029.x. [DOI] [PubMed] [Google Scholar]
- Bastidas RJ, Elwell CA, Engel JN, Valdivia RH. Chlamydial intracellular survival strategies. Cold Spring Harbor perspectives in medicine. 2013;3:a010256. doi: 10.1101/cshperspect.a010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer MF, Hofmann S, Neupert W, Brunner M. Protein translocation into mitochondria: the role of TIM complexes. Trends in cell biology. 2000;10:25–31. doi: 10.1016/s0962-8924(99)01684-0. [DOI] [PubMed] [Google Scholar]
- Betts-Hampikian HJ, Fields KA. The Chlamydial Type III Secretion Mechanism: Revealing Cracks in a Tough Nut. Frontiers in microbiology. 2010;1:114. doi: 10.3389/fmicb.2010.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujny MV, Ewels PA, Humphrey S, Attar N, Jepson MA, Cullen PJ. Sorting nexin-1 defines an early phase of Salmonella-containing vacuole-remodeling during Salmonella infection. J Cell Sci. 2008;121:2027–2036. doi: 10.1242/jcs.018432. [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Carlton JG. Phosphoinositides in the Mammalian Endo-lysosomal Network. Subcell Biochem. 2012;59:65–110. doi: 10.1007/978-94-007-3015-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Korswagen HC. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol. 2012;14:29–37. doi: 10.1038/ncb2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ZH, Verschueren E, Jang GM, Kleffman K, Johnson JR, Park J, Von Dollen J, Maher MC, Johnson T, Newton W, et al. Global Mapping of Herpesvirus-Host Protein Complexes Reveals a Transcription Strategy for Late Genes. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehoux P, Flores R, Dauga C, Zhong G, Subtil A. Multi-genome identification and characterization of chlamydiae-specific type III secretion substrates: the Inc proteins. BMC Genomics. 2011;12:109. doi: 10.1186/1471-2164-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C, Nilges M, Dehoux P, Paumet F, Perrinet S, Dautry-Varsat A, Subtil A. SNARE protein mimicry by an intracellular bacterium. PLoS Pathog. 2008;4:e1000022. doi: 10.1371/journal.ppat.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derre I, Swiss R, Agaisse H. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog. 2011;7:e1002092. doi: 10.1371/journal.ppat.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, Melancon P, Engel JN. Chlamydia trachomatis Co-opts GBF1 and CERT to Acquire Host Sphingomyelin for Distinct Roles during Intracellular Development. PLoS Pathog. 2011;7:e1002198. doi: 10.1371/journal.ppat.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsel I, Ragaz C, Hoffmann C, Harrison CF, Weber S, van Rahden VA, Johannes L, Hilbi H. The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell host & microbe. 2013;14:38–50. doi: 10.1016/j.chom.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic acids research. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell. 2008;132:807–817. doi: 10.1016/j.cell.2007.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauliard E, Ouellette SP, Rueden KJ, Ladant D. Characterization of interactions between inclusion membrane proteins from Chlamydia trachomatis. Frontiers in cellular and infection microbiology. 2015;5:13. doi: 10.3389/fcimb.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths E, Ventresca MS, Gupta RS. BLAST screening of chlamydial genomes to identify signature proteins that are unique for the Chlamydiales, Chlamydiaceae, Chlamydophila and Chlamydia groups of species. BMC Genomics. 2006;7:14. doi: 10.1186/1471-2164-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groppelli E, Len AC, Granger LA, Jolly C. Retromer regulates HIV-1 envelope glycoprotein trafficking and incorporation into virions. PLoS Pathog. 2014;10:e1004518. doi: 10.1371/journal.ppat.1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Yang Y, Zhang C, Niu Y, Li K, Zhao X, Liu JJ. The retromer component SNX6 interacts with dynactin p150(Glued) and mediates endosome-to-TGN transport. Cell Res. 2009;19:1334–1349. doi: 10.1038/cr.2009.130. [DOI] [PubMed] [Google Scholar]
- Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV-human protein complexes. Nature. 2011;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Fisher DJ. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS One. 2013;8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koharudin LM, Furey W, Liu H, Liu YJ, Gronenborn AM. The phox domain of sorting nexin 5 lacks phosphatidylinositol 3-phosphate (PtdIns(3)P) specificity and preferentially binds to phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) J Biol Chem. 2009;284:23697–23707. doi: 10.1074/jbc.M109.008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffer BE, Melero C, Temkin P, Lei C, Hong W, Kortemme T, von Zastrow M. SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. J Cell Biol. 2010;190:565–574. doi: 10.1083/jcb.201004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CA, Borel N. Chronic Chlamydial Diseases: From Atherosclerosis to Urogenital Infections. Curr Clin Micro Rpt. 2014;1:61–72. [Google Scholar]
- Li Z, Chen C, Chen D, Wu Y, Zhong Y, Zhong G. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect Immun. 2008;76:2746–2757. doi: 10.1128/IAI.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie MA, Poffenberger AC, Nelson CD, Atwood WJ, et al. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A. 2013;110:7452–7457. doi: 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter EI, Barger AC, Nair V, Hackstadt T. Chlamydia trachomatis inclusion membrane protein CT228 recruits elements of the myosin phosphatase pathway to regulate release mechanisms. Cell reports. 2013;3:1921–1931. doi: 10.1016/j.celrep.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter EI, Martens C, Hackstadt T. Evolution and conservation of predicted inclusion membrane proteins in chlamydiae. Comparative and functional genomics. 2012;2012:362104. doi: 10.1155/2012/362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell GL, Bennett JE, Dolin R. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. Philadelphia, PA: Churchill Livingstone/Elsevier; 2010. p. 1. online resource (2 v. (cl, 4028, xcvii p.)) [Google Scholar]
- Marshansky V, Futai M. The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Current opinion in cell biology. 2008;20:415–426. doi: 10.1016/j.ceb.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough JA, Newton HJ, Klum S, Swiss R, Agaisse H, Roy CR. Host pathways important for Coxiella burnetii infection revealed by genome-wide RNA interference screening. mBio. 2013;4:e00606–00612. doi: 10.1128/mBio.00606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mital J, Miller NJ, Dorward DW, Dooley CA, Hackstadt T. Role for chlamydial inclusion membrane proteins in inclusion membrane structure and biogenesis. PLoS One. 2013;8:e63426. doi: 10.1371/journal.pone.0063426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mital J, Miller NJ, Fischer ER, Hackstadt T. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell Microbiol. 2010 doi: 10.1111/j.1462-5822.2010.01465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore ER, Ouellette SP. Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins. Frontiers in cellular and infection microbiology. 2014;4:157. doi: 10.3389/fcimb.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive AJ, Haff MG, Emanuele MJ, Sack LM, Barker JR, Elledge SJ, Starnbach MN. Chlamydia trachomatis-induced alterations in the host cell proteome are required for intracellular growth. Cell host & microbe. 2014;15:113–124. doi: 10.1016/j.chom.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage HR, Kumar GR, Verschueren E, Johnson JR, Von Dollen J, Johnson T, Newton B, Shah P, Horner J, Krogan NJ, et al. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol Cell. 2015;57:329–340. doi: 10.1016/j.molcel.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roan NR, Starnbach MN. Antigen-specific CD8+ T cells respond to Chlamydia trachomatis in the genital mucosa. J Immunol. 2006;177:7974–7979. doi: 10.4049/jimmunol.177.11.7974. [DOI] [PubMed] [Google Scholar]
- Rockey DD, Scidmore MA, Bannantine JP, Brown WJ. Proteins in the chlamydial inclusion membrane. Microbes Infect. 2002;4:333–340. doi: 10.1016/s1286-4579(02)01546-0. [DOI] [PubMed] [Google Scholar]
- Rojas R, van Vlijmen T, Mardones GA, Prabhu Y, Rojas AL, Mohammed S, Heck AJ, Raposo G, van der Sluijs P, Bonifacino JS. Regulation of retromer recruitment to endosomes by sequential action of Rab5 and Rab7. J Cell Biol. 2008;183:513–526. doi: 10.1083/jcb.200804048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic acids research. 2010;38:D497–501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzomp KA, Moorhead AR, Scidmore MA. The GTPase Rab4 interacts with Chlamydia trachomatis inclusion membrane protein CT229. Infect Immun. 2006;74:5362–5373. doi: 10.1128/IAI.00539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scidmore MA, Hackstadt T. Mammalian 14-3-3beta associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol Microbiol. 2001;39:1638–1650. doi: 10.1046/j.1365-2958.2001.02355.x. [DOI] [PubMed] [Google Scholar]
- Scidmore-Carlson MA, Shaw EI, Dooley CA, Fischer ER, Hackstadt T. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol Microbiol. 1999;33:753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- Seaman MN. The retromer complex - endosomal protein recycling and beyond. J Cell Sci. 2012;125:4693–4702. doi: 10.1242/jcs.103440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisko JL, Spaeth K, Kumar Y, Valdivia RH. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol Microbiol. 2006;60:51–66. doi: 10.1111/j.1365-2958.2006.05074.x. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchland RJ, Rockey DD, Weeks SK, Alzhanov DT, Stamm WE. Development of secondary inclusions in cells infected by Chlamydia trachomatis. Infect Immun. 2005;73:3954–3962. doi: 10.1128/IAI.73.7.3954-3962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale RD, Collins BM. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem J. 2012;441:39–59. doi: 10.1042/BJ20111226. [DOI] [PubMed] [Google Scholar]
- van Ooij C, Apodaca G, Engel J. Characterization of the Chlamydia trachomatis vacuole and its interaction with the host endocytic pathway in HeLa cells. Infect Immun. 1997;65:758–766. doi: 10.1128/iai.65.2.758-766.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren E, Von Dollen J, Cimermancic P, Gulbahce N, Sali A, Krogan NJ. Scoring Large-Scale Affinity Purification Mass Spectrometry Datasets with MiST. Curr Protoc Bioinformatics. 2015;49:8. 19 11–18 19 16. doi: 10.1002/0471250953.bi0819s49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Persson K, Bjartling C, Clarke IN. Genetic transformation of a clinical (genital tract), plasmid-free isolate of Chlamydia trachomatis: engineering the plasmid as a cloning vector. PLoS One. 2013;8:e59195. doi: 10.1371/journal.pone.0059195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmer T, Attar N, Bujny MV, Oakley J, Traer CJ, Cullen PJ. A loss-of-function screen reveals SNX5 and SNX6 as potential components of the mammalian retromer. J Cell Sci. 2007;120:45–54. doi: 10.1242/jcs.03302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.