Abstract
The rimJ gene, which codes for a crotonyl-CoA carboxylase/reductase, lies within the biosynthetic gene cluster for two polyketides belonging to the polyene macrolide group (CE-108 and rimocidin) produced by Streptomyces diastaticus var. 108. Disruption of rimJ by insertional inactivation gave rise to a recombinant strain overproducing new polyene derivatives besides the parental CE-108 (2a) and rimocidin (4a). The structure elucidation of one of them, CE-108D (3a), confirmed the incorporation of an alternative extender unit for elongation step 13. Other compounds were also overproduced in the fermentation broth of rimJ disruptant. The new compounds are in vivo substrates for the previously described polyene carboxamide synthase PcsA. The rimJ disruptant strain, constitutively expressing the pcsA gene, allowed the overproduction of CE-108E (3b), the corresponding carboxamide derivative of CE-108D (3a), with improved pharmacological properties.
Introduction
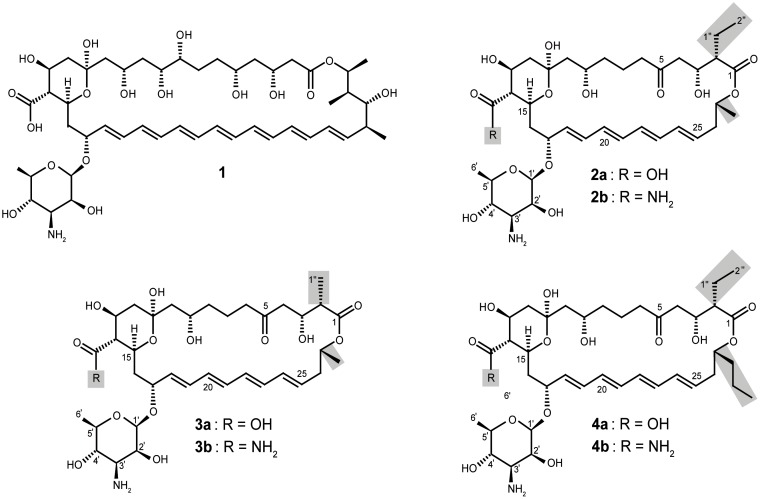
The polyene-macrolides are a group of polyketides that are commercially important because of their antifungal properties. This group includes well-known drugs such as amphotericin B (1), pimaricin, nystatin, 4a, and candicidin. They consist of a macrolactone ring containing several conjugated double bonds, which are in part responsible for the physical and chemical properties of these compounds (strong light sensitivity and low solubility in water). A sugar moiety (typically mycosamine) and a free carboxyl group are usually found on the macrocycle. The biological activity of these polyenes is rather specific for fungi, due to their preferred affinity toward ergosterol-containing membranes (fungal membranes and the membranes of some parasites such as Trypanosoma, Leishmania, etc.) rather than cholesterol-containing membranes. This interaction seems to affect some physico-chemical properties of the membranes, leading to changes on their ionic permeability, pore formation, loss of ions and thus cell death. In contrast to other antifungal drugs, the rate of appearance of resistant forms of the target microorganisms after treatment with polyenes is very low.
Undoubtedly the most important drawback for the clinical use of some polyenes like amphotericin B (1) (Fig 1) are the undesirable side effects during treatment of systemic fungal infections, particularly nephrotoxicity and hepatotoxicity. These toxicities seem to be associated with the interactions between 1 and cell membranes [1]. The need for new alternative antifungals with improved pharmacological properties has given rise to semi-synthetic derivatives of 1 designed to reduce the undesirable side effects while retaining antifungal activity. These attempts yielded new derivatives with good prospects as drugs and, more importantly, have increased knowledge of structure-activity relationships. Thus, exchanging several functional groups of 1, such as the carboxyl group of the macrolactone ring and/or the amino group of the mycosamine sugar or the polyol chain, seems to be crucial for the improvement of pharmacological properties (lower toxicity and increased antifungal activity and water solubility) [2–4]. Semisynthetic derivatives of other polyene macrolides have been developed with similar results [5,6]. Polyenes, like all macrolides, are produced through the action of type I modular polyketide synthases (PKSs). These large multifunctional enzymes consist of distinct domains, each of which catalyzes one non-iterative polyketide chain elongation step [7]. The availability of biosynthetic genes for several polyene macrolide pathways [8] has provided additional tools for in vivo biosynthesis of new chemical entities. In this way, new derivatives were successfully obtained by genetic engineering of polyene producer strains [9–18]. Due to the chemical complexity of the polyene macrolides, the biosynthetic route seemed to be a highly promising alternative for affording chemical modification of the macrolactone ring and thus generation of derivatives with improved pharmacological properties.
Fig 1. Chemical structures of polyenes cited in the text.
1: Amphotericin B; 2a: CE-108; 2b: CE-108B; 3a: CE-108D; 3b: CE-108E; 4a: Rimocidin; 4b: Rimocidin B. Note that the depicted stereochemistry of 2a, 2b, 3a, 3b and 4b was deduced from the known stereoschemistry of 4a and has not been experimentally established.
We have previously characterized a chromosomal region of Streptomyces diastaticus var. 108 [19] encoding the biosynthetic machinery for two related tetraenes: 4a and 2a (Fig 1). Based on their chemical structures, we proposed a model for the biosynthetic pathway [20]. The incorporation of a malonyl-CoA or ethylmalonyl-CoA starter unit determines the formation of 2a and 4a, respectively. RimA (module 0), the loading PKS, exhibits the domain structure carboxylic acid:CoA ligase-ACP-KSS-AT-ACP, in which the KSS domain (ketosynthase with a serine residue in place of the conserved active site cysteine) would not catalyze a condensation reaction, but rather select and possibly decarboxylate the starter unit, before transferring it to the C-terminal ACP. This peculiar KSS is also present in other polyene loading PKSs [21–23]. It is known that KS domains are converted to potent decarboxylases when the active site cysteine is replaced with glutamine [24,25] so this domain could function as a decarboxylase that acts on malonyl-CoA or ethylmalonyl-CoA to generate acetyl-CoA and butyryl-CoA starter units, respectively. Site-directed mutagenesis of the KSS domain of NysA, the loading PKS for nystatin biosynthesis, surprising suggested that the conserved serine residue S413 may be important for the decarboxylation of the malonyl-CoA starter unit, rater than S170 that sits in place of the active site cysteine. Since the NysA-KSS-S413N mutant retained some activity, it was further proposed that acetyl-CoA can be used as starter unit too. Although this residue is not present in RimA, we cannot rule out the possibility that acetyl-CoA and butyryl-CoA could also be loaded directly by RimA for the biosynthesis of 2a and 4a respectively.
The remaining 13 Type I PKS elongation modules are responsible for the correct formation of the polyketide chain for 2a and 4a biosynthesis. Acetate would be incorporated as elongation unit in all modules by decarboxylative condensation of malonyl-CoA, except modules 7 and 13, which would incorporate propionate and butyrate by decarboxylative condensation of methylmalonyl-CoA or ethylmalonyl-CoA units, respectively. The incorporation of methylmalonyl-CoA and ethylmalonyl-CoA to the growing polyketide yields methyl and ethyl side chains, respectively on the macrolactone ring.
Several routes have been proposed for providing the methylmalonyl-CoA precursor for polyketide biosynthesis: (i) the isomerization of succinyl-CoA, catalyzed by the coenzyme B12-dependent methylmalonyl-CoA mutase (MCM); (ii) carboxylation of propionyl-CoA, catalyzed by the propionyl-CoA carboxylase (PCC); (iii) methylmalonyl-CoA ligase (MatB) [26]. In the case of ethylmalonyl-CoA, two pathways have been described: catabolism of valine and the butyryl-CoA pathway [27]. Crotonyl-CoA reductase (CCR) was described as the key enzyme in the butyryl-CoA pathway, catalyzing the last step in the conversion of two acetyl-CoA molecules into butyryl-CoA. Besides the hydrogenation of crotonyl-CoA to butyryl-CoA, it has been recently reported that CCR also catalyzes the reductive carboxylation of crotonyl-CoA to ethylmalonyl-CoA, this last reaction being the physiologically favored reaction [28,29]. This reaction catalyzed by CCR is the key step in the recently discovered ethylmalonyl-CoA pathway, involved in assimilation of acetate in several bacterial species [28] and in the biosynthesis of different polyketides [30,31].
A crotonyl-CoA carboxylase/reductase encoded by rimJ, previously described as a crotonyl-CoA reductase, was found within the 4a and 2a gene cluster [20]. Interestingly, disruption of rimJ did not abolish 4a production and, in addition, a new compound with a typical tetraene spectrum was detected along with 2a [20]. At that time we hypothesized that rimJ mutant might unbalance the incorporation of starter or/and extender units by the loading module RimA or elongation step 13. In this work we aimed at a deeper characterization of the profile of polyene being produced by a rimJ mutant with reduced availability of some metabolic building blocks for polyene production. The versatility of RimA of S. diastaticus var. 108 towards starter units encouraged this approach for isolating recombinant strains producing different chemical structures.
We recently described the isolation of the pcsA gene, unlinked to the polyene biosynthetic cluster in S. diastaticus var. 108. This gene encodes a polyene carboxamide synthase, PcsA, involved in tailoring of the exocyclic carboxyl group of 2a and 4a into their carboxamide derivatives [32]. PcsA shows glutamine amidotransferase activity and belongs to the asparagine synthases B (Class II amidotransferases) [33,34], which recognizes the final polyenes 2a and 4a as substrates. Moreover, PcsA can also convert in vivo and in vitro not only 2a and 4a into their corresponding amides, but also pimaricin (a heterologous substrate) [17], revealing interestingly reduced selectivity of this enzymatic activity for polyene macrolides. A similar gene, pcsB, encoding another polyene carboxamide synthase, PcsB, but specific for converting pimaricin into its corresponding amide (AB-400), was recently reported [35]. Polyene amide derivatives showed higher antimicrobial activity than their parent compounds without increasing the haemolytic rate, suggesting increased selective toxicity against fungal membranes. Thus, use of this tailoring gene with reduced substrate selectivity within a genetic background, such as S. diastaticus var. 108, makes this strain a promising system for the attempted generation of new molecules with improved biological activity.
Material and Methods
Bacterial strains, cloning vectors and growth conditions
Bacterial strains and plasmids are described in Table 1. S. diastaticus var. 108 and its engineered derivatives were cultured in SYM2 medium [36] for tetraene production analysis, and liquid TSB (Oxoid) for plasmid and total DNA extraction. Streptomyces lividans TK21 [37] was used as a general cloning host and grown on solid R5 medium and in liquid YEME medium [37]. E. coli JM101 [38] was grown on Luria-Bertani (LB) agar or in LB broth [39]. P. chrysogenum, C. krusei, A. niger, C. albicans and F. neoformans, used for testing antifungal activity, were grown in RPMI-1640 medium (Sigma, catalog. No N-3503) supplemented with 0.165M MOPS (morpholinepropanesulfonic acid) buffer. The pH was adjusted to 6.9–7. pGAe-1 [16], pIJ2925 [37], PM1 phage [40], pHJL401 [41], pIJ4090 [37] and pIJ922 [42] vectors were used for cloning.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Properties | Reference |
|---|---|---|
| S. diastaticus var. 108 | Wild-type (WT); CE-108 and rimocidin producer. | [19] |
| S. diastaticus var. 108/PM1-709B | WT derivative with rimJ disrupted by integration of PM1-709B. | This work |
| S. diastaticus var. 108::PM1-709B/860 | WT derivative with rimJ disrupted by integration of PM1-709B and transformed with pSM860. | This work |
| S. diastaticus var. 108/780 | WT derivative by transformation with pSM780. CE-108 and rimocidin producer. | This work |
| S. diastaticus var. 108/781 | WT derivative by transformation with pSM781. Rimocidin producer as majority polyene. | This work |
| S. lividans TK21 | General cloning host | [37] |
| S. lividans TK21/pSM858 | S. lividans TK21 WT derivative transformed with pSM858 plasmid | [32] |
| E. coli JM101 | General cloning host | [38] |
| Penicillium chrysogenum ATCC10003 | Antifungal activity assays | ATCC |
| Aspergillus niger ATCC1004 | Antifungal activity assays | ATCC |
| Issatchenkia orientalis CECT 1688 | Antifungal activity assays | CECT |
| Filobasidiella neoformans CECT 1078 | Antifungal activity assays | CECT |
| PM1-709B | 1.8 kb XhoI-BglII fragment from pGAe-1 [16] carrying the ermE gene and 840 bp BamHI-SacI internal fragment of rimJ cloned into the XhoI/SacI sites of PM1 phage [40]. | This work |
| pSM859 | HindIII-EcoRI fragment from pSM858 (carrying oriT and pcsA under the control of ermE P*) cloned into the HindIII/EcoRI sites of pIJ2925 [37]. | This work |
| pSM860 | BglII-EcoRI fragment from pSM859 (carrying oriT and pcsA under the control of ermE P*) cloned into the BamHI/EcoRI sites of pIJ922 [42]. | This work |
| pSM780 | pHJL401 [41] derived vector carrying the ermE P* promoter from pIJ4090 [37] and oriT. | [32] |
| pSM781 | 17,866–16,005 bp fragment from the sequence deposited under accession number AY442225 isolated as BamHI present in the oligonucleotide CCR-D (CGGGATCCCGCCTTTTCCGGAGGC, 17849–17866 bp from AY442225) and Ecl136II of the chromosome cloned into the BamHI-Ecl136II sites of pSM780. This plasmid carries rimJ under the control of the ermE P* promoter. | This work |
ermE P*: constitutive erythromycin-resistance promoter where the asterisk signifies the presence of a one-base-pair mutation [43].
Genetic procedures
E. coli JM101 was grown and transformed as described elsewhere [39]. Streptomyces strains were manipulated as previously described [37]. Intraspecific conjugation was carried out as previously described [16]. DNA manipulations were performed as described by Maniatis et al. [39].
Assay for tetraene production
0.2 ml of total culture was extracted with 0.8 ml of methanol. The extracts were filtered and 20 μl were applied to an HPLC with a Waters 600S Controller, equipped with a Waters 996 PDA. The chromatographic parameters and the mobile phases were: 2 min with 100% of B (ammonium acetate 20 mM pH 5, ethanol 20%), 2 min of a binary gradient up to 50% of A (methanol) and 50% of B (curve 6); 6 min of a binary gradient up to 100 of A (curve 9), and a constant flow of 0.7 ml/min. The chromatograms were monitored at a wavelength of 304 nm.
HPLC-MS Assays
The mass spectra were determined in a 1100MSD HPLC connected to a quadrupole Agilent Technology Detector using electrospray as source and a positive ionization mode. The chromatographic conditions were the same as described above and the applied voltage was 150V.
Spectroscopic and spectrometric measurements
UV/vis spectra were recorded on a Perkin-Elmer Lambda 15 UV/vis spectrometer. NMR spectra were measured on a Varian Inova 600 (600.7 MHz) spectrometer. ESIMS was recorded on a Finnigan LCQ with a Rheos 4000 (Flux Instrument) quaternary pump. HRMS was recorded by ESI MS on an Apex IV 7 Tesla Fourier-Transform Ion Cyclotron Resonance Mass Spectrometer (Bruker Daltonics, Billerica, MA, USA). Reserpine (MW 608) and leucine enkephalin (MW 555) were used as standards in positive and negative mode.
Purification of polyenes
2a, 2b, 4a and 4b were purified as previously described [16] by a similar protocol as described below for 3a and 3b. 1 was purchased from Sigma (catalog. No N-3503).
3a was purified from Streptomyces diastaticus var. 108::PM1-709B which was grown on solid SYM2 medium supplemented with erythromycin (25 μg/ml) for the selection of chromosomal insertions. Four plates of 24 x 24 cm were used for purification and the yield was up to 40 mg of tetraene-containing sample per plate. After 6 days, the whole solid medium was fragmented through a syringe, extracted with four volumes of methanol and 25 mM formic acid, stirred for 1 hr and centrifuged at 5000 x g for 20 min to remove solid particles. The clear supernatant was concentrated by rotaevaporation to 10–20 x 106 U/μl measured at a wavelength of 304 nm. The sample was stored in 80% methanol/water until use. The methanol-extracted samples were brought to 20% methanol with water and filtered to remove precipitated material. An Omnifit column (250 x 25 mm, Supelco Catalog No. 56010) packed with SP-Sepharose Fast Flow (GE Healthcare) was equilibrated in the same solution. 3a and the rest of the carboxylated polyenes were eluted with the flowthrough whereas minority polyene amides were completely retained in the column. A mixture containing 3a was applied to a semipreparative column (Supelcosil PLC-8, 250.0 x 21.2 mm). The chromatographic parameters and the mobile phases, controlled with a Waters Automated Gradients Controller, were: 12 min with 100% of B (ammonium acetate 20 mM pH 5, ethanol 20%), 43 min of a binary gradient up to 50% of A (methanol) and 50% of B (curve 6); 35 min of a binary gradient up to 100 of A (curve 8), and a constant flow of 5 ml/min. Fractions were collected at regular intervals (5 ml per fraction) and those carrying the purified compounds were pooled and subjected to an additional desalting step, as above, and finally freeze-dried twice.
3b was purified from S. diastaticus var. 108::PM1-709B/860 which was grown on solid SYM2 medium supplemented with erythromycin (25 μg/ml) and thiostrepton (50 μg/ml) for the selection of chromosomal insertions and plasmid markers, respectively. The solid medium was processed as described above. An Omnifit column (250 x 25 mm, Supelco Catalog No. 56010) packed with SP-Sepharose Fast Flow (GE Healthcare) was used as described for 3a compound. 3b and the rest of the polyene amides were retained in the column, which was exhaustively washed with the same solution and the polyene amides were eluted with 300 mM ammonium acetate pH 5 in 20% methanol. The fractions containing the mixtures of polyene amides were desalted and 3b was separated by using a semipreparative column (Supelcosil PLC-8, 250.0 x 21.2 mm) as described above. 3b compound was pooled, subjected to an additional desalting step and finally freeze-dried twice.
3a: UV absorbing, pale yellow solid (6.3 mg). NMR data see Table 2 and S1–S5 Figs (+)-ESI MS: m/z (%) = 726 ([M+H]+, 100). (-)-ESI MS: m/z (%) = 724 ([M-H]-, 100). (+)-ESI HRMS: m/z = 726.36981 [M+H]+, (calcd 726.37006 for C36H56NO14).
Table 2. 1H NMR and 13C NMR Data of 3a and 3b in DMSO-d 6.
| 3a | 3b | |||
|---|---|---|---|---|
| Position | 1H (Int., mult, J [Hz]) a) | 13C b) | 1H (Int., mult., J [Hz]) c) | 13C b) |
| 1 | - | 173.0 | - | 173.0 |
| 2 | 2.20 (1H, m) | 47.0 | 2.20 (1H, m) | 47.0 |
| 2-Me | 1.08 (3H, d, 10.8) | 13.2 | 1.08 (3H, d, 7.0) | 13.1 |
| 3 | 4.03 (1H, m) | 67.7 | 4.03 (1H, m) | 67.8 |
| 4 | 2.36, 2.28 (2H, m) | 48.1 | 2.36, 2.30 (2H, m) | 48.1 |
| 5 | - | 208.5 | - | 208.6 |
| 6 | 2.39, 2.24 (2H, m) | 43.1 | 2.43, 2.24 (2H, m) | 43.1 |
| 7 | 1.49, 1.26 (2H, m) 2.06 | 19.3 | 1.53, 1.28 (2H, m) | 19.3 |
| 8 | 1.28, 1.20 (2H, m) | 37.4 | 1.28, 1.20 (2H, m) | 37.4 |
| 9 | 3.98 (1H, m) | 67.6 | 3.98 (1H, m) | 67.6 |
| 10 | 1.48 (2H, m) | 45.6 | 1.45 (2H, m) | 45.6 |
| 11 | - | 96.9 | - | 96.9 |
| 12 | 1.82, 1.11 (2H, m) | 44.4 | 1.89, 1.12 (2H, m) | 44.7 |
| 13 | 4.00 (1H, m) | 65.4 | 4.02 (1H, m) | 64.7 |
| 14 | 1.84 (1H, m) | 57.8 | 1.92 (1H, t, 10.3) | 56.6 |
| CONH2 | - | 176.4 | - | 174.2 |
| 15 | 4.16 (1H, t, 8.4) | 65.4 | 4.17 (1H, t, 9.6) | 65.2 |
| 16 | 2.16, 1.53 (2H, m) | 36.7 | 2.06, 1.51 (2H, m) | 36.6 |
| 17 | 4.38 (1H, m) | 74.1 | 4.37 (1H, m) | 74.4 |
| 18 | 5.89 (1H, dd, 15.2, 8.2) | 136.4 | 5.87 (1H, dd, 15.3, 8.4) | 136.3 |
| 19 | 6.06 (1H, dd, 15.2, 10.7) | 128.5 | 6.06 (1H, m) | 128.5 |
| 20 | 6.32 (1H, m) | 132.2 | 6.31(1H, dd, 13.9, 10.7) | 132.9 |
| 21 | 6.13 (1H, m) | 131.5 | 6.13 (1H, m) | 131.2 |
| 22 | 6.13 (1H, m) | 131.9 | 6.13 (1H, m) | 131.9 |
| 23 | 6.13 (1H, m) | 131.7 | 6.13 (1H, m) | 131.8 |
| 24 | 6.11 (1H, m) | 133.0 | 6.13 (1H, m) | 133.2 |
| 25 | 5.60 (1H, m) | 130.4 | 5.61 (1H, m) | 130.4 |
| 26 | 2.39, 2.24 (2H, m) | 39.0 | 2.41, 2.29 (2H, m) | 39.0 |
| 27 | 4.88 (1H, m) | 69.5 | 4.88 (1H, m) | 69.5 |
| 28 | 1.17 (3H, d, 6.1) | 20.2 | 1.16 (3H, d, 6.7) | 20.2 |
| Sugar | ||||
| 1' | 4.53 (1H, s) | 95.9 | 4.39 (1H, s) | 96.4 |
| 2' | 3.75 (1H, d, 1.7) | 68.0 | 3.69 (1H, d, 1.6) | 68.5 |
| 3' | 2.81 (1H, d, 4.7) | 56.0 | 2.62 (1H, m) | 56.0 |
| 4' | 3.16 (1H, dd, 9.6, 8.9) | 70.1 | 3.06 (1H, m) | 70.9 |
| 5' | 3.24 (1H, m) | 72.7 | 3.12 (1H, m) | 72.9 |
| 6' | 1.17 (3H, d, 6.1) | 17.8 | 1.16 (3H, d, 6.7) | 17.8 |
| OH/NH | 7.18, 5.20 (brs) | - | 7.32, 6.83 (brs) | - |
a) 300 MHz;
b) 125 MHz;
c) 600 MHz. Chemical shifts (δ) is expressed in ppm.
3b: UV absorbing, pale yellow solid (8.2 mg). UV/Vis (0.1 mg/ml MeOH): λmax (log ε) = 317 (4.09), 302 (4.14), 287 (4.10) nm. NMR data see Table 2 and S6 Fig. (-)-ESI MS: m/z (%) = 769 ([M+HCOO]-, 100). –ESIHR MS: m/z = 725.38606 [M+H]+, (calcd 725.38605 for C36H57N2O13).
Antifungal susceptibility testing
MICs were determined according to NCCLS document M27-A [44] and M38-P [45] for yeast and conidia-forming filamentous fungi, respectively. Due to the nature of the assayed compounds, the dilutions were made according to the indications for insoluble antibiotics in water. The transmittance of the cultures was adjusted with 0.82% NaCl up to values between 75–80% for Filobasidiella neoformans and Issatchenkia orientalis, 78–82% for Aspergillus and 74–76% for Penicillium. Each suspension was diluted in RPMI-1640 medium 1:100 for Aspergillus and Penicillium, and 1:2000 for F. neoformans and I. orientalis. 1, 1a and 4b were dissolved in 2 g/L DMSO and, 2a, 2b and 3a in 6 g/L DMSO. Each drug was serially diluted twofold in RPMI-1640 medium and the appropriate dilutions were finally diluted 1:50. The assays were performed in multi-well plates by mixing 50 μl of the polyene dilutions with 50 μl of the cellular suspension. MICs were interpreted after 24 hours incubation at 37°C for Issatchenkia orientalis and Aspergillus niger and 48 hours for Filobasidiella neoformans and Penicillium chrysogenum. For all the polyenes assayed, the endpoint was defined at the lowest concentration that completely inhibited growth. The polyenes used in the study were purified as described above.
Hemolytic activity assay
The different polyenes were dissolved in DMSO at 5 nmol/μl for 2b and 4b, 20 nmol/μl for 3b, and 0.05 nmol/μl for 1. Increasing quantities of the different polyenes were brought to a final volume of 25 μl of DMSO and mixed by gently shaking with 125 μl of PBS buffer containing 2.5% of horse blood. After incubation at 37°C for 30 min without agitation, cells were pelleted by centrifugation. The hemolysis was evaluated by measuring the absorbance at 545 nm. The values corresponding to total hemolysis were estimated with a suspension of 2.5% horse blood in distiller water. Horse blood was from Oxoid (defibrinated blood).
Results
Production of a new CE-108 derivative, CE-108D (3a), by genetic engineering
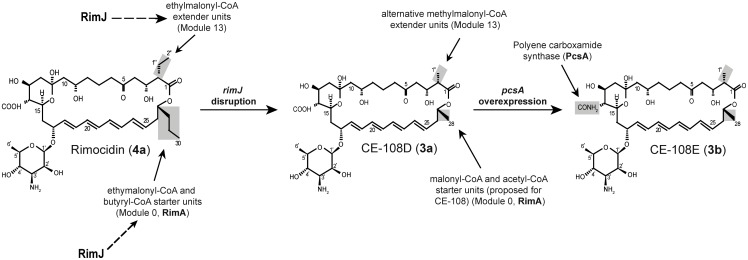
According to our previous findings [20], disruption of the rimJ gene, encoding a crotonyl-CoA carboxylase/reductase belonging to the rimocidin/CE-108 gene cluster, caused some interesting differences in the chromatographic profile of the produced polyenes compared to the wild type strain. The observed chromatographic profile of polyene production is consistent with the putative role of rimJ in controlling the cellular balance of ethylmalonyl-CoA (or possibly ethylmalonyl-CoA and butyryl-CoA) needed as starter unit for rimocidin production. Bearing in mind that ethylmalonyl-CoA is also needed for elongation step 13 in rimocidin/CE-108 biosynthesis, an altered intracellular concentration of ethylmalonyl-CoA units might be targeting two biosynthetic steps of the rimocidin/CE-108 pathway: the loading process and elongation step 13.
To further explore the in vivo specificity of PcsA toward the new compounds, we first had to design a new rimJ disruptant, as it shared the same thiostrepton resistance marker with the PcsA expression plasmid. We therefore exchanged the thiostrepton ressistance marker for erythromycin, which subsequently allowed the transformation with the PcsA expression plasmid (as discussed later). Thus, the ermE gene and an internal fragment of rimJ (positions 16,691 to 17,530 bp from the previously determined sequence, Accession Number AY442225) were cloned together into the XhoI/SacI sites of the actinophage PM1 [40], replacing the thiostrepton resistance gene and part of the hygromycin resistance gene, as described in Table 1. The resulting recombinant phage carrying the erythromycin resistance gene, named PM1-709B, was used to infect S. diastaticus var. 108. The correct rimJ disruptant (S. diastaticus var. 108::PM1-709B) was confirmed by Southern blotting. HPLC analysis of the fermentation broth of this recombinant showed a clear decrease in 4a production, an increase in 2a and the overproduction of new compounds with typical tetraene spectra (Fig 2A). One of them, with a retention time lower than that of 2a was called 3a and two other compounds, with a retention time between 2a and 4a were additionally found, but were not fully elucidated. HPLC-MS analysis of these compounds revealed preliminary masses of 725 Dalton for 3a and 753 Dalton for both new overproduced compounds (marked as 753 in Fig 2A). The loss of 14 units in 3a compared with 2a and the same loss for the other two compounds with respect to 4a is consistent with the incorporation of an alternative unit by the loading module RimA and/or the extender module 13. The two compounds with a deduced mass of 753 Dalton could not be efficiently separated by HPLC and were thus not further investigated. Hence, we focused on the characterization of 3a.
Fig 2. rimJ recombinants.
(A) HPLC analysis of the fermentation broth of S. diastaticus var. 108/PM1-709B (rimJ disruptant). (B) HPLC analysis of the fermentation broth of the rimJ disruptant carrying pcsA under the control of the constitutive promoter ermE P* (S. diastaticus var. 108::PM1-709B/860); the numbers on the peaks in A and B correspond to the polyenes shown in Fig 1. Peaks marked with 753 and 752 are cited in the text. (C) Percentage of CE-108 and rimocidin production. Polyenes production was measured from liquid cultures by HPLC as described in Material and Methods. The data shown are the mean of three independent experiments. The standard deviation of the mean is indicated by error bars. 1, WT control S. diastaticus var. 108/780 carrying the empty vector; 2, WT derivative carrying rimJ under the control of the ermE P* promoter.
Also, we isolated rimJ and expressed it under the control of a constitutive promoter in the wild type strain. A DNA fragment from the rim cluster containing rimJ and the 5´end of rimK (17,866–16,005 bp of the sequence at accession number AY442225) and flanked by BamHI/EheI sites, was fused with the ermEp* promoter [43] in the pHJL401 vector by several steps detailed in Table 1 (see also Material and Methods). This recombinant plasmid, named pSM781, carrying extra copies of recombinant rimJ, was introduced by transformation into S. diastaticus var. 108 to give rise to S. diastaticus var. 108/781. The presence of the plasmid was confirmed by direct plasmid extraction. The fermentation broth of this recombinant strain was tested for tetraene production by HPLC analysis and compared to the wild type control (S. diastaticus var. 108/780); the chromatograms showed that the production of 2a was substantially reduced compared with the wild type in favour of an increase of 4a (Fig 2C). This result confirms that the ethylmalonyl-CoA precursor required as starter and extender units (module 13) acts as a limiting factor in 4a production by the wild type strain.
3a is a new substrate for the carboxamide synthase PcsA
In our previous report [32], PcsA was shown to exhibit a broad substrate specificity and converts the exoxyclic carboxyl group of 2a, 4a and pimaricin into the corresponding amides 2b, 4b and AB-400, respectively. Because the carboxamide polyenes were improved in some pharmacological properties, we were interested in knowing if the polyene derivative 3a isolated from the rimJ mutant strain can be also be recognized by PcsA, and if so, if the carboxamide derivative has improved antimicrobial properties.
To answer these questions, the engineered tetraene 3a was purified as described in Material and Methods and used as substrate for in vitro amidotransferase assays performed using cell-free extracts from S. lividans TK21/pSM858 as previously described [32]. The reaction products were analyzed by HPLC. A clear conversion of 3a into another peak was observed (data not shown). The identity of the new peak in the amidation reaction was confirmed by HPLC-MS analysis, with an experimental mass for the reaction product of 724 Dalton for the putative amide of 3a. The loss of 1 unit in the mass of 3a strongly suggested that conversion of a carboxyl group into the amide took place, indicating that the new carboxyl polyene 3a is a new substrate of PcsA activity. The new polyene amide was called 3b.
Suitably engineered S. diastaticus var. 108 overproduces the new carboxylated and amidated tetraene
The first step towards the characterization of 3a and its corresponding amide 3b was to generate a strain able to produce these new polyenes. For this purpose, a double recombinant strain was generated. Protoplasts of the lysogen S. diastaticus var. 108/PM1-709B (rimJ disruptant) were transformed with a construct carrying pcsA under the control of ermE P * in the low copy number plasmid pSM860 (Table 1). Several colonies were selected with erythromycin and thiostrepton. The correct rimJ disruption and the presence of the plasmid pSM860 were both confirmed by Southern blotting and direct plasmid extraction, respectively.
As expected, HPLC analysis of the fermentation broth of the double recombinant S. diastaticus var. 108::PM1-709B/pSM860 clearly revealed overproduction of the expected polyene amide 3b (Fig 2B). Other new peaks with retention times between 2a and 4b with masses of 752 Dalton deduced by HPLC-MS analysis could be the respective amides of the 753 Dalton carboxylated polyenes described above.
Characterization of the new tetraenes
3a and 3b were isolated from the recombinant strains S. diastaticus var. 108::PM1-709B and S. diastaticus var. 108::PM1-709B/pSM860, respectively, as described in Material and Methods.
Chemical structure elucidation of 3a and 3b
Both polyene macrolides were faint yellow solids. Compound 3a was sparingly soluble in methanol and readily soluble in DMSO and pyridine, while 3b was well soluble in methanol and DMSO.
The HR-ESI mass spectrum of the pale yellow powdery 3a showed a pseudomolecular ion peak at m/z = 726.36927 [M+H]+, which corresponds to the ion formula C36H56NO14, and fits only with structure 3a amongst the alternatives listed above. The 1H NMR spectrum showed very close similarity to those of 2a, 2b, 4a, 4b, CE-108C, and rimocidin C [16,17,19].
The 1H NMR spectrum displayed four signals in the sp 2 region at δ 6.32 (1H, m), between δ 6.05–6.15 (5H, m), at δ 5.89 (1H, dd), and at δ 5.65 (1H, m), with integration of eight protons in total. Two exchangeable signals appeared at δ 7.18 and 5.20 as broad singlets. One oxygenated methine at δ 4.53 was due to an anomeric proton, as the HSQC spectrum displayed. The other protons of the sugar moiety appeared in the range of δ 4.62–3.25; a methine group at δ 2.81 was possibly connected to a nitrogen atom. In the aliphatic region between δ 2.60–1.40, the spectrum showed a complex multiplet pattern, in addition to three methyl doublets: two at δ 1.17 and the third at δ 1.08 were present (S1 Fig).
In the 13C NMR spectrum, 36 carbon signals were observed, which is in agreement with the HR-ESI mass spectrum. These carbon signals could be classified as three carbonyls: one ketone CO at δ 208.5, two CO signals corresponding to an acid, amid or ester at δ 173.0 and 176.4. Eight sp 2 carbon signals were in the range of δ 136.4–128.4, two anomeric carbons gave signals at δ 96.9 (Cq) and δ 95.9 (CH). Oxygenated carbons were observed between δ 74.1 to 65.5, which were attributed to C-3, 9, 13, 15, 17, 27, 2', 4' and 5'. Finally three methyls were present at δ 20.2, 17.8, and 13.2, respectively (S2 Fig).
To confirm the structure of 3a, it was subjected to 2D NMR measurements. The H,H COSY spectrum showed a correlation series beginning with the methine carbinol at δ 4.38 assigned to H-17, which was coupled with one of the methylene protons at δ 2.16 assigned to H-16 and with the sp 2 methine doublet of doublets at δ 5.89 (H-18). The latter proton correlated with H-19 at δ 6.06, which in turn coupled with another sp 2 methine proton H-20 at δ 6.32. The signal at δ 2.81 (H-3') was correlated to the proton at δ 3.75 (H-2') and also with the methine proton at δ 3.16 (H-4'), which correlated to another methine at δ 3.24 to construct a part of the sugar moiety. The methylene protons at δ 2.15 and 1.53 assigned to H-16 coupled with the methine carbinol at δ 4.16 (H-15) (see Table 2; S3 and S4 Fig). The COSY and HMBC correlation confirmed the southern hemisphere and the structure of the amino sugar clearly. The anomeric proton H-1' (4.53) exhibited a 3 J correlation with C-17 (74.1), which confirmed the position of the sugar.
The interpretation of couplings in the northern part of the molecule was difficult, due to strong signal overlapping. The HMBC 3 J coupling of the proton at δ 4.88 (C-27) to the carbonyl at δ 173.0 (C-1) confirmed the lactone. The methyl protons of 2-Me displayed 3 J couplings with the lactone carbonyl, the carbinol carbon C-3 (67.7) and a 2 J coupling with C-2 (47.0). The methyl protons (2-Me) showed an additional COSY correlation with the methine at δ 2.20 (H-2), which in turn correlated to the carbinol methine at δ 4.03 (H-3). The HMBC correlations (S5 and S6 Figs) confirmed those relations observed in the COSY spectrum, which led to the complete elucidation of the structure of 3a as shown in Fig 1. It should be mentioned, however, that there were no COSY or HMBC correlation visible between CH2-8 and CH2-9, and also not between the C-14-carboxyl group and H-13 or H-15, respectively, in both 3a and 3b.
Compound 3b was obtained as a pale yellow powder and showed a typical tetraene UV spectrum with λmax at 317, 302 and 287 nm similar to that of 3a. The combined data of HR ESI MS (m/z = 725.38543 [M+H]+), 13C NMR and 1H NMR data (see Table 2) of 3b delivered the molecular formula C36H56N2O13. The 1H NMR spectrum was very similar to that of 3a. By comparing the molecular formula of 3b and 3a, the former one must have an amide group (CONH2) instead of the carboxylic group (COOH) in the latter one. The 2D NMR experiments showed the same correlations like 3a, so that the structure 3b is fully confirmed (Fig 1).
Antifungal properties of the recombinant polyenes
MICs were measured for 3a, 3b, and the previously characterized compounds 2a, 2b, 4a and 4b. Amphotericin B (1) was also included for reference. Broth microdilution MICs were determined according to NCCLS document M27-A [44] for F. neoformans, C. krusei and according to document M38-P [45] for A. niger and P. chrysogenum as described in Material and Methods.
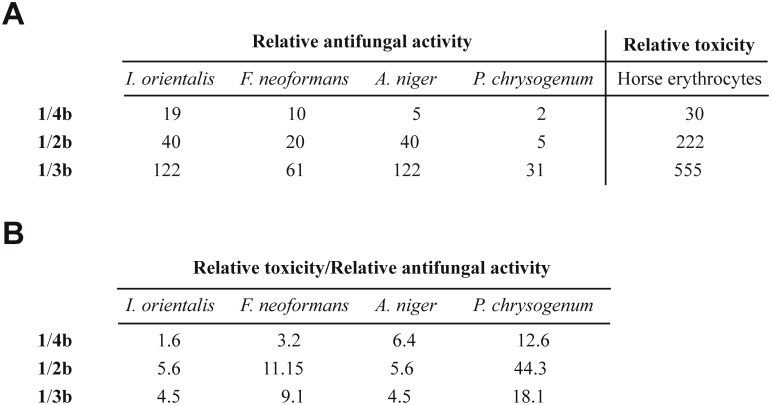
The results revealed a low biological activity for the carboxylated polyene 3a against all the microorganisms tested. The conversion of the side chain carboxyl group of 3a into an amide group present in 3b led to an increasing biological activity against all fungi tested (Fig 3A). As expected from previous data [16], the MIC values for the polyene amides 2b and 4b are substantially lower than those of the corresponding carboxyl polyene compounds against all fungi tested (Fig 3A).
Fig 3. Biological activities for the polyenes tested.
(A) In vitro susceptibilities of Issatchenkia orientalis, Filobasidiella neoformans, Aspergillus niger and Penicillium chrysogenum to seven antifungal agents. Minimal and maximum values of the range of concentrations for the MICs (μM) are detailed. Minimal values correspond to the lowest concentration of polyene in which growth appeared and maxima values were the lowest concentration that completely inhibited growth. Data are shown at 24 hours for I. orientalis and A. niger and at 48 hours for F. neoformans and P. chrysogenum. (B) Hemolytic activity of 1, 4b, 2b and 3b. Concentration in μM of the polyenes for 50% and 100% hemolysis is expressed. The standard deviation is shown in parentheses.
For the amide derivatives, we observed that the biological activity of 3b is lower than that of 2b. We conclude from this that the substitution of an ethyl group at C-2 by a methyl residue led to a reduction in biological activity. In this sense, 4a is more bioactive than 2a and 3a because it presents a lateral ethyl chain at C-2 besides a propyl chain at C-27 instead of a methyl group present in both 2a and 3a. So, interestingly, a decrease of biological activity occurs when the number of carbons in side chains at C-2 or C-27 is reduced.
Toxicity assays
Horse erythrocytes were used as a cellular model for this study since similar hemolytic activities have been reported for horse and human blood [16]. Since we had previously found that conversion of the free carboxyl group into an amide did not alter hemolytic activity, we confined the toxicity assays to the polyene amides due to their major antifungal activity. The hemolytic activity assays of 3b were evaluated versus 1, 2b and 4b as described in Material and Methods. Concentrations expressed in μM of the different tetraenes producing 50% and 100% of hemolysis are detailed in Fig 3B. Maximum toxicity was found for 1. It is noteworthy that, as occurred for the antifungal activity, the toxicity of the polyene amides assayed was directly related to the number of carbons in the side chains at C-2 and C-27, with toxicity being higher for the polyenes having longer side chains.
To correlate the antifungal activity and toxicity of these new compounds, Fig 4A shows the number of times that 1 is more active in terms of both antifungal activity and toxicity than the assayed polyenes. On a comparable level of inhibition against the tested fungi, all analyzed polyene amides showed lower toxicity than 1 (Fig 4B).
Fig 4. Relative antifungal activity and toxicity.
(A) Number of times that 1 is more active against all the fungi tested and toxic in terms of hemolytic activity than the polyenes 4b, 2b and 3b. (B) Ratio between relative toxicity and relative antifungal activities. The values indicate the times that the polyene is less toxic than 1 for the same level of inhibition against the fungi tested.
Discussion
Here we report the in vivo biosynthesis of new rimocidin analogues through inactivation of the gene rimJ, which encodes a predicted crotonyl-CoA carboxylase/reductase in the rimocidin biosynthetic gene cluster in S. diastaticus var. 108. The newly produced macrolide antibiotics were furthermore shown to be substrates for the heterologously expressed carboxamide synthase PscA, which allowed the in vivo and in vitro generation of further rimocidin derivatives with increased bioactivity.
3a and 3b are structural derivatives of 2a and 2b, respectively, by substitution of an ethyl side chain with a methyl side chain at C-2. During 3a formation, the elongation module 13 incorporates acetate (derived from methylmalonyl-CoA) rather than the naturally observed butyrate (derived from ethylmalonyl) in the biosynthesis of 2a and 4a [20], indicating a reduced selectivity for the PKS involved in elongation module 13, which can incorporate methylmalonyl-CoA units when the availability of ethylmalonyl-CoA units is reduced as a consequence of rimJ disruption. Once the polyketide chain is completed and cyclized, two tailoring reactions occur: the conversion of the side chain methyl group at C-14 into a free carboxyl group by cytochrome P450 monooxygenase RimG; and the incorporation of mycosamine at C-17 by glycosyltransferase RimE [20]. The carboxylated compound 3a is the product of this biosynthetic pathway but can be a substrate for the polyene carboxamide synthase PcsA activity, as we have confirmed here through in vitro amidation assays, giving rise to 3b (Fig 5).
Fig 5. Proposed model for 3a and 3b biosynthesis.
Besides this new characterized compound, other uncharacterized peaks with typical tetraene spectra were found in the fermentation broth of the rimJ recombinant whose deduced masses of 753 Dalton in both cases are in accordance with rimocidin (4a) derivatives carrying: (i) an ethyl side chain at C-27 by incorporation of methylmalonyl-CoA, or possibly methylmalonyl-CoA and propionyl-CoA, in the loading module; (ii) a methyl side chain at C-2 by incorporation of methylmalonyl-CoA as extender unit in elongation module 13 as described above for 3a biosynthesis. Thus, we expect the presence of both rimocidin derivatives in this recombinant strain and probably a new derivative carrying both chemical modifications.
As we report here, the availability of ethylmalonyl-CoA units constitutes a limiting factor in 4a production, so that all the other carboxylated tetraene derivatives produced by this strain are a consequence of a low availability of these precursors. The presence of 4a and 2a in the fermentation broth of the rimJ mutant suggests the presence of at least one other CCR activity related to primary metabolism in S. diastaticus var. 108; the corresponding gene and others related to valine catabolism also involved in formation of these carboxylic acids [46] could be good tools for trying to overproduce new interesting compounds by gene disruption. An appropriated genetic manipulation of the producer strain S. diastaticus var. 108, with a highly flexible PKS, would favor the overproduction of these new carboxylated tetraenes.
Loading module RimA could have a higher flexibility than described for 2a and 4a recognizing not only acetyl-CoA and butyryl-CoA but probably propionyl-CoA, making it a good tool for obtaining polyketide derivatives by combinatorial biosynthesis. PimS0, the loading module for pimaricin biosynthesis, has a high degree of identity with RimA (70%), but PimS0 cannot incorporate either propionyl-CoA or butyryl-CoA as starter units in the same genetic context as RimA [47].
The new carboxylated compound 3a turned out to be a new substrate for PcsA activity, converting it into its amide 3b. However, as occurred for 4a and 2a, 3a was not recognized by the polyene carboxamide synthase PcsB involved in the conversion of pimaricin to its corresponding amide [35]. The improved pharmacological properties of the polyene amide 3b) compared to the carboxylated compound 3a suggests the interesting possibility of extending this chemical modification to other polyenes.
The major compounds produced by a strain are not necessarily the most interesting. It is worth seeking also minor components in the fermentation broth of the producer strain to obtain new metabolites with improved pharmacological properties. A new CE-108 derivative produced by incorporation of malonyl-CoA as alternative extender unit by elongation module 13 is also possible. HPLC-MS analysis confirmed the presence of other uncharacterized compounds with typical tetraene spectra and minor retention times in the fermentation broth of S. diastaticus var. 108. whose masses of 711 and 710 are consistent with this new derivative and its corresponding amide. Here we show a clear correlation between the number of carbons in the side chains at C-2 and C-27 and the antifungal activity, but the loss of antifungal activity when the number of carbons is reduced is compensated by a substantial reduction in toxicity (Fig 4). This suggests these compounds as potential alternatives to 1 in the treatment of systemic infections.
Conclusions
In a previous study [32], we described a polyene carboxamide synthase, PcsA, from S. diastaticus var. 108 involved in a post-PKS activity converting the free carboxyl group widespread in most natural polyene macrolides into an amide group. PcsA not only converts the carboxyl group of 2a and 4a (the predominant polyenes produced by this strain) into their respective polyene amides 2b and 4b, but also the heterologous substrate pimaricin, leading to an increase in selectivity towards fungal membranes in all cases [16]. Importantly, the high flexibility of the PKS involved in polyene macrolide biosynthesis in this strain allowed the overproduction of new polyene derivatives by disrupting a gene encoding a crotonyl-CoA carboxylase/reductase (rimJ) in the polyene biosynthetic gene cluster. One of them, 3a, obtained by incorporation of an alternative extender unit in elongation step 13 of the biosynthetic pathway, turned out to be a new substrate for PcsA. 3b, the corresponding carboxamide derivative of 3a, showed improved pharmacological properties with respect to the parental products. The reduced selectivity of this enzymatic activity plus the high flexibility of the S. diastaticus var. 108 PKS are useful tools for generating new antifungal drugs.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
This work was supported by grants to F.M. from the Spanish Ministerio de Ciencia e Innovación BIO2005-02785 and BIO2008-03683, and to J.C.A. from the Spanish Ministerio de Economía y Competitividad BFU2012-39879-C02-01. The German Academic Exchange Service (DAAD) supported M.A.R. with a Ph.D. grant. We thank Prof. D.A. Hopwood for critical reading and suggestions, J.C.A. for his interest in the project and T. Cuesta for excellent technical help.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants to F.M. from the Spanish Ministerio de Ciencia e Innovación BIO2005-02785 and BIO2008-03683, and to J.C.A. from Spanish Ministerio de Economía y Competitividad BFU2012-39879-C02-01. The German Academic Exchange Service (DAAD) supported M.A.R. with a Ph.D. grant.
References
- 1. Goldman RD, Ong M, Wolpin J, Doyle J, Parshuram C, et al. (2007) Pharmacological risk factors for amphotericin B nephrotoxicity in children. J Clin Pharmacol 47: 1049–1054. [DOI] [PubMed] [Google Scholar]
- 2. Cybulska B, Bolard J, Seksek O, Czerwinski A, Borowski E (1995) Identification of the structural elements of amphotericin B and other polyene macrolide antibiotics of the hepteane group influencing the ionic selectivity of the permeability pathways formed in the red cell membrane. Biochim Biophys Acta 1240: 167–178. [DOI] [PubMed] [Google Scholar]
- 3. Paquet V, Volmer AA, Carreira EM (2008) Synthesis and in vitro biological properties of novel cationic derivatives of amphotericin B. Chemistry 14: 2465–2481. 10.1002/chem.200701237 [DOI] [PubMed] [Google Scholar]
- 4. Szlinder-Richert J, Mazerski J, Cybulska B, Grzybowska J, Borowski E (2001) MFAME, N-methyl-N-D-fructosyl amphotericin B methyl ester, a new amphotericin B derivative of low toxicity: relationship between self-association and effects on red blood cells. Biochim Biophys Acta 1528: 15–24. [DOI] [PubMed] [Google Scholar]
- 5. Bruzzese T, Rimaroli C, Bonabello A (1996) Amide derivatives of partricin A with potent antifungal activity. Eur J Med Chem 31: 965–972. [Google Scholar]
- 6. Preobrazhenskaya MN, Olsufyeva EN, Solovieva SE, Tevyashova AN, Reznikova MI, et al. (2009) Chemical modification and biological evaluation of new semisynthetic derivatives of 28,29-Didehydronystatin A1 (S44HP), a genetically engineered antifungal polyene macrolide antibiotic. J Med Chem 52: 189–196. 10.1021/jm800695k [DOI] [PubMed] [Google Scholar]
- 7. Donadio S, Staver MJ, McAlpine JB, Swanson SJ, Katz L (1991) Modular organization of genes required for complex polyketide biosynthesis. Science 252: 675–679. [DOI] [PubMed] [Google Scholar]
- 8. Aparicio JF, Caffrey P, Gil JA, Zotchev SB (2003) Polyene antibiotic biosynthesis gene clusters. Appl Microbiol Biotechnol 61: 179–188. [DOI] [PubMed] [Google Scholar]
- 9. Borgos SE, Tsan P, Sletta H, Ellingsen TE, Lancelin JM, et al. (2006) Probing the structure-function relationship of polyene macrolides: engineered biosynthesis of soluble nystatin analogues. J Med Chem 49: 2431–2439. [DOI] [PubMed] [Google Scholar]
- 10. Brautaset T, Bruheim P, Sletta H, Hagen L, Ellingsen TE, et al. (2002) Hexaene derivatives of nystatin produced as a result of an induced rearrangement within the nysC polyketide synthase gene in S. noursei ATCC 11455. Chem Biol 9: 367–373. [DOI] [PubMed] [Google Scholar]
- 11. Caffrey P, Aparicio JF, Malpartida F, Zotchev SB (2008) Biosynthetic engineering of polyene macrolides towards generation of improved antifungal and antiparasitic agents. Curr Top Med Chem 8: 639–653. [DOI] [PubMed] [Google Scholar]
- 12. Carmody M, Murphy B, Byrne B, Power P, Rai D, et al. (2005) Biosynthesis of amphotericin derivatives lacking exocyclic carboxyl groups. J Biol Chem 280: 34420–34426. [DOI] [PubMed] [Google Scholar]
- 13. Mendes MV, Recio E, Fouces R, Luiten R, Martin JF, et al. (2001) Engineered biosynthesis of novel polyenes: a pimaricin derivative produced by targeted gene disruption in Streptomyces natalensis . Chem Biol 8: 635–644. [DOI] [PubMed] [Google Scholar]
- 14. Power P, Dunne T, Murphy B, Nic Lochlainn L, Rai D, et al. (2008) Engineered synthesis of 7-oxo- and 15-deoxy-15-oxo-amphotericins: insights into structure-activity relationships in polyene antibiotics. Chem Biol 15: 78–86. [DOI] [PubMed] [Google Scholar]
- 15. Brautaset T, Sletta H, Nedal A, Borgos SE, Degnes KF, et al. (2008) Improved antifungal polyene macrolides via engineering of the nystatin biosynthetic genes in Streptomyces noursei . Chem Biol 15: 1198–1206. 10.1016/j.chembiol.2008.08.009 [DOI] [PubMed] [Google Scholar]
- 16. Seco EM, Cuesta T, Fotso S, Laatsch H, Malpartida F (2005) Two polyene amides produced by genetically modified Streptomyces diastaticus var. 108. Chem Biol 12: 535–543. [DOI] [PubMed] [Google Scholar]
- 17. Seco EM, Fotso S, Laatsch H, Malpartida F (2005) A tailoring activity is responsible for generating polyene amide derivatives in Streptomyces diastaticus var. 108. Chem Biol 12: 1093–1101. [DOI] [PubMed] [Google Scholar]
- 18. Tevyashova AN, Olsufyeva EN, Solovieva SE, Printsevskaya SS, Reznikova MI, et al. (2013) Structure-antifungal activity relationships of polyene antibiotics of the amphotericin B group. Antimicrob Agents Chemother 57: 3815–3822. 10.1128/AAC.00270-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pérez-Zúñiga FJ, Seco EM, Cuesta T, Degenhardt F, Rohr J, et al. (2004) CE-108, a new macrolide tetraene antibiotic. J Antibiot (Tokyo) 57: 197–204. [DOI] [PubMed] [Google Scholar]
- 20. Seco EM, Perez-Zuniga FJ, Rolon MS, Malpartida F (2004) Starter unit choice determines the production of two tetraene macrolides, rimocidin and CE-108, in Streptomyces diastaticus var. 108. Chem Biol 11: 357–366. [DOI] [PubMed] [Google Scholar]
- 21. Aparicio JF, Colina AJ, Ceballos E, Martin JF (1999) The biosynthetic gene cluster for the 26-membered ring polyene macrolide pimaricin. A new polyketide synthase organization encoded by two subclusters separated by functionalization genes. J Biol Chem 274: 10133–10139. [DOI] [PubMed] [Google Scholar]
- 22. Brautaset T, Sekurova ON, Sletta H, Ellingsen TE, StrLm AR, et al. (2000) Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem Biol 7: 395–403. [DOI] [PubMed] [Google Scholar]
- 23. Caffrey P, Lynch S, Flood E, Finnan S, Oliynyk M (2001) Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem Biol 8: 713–723. [DOI] [PubMed] [Google Scholar]
- 24. Bisang C, Long PF, Cortes J, Westcott J, Crosby J, et al. (1999) A chain initiation factor common to both modular and aromatic polyketide synthases. Nature 401: 502–505. [DOI] [PubMed] [Google Scholar]
- 25. Witkowski A, Joshi AK, Lindqvist Y, Smith S (1999) Conversion of a beta-ketoacyl synthase to a malonyl decarboxylase by replacement of the active-site cysteine with glutamine. Biochemistry 38: 11643–11650. [DOI] [PubMed] [Google Scholar]
- 26. Jung WS, Yoo YJ, Park JW, Park SR, Han AR, et al. (2011) A combined approach of classical mutagenesis and rational metabolic engineering improves rapamycin biosynthesis and provides insights into methylmalonyl-CoA precursor supply pathway in Streptomyces hygroscopicus ATCC 29253. Appl Microbiol Biotechnol 91: 1389–1397. 10.1007/s00253-011-3348-6 [DOI] [PubMed] [Google Scholar]
- 27. Wallace KK, Zhao B, McArthur HA, Reynolds KA (1995) In vivo analysis of straight-chain and branched-chain fatty acid biosynthesis in three actinomycetes. FEMS Microbiol Lett 131: 227–234. [DOI] [PubMed] [Google Scholar]
- 28. Erb TJ, Berg IA, Brecht V, Muller M, Fuchs G, et al. (2007) Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc Natl Acad Sci U S A 104: 10631–10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Erb T, Brecht V, Fuchs G, Muller M, Alber B (2009) Carboxylation mechanism and stereochemistry of crotonyl-CoA carboxylase/reductase, a carboxylating enoyl-thioester reductase. Proc Natl Acad Sci U S A 106: 8871–8876. 10.1073/pnas.0903939106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Hazzard C, Eustaquio AS, Reynolds KA, Moore BS (2009) Biosynthesis of salinosporamides from alpha,beta-unsaturated fatty acids: implications for extending polyketide synthase diversity. J Am Chem Soc 131: 10376–10377. 10.1021/ja9042824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kosec G, Goranovic D, Mrak P, Fujs S, Kuscer E, et al. (2012) Novel chemobiosynthetic approach for exclusive production of FK506. Metab Eng 14: 39–46. 10.1016/j.ymben.2011.11.003 [DOI] [PubMed] [Google Scholar]
- 32. Seco EM, Miranzo D, Nieto C, Malpartida F (2010) The pcsA gene from Streptomyces diastaticus var. 108 encodes a polyene carboxamide synthase with broad substrate specificity for polyene amides biosynthesis. Appl Microbiol Biotechnol 85: 1797–1807. 10.1007/s00253-009-2193-3 [DOI] [PubMed] [Google Scholar]
- 33. Massiere F, Badet-Denisot MA (1998) The mechanism of glutamine-dependent amidotransferases. Cell Mol Life Sci 54: 205–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zalkin H, Smith JL (1998) Enzymes utilizing glutamine as an amide donor. Adv Enzymol Relat Areas Mol Biol 72: 87–144. [DOI] [PubMed] [Google Scholar]
- 35. Miranzo D, Seco EM, Cuesta T, Malpartida F (2010) Isolation and characterization of pcsB, the gene for a polyene carboxamide synthase that tailors pimaricin into AB-400. Appl Microbiol Biotechnol 85: 1809–1819. 10.1007/s00253-009-2195-1 [DOI] [PubMed] [Google Scholar]
- 36.Atlas RM (1993) Microbiological media. Florida.
- 37. Kieser T, Bibb MJ, Buttner MJ (2000) Practical Streptomyces Genetics; Foundation JI, editor. Norwich. [Google Scholar]
- 38. Yanisch-Perron C, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33: 103–119. [DOI] [PubMed] [Google Scholar]
- 39. Maniatis T, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual. New York. [Google Scholar]
- 40. Malpartida F, Hopwood DA (1986) Physical and genetic characterisation of the gene cluster for the antibiotic actinorhodin in Streptomyces coelicolor A3(2). Mol Gen Genet 205: 66–73. [DOI] [PubMed] [Google Scholar]
- 41. Larson JL, Hershberger CL (1986) The minimal replicon of a streptomycete plasmid produces an ultrahigh level of plasmid DNA. Plasmid 15: 199–209. [DOI] [PubMed] [Google Scholar]
- 42. Lydiate DJ, Malpartida F, Hopwood DA (1985) The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene 35: 223–235. [DOI] [PubMed] [Google Scholar]
- 43. Bibb MJ, Janssen GR, Ward JM (1985) Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene 38: 215–226. [DOI] [PubMed] [Google Scholar]
- 44.M27-A. NCfCLS (1997) Reference method for broth dilution antifungal susceptibility testing of yeast: proposed standard 1997.
- 45.Standards NCfCL (1998) Reference method for broth dilution antifungal susceptibility testing of conidium-forming filamentous fungi. M38-P, vol18, No 13. Wayne, Pa.
- 46. Wallace KK, Bao ZY, Dai H, Digate R, Schuler G, et al. (1995) Purification of crotonyl-CoA reductase from Streptomyces collinus and cloning, sequencing and expression of the corresponding gene in Escherichia coli. Eur J Biochem 233: 954–962. [DOI] [PubMed] [Google Scholar]
- 47. Heia S, Borgos SE, Sletta H, Escudero L, Seco EM, et al. (2011) Initiation of polyene macrolide biosynthesis: interplay between polyketide synthase domains and modules as revealed via domain swapping, mutagenesis, and heterologous complementation. Appl Environ Microbiol 77: 6982–6990. 10.1128/AEM.05781-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.