Abstract
The diverse microbial populations constituting the intestinal microbiota promote immune development and differentiation, but because of their complex metabolic requirements and the consequent difficulty culturing them, they remained, until recently, largely uncharacterized and mysterious. In the last decade, deep nucleic acid sequencing platforms, new computational and bioinformatics tools, and full-genome characterization of several hundred commensal bacterial species facilitated studies of the microbiota and revealed that differences in microbiota composition can be associated with inflammatory, metabolic, and infectious diseases, that each human is colonized by a distinct bacterial flora, and that the microbiota can be manipulated to reduce and even cure some diseases. Different bacterial species induce distinct immune cell populations that can play pro- and anti-inflammatory roles, and thus the composition of the microbiota determines, in part, the level of resistance to infection and susceptibility to inflammatory diseases. This review summarizes recent work characterizing commensal microbes that contribute to the antimicrobial defense/inflammation axis.
Keywords: commensals, mucins, TLR signaling, antimicrobial molecules, dendritic cells
INTRODUCTION
The development in metazoans of a tubular tract for food digestion, nutrient absorption, and waste expulsion is believed to have occurred over 500 million years ago (1, 2). The evolution of the intestinal tract created a niche for microbial symbionts, with resulting complex microbial populations that are uniquely adapted to life in this environment and that, to greater and lesser extents, depend on each other for survival. Although these microbial populations have, for hundreds of years, provoked curiosity among microscopists, microbiologists, and physiologists, they remained largely uncultured and therefore uncharacterized, in part because they generally do not cause disease. The last decade, however, has seen remarkable progress in our understanding of the mammalian intestinal microbiota and their impact on immune system development and function (3, 4). Whereas much of the focus of microbiota and microbiome studies has been on microbiota composition of healthy individuals (5–7), parallel studies have correlated microbiota composition with a wide range of diseases, which are outlined in this review. The major messages emerging from this growing body of work are that healthy humans are colonized with diverse microbial populations that differ between individuals; that different microbes can have distinct interactions with the innate and adaptive immune systems of their mammalian host; and that exogenous factors, in particular medical interventions such as antibiotic administration, can lead to marked changes in the microbiota with downstream implications for health.
Variation in microbiota composition within healthy human populations has led to the enterotype hypothesis, which posits that humans can be divided into three groups on the basis of their fecal microbiota composition (7). The fecal microbiota of healthy individuals could be grouped into three enterotypes, the first predominated by Prevotella, the second by Bacteroides, and the third by Ruminococcus. Other studies suggest that microbiota differences between individuals occur over a wide and continuous range of compositions with indistinct boundaries (8). Further analysis of sequence data obtained from healthy subjects established associations between microbiota composition and breast-feeding, gender, and level of education. These studies also revealed correlations between microbial composition at different anatomic sites and demonstrated that the gut and vaginal microbiota are more stable than the oral microbiota (9).
Our understanding of how microbiota complexity is maintained and how the individual bacterial taxa compete with and support each other is rudimentary, but important new insights have emerged. Metabolism of host-derived polysaccharides by Bacteroides fragilis, an obligately anaerobic bacterial species that inhabits the human colon, is required for intestinal colonization and persistence, and genetic deletion of the polysaccharide utilization locus markedly reduces competitiveness in the gut (10). Commensal microbiota–mediated metabolism of host polysaccharides, for example the cleavage of sialic acid or fucose from host mucin, can promote infection with enterohemorrhagic Escherichia coli, Salmonella typhimurium, and Clostridium difficile by promoting virulence gene expression (11) or by enhancing growth (12). Competition between different microbial species and strains can also be mediated by distinct susceptibilities and resistances to phage-mediated lysis, a mechanism that has been shown to facilitate colonization of the gut with some strains of Enterococcus faecalis (13).
The interactions between bacterial taxa in the gut can be direct (e.g., bacterial species B inhibits or promotes bacterial species A) or indirect (e.g., bacterial species B modifies immunologic or physiologic host factors, which then either inhibit or promote colonization by species A). Studies of these interactions are greatly facilitated by isolation, growth, and characterization of the wide array of commensal bacterial species, a critical step that is both technically challenging and, given the marked genomic differences between bacterial strains belonging to the same species, daunting in terms of the massive number of potential strains to be studied. The importance of characterizing multiple strains was demonstrated in a study of four Bifidobacterium longum strains, of which only two provided resistance against an intestinal pathogen (14). Recent studies demonstrate that many colon-derived bacterial species can be cultured in vitro (15), including bacterial species that drive in vivo T cell differentiation (16, 17). The immunologic impact of microbiota composition is increasingly recognized as important; some bacterial taxa drive intestinal T regulatory cell (Treg) development, whereas others induce Th17 T cell development (16, 18). Microbial populations associated with specific mammalian host species have evolved to optimally promote their respective hosts’ immune system maturation (19).
BIOINFORMATIC AND COMPUTATIONAL PLATFORMS FOR MICROBIOTA/MICROBIOME ANALYSIS
Multiparallel nucleic acid sequencing has greatly enhanced our understanding of commensal bacterial populations. Microbiota composition is generally determined by sequencing PCR-amplified bacterial 16S ribosomal RNA genes, and the microbiome is determined by shotgun sequencing of randomly generated DNA fragments obtained by shearing DNA isolated from fecal or other samples (5). These approaches generated massive amounts of sequence data that required the development of bioinformatic programs to facilitate analysis. A number of platforms, including mothur (20) and QIIME (21), have been developed to organize sequence data and to assign taxonomic labels to each sequence. Other methods, such as UniFrac, enable investigators to compare complex samples and to correlate microbiota composition with specific experimental or clinical scenarios (22). Another method that has enabled investigators to identify bacterial taxa that differ between samples is LEfSe (linear discriminant analysis effect size), which supports high-dimensional class comparison between microbiomes obtained from different groups (for example, colitis versus normal control samples) (23). Programs such as MetaPhlAn (24) facilitate the determination of bacterial taxon prevalence in samples that have been shotgun sequenced, whereas PICRUSt enables investigators to estimate the representation of microbial metabolic pathways on the basis of 16S rRNA taxonomy (25). These platforms are well established and are commonly used for microbiota and microbiome analyses.
More recently, mathematical models have been used to predict shifts in microbiota composition following different perturbations and to identify interactions between distinct bacterial taxa. Using modified Lotka-Volterra equations, which were originally derived to mathematically model predator-prey dynamics, one mathematical approach incorporates the growth rates of different bacterial taxa, their susceptibilities to specific perturbations (such as antibiotic administration), and their impacts on each other. If provided with quantitative data on the densities of specific bacterial populations and knowledge of their growth rates and susceptibilities to a specific perturbation, one can calculate the strength of interactions between bacterial populations (26). This approach was used to identify a subnetwork of bacterial groups that appears to confer protection against C. difficile infection. Another mathematical approach to study reconstitution of germfree mice with complex flora suggested that interactions between bacterial species range from competitive to parasitic, with the surprising result that mutualistic interactions were not detectable (27).
INTESTINAL EPITHELIAL BARRIER AND INNATE IMMUNE DEFENSES
Mucin and Epithelial Cell–Mediated Barriers Against Infection
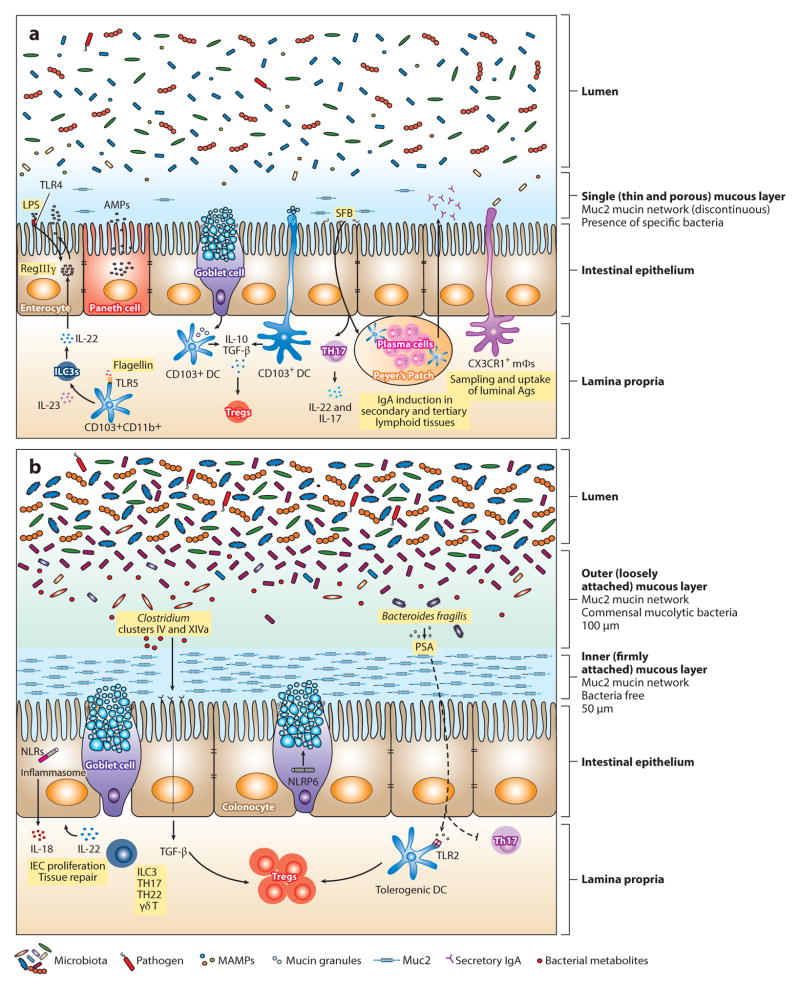
Although blood draining from the gut flows to the liver, which protects the systemic circulation by filtering microbes that traverse the intestinal epithelium (28), the most important barrier to bacterial entry of deeper tissues is the intestinal epithelium. The mucin layer that coats luminal epithelial cell surfaces physically excludes microbes inhabiting the intestinal lumen. Direct bacterial contact with luminal epithelial cell surfaces can occur in the absence of intestinal mucin layers or when specific microbes can penetrate mucin. The predominant mucin secreted by goblet cells is Muc2, which is a large, highly glycosylated multidomain protein that forms disulfide-bonded trimers that are partially resistant to trypsin. Muc2 forms a dense, approximately 50-μm-thick mucin layer in the colon that is attached to underlying epithelial cells and a more dispersed outer layer (29). Some pathogens and commensal bacteria can penetrate the dense mucin layer by proteolytic degradation (30, 31), and a subset of commensal bacteria can penetrate the outer mucin layer and break down and metabolize the O-linked glycans attached to Muc2 (32–34) (Figure 1). Deficient O-linked glycosylation of Muc2 by epithelial cells increases susceptibility to dextran sodium sulfate (DSS)-induced colitis and also shifts the composition of the microbiota (35, 36). DSS administration to mice is associated with increased bacterial penetration of the inner mucous layer by commensal bacteria, potentially leading to subsequent inflammation (37).
Figure 1.
Maintenance of intestinal homeostasis in the gastrointestinal tract. The intestinal epithelial surface is coated with a layer of mucus that has a pivotal role in intestinal barrier function. This mucous layer is organized by Muc2 mucin glycoproteins that polymerize into a gel-like structure, preventing luminal bacteria from coming into contact with epithelial cells. (b) In the large intestine, two mucous layers protect the colonic epithelium: a bacteria-free, dense, inner layer followed by an outer layer that harbors mucus-degrading bacteria. (a) In contrast, a single, loosely attached layer of mucus lines the small intestine. Mucins are produced by goblet cells and stored in secretory granules until appropriate stimulation, such as signaling through the Nod-like receptor pyrin domain 6 (NLRP6) inflammasome, prompts their release. Consistent with abundant mucus production in the colon, the number of goblet cells is much greater in the large intestine compared with the small bowel. The density and composition of luminal bacteria also differ between these two compartments. The small intestine harbors ~108 bacteria per gram of content. In mice, segmented filamentous bacteria (SFB) adhere to the intestinal surface and enhance the development of Th17 cells and the production of IgA by B cells. Microbial molecules stimulate the production of antimicrobial peptides (AMPs) from epithelial and Paneth cells through activation of innate immune receptors. For example, induction of regenerating islet-derived 3 γ(RegIIIγ), an antimicrobial protein that targets gram-positive bacteria and maintains host-bacterial segregation, is mediated through lipopolysaccharide (LPS) and flagellin stimulation of TLR4+ (Toll-like receptor 4) radioresistant cells and TLR5+CD103+CD11b+ dendritic cells (DCs), respectively. DCs and macrophages sample luminal antigens and stimulate cytokine production and T cell differentiation. Although CX3CR1+ macrophages are the major antigen-sampling mononuclear phagocytes in the small intestine, CD103+ DCs play a role as well. Nearly 1011–1012 bacteria reside in the large intestine. The microbiota are diverse, mainly comprising Lachnospiraceae, Bacteroides, and Clostridium groups IV and XIV. Tissue repair and tolerance to commensal and food antigens are key factors for maintaining homeostasis in the gut. IL-22 promotes epithelial cell proliferation and repair; however, excessive and aberrant signaling can lead to inflammation and colitis. Tolerance is accomplished by the induction of regulatory T cells (Tregs) through several mechanisms. In the small intestine, CD103+ DCs take up mucus and stimulate Tregs through IL-10 and TGF-β. Although the mechanism has not been elucidated, it is possible that DCs take up mucus directly by extending dendrites into the lumen or that goblet cells transfer mucus to DCs, as has been shown with antigen. In the colon, specific Treg-inducing bacteria have been identified. For instance, Clostridia spp. generate metabolites that upon receptor binding stimulate the production of TGF-βby epithelial cells. Polysaccharide A (PSA) from Bacteroides fragilis enhances Treg function (either through direct stimulation of TLR2+ Tregs or through a TLR2+ DC intermediate) while inhibiting proinflammatory Th17 responses. Abbreviations: ILC3, type 3 innate lymphoid cell; MAMPs, microbe-associated molecular patterns; mΦ, macrophage.
Under normal circumstances, bacterial penetration of the dense inner mucin layer is limited (38), presumably because of the density of the mucin but also because of mucin association with the antimicrobial C-type lectin, regenerating islet-derived 3 γ (RegIIIγ) (39, 40). Commensal microbes enhance mucin production by goblet cells in a MyD88-dependent fashion, and germfree or antibiotic-treated mice have markedly reduced levels of mucin. The thickness, density, and composition of mucin differ between the stomach, small intestine, and colon (41, 42), and the ability of bacteria to penetrate the mucous layers along the gut differs, with greater mucus penetration in the small intestine than in either the stomach or the colon (41). Muc1 expression in the stomach and small intestine has been implicated in resistance to Campylobacter jejuni and Helicobacter pylori infection (43, 44), whereas Muc2 provides resistance to enteric S. typhimurium infection (45). In addition to functioning as a barrier against penetration by luminal bacteria, Muc2 conditions lamina propria dendritic cells to become tolerizing, thereby reducing inflammatory responses to absorbed substances (46). Goblet cells have recently been shown to deliver antigens to lamina propria dendritic cells, providing a potential delivery pathway to facilitate mucin-mediated tolerization of antigen-presenting cells (47).
Below the mucin layer, intestinal epithelial cells form a continuous barrier that is one cell layer thick. Intestinal epithelial cells absorb a wide range of nutrients, including proteins, carbohydrates, and fats but excluding bacteria. Achieving the right balance between nutrient absorption and microbial exclusion is critical to optimal health, and deviation from this balance is deleterious to the host. Intestinal epithelial cells express innate immune receptors, but receptor positioning (i.e., apical versus basal) minimizes unnecessary activation by harmless commensals in the intestinal lumen while enabling rapid defensive responses when the barrier is breached. For example, Toll-like receptor 5 (TLR5), the innate immune receptor specific for bacterial flagellin, is not expressed on the apical surface of epithelial cells, where flagellin is presumably present at high concentrations (48). Deletion of MyD88 in intestinal epithelial cells reduces production of mucin and antibacterial peptides and increases susceptibility to infection (49). Certain components of the inflammasome, such as Nod-like receptor pyrin domain 6 (NLRP6), are selectively expressed by intestinal epithelial cells and can influence the composition of the intestinal microbiota by inducing IL-18 expression (50) (Figure 1). Nod2 is also expressed in intestinal epithelial cells and has been implicated in the regulation of defensin expression (51). Selective deletion of IKKβ in the intestinal epithelial cells prevents NF-κB activation and is associated with impaired lamina propria dendritic cell conditioning and markedly reduced Th2 CD4 T cell responses to intestinal worm infection but enhanced inflammatory Th1 responses (52).
Intestinal epithelial cells can also express CD1d, an MHC-like molecule that presents glycolipids to natural killer (NK) T cells (see below). Cross-linking of CD1d, however, activates STAT3 and induces intestinal epithelial cells to produce IL-10. Deletion of either CD1d or IL-10 in intestinal epithelial cells results in enhanced NK T cell–mediated intestinal inflammation (53). Lymphocytes, in particular T cells expressing the γδ T cell receptor, infiltrate the intestinal epithelial cell layer and receive signals from epithelial cells that induce expression of antimicrobial factors (54).
Lamina Propria Macrophages and Dendritic Cells
Immediately below the intestinal epithelial cell layer, complex cell populations make up the lamina propria. The lamina propria is a well-vascularized and lymph-drained tissue whose functions include facilitating systemic nutrient distribution, establishing immune tolerance toward innocuous antigens and microbes, and providing immune defense against potentially pathogenic microbes that emerge from bacterial populations inhabiting the intestinal lumen. Mesenchymal stromal cells of the intestinal lamina propria help orchestrate immune responses by responding to microbe-derived molecules that trigger MyD88-mediated signals and potentially other innate immune signaling pathways (55). Although the role of stromal cells in the lamina propria is likely underappreciated and underinvestigated, great progress has been made in the last decade in our understanding of the macrophage and dendritic cell populations of the gut (56, 57).
Given the density and complexity of microbial populations in the intestinal lumen and the obvious benefits of mucus and epithelial cell–mediated segregation of these organisms from underlying tissues, the initial revelation that myeloid cells disrupt interepithelial cell tight junctions and extend processes into the intestinal lumen was both astounding and disquieting (58, 59). Subsequent studies implicated CX3CR1-expressing bone marrow–derived cells as the key population accessing the intestinal lumen and taking up pathogenic organisms such as S. typhimurium (59). Extension of processes into the intestinal lumen is dependent on MyD88-mediated signaling in CX3CR1-expressing mononuclear cells (60). CD103+ dendritic cells of the lamina propria, which have been implicated in the transport of bacteria and antigens to draining mesenteric lymph nodes and in the induction of tolerance by production of retinoic acid (61), have also been found to patrol between intestinal epithelial cells and extend dendrites into the intestinal lumen, taking up pathogenic bacteria such as S. typhimurium and nonpathogenic commensal bacteria and transporting them to draining lymph nodes (62) (Figure 1). Diehl et al. (63) investigated the contribution of the commensal flora to microbial transport by lamina propria–residing, bone marrow–derived cells and found that antibiotic treatment or MyD88 deficiency increased the delivery of bacteria to mesenteric lymph nodes and enhanced specific T cell and antibody responses.
The identity of the cell populations capable of delivering bacteria to draining lymph nodes remains somewhat controversial. CX3CR1+ mononuclear phagocytes have been shown to transport bacteria (63), while other studies implicate CD103+ dendritic cells (64). It is likely that this controversy in part stems from varied dendritic cell definitions and the plasticity of inflammatory monocytes infiltrating the intestinal lamina propria, particularly during intestinal inflammation. For example, CCR2+Ly6Chi monocytes give rise to CX3CR1+ mononuclear phagocytes, and once differentiated, they appear to become nonmigratory. However, under inflammatory conditions, CCR2+Ly6Chi monocytes can differentiate into migratory dendritic cells and prime T lymphocytes (65, 66). In other organs, such as the lung, CCR2+Ly6Chi monocytes pick up bacteria and particulate antigens and deliver them to draining lymph nodes, in the process differentiating and acquiring markers that overlap with those of dendritic cells and nonmigratory CX3CR1+ cells (67). An additional mechanism by which antigen can make its way from the lumen to the draining lymph node is uptake by CX3CR1+ mononuclear phagocytes followed by connexin 43–dependent transfer to CD103+ dendritic cells, which then traffic to the draining lymph node (68). Deletion of connexin 43 demonstrated that antigen transfer to CD103+ dendritic cells was necessary to establish immune tolerance. A small population of CX3CR1+ mononuclear phagocytes that are not monocyte derived exists in the intestine. These cells lack monocyte/macrophage surface markers and are thought to be bona fide dendritic cells. Although not much is known about their function, they have recently been implicated in the differentiation of Th1 and Th17 cells (69).
Bone marrow–derived neutrophils and monocytes play central roles in innate immune responses to intestinal infection, in some cases reducing potentially deleterious inflammation and in other cases increasing inflammation-mediated antimicrobial defenses. For example, during intestinal infection with Toxoplasma gondii, inflammatory monocytes are recruited to the lamina propria and, in response to commensal bacteria, produce prostaglandin E2, which reduces neutrophil-mediated tissue damage (70). Inflammatory monocyte–derived lamina propria mononuclear phagocytes, in response to stimulation by microbe-derived molecules, also contribute to intestinal tolerance by producing IL-1β, which induces RORγt-expressing innate lymphocytes to produce granulocyte-macrophage colony-stimulating factor (GM-CSF), which feeds back to mononuclear phagocytes and enhances differentiation of Tregs (71). Intestinal phagocytes can also produce IL-1β in response to infection with gram-negative pathogens such as S. typhimurium and Pseudomonas aeruginosa following induction of NLRC4 inflammasome signaling (72). In other settings, however, lamina propria mononuclear phagocytes and dendritic cells contribute to innate immune defense against epithelial barrier penetration by rapidly producing inflammatory cytokines and chemokines and orchestrating the recruitment of bactericidal cell populations. Production of IL-23 by macrophages and dendritic cells is essential for innate immune defense against infection with the murine pathogen Citrobacter rodentium (73). CD103+ dendritic cells in the lamina propria can be divided into a large population that expresses CD11b and a less frequent, Batf3-dependent population, which probably resides within lymphoid aggregates and Peyer’s Patches (56, 74). The CD11b+CD103+ dendritic cell subset, which expresses TLR5, is the major IL-23-producing cell population following flagellin administration (75) and also following C. rodentium infection (76). Batf3-dependent CD11b−CD103+ dendritic cells, though, have been shown to prime the development of CD8+ effector T cells and stimulate the production of antigen-specific IgG upon TLR activation (77–79). Dendritic cells of the cecum-draining lymph node can harbor live bacteria that are relatively unresponsive to antibiotics, suggesting that some intestinal bacteria have evolved mechanisms to access subepithelial tissues for long-term persistence (80). CD11c+CD103−CD11b− plasmacytoid dendritic cells (pDCs) are a minor dendritic cell population in the intestine. Derived from the same common dendritic cell progenitor (CDP) as classical dendritic cells, pDCs are unique in their ability to secrete copious amounts of type I IFNs in response to viral and bacterial antigens (81).
Innate Immune Signaling in the Intestinal Wall
Activation of the innate immune system by microbial stimulation of innate immune receptors is an essential early step in defense against infection by pathogenic organisms. However, innocuous and potentially beneficial colonizing bacteria are composed of molecules that can bind innate immune receptors and potentially trigger inflammatory responses. Triggering of some innate immune receptors, however, can also induce tolerance by reducing inflammatory responses. Indeed, development of the mucosal immune system is in part dependent on innate immune receptor triggering by commensal microbes in the intestinal lumen (82). Colonization of the murine gut with the anaerobic bacterium B. fragilis and its production of polysaccharide A (PSA) enhance T cell development and differentiation (83). PSA is believed to promote Treg development and inhibit Th17 responses by stimulating TLR2 (84) (Figure 1). In this setting, TLR2 signaling occurs in part via Gadd45α in intestinal dendritic cells upon exposure to B. fragilis outer membrane vesicles containing PSA (85). Although the specific dendritic cell population that responds to PSA stimulation in the colon has not been identified yet, a recent study suggests that pDCs might have a role, given that transfer of TLR2−/− bone marrow–derived pDCs rendered mice susceptible to colitis (86). It is likely that other monocytes/DCs also contribute to Th17 development.
Intestinal epithelial cells express antimicrobial molecules, including the bactericidal C-type lectin RegIIIγ, in response to gut colonization by commensal bacteria (87). Microbe-derived molecules drive RegIIIγ expression by stimulating TLRs (88, 89). RegIIIγ expression reduces bacterial colonization of the dense mucous layer associated with intestinal epithelial cells, thereby reducing microbial contact with the epithelium (40, 90). RegIIIγ binds to peptidoglycan by its lectin domain, and more recent studies of closely related RegIIIα demonstrated that bactericidal activity results from association with bacterial phospholipids and the formation of a hexameric structure that produces a pore in the bacterial membrane (91). In contrast to RegIIIα, which is inhibited by gram-negative lipopolysaccharide (LPS), RegIIIβ has bactericidal activity against gram-negative bacteria by lectin-mediated association with LPS and, at least in part, pore formation in the gram-negative outer membrane (92, 93).
Which TLRs contribute to immune defense versus tolerance induction in the gut remains incompletely resolved. TLR5 has been investigated most extensively. Whereas early studies suggested that TLR5 is expressed on the basal surface of intestinal epithelial cells, subsequent studies demonstrated that TLR5 is predominantly expressed on lamina propria dendritic cells and that TLR5 expression on lamina propria dendritic cells facilitates the development of Th17 T cells and IgA-producing cells (94, 95). Systemic administration of bacterial flagellin, the microbial ligand for TLR5, induces the expression of RegIIIγ by intestinal epithelial cells along the entire length of the small intestine and provides significant protection against intestinal colonization by vancomycin-resistant Enterococcus (VRE) (96) and infection by C. difficile (97). Systemic flagellin rapidly induces expression of IL-23 by CD103+CD11b+ lamina propria dendritic cells, as noted above, which in turn stimulates IL-22 production by type 3 innate lymphoid cells (ILC3s), which then leads to RegIIIγ expression by intestinal epithelial cells (75) (Figure 1).
Although TLR5 stimulation can promote innate immune defenses against intestinal infection, TLR5 signaling, under homeostatic conditions, reduces gut inflammation and metabolic syndrome (98). In some mouse colonies, TLR5 deficiency is associated with development of spontaneous colitis and massive weight gain with parallel development of metabolic syndrome. This association has been attributed to alteration of the intestinal microbiota, including expansion of proteobacterial populations, resulting from deficient TLR5 signaling (99). In the absence of TLR5, flagellin-specific-IgA levels are reduced, which appears to increase the expression of flagellin and invasion of flagellated bacteria into subepithelial tissues (100). Although some studies have demonstrated that TLR5 deficiency results in the development of obesity and metabolic syndrome (98), this finding is not replicated in all TLR5-deficient mouse colonies (101).
Flagellin and TLR5, although contributing to intestinal defense against microbial pathogens, are only part of the story. Oral administration of DNA isolated from the gut flora, for example, corrects innate immune antimicrobial responses in antibiotic-treated mice (102), and oral administration of LPS corrects antibiotic-induced innate immune deficiency in the gut and enhances resistance against VRE (103). The extent to which TLR-mediated signals influence the composition of the intestinal microbiota is controversial. Comparison of wild-type C57BL/6 mice with littermate control TLR2-, TLR4-, TLR5-, TLR9-, or MyD88-deficient mice did not detect differences in their luminal or mucosa-associated ileal, cecal, or fecal microbiota under homeostatic conditions and following recovery from antibiotic treatment (104). This study demonstrated that microbiota composition is largely determined by familial transmission of the microbiota and that the composition of bacterial populations inhabiting the gut can drift over time as distinct breeding colonies are kept in isolation from each other.
Defects in innate immune signaling have also been implicated in microbiota-mediated development of inflammatory bowel disease, obesity, and obesity-associated steatohepatitis (105). Deficiency of the NLRP6 inflammasome subunit in mice is associated with a shift in colonic microbiota composition that leads to inflammatory bowel disease and steatohepatitis (50, 106). Transfer of the aberrant microbiota from NLRP6 knockout mice to wild-type mice resulted in the development of these inflammatory diseases, suggesting that loss of NLRP6 alters the mi-crobiota to be proinflammatory, or in other words, that NLRP6 signaling in intestinal epithelial cells controls the composition of the microbiota and steers it to an anti-inflammatory state. This process is in part regulated by IL-18-mediated signaling (50). Another important mechanism by which NLRP6 modulates microbiota composition is regulation of intestinal goblet cell secretion of mucin (Figure 1). NLRP6 facilitates goblet cell autophagy and also exocytosis of mucin granules, and mucin production is markedly reduced in its absence (107). Reduced mucin production renders mice more susceptible to infection by intestinal pathogens, as noted above.
Whereas inflammasome signaling in intestinal epithelial cells contributes to antimicrobial defense, inflammasome signaling following systemic infection can have adverse effects. For example, mortality following systemic spread of a pathogenic strain of E. coli was mediated in part by the Naip5/NLRC4 inflammasome (108). In contrast, systemic absorption of microbial molecules from the gut, such as peptidoglycan fragments, can trigger Nod2 signaling and activate circulating neutrophils, thereby enhancing resistance against infection by Streptococcus pneumoniae and Staphylococcus aureus (109). Nod2 signaling in the gut also acts locally, by promoting inflammatory monocyte recruitment into the gut during C. rodentium infection (110). Studies with germfree mice demonstrated a systemic effect of microbial colonization on mononuclear phagocytes. Specifically, mononuclear phagocytes from germfree mice were hyporesponsive to microbial stimulation, a result of aberrant chromatin modification of innate immune response genes (111). These studies are revealing the complexity of innate immune recognition and responses in mucosal tissues, and it is likely that many more examples of proinflammatory antimicrobial responses and contrasting anti-inflammatory or tolerizing responses will be discovered in the coming years.
Innate Lymphocytes and the Intestinal Microbiota
Innate lymphocytes derive from common lymphoid progenitors, depend on common γ-chain and IL-7R signaling and the Id2 transcriptional regulator, and differentiate under the control of specific transcription factors that are shared with T lymphocytes into an array of phenotypes that are categorized as ILC1 (T-bet dependent and IFN-γ producing), ILC2 (GATA-3 dependent and IL-4 and IL-13 producing), and ILC3 (RORγt dependent and IL-17 and IL-22 producing). ILCs represent an important arm of the innate immune defense system, at the level of orchestrating immune tissue development and, more directly, antimicrobial defenses (112).
A subset of ILC3 cells contributes to innate immune tissue development in the gut by producing lymphotoxin α and lymphotoxin β. The ILC3 cells involved in this process are referred to as lymphoid tissue inducer (LTi) cells, and their existence depends on the RORγt transcription factor. Soluble lymphotoxin α facilitates the production of T cell–dependent IgA in the gut, whereas lymphotoxin β on the surface of ILC3s promotes T cell–independent IgA production (113). In utero development of LTi cells depends on maternal retinoid intake, and retinoic acid in the developing fetus is required for RORγt induction and LTi development (114).
ILCs play essential roles in defense against intestinal bacteria and also nonpathogenic bacteria that cross the intestinal epithelial barrier and reside in gut-associated lymphoid tissues. Upon depletion of IL-22-producing ILC3s, lymphoid tissue–residing bacteria such as Alcaligenes can disseminate and drive systemic flares of inflammation (115). Because ILCs and differentiated T lymphocytes can express overlapping phenotypes, and because depletion of ILCs often also results in depletion of T lymphocytes, it has been challenging to distinguish the roles of these cell populations in the induction of inflammation and during defense against infection. Using ILC-specific depletion strategies, Hepworth et al. (116) found that the absence of ILC3s resulted in dysregulated T cell responses to the commensal flora. Furthermore, this process required MHC class II expression by ILC3s (116). Although this finding suggests that responding T lymphocytes may interact directly with MHC class II–expressing ILC3s, the role of ILC3-mediated antigen presentation remains unclear. IL-22 expression by ILC3s is promoted by the aryl hydrocarbon receptor (Ahr), and mice lacking Ahr have markedly reduced numbers of IL-22-producing ILC3s. Surprisingly, Ahr deficiency results in increased numbers of Th17 cells, potentially the result of increased intestinal colonization by segmented filamentous bacteria (SFB) (117), and also increases the risk of spontaneous colitis. ILC3s were recently shown to be a major source of GM-CSF following stimulation by IL-1β, which is produced by mononuclear phagocytes upon stimulation by microbial molecules. In turn, GM-CSF acts on mononuclear phagocytes to enhance their effector functions (71). The interactions between myeloid cells, ILCs, T lymphocytes, and B lymphocytes are complex, and our knowledge of these relationships is far from complete. Indeed, novel cell populations, such as populations of CD71+ erythroid cells, which were recently demonstrated to attenuate inflammatory responses against newly colonizing intestinal bacteria in the neonate by producing arginase-2, are being discovered and assigned functional roles in the relationship between the host immune system and colonizing microbes (118).
ADAPTIVE IMMUNE DEFENSES AND THE INTESTINAL MICROBIOTA
Antibody- and T lymphocyte–mediated recognition of intestinal bacteria contributes to immune defense but also to the pathogenesis of intestinal and systemic inflammatory diseases (119). In parallel with advances in our understanding of the complex microbial populations inhabiting the gut and the more detailed characterization of individual members of the microbiota, recent studies have begun to define the specificity of antibody and T cell responses to intestinal microbes and the mechanisms driving microbe-mediated differentiation of immune cells.
Secretory IgA
Whereas the intestinal microbiota influence systemic antibody responses, as demonstrated by increased serum IgE levels in germfree mice (120), induction of secreted IgA in response to intestinal bacteria is particularly important for antimicrobial defense and intestinal microbiota homeostasis. Secretory IgA provides defense against intestinal pathogens, and its absence has been implicated in the expansion of some commensal bacterial populations such as SFB (121) and in the maturation of the intestinal microbiota during neonatal life (122). Colonization of the murine small intestine with SFB and its ability to induce IgA were established by studies over 15 years ago (123), and recent studies demonstrate that SFB induce Peyer’s Patch development and IgA secretion (124) (Figure 1).
The PD-1 inhibitory receptor regulates the development of T follicular helper cells in Peyer’s Patches, and its absence promotes precursor cells that produce IgA with lower affinity for intestinal bacteria, ultimately affecting the composition of the microbiota (125). Kinetic analyses of intestinal IgA responses against commensal bacteria suggest that specific antibodies can be secreted for prolonged periods but also that they are dynamic and that new IgA antibodies are generated following shifts in microbiota composition (126). Deep sequencing of the IgA-encoding genes of mice revealed a very broad repertoire with many distinct specificities and increasing diversity with aging (127). Whereas secreted IgA, by virtue of its specificity, influences composition of the microbiota, some IgA-producing plasma cells also produce TNF and iNOS, and depletion of these cells alters microbiota composition and decreases resistance against infection (128). Thus, IgA-producing plasma cells may augment defensive strategies that have largely been attributed to myeloid cell populations.
Th17 Cells
SFB also contribute to the differentiation of Th17 cells in the murine small intestinal lamina propria (18, 129). SFB appear to have a unique ability to penetrate the small intestinal mucous layer and directly contact the surface of intestinal epithelial cells, a characteristic that likely contributes to their ability to drive the differentiation of Th17 T cells (Figure 1). Whole-genome sequencing of SFB revealed a reduced genome size compared with other Clostridiales and its dependence on exogenous amino acids, suggesting that this organism is highly adapted to and dependent on its mammalian host (130). Two recent studies investigated the antigen specificity of Th17 cells induced by SFB monocolonization and discovered that the majority of T cells belonging to this subset are specific for SFB. Thus, without apparent invasion of the lamina propria, SFB induce a broad Th17 response that is directed against several SFB-encoded antigens (131) and that depends on MHC class II presentation by lamina propria dendritic cells and is independent of secondary lymphoid tissues (132). Whereas T cell responses to commensal bacteria can be induced by intestinal infections that destroy the epithelial cell layer and induce potent inflammatory responses (133), the response to SFB is distinct and occurs in the presence of an intact epithelial barrier and in the absence of apparent inflammatory cell infiltration of the lamina propria.
Gut microbiota, including SFB, induce IL-1β in intestinal macrophages (also known as mononuclear phagocytes), and it has been suggested that the IL-β then binds to T cell–expressed IL-1 receptor to induce Th17 T cells (134). A recent study has demonstrated that light-cycle disruption in mouse colonies can promote the differentiation of Th17 cells (135). This study demonstrated that the REV-ERBα transcription factor, which is implicated in regulation of the circadian clock, enhances NFIL3 activity, which in turn interferes with RORγt binding to its promoter. Thus, interference with REV-ERBα expression reduces NFIL3 activity and enhances Th17 development. This finding suggests that sleep abnormalities or frequent challenges to the baseline circadian rhythm of mammals may incrementally increase Th17 development and increase the risk of inflammatory diseases.
Regulatory T Cells
The intestinal lumen contains a wide range of microbial inhabitants and ingested antigens that are potential inducers and targets of T cell responses. Tregs limit naive T cell differentiation into effector cells that can target many of the innocuous antigens that are present in the intestinal lumen. In the absence of Tregs, inflammatory diseases involving the gut occur rapidly and can be lethal. Recent studies have identified bacterial species that drive intestinal Treg development. Colonization of mice with altered Schaedler flora, which consists of eight distinct bacterial species, induced colonic Tregs in germfree mice (136) and prevented colitis. Characterization of colonic Tregs induced by commensal bacteria revealed that their repertoire of T cell receptors was distinct from that of Tregs at other sites, suggesting that colonic Tregs are derived from circulating naive CD4 T cells (137). Remarkably, induction of colonic Tregs can be driven by colonization with a single bacterial strain of B. fragilis that expresses PSA (138). The ability of bacteria belonging to the Bacteroidetes phylum to drive Treg differentiation was also demonstrated by colonizing germfree mice with different mixtures of human fecal bacterial cultures that included Bacteroides caccae, B. thetaiotaomicron, B. vulgatus, B. massiliensis, and Parabacteroides distasonis (139). Other studies have demonstrated that murine- and human-derived fecal microbes that belong to Clostridium clusters IV and XIVa drive Treg development in the murine colon (17), in part by inducing TGF-β production by intestinal epithelial cells (Figure 1). Optimal Treg development in mice can be induced by colonization with 17 human-derived strains belonging to these Clostridia groups (16, 140). Trafficking of Tregs to colonic lamina propria is mediated by the G protein–coupled receptor 15 (GPR15) heterotrimeric guanine nucleotide–binding protein–coupled receptor, the expression of which is modulated by the gut microbiota (141). One mechanism by which Tregs are induced involves the intestinal microbiota upregulation of Uhrf1 expression. Uhrf1, in turn, DNA methylates the cyclin-dependent kinase inhibitor p21 promoter, thereby inhibiting its transcription and promoting Treg proliferation (142).
Natural Killer T Cells and Mucosal-Associated Invariant T Cells
NK T cells are present in the intestinal lamina propria and have been implicated in inflammatory bowel disease. The number of NK T cells in the colonic lamina propria is increased in germfree mice and can be normalized by administering microbiota to neonatal but not adult mice (143). Other studies have demonstrated that NK T cells isolated from germfree mice are hyporesponsive to antigen and that colonization of mice with Sphingomonas, but not E. coli, normalized NK T cell responsiveness to antigen (144). Recent studies have demonstrated that, in addition to Sphingomonas, other bacterial inhabitants of the gut produce glycolipids that are bound by CD1d and presented to NK T cells. B. fragilis, for example, encodes a serine palmitoyltransferase that catalyzes synthesis of sphingolipids (145), including a B. fragilis–produced α-galactosylceramide (α-GalCer) closely related to α-GalCer produced by a marine sponge; the latter has been extensively used to characterize NK T cells in experimental systems and to augment NK T cell responses in vivo. Studies of B. fragilis–derived glycosphingolipids have identified variants that reduce lamina propria NK T cell numbers in germfree mice and enhance resistance to oxazolone-induced colitis (146).
Mucosal-associated invariant T (MAIT) cells are prevalent in the gut and respond to the MHC class I–like molecule MR1. Recent studies have demonstrated that MAIT cells can respond to intestinal epithelial cells that are infected with Shigella flexneri but not S. typhimurium (147). The ligands that are bound by MR1 and their role during in vivo antimicrobial defense warrant further study.
DIET, VITAMINS, METABOLISM, IMMUNITY, AND THE INTESTINAL METABOLOME
Food digestion, nutrient and vitamin absorption, immune defense, and bacterial and host metabolism are interrelated and interdependent processes occurring in the gut. The implications of these interactions for the mammalian host are profound. The mechanisms by which commensal bacteria contribute to host metabolism are increasingly being defined (148). Given the complexity of intestinal microbial populations, one approach to determine the impact of specific bacterial species on host metabolism has been to colonize germfree mice with individual bacterial strains and measure the impact on host gene expression (149). A study taking this approach demonstrated that two bacterial species, B. longum and B. thetaiotaomicron, have distinct effects on gene expression by intestinal epithelial cells but also influence each other’s gene expression patterns. Ingestion of different dietary carbohydrates, such as fructose-based polysaccharides, can drive the selective expansion of bacterial species that encode the appropriate hydrolytic enzymes and can thereby alter microbiota composition (150). The impact of altered microbiota composition can be profound, in some cases leading to obesity that can be reversed by transfer and expansion of specific members of the Bacteroidetes phylum (151). Changes in diet can rapidly alter the composition of the human fecal microbiota (152), with animal-based diets reducing the density of bacteria belonging to the Firmicutes phylum and increasing organisms associated with bile tolerance. Pregnancy has also been associated with changes in the intestinal microbiota toward a composition that, upon transfer into germfree mice, increases adiposity and reduces insulin sensitivity (153).
Vitamin A is an essential dietary component and is metabolized into retinoic acid, which plays an important role in T cell differentiation, including development of Tregs (154–156). Ingested vitamins, such as vitamin B12, are also of benefit to the microbiota, and recent studies revealed that the intestinal symbiont B. thetaiotaomicron expresses three nonredundant vitamin B12 receptors that can each confer competitive advantages to the bacterial strain, depending on the presence of distinct vitamin B12 analogs (157). Intestinal bacteria can also produce vitamins, such as vitamin B3 (also known as niacin), and signal via GPR109A and reduce intestinal inflammation (158). Recent studies have demonstrated that a dietary switch from carbohydrates to tryptophan leads to production of tryptophan metabolites by intestinal microbes that enhance signaling via the aryl hydrocarbon receptor, in turn leading to greater IL-22 production (159). The impact of microbiota-mediated metabolism of ingested nutrients on host health can also be negative, as demonstrated by studies of L-carnitine, a trimethylamine that is acquired by meat ingestion. Conversion of L-carnitine to trimethylamine-N-oxide by microbiota members that are expanded in meat-eating individuals led to increased progression of atherosclerosis in mice (160).
It is clear that intestinal microbes can dramatically alter the absorption and metabolism of orally administered medications. Recent studies have demonstrated, for example, that some strains of Eggerthella lenta, a member of the human microbiota, can inactivate the cardiac drug digoxin and that changes in diet can alter in vivo metabolism of the drug, leading to different drug levels (161). For a drug like digoxin, which has a relatively narrow therapeutic window of activity and potentially lethal toxicities, changes in its metabolism by intestinal microbes can have serious clinical implications.
Although most studies of the microbiota have focused on microbiota composition, determined either by deep bacterial 16S rRNA gene sequencing or by shotgun sequencing of fragmented bacterial DNA, recent studies have started to investigate the bacterial transcriptome and metabolome. Analysis of the bacterial transcriptome in the gut demonstrated that many genes and their respective transcripts are similarly abundant. However, genes associated with sporulation and amino acid synthesis are often downregulated, whereas genes involved in ribosome biogenesis and methanogenesis are upregulated (162). Administration of xenobiotics to mice can dramatically alter gene expression by intestinal microbes, with transcript fluctuations greatly exceeding the quantitative changes in the representation of different bacterial taxa (163). Beyond determination of the microbiota transcriptome, mass spectrometric analysis of microbially produced or modified metabolic products promises to provide new insights into the role of intestinal microbes in health and disease (164). The impact of the microbiota on a wide range of metabolites was revealed by treatment of mice with antibiotics; over 87% of metabolites, including bile salts, eicosanoids, and steroid hormones, were affected by antibiotic-induced shifts in the microbiota (165). The metabolomic changes induced by antibiotic administration have been implicated in the development of adiposity and have been suggested to contribute to the ongoing obesity epidemic (166). Administration of fecal microbiota from different humans to germfree mice demonstrated conserved metabolite differences that could be detected by mass spectrometry–based metabolomic assays of urine samples (167).
Among the metabolites that are being characterized in the intestinal lumen, short-chain fatty acids and bile salts stand out as having clearly demonstrable roles in immune development and defense against infection. Short-chain fatty acids include acetate, butyrate, and propionate and are produced by fermentation of fiber and other polysaccharides by a range of intestinal bacteria (168). Bacteria belonging to the Bifidobacterium genus can produce acetate, which reduces the susceptibility of the colonic epithelium to damage caused by the E. coli O157:H7 Shiga toxin (14). In this setting, a Bifidobacterium-expressed transporter pumps acetate into the intestinal lumen (169). Short-chain fatty acids produced by intestinal microbes stimulate GPR43 expressed by host inflammatory cells and reduce inflammation, and mice with a deletion of the gene encoding GPR43 have enhanced colitis and arthritis upon induction of inflammation (170). Short-chain fatty acids, and butyrate in particular, have recently been demonstrated to regulate extrathymic Treg development in a GPR43-dependent manner (171–173). Butyrate and propionate administration enhances histone acetylation in the promoter of the Foxp3-encoding gene, potentially by inhibiting histone deacetylase function.
On the one hand, bile salts released into the intestinal lumen can affect the composition of the microbiota; on the other hand, commensal microbes can alter the composition of intestinal bile salts. Ingestion of milk fats, for example, increases production of taurocholate by the liver, which in turn leads to expansion of the bacterial species Bilophila wadsworthia (174). B. wadsworthia can enhance the development of colitis in IL-10-deficient mice, demonstrating the indirect mechanism by which dietary changes can alter the risk of inflammatory diseases. Antibiotic treatment, on the other hand, can alter the gut microbiota and eliminate microbes that deconjugate primary bile salts and also dehydroxylate primary bile salts to produce secondary bile salts, which have been implicated not only in the induction of colitis but also in defense against a subset of intestinal pathogens (175).
AUTOIMMUNITY, INFLAMMATORY DISEASES, AND THE MICROBIOTA
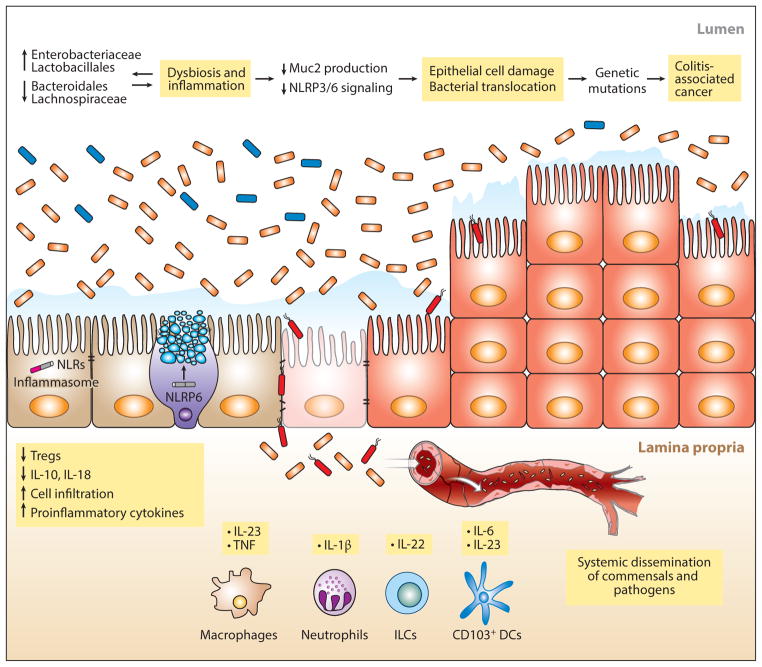
Mouse models have provided important insights into the role of different bacterial species in the development of colitis. The role of Muc2 and the mucin layer in protection against pathogen-associated colitis was demonstrated in a study of C. rodentium infection in mice deficient in Muc2 (176). Even in the absence of pathogens, loss of O-linked glycans on mucin can lead to colitis associated with myeloid cell infiltration of the colonic lamina propria and increased TNF production (35). Colitis in mice lacking core 1-derived O-glycans was associated with increases in the frequency of Lactobacillus and Clostridium species and a decrease in bacteria belonging to the Ruminococcaceae and Lachnospiraceae families (177) (Figure 2). The development of colitis is associated with mucous layer penetration by luminal bacteria that are normally kept at a distance from epithelial cells and presumably also dendritic cells and mononuclear phagocytes inhabiting the underlying lamina propria (178). In mice, Helicobacter hepaticus is associated with the development of colitis, but this colitis can be ameliorated by PSA, which is produced by some strains of B. fragilis (179). Establishing causative relationships between the intestinal microbiota composition and development of colitis is challenging because intestinal inflammation alters the composition of the microbiota (180–182). Thus, changes in microbiota composition in individuals with established intestinal inflammation might be causative, or they may simply reflect the presence of inflammation. In a mouse model, the potential impact of carbohydrate metabolism and even the presence of substances like dopamine on the microbiome and metabolome varied with colitis flares and remissions (183). A recent study characterized the microbiome of people with newly diagnosed and untreated Crohn’s disease and found increased prevalence of Enterobacteriaceae and decreased abundance of Bacteroidales in patients with active disease (184) (Figure 2).
Figure 2.
Inflammation, colitis, and cancer. Shifts in microbiota composition, in particular the expansion of γ-proteobacteria belonging to the Enterobacteriaceae family (orange), have been associated with colitis. These changes in bacterial composition are driven by factors such as infection, inflammation, antibiotic treatment, and deficiency of genes involved in the host-microbial response. Although specific bacteria that confer a colitogenic phenotype on normal hosts have been identified (188, 189), whether altered gut microbiota are the culprit behind colitis rather than a result of inflammation is still a matter of debate. Thinning of the mucous layer makes the intestine vulnerable to invasion by bacteria that may spread to other organs through the bloodstream. The continued presence of a colitogenic flora and production of proinflammatory cytokines plus alterations in the host genome perpetuate a state of inflammation that can eventually progress to cancer. Abbreviations: DC, dendritic cell; ILC, innate lymphoid cell; NLRP6, Nod-like receptor pyrin domain 6; Treg, T regulatory cell.
Deficiency of IL-22 can result in changes in the intestinal microbiota that increase susceptibility to experimental colitis (185). Resistance to colitis can be enhanced by retinoic acid administration, which, at least in part, acts by increasing IL-22 production by γδ T cells (186). Mice deficient in B and T cells and also deficient in the T-bet transcription factor develop colitis that is characterized by high levels of TNF production and microbiota that, upon transfer into wild-type mice, induce colitis (187). The microbiota of these mice had increased prevalence of Klebsiella pneumoniae and Proteus mirabilis, although these two bacterial species did not transmit colitis in the absence of other microbiota members (188). Subsequent studies identified Helicobacter typhlonius as the cause of colitis in this model and implicated TNF and IL-23 production by lamina propria dendritic cells followed by IL-17 production by ILC3s (189). Deficiency of NOD2 has also been associated with a shift in microbiota composition that predisposes mice to the development of colitis (190). The RIG-I-MAVS pathway, which is an important innate immune signaling system for defense against viruses, also contributes to defense against colitis that may result from stimulation by bacterial RNA and the induction of IFN-β and RegIIIγ (191).
The contribution of the intestinal microbiota to other autoimmune diseases has also been investigated, revealing additional interesting associations. Gender-associated differences in the intestinal microbiota have been implicated in the development of diabetes in nonobese mice (192, 193). Development of diabetes in these mice is dependent on MyD88-mediated signaling in response to the colonizing microbiota (194). Development of obesity and associated type 2 diabetes has been associated with reduced prevalence of a specific commensal bacterial species, Akkermansia muciniphila (195). In contrast, new-onset rheumatoid arthritis in humans was associated with increased prevalence of Prevotella copri in fecal samples, and colonization of mice with this organism increased susceptibility to colitis (196). Positive correlations between gut microbes belonging to the genus Collinsella and atherosclerosis (197) and negative correlations between Lactobacillales and the development of graft-versus-host disease following allogeneic hematopoietic stem cell transplantation have been found and suggest that the impact of microbiota composition can extend beyond the gut (198). Indeed, colonization with SFB and induction of Th17 responses promote the development and progression of arthritis and inflammatory disorders of the central nervous system (199, 200). Furthermore, in a murine model of autism spectrum disorder, colonization of susceptible mice with B. fragilis reduced the development of abnormal anxiety and sensorimotor disorders (201).
THE INTESTINAL MICROBIOTA AND CANCER
Although it is widely accepted that chronic inflammation can lead to cancer, increasing evidence is indicating that colonizing microbes can drive cancer development and progression by direct or indirect effects on host tissues (202–204). Fusobacterium nucleatum is an obligate anaerobic gram-negative bacterium that is associated with colon tumors and adenomas. In the ApcMin/+ mouse model of intestinal tumorigenesis, exposure to F. nucleatum increased myeloid cell recruitment to tumors, suggesting that some microbes in the intestinal lumen mediate tumor development or progression by orchestrating recruitment of suppressive cells to the tumor (205). The FadA adhesin of F. nucleatum has also been demonstrated to bind to E-cadherin and to activate β-catenin signaling in epithelial cells, providing a potential mechanism to enhance immune cell recruitment and activate oncogenic pathways within tumors and adenomas (206). B. fragilis, specifically a strain expressing an enterotoxin, has also been associated with intestinal cancer progression in multiple intestinal neoplasia (Min) mice. In this model, Stat3 signaling and the induction of IL-23 and IL-17 were essential steps for tumor progression (207).
Inflammasome components have been implicated in resistance to tumor development. Caspase-1 is required for IL-1 and IL-18 activation, and caspase-1 deficiency enhances tumor development (208). Functional caspase-1 promotes intestinal epithelial cell apoptosis and reduces epithelial cell proliferation, thereby reducing tumor development. NLRP3 deficiency, but not NLRC4 deficiency, in mice leads to increased colitis-associated cancer development (209). However, there is also evidence suggesting that overactivation of the inflammasome can lead to, rather than protect from, cancer progression. In this model, commensal-derived LPS stimulates the production of IL-1β from neutrophils during inflammation, which in turn stimulates the production of IL-6 from intestinal mononuclear phagocytes, leading to colitis-associated colorectal cancer (210) (Figure 2). Progression of colitis to cancer has also been suggested to result from expansion of bacterial species, such as potentially genotoxic strains of E. coli, that thrive in the presence of inflammation (211). Deletion of the NLRP6 inflammasome component led to the development of colitis and progression to colon cancer (50, 212). NLRP6 deficiency results in defective tissue repair following epithelial damage, enhancing inflammatory responses (213). In the case of NLRP6 deficiency, the microbiota composition changes and the microbiota acquire the ability to induce colitis upon transfer into wild-type mice, suggesting that NLRP6, under normal circumstances, pushes the microbiota composition toward a noninflammatory state. Along similar lines, deletion of NOD2 or RIP2 leads to alterations in the microbiota that increase the incidence of colitis-associated cancer, a phenotype that is transmissible to wild-type mice and that can be ameliorated by antibiotic administration (190). IL-18 deficiency in mice results in a phenotype very similar to NLRP6 deficiency, indicating that IL-18-mediated signaling is a key step in this model of cancer development (214).
Increased growth of colon tumors has been associated with increased IL-23 production by tumor-infiltrating myeloid cells in response to stimulation by microbial molecules that access the lamina propria via discontinuities in the epithelial cell layer (215). IL-23 induces expression of IL-22 by innate lymphocytes, which plays an important role in promoting epithelial cell proliferation when the epithelium is damaged but can also enhance tumor growth (216, 217) (Figure 2). The potential impact of microbiota composition is not limited to the intestinal epithelium but extends to the liver, where changes in the intestinal microbiota composition associated with NLRP6, NLRP3, or IL-18 deficiency promote progression to steatohepatitis, an inflammatory disease of the liver that, in susceptible mice, is a precursor for hepatocellular carcinoma (106). Another mechanism by which the intestinal microbiota can promote hepatic inflammation and progression to hepatocellular carcinoma involves the bacterially mediated conversion of primary bile acids to the secondary bile acid, deoxycholic acid, which can cause DNA damage and induce the production of inflammatory cytokines and tumor-promoting factors by hepatic stellate cells (218).
Recent studies in murine models have also implicated the intestinal microbiota in responses to cancer chemotherapy by two distinct immunologically mediated mechanisms. Iida et al. found that in vivo responses of lymphoma, colon carcinoma, and melanoma to immunotherapy were reduced in mice with absent or antibiotic-depleted microbiota, as reflected by reduced TNF production by tumor-infiltrating myeloid cells (219). In this study, the response of a lymphoma to platinum chemotherapy was also reduced in the absence of a complete microbiota. In a parallel study, administration of cyclophosphamide was found to reduce intestinal epithelial integrity, alter the intestinal microbiota composition, and increase bacterial translocation from the intestinal lumen to secondary lymphoid organs, which resulted in enhanced populations of CD4 T cells that expressed both IFN-γ and IL-17. Responses of a mastocytoma and sarcoma to cyclophosphamide chemotherapy were reduced in mice with an antibiotic-damaged microbiota, a defect that was corrected by administration of IFN-γ/IL-17-producing T cells (220). These results demonstrate the complex interactions between the intestinal microbiota, tumors, and chemotherapeutic agents and suggest that increased study of microbiota composition in the setting of cancer treatment is warranted.
MICROBIOTA-MEDIATED RESISTANCE TO INFECTION
The ability of the normal microbiota to confer resistance to infection became apparent in the decades following the introduction of broad-spectrum antibiotics, with the discovery that antibiotic-treatment could render mice much more susceptible to a range of intestinal infections (221–224). In the last decade, significant progress has been made in our understanding of the mechanism by which the microbiota provide colonization resistance (225–227). Studies are revealing that colonization resistance can be mediated by direct interactions between different bacterial species or by indirect mechanisms that involve either stimulation of the host’s innate immune system, which induces expression of antimicrobial factors, or modification of host metabolic products into molecules that inhibit pathogenic organisms.
Antibiotics can induce dramatic and long-lasting changes to the microbiota, with expansion of some species and loss of others (228–231). Antibiotic treatment enhances susceptibility to infection by S. typhimurium and C. rodentium, and resistance to these infections has been associated with the presence of obligate anaerobes belonging to the phylum Bacteroidetes (232–234). Administration of a normal microbiota can clear pathogens from the intestinal lumen (235, 236).
The emergence of highly antibiotic-resistant bacterial pathogens is a growing clinical problem. Many of the most antibiotic-resistant pathogens densely colonize the intestinal tract of patients following treatment with antibiotics (230, 237). VRE are one group of such intestinal colonizers and can achieve very high densities in the small intestine and colon following antibiotic-mediated depletion of the microbiota. Administration of diverse and antibiotic-naive microbiota to VRE-dominated mice eliminates VRE from the gut, demonstrating that colonization resistance can be reestablished and bacteria can be cleared with administration of normal flora. Obligate anaerobes are essential for clearance, as demonstrated by fractionation of the microbiota by anaerobic or aerobic culture and treatment with chloroform to eliminate non-spore-forming organisms. Bacteria belonging to the genus Barnesiella were strongly associated with resistance to VRE colonization and with VRE clearance from the gut (236). Innate immune activation via TLR4 or TLR5 stimulation can enhance resistance to VRE colonization, in part by stimulating the production of RegIIIγ expression in the small intestine (96, 103).
Antibiotic treatment also predisposes individuals to infection with C. difficile (238). In the case of C. difficile infection, TLR signaling is essential for short-term defense, and mice lacking the adaptor protein MyD88 have markedly increased mortality (239) that results from severely attenuated neutrophil recruitment to the intestinal lamina propria and damaged epithelium (240). MyD88 facilitates signaling through most TLRs but also through the IL-1 and IL-18 receptors. Administration of flagellin to antibiotic-treated mice enhances resistance against C. difficile infection (97). Although treatment of mice with antibiotics reduces microbiota diversity and markedly increases susceptibility to C. difficile infection, it is unclear which organisms are most important for resistance, and their mechanisms of providing resistance are largely undefined (231, 241, 242). The composition of the microbiota in patients with C. difficile is distinct from the composition of the microbiota in patients with other causes of diarrhea (243). One mechanism by which C. difficile growth in vivo is enhanced is release of sialic acid from host mucus, which transiently increases following antibiotic treatment (12).
Fecal transplantation is one method to reestablish a complex microbiota and has been used in patients with recurrent C. difficile infection (244). This procedure, which involves obtaining fecal samples from healthy donors and administering them to individuals with recurrent C. difficile infection, is remarkably effective and reestablishes a normal microbiota that confers resistance to infection in the recipient. An important and obvious concern with fecal transplantation is the potential risk of transmitting unknown or uncharacterized infections from a resistant donor to a susceptible recipient (245). This concern is not easily eliminated, because the complete composition of a fecal sample is difficult to determine. Although it is likely that well-defined mixtures of bacterial populations will be developed for administration to individuals with specific intestinal infections (246–248), further work is required to precisely identify and characterize the bacterial strains that confer resistance to C. difficile and potentially other infections and inflammatory diseases.
SUMMARY
The role of the mucosal immune system in human health and resistance to infectious and inflammatory diseases is being defined with experimental studies of mice and clinical studies of patients with a wide range of diseases. Characterization of microbial populations associated with mucosal surfaces, particularly beneficial commensal bacterial species, has added a new dimension to the study of mammalian immune development, tolerance induction, and antimicrobial defense. Countless individuals have been protected from death and disease by vaccines that exploit the immune system to induce resistance to pathogens, and recent successes with immunotherapy for cancer suggest that manipulation of the immune system will continue to enhance human health and enhance longevity. Antibiotics have been essential in controlling infections, but injudicious use of antibiotics has accelerated development of antibiotic resistance in pathogenic organisms and, as we are now learning, changes the microbiome in ways that are deleterious to the host. Whereas the field of immunology began with the study of resistance to pathogen-induced infections, and thus has deep roots in microbiology, defining the mammalian host’s relationship with commensal, nonpathogenic bacterial colonizers and fully characterizing the harmless microbial species that provide resistance to infections and ameliorate inflammation are new challenges. In addition to providing fascinating terrain for the exploration of immunologic and microbiologic mechanisms, this new frontier has enormous potential for enhancing resistance to a wide range of human diseases.
Glossary
- Mucin
a cell surface or secreted glycoprotein characterized by the presence of large, densely O-glycosylated filamentous domains
- Goblet cell
a specialized cell that produces, stores, and secretes mucins
- Inflammasomes
large intracellular multiprotein complexes consisting of the ASC/PYCARD adaptor and NLR proteins
- Polysaccharide A (PSA)
polymeric carbohydrate molecule with a zwitterionic motif that is a component of the capsule of some strains of Bacteroides fragilis
- Innate lymphoid cells (ILCs)
lymphoid lineage cells that lack antigen-specific receptors
- Gut microbiota (flora)
the population of microbes living in our intestines
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Hejnol A, Martindale MQ. Acoel development indicates the independent evolution of the bilaterian mouth and anus. Nature. 2008;456:382–86. doi: 10.1038/nature07309. [DOI] [PubMed] [Google Scholar]
- 2.Maxmen A. Evolution: a can of worms. Nature. 2011;470:161–62. doi: 10.1038/470161a. [DOI] [PubMed] [Google Scholar]
- 3.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–95. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hum. Microbiome Proj. Consort. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–38. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, et al. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLOS Comput Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–60. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–29. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–17. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. A composite bacteriophage alters colonization by an intestinal commensal bacterium. PNAS. 2012;109:17621–26. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–47. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 15.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. PNAS. 2011;108:6252–57. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–36. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 17.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–41. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–98. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–93. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–36. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–14. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein RR, Bucci V, Toussaint NC, Buffie CG, Ratsch G, et al. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLOS Comput Biol. 2013;9:e1003388. doi: 10.1371/journal.pcbi.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marino S, Baxter NT, Huffnagle GB, Petrosino JF, Schloss PD. Mathematical modeling of primary succession of murine intestinal microbiota. PNAS. 2014;111:439–44. doi: 10.1073/pnas.1311322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balmer ML, Slack E, de Gottardi A, Lawson MA, Hapfelmeier S, et al. The liver may act as a firewall mediating mutualism between the host and its gut commensal microbiota. Sci Transl Med. 2014;6:237ra66. doi: 10.1126/scitranslmed.3008618. [DOI] [PubMed] [Google Scholar]
- 29.Godl K, Johansson ME, Lidell ME, Morgelin M, Karlsson H, et al. The N terminus of the MUC2 mucin forms trimers that are held together within a trypsin-resistant core fragment. J Biol Chem. 2002;277:47248–56. doi: 10.1074/jbc.M208483200. [DOI] [PubMed] [Google Scholar]
- 30.Lidell ME, Moncada DM, Chadee K, Hansson GC. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. PNAS. 2006;103:9298–303. doi: 10.1073/pnas.0600623103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Post S, Subramani DB, Backstrom M, Johansson ME, Vester-Christensen MB, et al. Site-specific O-glycosylation on the MUC2 mucin protein inhibits cleavage by the Porphyromonas gingivalis secreted cysteine protease (RgpB) J Biol Chem. 2013;288:14636–46. doi: 10.1074/jbc.M113.459479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–57. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem. 2009;284:18445–57. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. PNAS. 2011;108(Suppl 1):4659–65. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu J, Wei B, Wen T, Johansson ME, Liu X, et al. Loss of intestinal core 1–derived O-glycans causes spontaneous colitis in mice. J Clin Investig. 2011;121:1657–66. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sommer F, Adam N, Johansson ME, Xia L, Hansson GC, Backhed F. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PLOS ONE. 2014;9:e85254. doi: 10.1371/journal.pone.0085254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson ME, Gustafsson JK, Sjoberg KE, Petersson J, Holm L, et al. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLOS ONE. 2010;5:e12238. doi: 10.1371/journal.pone.0012238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. PNAS. 2008;105:15064–9. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansson GC, Johansson ME. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut Microbes. 2010;1:51–54. doi: 10.4161/gmic.1.1.10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–58. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointest Liver Physiol. 2013;305:G341–47. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmen Larsson JM, Thomsson KA, Rodriguez-Pineiro AM, Karlsson H, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. III Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am J Physiol Gastrointest Liver Physiol. 2013;305:G357–63. doi: 10.1152/ajpgi.00048.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McAuley JL, Linden SK, Png CW, King RM, Pennington HL, et al. MUC1 cell surface mucin is a critical element of the mucosal barrier to infection. J Clin Investig. 2007;117:2313–24. doi: 10.1172/JCI26705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linden SK, Sheng YH, Every AL, Miles KM, Skoog EC, et al. MUC1 limits Helicobacter pylori infection both by steric hindrance and by acting as a releasable decoy. PLOS Pathog. 2009;5:e1000617. doi: 10.1371/journal.ppat.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zarepour M, Bhullar K, Montero M, Ma C, Huang T, et al. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect Immun. 2013;81:3672–83. doi: 10.1128/IAI.00854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342:447–53. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDole JR, Wheeler LW, McDonald KG, Wang B, Konjufca V, et al. Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature. 2012;483:345–49. doi: 10.1038/nature10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–85. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 49.Frantz AL, Rogier EW, Weber CR, Shen L, Cohen DA, et al. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 2012;5:501–12. doi: 10.1038/mi.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–34. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 52.Zaph C, Troy AE, Taylor BC, Berman-Booty LD, Guild KJ, et al. Epithelial-cell-intrinsic IKK-β expression regulates intestinal immune homeostasis. Nature. 2007;446:552–56. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 53.Olszak T, Neves JF, Dowds CM, Baker K, Glickman J, et al. Protective mucosal immunity mediated by epithelial CD1d and IL-10. Nature. 2014;509:497–502. doi: 10.1038/nature13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, et al. γδintraepithelial lymphocytes are essential mediators of host-microbial homeostasis at the intestinal mucosal surface. PNAS. 2011;108:8743–48. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown SL, Riehl TE, Walker MR, Geske MJ, Doherty JM, et al. Myd88-dependent positioning of Ptgs2-expressing stromal cells maintains colonic epithelial proliferation during injury. J Clin Investig. 2007;117:258–69. doi: 10.1172/JCI29159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varol C, Zigmond E, Jung S. Securing the immune tightrope: mononuclear phagocytes in the intestinal lamina propria. Nat Rev Immunol. 2010;10:415–26. doi: 10.1038/nri2778. [DOI] [PubMed] [Google Scholar]
- 57.Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Kc W, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490:502–7. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, et al. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2:361–67. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 59.Niess JH, Brand S, Gu X, Landsman L, Jung S, et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–58. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 60.Chieppa M, Rescigno M, Huang AY, Germain RN. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J Exp Med. 2006;203:2841–52. doi: 10.1084/jem.20061884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid–dependent mechanism. J Exp Med. 2007;204:1757–64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farache J, Koren I, Milo I, Gurevich I, Kim KW, et al. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity. 2013;38:581–95. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diehl GE, Longman RS, Zhang JX, Breart B, Galan C, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature. 2013;494:116–20. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206:3101–14. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zigmond E, Varol C, Farache J, Elmaliah E, Satpathy AT, et al. Ly6Chi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity. 2012;37:1076–90. doi: 10.1016/j.immuni.2012.08.026. [DOI] [PubMed] [Google Scholar]
- 66.Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14:571–78. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Samstein M, Schreiber HA, Leiner IM, Susac B, Glickman MS, Pamer EG. Essential yet limited role for CCR2+ inflammatory monocytes during Mycobacterium tuberculosis–specific T cell priming. eLife. 2013;2:e01086. doi: 10.7554/eLife.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mazzini E, Massimiliano L, Penna G, Rescigno M. Oral tolerance can be established via gap junction transfer of fed antigens from CX3CR1+ macrophages to CD103+ dendritic cells. Immunity. 2014;40:248–61. doi: 10.1016/j.immuni.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 69.Cerovic V, Houston SA, Scott CL, Aumeunier A, Yrlid U, et al. Intestinal CD103− dendritic cells migrate in lymph and prime effector T cells. Mucosal Immunol. 2013;6:104–13. doi: 10.1038/mi.2012.53. [DOI] [PubMed] [Google Scholar]
- 70.Grainger JR, Wohlfert EA, Fuss IJ, Bouladoux N, Askenase MH, et al. Inflammatory monocytes regulate pathologic responses to commensals during acute gastrointestinal infection. Nat Med. 2013;19:713–21. doi: 10.1038/nm.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014;343:1249288. doi: 10.1126/science.1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, et al. NLRC4-driven production of IL-1βdiscriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13:449–56. doi: 10.1038/ni.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–89. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 74.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kinnebrew MA, Buffie CG, Diehl GE, Zenewicz LA, Leiner I, et al. Interleukin 23 production by intestinal CD103+CD11b+ dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–87. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Satpathy AT, Briseno CG, Lee JS, Ng D, Manieri NA, et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol. 2013;14:937–48. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujimoto K, Karuppuchamy T, Takemura N, Shimohigoshi M, Machida T, et al. A new subset of CD103+CD8α+ dendritic cells in the small intestine expresses TLR3, TLR7, and TLR9 and induces Th1 response and CTL activity. J Immunol. 2011;186:6287–95. doi: 10.4049/jimmunol.1004036. [DOI] [PubMed] [Google Scholar]
- 78.Cerovic V, Bain CC, Mowat AM, Milling SW. Intestinal macrophages and dendritic cells: What’s the difference? Trends Immunol. 2014;35:270–77. doi: 10.1016/j.it.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 79.Bekiaris V, Persson EK, Agace WW. Intestinal dendritic cells in the regulation of mucosal immunity. Immunol Rev. 2014;260:86–101. doi: 10.1111/imr.12194. [DOI] [PubMed] [Google Scholar]
- 80.Kaiser P, Regoes RR, Dolowschiak T, Wotzka SY, Lengefeld J, et al. Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLOS Biol. 2014;12:e1001793. doi: 10.1371/journal.pbio.1001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yrlid U, Milling SW, Miller JL, Cartland S, Jenkins CD, MacPherson GG. Regulation of intestinal dendritic cell migration and activation by plasmacytoid dendritic cells, TNF-alpha and type 1 IFNs after feeding a TLR7/8 ligand. J Immunol. 2006;176:5205–12. doi: 10.4049/jimmunol.176.9.5205. [DOI] [PubMed] [Google Scholar]
- 82.Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 2013;14:668–75. doi: 10.1038/ni.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 84.Round JL, Lee SM, Li J, Tran G, Jabri B, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–77. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–20. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dasgupta S, Erturk-Hasdemir D, Ochoa-Reparaz J, Reinecker HC, Kasper DL. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe. 2014;15:413–23. doi: 10.1016/j.chom.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–30. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIIIγ and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204:1891–900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. PNAS. 2008;105:20858–63. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Loonen LM, Stolte EH, Jaklofsky MT, Meijerink M, Dekker J, et al. REG3γ-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 2014;7:939–47. doi: 10.1038/mi.2013.109. [DOI] [PubMed] [Google Scholar]
- 91.Mukherjee S, Zheng H, Derebe MG, Callenberg KM, Partch CL, et al. Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature. 2014;505:103–7. doi: 10.1038/nature12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miki T, Holst O, Hardt WD. The bactericidal activity of the C-type lectin RegIIIβ against gram-negative bacteria involves binding to lipid A. J Biol Chem. 2012;287:34844–55. doi: 10.1074/jbc.M112.399998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miki T, Hardt WD. Outer membrane permeabilization is an essential step in the killing of gram-negative bacteria by the lectin RegIIIβ. PLOS ONE. 2013;8:e69901. doi: 10.1371/journal.pone.0069901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Uematsu S, Jang MH, Chevrier N, Guo Z, Kumagai Y, et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat Immunol. 2006;7:868–74. doi: 10.1038/ni1362. [DOI] [PubMed] [Google Scholar]
- 95.Uematsu S, Fujimoto K, Jang MH, Yang BG, Jung YJ, et al. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat Immunol. 2008;9:769–76. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- 96.Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5–dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534–43. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jarchum I, Liu M, Lipuma L, Pamer EG. Toll-like receptor 5 stimulation protects mice from acute Clostridium difficile colitis. Infect Immun. 2011;79:1498–503. doi: 10.1128/IAI.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–52. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, et al. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571–81. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Letran SE, Lee SJ, Atif SM, Flores-Langarica A, Uematsu S, et al. TLR5-deficient mice lack basal inflammatory and metabolic defects but exhibit impaired CD4 T cell responses to a flagellated pathogen. J Immunol. 2011;186:5406–12. doi: 10.4049/jimmunol.1003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–49. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–7. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209:1445–56. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jin C, Henao-Mejia J, Flavell RA. Innate immune receptors: key regulators of metabolic disease progression. Cell Metab. 2013;17:873–82. doi: 10.1016/j.cmet.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 106.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–85. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045–59. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ayres JS, Trinidad NJ, Vance RE. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat Med. 2012;18:799–806. doi: 10.1038/nm.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–31. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim YG, Kamada N, Shaw MH, Warner N, Chen GY, et al. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity. 2011;34:769–80. doi: 10.1016/j.immuni.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–86. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 112.Sonnenberg GF, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601–10. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kruglov AA, Grivennikov SI, Kuprash DV, Winsauer C, Prepens S, et al. Nonredundant function of soluble LTα3 produced by innate lymphoid cells in intestinal homeostasis. Science. 2013;342:1243–46. doi: 10.1126/science.1243364. [DOI] [PubMed] [Google Scholar]
- 114.van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 2014;508:123–27. doi: 10.1038/nature13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–25. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–17. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–99. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Elahi S, Ertelt JM, Kinder JM, Jiang TT, Zhang X, et al. Immunosuppressive CD71+ erythroid cells compromise neonatal host defence against infection. Nature. 2013;504:158–62. doi: 10.1038/nature12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–70. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. PNAS. 2004;101:1981–86. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. 2014;5:28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect Immun. 1999;67:3504–11. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–20. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 125.Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, et al. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485–89. doi: 10.1126/science.1217718. [DOI] [PubMed] [Google Scholar]
- 126.Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–9. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lindner C, Wahl B, Fohse L, Suerbaum S, Macpherson AJ, et al. Age, microbiota, and T cells shape diverse individual IgA repertoires in the intestine. J Exp Med. 2012;209:365–77. doi: 10.1084/jem.20111980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fritz JH, Rojas OL, Simard N, McCarthy DD, Hapfelmeier S, et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature. 2012;481:199–203. doi: 10.1038/nature10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–89. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 130.Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, et al. The genome of Th17 cell–inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. 2011;10:260–72. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–56. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Goto Y, Panea C, Nakato G, Cebula A, Lee C, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hand TW, Dos Santos LM, Bouladoux N, Molloy MJ, Pagan AJ, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–56. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shaw MH, Kamada N, Kim YG, Nunez G. Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209:251–58. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, et al. TH17 cell differentiation is regulated by the circadian clock. Science. 2013;342:727–30. doi: 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 137.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–54. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. PNAS. 2010;107:12204–9. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Narushima S, Sugiura Y, Oshima K, Atarashi K, Hattori M, et al. Characterization of the 17 strains of regulatory T cell–inducing human-derived Clostridia. Gut Microbes. 2014;5:333–39. doi: 10.4161/gmic.28572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim SV, Xiang WV, Kwak C, Yang Y, Lin XW, et al. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science. 2013;340:1456–59. doi: 10.1126/science.1237013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Obata Y, Furusawa Y, Endo TA, Sharif J, Takahashi D, et al. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol. 2014;15:571–79. doi: 10.1038/ni.2886. [DOI] [PubMed] [Google Scholar]
- 143.Olszak T, An D, Zeissig S, Vera MP, Richter J, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wingender G, Stepniak D, Krebs P, Lin L, McBride S, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143:418–28. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wieland Brown LC, Penaranda C, Kashyap PC, Williams BB, Clardy J, et al. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLOS Biol. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.An D, Oh SF, Olszak T, Neves JF, Avci FY, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. 2014;156:123–33. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Le Bourhis L, Dusseaux M, Bohineust A, Bessoles S, Martin E, et al. MAIT cells detect and efficiently lyse bacterially-infected epithelial cells. PLOS Pathog. 2013;9:e1003681. doi: 10.1371/journal.ppat.1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–84. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLOS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, et al. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–52. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–80. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hall JA, Grainger JR, Spencer SP, Belkaid Y. The role of retinoic acid in tolerance and immunity. Immunity. 2011;35:13–22. doi: 10.1016/j.immuni.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hall JA, Cannons JL, Grainger JR, Dos Santos LM, Hand TW, et al. Essential role for retinoic acid in the promotion of CD4+ T cell effector responses via retinoic acid receptor alpha. Immunity. 2011;34:435–47. doi: 10.1016/j.immuni.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Spencer SP, Wilhelm C, Yang Q, Hall JA, Bouladoux N, et al. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science. 2014;343:432–37. doi: 10.1126/science.1247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell Host Microbe. 2014;15:47–57. doi: 10.1016/j.chom.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–39. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–85. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 160.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–85. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–98. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, et al. Relating the metatranscriptome and metagenome of the human gut. PNAS. 2014;111:E2329–38. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ursell LK, Haiser HJ, Van Treuren W, Garg N, Reddivari L, et al. The intestinal metabolome: an intersection between microbiota and host. Gastroenterology. 2014;146:1470–76. doi: 10.1053/j.gastro.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Antunes LC, Han J, Ferreira RB, Lolic P, Borchers CH, Finlay BB. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother. 2011;55:1494–503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cho I, Yamanishi S, Cox L, Methe BA, Zavadil J, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–26. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–43. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 169.Fukuda S, Toh H, Taylor TD, Ohno H, Hattori M. Acetate-producing bifidobacteria protect the host from enteropathogenic infection via carbohydrate transporters. Gut Microbes. 2012;3:449–54. doi: 10.4161/gmic.21214. [DOI] [PubMed] [Google Scholar]
- 170.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–86. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–50. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 173.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–55. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Vrieze A, Out C, Fuentes S, Jonker L, Reuling I, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol. 2014;60:824–31. doi: 10.1016/j.jhep.2013.11.034. [DOI] [PubMed] [Google Scholar]
- 176.Bergstrom KS, Kissoon-Singh V, Gibson DL, Ma C, Montero M, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLOS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Perez-Munoz ME, Bergstrom K, Peng V, Schmaltz R, Jimenez-Cardona R, et al. Discordance between changes in the gut microbiota and pathogenicity in a mouse model of spontaneous colitis. Gut Microbes. 2014;5:286–95. doi: 10.4161/gmic.28622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–91. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–25. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 180.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–29. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 181.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–29. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–11. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J. 2014;8:1403–17. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–92. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, et al. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol. 2013;190:5306–12. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mielke LA, Jones SA, Raverdeau M, Higgs R, Stefanska A, et al. Retinoic acid expression associates with enhanced IL-22 production by γδ T cells and innate lymphoid cells and attenuation of intestinal inflammation. J Exp Med. 2013;210:1117–24. doi: 10.1084/jem.20121588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Powell N, Walker AW, Stolarczyk E, Canavan JB, Gokmen MR, et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity. 2012;37:674–84. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Investig. 2013;123:700–11. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li XD, Chiu YH, Ismail AS, Behrendt CL, Wight-Carter M, et al. Mitochondrial antiviral signaling protein (MAVS) monitors commensal bacteria and induces an immune response that prevents experimental colitis. PNAS. 2011;108:17390–95. doi: 10.1073/pnas.1107114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–12. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–88. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 194.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. PNAS. 2013;110:9066–71. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Karlsson FH, Fak F, Nookaew I, Tremaroli V, Fagerberg B, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245. doi: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jenq RR, Ubeda C, Taur Y, Menezes CC, Khanin R, et al. Regulation of intestinal inflammation by microbiota following allogeneic bone marrow transplantation. J Exp Med. 2012;209:903–11. doi: 10.1084/jem.20112408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–27. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. PNAS. 2011;108(Suppl 1):4615–22. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–63. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nat Rev Cancer. 2009;9:57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- 203.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–28. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gagliani N, Hu B, Huber S, Elinav E, Flavell RA. The fire within: Microbes inflame tumors. Cell. 2014;157:776–83. doi: 10.1016/j.cell.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 205.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–15. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–22. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hu B, Elinav E, Huber S, Booth CJ, Strowig T, et al. Inflammation-induced tumorigenesis in the colon is regulated by caspase-1 and NLRC4. PNAS. 2010;107:21635–40. doi: 10.1073/pnas.1016814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Allen IC, TeKippe EM, Woodford RM, Uronis JM, Holl EK, et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J Exp Med. 2010;207:1045–56. doi: 10.1084/jem.20100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang Y, Wang K, Han GC, Wang RX, Xiao H, et al. Neutrophil infiltration favors colitis-associated tumorigenesis by activating the interleukin-1 (IL-1)/IL-6 axis. Mucosal Immunol. 2014;7:1106–15. doi: 10.1038/mi.2013.126. [DOI] [PubMed] [Google Scholar]
- 211.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–23. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chen GY, Liu M, Wang F, Bertin J, Nunez G. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol. 2011;186:7187–94. doi: 10.4049/jimmunol.1100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Normand S, Delanoye-Crespin A, Bressenot A, Huot L, Grandjean T, et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. PNAS. 2011;108:9601–6. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hu B, Elinav E, Huber S, Strowig T, Hao L, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. PNAS. 2013;110:9862–67. doi: 10.1073/pnas.1307575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491:254–58. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Huber S, Gagliani N, Zenewicz LA, Huber FJ, Bosurgi L, et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 2012;491:259–63. doi: 10.1038/nature11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med. 2013;210:917–31. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 219.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–70. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–76. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bohnhoff M, Drake BL, Miller CP. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med. 1954;86:132–37. doi: 10.3181/00379727-86-21030. [DOI] [PubMed] [Google Scholar]
- 222.Bohnhoff M, Miller CP. Enhanced susceptibility to Salmonella infection in streptomycin-treated mice. J Infect Dis. 1962;111:117–27. doi: 10.1093/infdis/111.2.117. [DOI] [PubMed] [Google Scholar]
- 223.Freter R. The fatal enteric cholera infection in the guinea pig, achieved by inhibition of normal enteric flora. J Infect Dis. 1955;97:57–65. doi: 10.1093/infdis/97.1.57. [DOI] [PubMed] [Google Scholar]
- 224.Freter R. In vivo and in vitro antagonism of intestinal bacteria against Shigella flexneri II The inhibitory mechanism. J Infect Dis. 1962;110:38–46. doi: 10.1093/infdis/110.1.38. [DOI] [PubMed] [Google Scholar]
- 225.Stecher B, Hardt WD. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol. 2011;14:82–91. doi: 10.1016/j.mib.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 226.Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–90. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLOS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Antunes LC, Finlay BB. A comparative analysis of the effect of antibiotic treatment and enteric infection on intestinal homeostasis. Gut Microbes. 2011;2:105–8. doi: 10.4161/gmic.2.2.15610. [DOI] [PubMed] [Google Scholar]
- 230.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Investig. 2010;120:4332–41. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile–induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bohnhoff M, Miller CP, Martin WR. Resistance of the mouse’s intestinal tract to experimental Salmonella infection. I Factors which interfere with the initiation of infection by oral inoculation. J Exp Med. 1964;120:805–16. doi: 10.1084/jem.120.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ferreira RB, Gill N, Willing BP, Antunes LC, Russell SL, et al. The intestinal microbiota plays a role in Salmonella-induced colitis independent of pathogen colonization. PLOS ONE. 2011;6:e20338. doi: 10.1371/journal.pone.0020338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium–induced colitis. Infect Immun. 2011;79:1536–45. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Endt K, Stecher B, Chaffron S, Slack E, Tchitchek N, et al. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLOS Pathog. 2010;6:e1001097. doi: 10.1371/journal.ppat.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun. 2013;81:965–73. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905–14. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526–36. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 239.Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77:3661–69. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jarchum I, Liu M, Shi C, Equinda M, Pamer EG. Critical role for MyD88-mediated neutrophil recruitment during Clostridium difficile colitis. Infect Immun. 2012;80:2989–96. doi: 10.1128/IAI.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Theriot CM, Young VB. Microbial and metabolic interactions between the gastrointestinal tract and Clostridium difficile infection. Gut Microbes. 2014;5:86–95. doi: 10.4161/gmic.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Schubert AM, Rogers MA, Ring C, Mogle J, Petrosino JP, et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. MBio. 2014;5:e01021–14. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.van Nood E, Dijkgraaf MG, Keller JJ. Duodenal infusion of feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:2145. doi: 10.1056/NEJMc1303919. [DOI] [PubMed] [Google Scholar]
- 245.Pamer EG. Fecal microbiota transplantation: effectiveness, complexities, and lingering concerns. Mucosal Immunol. 2014;7:210–14. doi: 10.1038/mi.2013.117. [DOI] [PubMed] [Google Scholar]
- 246.Tvede M, Rask-Madsen J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet. 1989;1:1156–60. doi: 10.1016/s0140-6736(89)92749-9. [DOI] [PubMed] [Google Scholar]
- 247.Emanuelsson F, Claesson BE, Ljungstrom L, Tvede M, Ung KA. Faecal microbiota transplantation and bacteriotherapy for recurrent Clostridium difficile infection: a retrospective evaluation of 31 patients. Scand J Infect Dis. 2013;46:89–97. doi: 10.3109/00365548.2013.858181. [DOI] [PubMed] [Google Scholar]
- 248.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLOS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]