Abstract
Galpha(i)-coupled receptors comprise a diverse family of receptors that induce transformation by largely unknown mechanisms. We previously found that the Galpha(i)-coupled dopamine-D2short (D2S) receptor transforms Balb-D2S cells via Gαi3. To identify new Gαi effectors, a yeast two-hybrid screen was done using constitutively active Gαi3-Q204L as bait, and tumor necrosis factor-alpha (TNFα)-induced protein 8 (TNFAIP8, SCC-S2/NDED/GG2-1) was identified. In contrast, TNFAIP8-related TIPE1 and TIPE2 showed a very weak interaction with Gαi3. In yeast mating, in vitro pull-down, co-immunoprecipitation and bioluminescence resonance energy transfer (BRET) assays, TNFAIP8 preferentially interacted with activated Gαi proteins, consistent with direct Gαi-TNFAIP8 coupling. Over-expression or depletion of TNFAIP8 using antisense constructs in Balb-D2S cells did not affect D2S-induced signaling to Gαi-dependent inhibition of cAMP. In contrast, antisense depletion of TNFAIP8 completely inhibited spontaneous and D2S-induced foci formation, consistent with a role for TNFAIP8 in Gαi-dependent transformation. To address possible mechanisms, the effect of D2S signaling via TNFAIP8 on TNFα action was examined. D2S receptor activation inhibited TNFα-induced cell death in Balb-D2S cells, but not in cells depleted of TNFAIP8. However, depletion of TNFAIP8 did not prevent D2S-induced inhibition of TNFα-mediated caspase activation, suggesting that D2S/TNFAIP8-induced protection from TNFα-induced cell death is caspase-independent. The data suggest that Gαi-TNFAIP8-mediated rescue of pre-oncogenic cells enhances progression to oncogenic transformation, providing a selective target to inhibit cellular transformation.
Heterotrimeric G proteins (composed of Gα and Gβγ subunits) mediate intracellular signaling of a wide variety of receptors (Bockaert et al., 2002; Fredriksson et al., 2003; Wise et al., 2004), but traditional effectors such as adenylyl cyclase (AC) do not explain all of their actions (Albert and Robillard, 2002; Siderovski and Willard, 2005; Dorsam and Gutkind, 2007; Dave et al., 2009). The “inhibitory” G proteins (Gi1, Gi2, Gi3, and Go) couple to inhibition of AC in nearly all cell types, yet can stimulate cell proliferation and transformation in mesenchymal cell types, but inhibit these processes in neuroendocrine cells (Albert and Robillard, 2002; Dorsam and Gutkind, 2007). In neuroendocrine cells, the Gi/Go-coupled dopamine-D2short (D2S) receptor inhibits AC, phospholipase C (PLC), and mitogen-activated protein kinase (MAPK; ERK1/2), leading to decreased hormone synthesis, secretion, and reduced cell proliferation (Albert, 1994, 2002; Albert et al., 1997; Banihashemi and Albert, 2002). Paradoxically, in mesenchymal cells such as Balb/c-3T3 fibroblasts, D2S receptors stimulate calcium mobilization and MAPK activation, leading to increased cell proliferation and transformation (Ghahremani et al., 2000; Albert and Robillard, 2002). These D2S-induced responses were blocked by pretreatment with pertussis toxin (PTX), which selectively inhibits Gi/Go proteins, implicating them in these D2S-mediated actions (Ghahremani et al., 2000; Banihashemi and Albert, 2002). However, the mechanisms involved in differential regulation of cell proliferation by Gi/Go signaling remain incompletely characterized. We recently used yeast two hybrid screening to identify a novel Gαi effector, the Ras GTPase activating protein RASA3, which mediates D2S-induced inactivation of MAPK in neuroendocrine cells (Nafisi et al., 2008). However, the Gαi effector(s) that mediate stimulation of cell transformation in Balb/c-3T3 cells are not known.
In order to further address mechanisms of Gi/Go-mediated stimulation of cell proliferation, non-transformed Balb/c-3T3 cells were stably transfected with the Gi/Go-coupled D2S receptor to generate Balb-D2S cells (Ghahremani et al., 2000; Albert and Robillard, 2002), since Balb/c-3T3 cells represent a well-studied model of two-stage transformation that closely replicates sensitivity to transformation in the whole animal (Group, 1985; Miura et al., 2006; Maeshima et al., 2009). To identify which G proteins were required for D2S actions, we stably transfected Balb-D2S cells with PTX-insensitive Gi/Go proteins (PTX-Gi2, PTX-Gi3, and PTX-Go) and examined D2S-induced responses upon pretreatment with PTX to inactivate endogenous Gi/Go proteins (Ghahremani et al., 2000). We found that PTX-Gi3 rescued D2S-induced transformation, but not MAPK or cell proliferation. Thus, Gi3 is implicated in D2S-induced transformation in Balb/c-3T3 cells, but through an unknown pathway that does not involve MAPK-induced cell proliferation.
We hypothesized that Gαi3 couples to an unknown effector in Balb-D2S cells to mediate transformation and have used an activated form of Gαi3 as bait in a yeast two-hybrid screen to identify tumor necrosis factor-alpha-induced protein 8 (TNFAIP8) as a novel Gαi-interacting protein. We find that over-expression of TNFAIP8 reduces D2S signaling to Gβγ—but not Gαi-mediated pathways. Depletion of TNFAIP8 inhibited D2S/Gαi-induced transformation of Balb-D2S cells. Importantly, we have uncovered a new action of the D2S receptor signaling to inhibit TNFα-induced cell death that was blocked upon depletion of TNFAIP8. This indicates a previously unknown mechanism of crosstalk between Gi protein-induced growth signaling and death receptor signaling. Our results also indicate a link between Gαi-induced inhibition of cell death and cellular transformation. Taken together, these results suggest a mechanism whereby D2S/Gi-induced signaling to TNFAIP8 rescues of pro-oncogenic cells to prevent their elimination by cell death, leading to oncogenic transformation. Based on the known function of TNFAIP8 as an anti-apoptotic and pro-oncogenic signaling molecule in immune and breast cancers (Kumar et al., 2000, 2004), these results suggest a role for Gαi signaling to TNF AIP8 in the cellular transformation of mesenchymal cells in these tissues.
Materials and Methods
Materials
Restriction enzymes, endonucleases, and DNA polymerases total p42/44 MAPK antiserum, antiphospho-p42/44 MAPK (T202/Y204) antibody were purchased from New England Biolabs (Pickering, ON, Canada). Apomorphine (APO), dopamine, EGTA, forskolin, 3-isobutyl-1-methylxanthine (IBMX), and PTX were from Sigma (St. Louis, MO.). Enhanced chemiluminescence (ECL, Roche, Laval, QC) detection kits, [3H]spiperone (125 Ci/mM) were obtained from GE Healthcare (Baie d’Urfe, QC). Fura-2-AM was obtained from Molecular Probes (Eugene, OR). Hygromycin B, anti-Gαi1-2, and anti-Gαi3 were from Calbiochem (San Diego, CA). [125I]-Succinyl cAMP (2200 Ci/mMol) and polyvinylidene difluoride (PVDF) membrane were purchased from NEN Life Science Products (Boston, MA). Anti-3′,5′-cAMP antibody was obtained from ICN Biomedicals (Aurora, OH). Anti-TNFAIP8 antibody was raised against KLH-conjugated peptide (TNFAIP8 amino acids 87–97) RNNQFNQDELC (Sigma–Genosys, Oakville, ON, Canada) and affinity purified using peptide-conjugated Sepharose. Sera, media, and Geneticin (G418) were obtained from Wisent (St.-Bruno, QC).
Yeast two-hybrid screening
Constitutively active human Gαi3-Q204L cDNA (cDNA Resource Center, Rolla, MO) was subcloned into EcoRI-cut pAS2-1 (Clontech, Mountain View, CA). A mouse NIH-3T3 cDNA library (Clontech CAT. #ML4009AH) was screened using pAS2-1-Gi3-Q204L as bait in AH109 yeast strain grown at 30°C for 5–7 days. Transformants were selected on SD-Leu−Trp−His− plates and screened by X-galactosidase assay. DNA from positive clones was extracted, re-transformed into MH6 cells and selected on M9-Leu− plates and DNA extracted for verification and sequence analysis.
Northern blot analysis
Total RNA was extracted from individual cell lines using the Trizol method. Total RNA (10 μg/lane) was loaded on a 1% formaldehyde agarose gel, separated by electrophoresis, transferred onto a Hybond-A membrane and hybridized with [32P]-labeled TNFAIP8 cDNA probe. The membrane was washed and exposed to X-ray film for 5 days at −80°C, with intensifying screen.
Quantitative β-gal assay
Gα subunits were subcloned in pAS2-1 and mouse TNFAIP8, TIPE1 (BC032199), and TIPE2 (BC055879; Open Biosystems, Huntsville, AL) were cloned into pACT2 vector. The C-terminal fragment of TNFAIP8 (aa 108–199) was amplified with two primers: one with EcoRI →CCG GAA TTC CAG CTT GCC ATG ACG GTC GT (at aa108) and one with XhoI →CCG CTC GAG TCA TAT GTT CTC TTC ATC C (at aa199); other TNFAIP8 constructs were made by incorporating stop codons at aa positions 110, 144, 163, 184, using Stratagene Mutagenesis protocol. Gα and TNFAIP8 constructs were transformed in Y187 and AH109 yeast strains and positive transformants selected on SD-Trp− and SD-Leu− plates, respectively. Resistant colonies were mated, selected on SD-Leu− Trp− His−, lysed by three freeze-thaw cycles and resuspended in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7.0). For quantitative assay, O-nitrophenyl β-D-galactopyranoside (ONPG) and yeast cell lysates were incubated at 30°C for 30 min or until color developed and A420 nm was measured and normalized to positive control pCL1.
In vitro pull down assay
Bacterially expressed S-His6-TNFAIP8 (pET, Novagen, Gibbstown, NJ) and GST-tagged Gαi3 or Gαi2 (pGEX; GE Healthcare, Piscataway, NJ) proteins were transformed into BL21 (DE3)-competent cells, colonies isolated, grown overnight and induced with 0.5 mg/ml isopropyl-β-D-thiogalactoside (3 h). Bacteria were harvested, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole), sonicated (6 sec × 10 sec) and centrifuged at 20,000g for 30 min at 4°C. Gαi2 and Gαi3 supernatants were incubated with 100 μM GTPγS or GDPβS (to activate or inactivate, respectively) and 10 mM MgCl2 on shaker (30°C, 30 min). The TNFAIP8 supernatant was incubated with Ni-NTA-agarose (Qiagen, Mississauga, ON, Canada) on shaker (1 h, 4°C), washed 3× in wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole) and Gαi2 or Gαi3 added and incubated 1 h, 4°C on shaker. The resin was washed, incubated with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole) for 15 min on shaker at 4°C and eluted. The proteins were resolved on 10% SDS–PAGE, transferred onto a polyvinylidene difluoride membrane, and blocked in 5% skim milk in Tris-buffered saline (22°C, 1 h). The membrane was blotted with anti-S antigen (Novagen) or anti-GST (Millipore, Billerica, MA) antibodies overnight at 4°C, and then horseradish peroxidase-linked anti-mouse secondary antibody was added to detect anti-GST antibody. The membrane was incubated with chemiluminescence substrate (Laval, QC, Canada) and exposed to Kodak BioMax MR film.
Co-immunoprecipitation assay
The pcDNA3-FLAG-TNFAIP8 construct was transiently co-transfected with wild-type or constitutively active Gαi3 or Gαi2 in pcDNA3 vector into HEK-293 cells using lipofectamine 2000. Alternately TE cells were used. After 48 h, the cells were scraped in Mg2+Lysis/Wash Buffer (125 mM HEPES, pH 7.5, 750 mM NaCl, 5% Igepal CA-630, 50 mM MgCl2, 5 mM EDTA, and 10% glycerol, 10 μg/ml aprotinin, and 10 μg/ml leupeptin), placed on shaker (3 h, 4°C), passed through 25G needle and centrifuged at 10,000g for 20 min at 4°C. The supernatants were incubated 1 h (4°C) with 40 μl of protein G-Sepharose, Fast Flow (Sigma) in a preclear phase, re-centrifuged, and the supernatants incubated with 40 μl of anti-FLAG M2-agarose affinity gel (Sigma) on a shaker at 4°C overnight, washed 3× with Mg2+Lysis/Wash Buffer, resuspended in 2× SDS-loading buffer, boiled for 5 min, resolved on 10% SDS–PAGE, and analyzed by Western blotting.
Cell culture and stable transfection
Balb/c-3T3 cells, Balb-D2S (clone 11, selected in 400 μg/ml hygromycin B) and derivative clones were cultured in Dulbecco’s modified Eagle’s medium (DMEM) plus 8% fetal bovine serum (FBS), and 100 μg/ml penicillin/streptomycin. Balb/c-D2S cells were transfected by calcium phosphate co-precipitation with 30 μg/plate of pcDNA3-FLAG-TNFAIP8, pcDNA3-FLAG-TNFAIP8-ΔNT, or pcDNA3-AS-TNFAIP8 and selected after adding 800 μg/ml G418 for 4–6 weeks. The colonies were harvested and screened by RT-PCR for TNFAIP8 RNA, and positive clones were confirmed by Western blot using anti-Flag or anti-TNFAIP8 antibody.
Bioluminescence resonance energy transfer (BRET2) assay
TNFAIP8 cDNA was subcloned in the EcoRI site of pGFP2-C2 vector generated N-terminal tagged GFP2-TNFAIP8; Gαi1-91rLuc was from Dr. Michel Bouvier (Univ. de Montreal). HEK293 cells cotransfected with Gαi1-91rLuc and GFP2-TNFAIP8 were washed twice and resuspended in PBS with 2 μg/ml aprotinin 106 cells/100 μl in 96-well format) with addition of TNFα (2.5 ng/ml) or vehicle followed by 5 μM DeepBlueC (Perkin Elmer, Woodbridge, ON, Canada), and read using Victor V3 plate reader (Perkin Elmer) using excitation 405/5 nm and emission filters 510/30 nm for GFP2 and emission filter 410/80 nm for RLuc, with non-transfected cells as control (Whitaker et al., 2008). BRET ratio was determined as GFP2 test – GFP2 control)/(RLuc test –RLuc control).
Ligand binding
Cell membranes were prepared as previously described, resuspended in TME buffer (75 mM Tris–HCl, pH 7.4, 12.5 mM MgCl2, 1 mM EDTA), and added to triplicate tubes (100 μg/tube) containing 200 μl TME and 10 nM [3H]-spiperone (119 Ci/mmol; GE Healthcare) without (total) or with (non-specific) APO (10 μM) for 30 min at 22°C. Reactions were terminated by filtration through GF/C (Whatmann) filters, washing with 3 ml × 4 ml of ice-cold buffer (50 mM Tris–HCl, pH 7.4) and radioactivity quantified by liquid scintillation counting; specific D2 binding was determined as total–non-specific. Protein was assayed with the Bio-Rad protein assay kit with bovine serum albumin as a standard.
cAMP assay
Measurement of cAMP was performed as described previously (Albert et al., 1999). In brief, cells were plated (8 × 105 cells/ml in six-well dishes) and after 24 h, washed and incubated in 1 ml/well DMEM/20 mM HEPES, pH 7.0/100 μM IBMX for 15 min at 37°C. Experimental compounds were added to triplicate wells, as indicated. Media were collected, centrifuged (13,000g × 0.5 min) and the supernatant assayed for cAMP by radioimmunoassay (MP Biomedicals, Irvine, CA).
Soft agar foci formation
Cell lines (passages 4–10) were plated 0.5 × 106 cells/10 cm dish prior to plating in soft agar and media was changed 24 h prior to replating six-well plates prepared with a 1.5 ml base layer of 0.5% agar/RPMI/15% FBS and a top layer containing 2.5 × 104 cells/well in 0.35% agar/RPMI/15% FBS (<42°C) with 100 μg/ml penicillin/streptomycin ± 1 μM of APO. Cells were grown for 3 weeks in soft agar and 200 μl treatments of 15% FBS-RPMI ± 1 μM APO were added every 2–3 days. After 3 weeks, cells were stained with 1 ml of 0.005% crystal violet for 1.5 h. Microscopy was performed using Olympus SZ61 dissection microscope at a magnification of 0.8× and photographed with PixeLink Mega Pixel Firewire Camera. Images were analyzed and foci automatically counted using NIH ImageJ software. In each six-well plate, triplicates of control and APO wells were grown for each cell line. These experiments were repeated independently three to four times/cell line.
Live–dead assay
Cell lines (passages 8–15) grown in six-well plates were serum-starved in DMEM/0.5% FBS for 20 h and then treated in DMEM with 20 μg/ml cycloheximide (CHX) ± drug treatments including 5 ng/ml mouse TNFα (Calbiochem), 0.3 μM APO (Sigma), or 1.0 μM spiperone (SPP; Sigma) as indicated for 3 h at 37°C, 5% CO2. The protocol described in the Invitrogen Live/Dead Assay Kit was then followed and microscopy was performed on Zeiss Axiovert S100 and photographed with Sony Power HAD 3 CCD color video camera. Cells were counted manually and the counter was blind to treatment group.
Active caspase staining assay
Cell lines (passages 4–10) were grown in six-well plates (2 × 104 cells/well) on coverslips for 24 h and then serum-starved in 1.5 ml of DMEM/0.5% FBS for 16 h and then treated for 2.5 h with CHX ± TNFα, APO, or SPP as described in the Live/Dead Assay. Media was then removed and replaced with 350 μl of media containing 1× FLICA (or FAM-DEVD-FMK, Chemicon Caspatag Assay Kit) for 1.5 h incubation. After incubation, media was removed and cells were washed twice in 1 ml of wash buffer (kit). Cover slips were then mounted on microscope slides with 40 μl of fixative (kit). Microscopy was performed on Zeiss Axioskop 2 MOT and photography was performed on Q-imaging QICAM FAST mono 12-bit camera. Image capturing and processing was performed with Northern Eclipse v6.0 software. Cells were counted manually and the counter was blind to treatment group.
Results
TNFAIP8 interacts with Gαi proteins
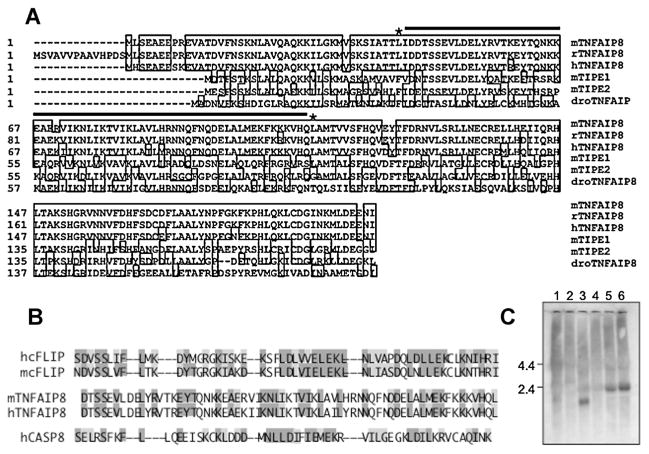
To identify new Gαi-dependent effectors, constitutively active Gαi3-Q204L was used as bait in a yeast two-hybrid screen of an NIH-3T3 cell cDNA library. Two screenings of 0.8 and 2.6 × 106 independent clones yielded positive clones for several known Gαi interacters, LGN (13/26), RGS19 (5/26), RGS20 (1/26), and AGS3 (1/26; Mochizuki et al., 1996; Takesono et al., 1999), and TNFAIP8 (4/26). TNFAIP8, (also known as SCC-S2, NDED, GG2-1, and MDC-3.13) was originally identified as TNFα-induced RNA transcript (Horrevoets et al., 1999). As shown in Figure 1A, mouse TNFAIP8 shares amino acid sequence identity with rat (98%), human (93%), and Drosophila melanogaster (42%) homologs, and with TNFAIP8-related proteins such as TIPE1 (56%) and TIPE2 (55%). Human TNFAIP8 (SCC-S2) has been characterized as an anti-apoptotic protein that is amplified in carcinoma (Kumar et al., 2000). TNFAIP8 contains a central region with homology to death effector domains (DED-like, Fig. 1B), particularly to cFLIP DED (43%) over the caspase 8 DED (31%). The TNFAIP8 DED-like domain is believed to confer anti-apoptotic activity, as observed for cFLIP (Kumar et al., 2000). To verify TNFAIP8 expression in NIH-3T3 and Balb/c-3T3 cells Northern blot for TNFAIP8 RNA was done and a single 2.4-kb murine species was detected (Fig. 1C), similar in size to the human TNFAIP8 transcript (Kumar et al., 2000). By contrast, in rat raphe RN46A and GH4C1 pituitary cells a smaller 1.8-kb transcript was identified. Thus, TNFAIP8 is expressed in both NIH-3T3 and Balb/c-3T3 cells.
Fig. 1.
A: Alignment and RNA expression of Gαi3-interacting mouse TNFAIP8 and homologs. Alignment of the predicted amino acid sequences of mouse TNFAIP8, rat (XP_225940), human TNFAIP8a (NP_055165), mouse TIPE1 (BC032199), mouse TIPE2 (BC055879), and Drosophila TNFAIP8 (NP_611820) was performed using DNAStar MegAlign software with % amino acid identity to mouse TNFAIP8 of 100%, 98.5%, 93.4%, 56.1%, 54.9%, and 42%, respectively. The putative death effector domain (DED) is indicated by the line between asterisks. B: Alignment of DED. human and mouse DED amino acid sequences (DED2) from human (hcFLIP, aa107–162) and mouse (mcFLIP, aa112–167). c-FLIP and human caspase 8 (hCASP8 aa115–170) are compared to mouse (aa44–109) and human TNFAIP8 putative DED (aa36–101). Dark shaded residues are identical in a majority of DED’s, light shaded residues are similar; the % similar amino acids was: TNFAIP8/cFLIP (43%); TNFAIP8/CASP8 (31%); cFLIP/CASP8 (51%). C: Expression ofTNFAIP8in rat and mouse cell lines. Total RNA was extracted from various cell lines (1, rat GH4C1 pituitary cells; 2, human HEK293 cells; 3, rat raphe RN46A cells; 4, rat X mouse septal SN48 cells; 5, murine Balb/c-3T3; and 6, NIH-3T3 cells) using Trizol method and analyzed by Northern blot. Equal loading was verified by ethidium bromide staining of 28S/18S RNA (not shown). Size markers correspond to 4.4 and 2.4 kb.
The Gα specificity of TNFAIP8 was examined by yeast mating and beta-galactosidase assays (Fig. 2). TNFAIP8 preferentially interacted with constitutively active Gαi family proteins, including Gαo, Gαi1, Gαi2, and Gαz proteins, but not with active Gαcone or Gαs (Fig. 2A); both wild-type and active Gαi3 interacted with TNFAIP8 in this assay. The interaction of Gαi3 or Gαo with TNFAIP8 (clone Gαi3-33; Mao et al., 2004) was not altered by the G184S mutation that inactivates Gα interactions with RGS proteins, indicating the specificity of the Gαi/o-TNFAIP8 interaction. By yeast mating assay, both TIPE1 and TIPE2 interacted with Gαi3 (Fig. 2B, inset), but much more weakly (1%) than TNFAIP8, indicating that the Gαi3 interaction is selective for TNFAIP8. A series of TNFAIP8 deletion constructs were examined to identify TNFAIP8 domains that mediate Gαi3 interaction. Only TNFAIP8 and N-terminal deleted TNFAIP8 (ΔNT) were positive (Fig. 2C,D), indicating that the TNFAIP8 N-terminal is dispensable for the interaction. Deletion of the last 15 amino acids of the C-terminal (ΔCT-D) was sufficient to prevent interaction with active Gαi3 (Fig. 2D), indicating that both the DED-like and extreme C-terminal are required for Gαi/o interaction.
Fig. 2.
TNFAIP8 interaction with Gα proteins in yeast. A: G-protein specificity. Mating of yeast transformed with pAS2-Gαi/o subunits wild-type or constitutively active Gαo-R179C or QL mutants and pACT2-TNFAIP8 was done, and transformants assayed by quantitative β-galactosidase assay. Active mutants of Gαz (Gz), Gαcone (Gc), and Gαs (Gs) were also assayed. Results are presented as normalized β-galactosidase activity (%pCL1-1, mean ± SE, N =3). B:TNFAIP8-like proteins. Yeast mating assay was done as in A, comparing activity of TNFAIP8 to TIPE1 and TIPE2; inset, enlarged scale of TIPE1/TIPE2 data. C: Specificity of Gαi–TNFAIP8 interaction. Interaction between wild-type (wt) or constitutively active Gαo-R179C or Gαi3-Q204L mutants was visualized by colonies in LTHA medium(arrows) and X-gal assay (dark-stained colonies). D: Summary of TNFAIP8 deletion mutants. The indicated TNFAIP8 deletion mutants (shown figuratively with amino acid sites indicated) were tested by yeast mating/X-gal assay and positive (+) and negative (−) interactions are shown. These experiments were repeated at least three times for each interaction.
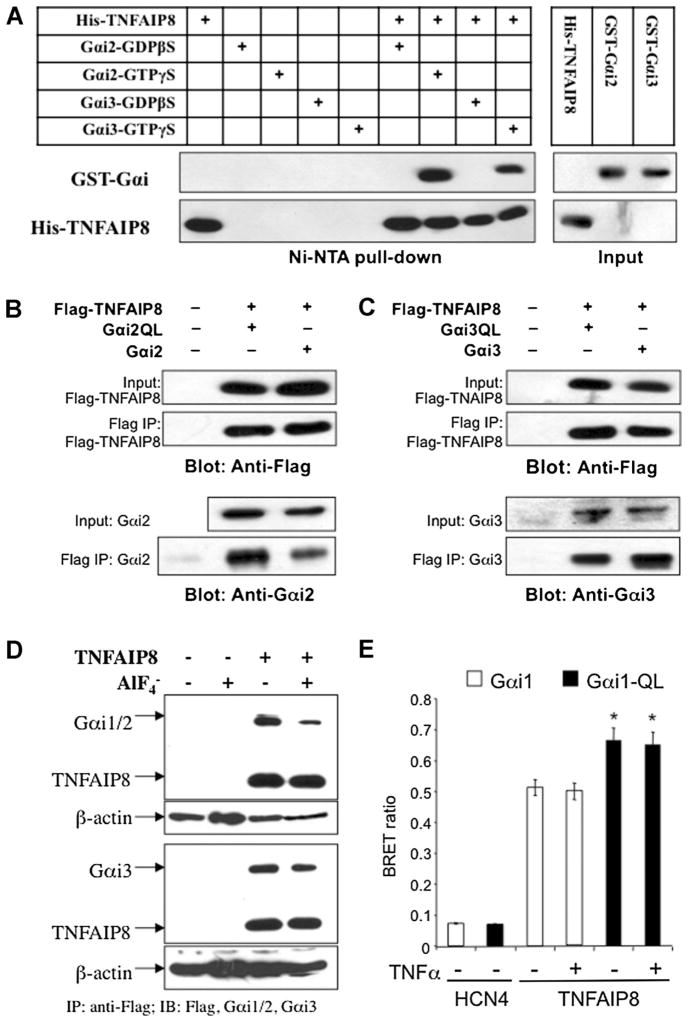
The Gαi-TNFAIP8 interaction was further verified by in vitro pull-down, co-immunoprecipitation, and BRET assays. For in vitro pull-down assay purified recombinant His-tagged TNFAIP8 and GST-tagged Gαi2 or Gαi3 proteins (Fig. 3) were pre-incubated with GDPβS or GTPγS for inactivation or activation, respectively (Fig. 3A). Upon pull-down of the His-tag, Gαi2, and Gαi3 were detected in the presence but not absence of His-TNFAIP8, indicating a direct interaction. A much greater pull-down of Gαi2 and especially Gαi3 was observed in the presence of GTPγS, consistent with a stronger TNFAIP8 interaction with activated G-proteins. To confirm that this interaction occurs in cells co-immunoprecipitation studies were done. HEK293 cells were transiently transfected with Flag-TNFAIP8 and either wild-type or constitutively active Gαi2- or Gαi3-Q204L point mutants (Fig. 3B,C). Upon immunoprecipitation of Flag-tag, both wild-type and active Gαi2 and Gαi3 mutants bound to TNFAIP8. To determine whether TNFAIP8 interacts with endogenous Gi/o proteins, an N-terminal tagged Flag-TNFAIP8 construct was stably transfected in Balb-D2S cells and immunoprecipitated using anti-Flag antibody (Fig. 3D). Western blot of the immunoprecipitate detected both non-activated and - activated Gαi2 and Gαi3, confirming the interaction of TNFAIP8 with these Gαi proteins. Taken together, these data indicate that TNFAIP8 directly and preferentially interacts with activated Gαi2 and Gαi3, consistent with Gαi/TNFAIP8 coupling. To address TNFAIP8–Gαi protein interaction in living cells we performed the BRET assay, which detects protein interaction within 100 Å (Hamdan et al., 2006). GFP2-TNFAIP8 and Gαi1-91rLuc (Gales et al., 2006) were co-transfected in HEK-293 cells and BRET ratio determined (Fig. 3E). TNFAIP8–GFP2 gave a strong BRET signal with wild-type or constitutively active (Gi1-QL) Gαi1-91rLuc, but was greater for the Gi1-QL suggesting a preferential interaction with activated Gαi1. The BRET signal was not altered by acute treatment with TNFα. By contrast, the potassium channel HCN4-GFP2, which does not interact with G-proteins (Whitaker and Accili, 2008), gave a weak background signal with Gαi1-91rLuc. These data support the preferential interaction of TNFAIP8 with activated Gαi protein in cells.
Fig. 3.
Interaction of TNFAIP8 with activated Gαi proteins in vitro and in cells. A: TNFAIP8 and Gαi2/Gαi3 interact in vitro. Recombinant His/S-tagged TNFAIP8 (His-TNFAIP8) and GST-tagged Gαi2 or Gαi3 in the presence of 100 μM GDPβS (to inactivate) or GTPγS (to activate) were combined and His/S tag-TNFAIP8 pulled down using Ni-NTA agarose, and the pull-down was analyzed by Western blot with anti-S tag (1:5,000) or anti-GST antibody (1:1,000). Input (10%) controls are shown at right. B,C: Interaction between TNFAIP8 and Gαi proteins in cells. HEK-293 cells were transiently cotransfected with Flag-TNFAIP8 and either vector or: B, Gαi2, wild-type or active Gαi2Q204L (Gαi2QL) or C, Gai3, wt Gαi3Q204L (Gαi3QL), as indicated. Cell extracts were immunoprecipitated using anti-Flag, and then 10% input or immunoprecipitates (Flag-IP) were analyzed by Western blot and probed using antibody to Flag to detect Flag-TNFAIP8 or to Gαi2 or Gαi3 as indicated. The bottom lanes show that Gαi2 and Gαi3 were co-immunoprecipitated with TNFAIP8. D: Co-immunoprecipitation of TNFAIP8 and Gαi proteins in Balb/c-3T3 cells. Balb-D2S cells were stably transfected with Flag-tagged TNFAIP8 (TE clone). The cells were lysed with NP-40 buffer and incubated with anti-FLAG antibody-agarose beads overnight at 4°C in the absence or presence of 10 mM NaF and 30 μM AlCl3 ( ). Immunoprecipitates examined by Western blot for TNFAIP8 (using anti-Flag) and endogenous Gi protein using anti-Gαi1/2 or anti-Gαi3 antibodies. Cell extracts were subjected to Western blot using anti-βactin as loading control. E: BRET studies of TNFAIP8/Gαi1 interaction in HEK293 cells. HEK293 cells were co-transfected with GFP2-TNFAIP8 and wt (Gαi1) or constitutively active (Gαi1-QL) Gαi1-91rLuc and were treated with TNFα (2.5 ng/ml) or vehicle and analyzed for BRET, with non-transfected cells as control (Whitaker and Accili, 2008). BRET ratio was determined as (GFP2test – GFP2 control)/(RLuc test – RLuc control). HCN4, which does not interact with G-proteins, was used as negative control to determine background fluorescent signal. *P < 0.05 by one-way ANOVA with Dunnett’s test.
D2S–Gαi signaling in TNFAIP8 clones
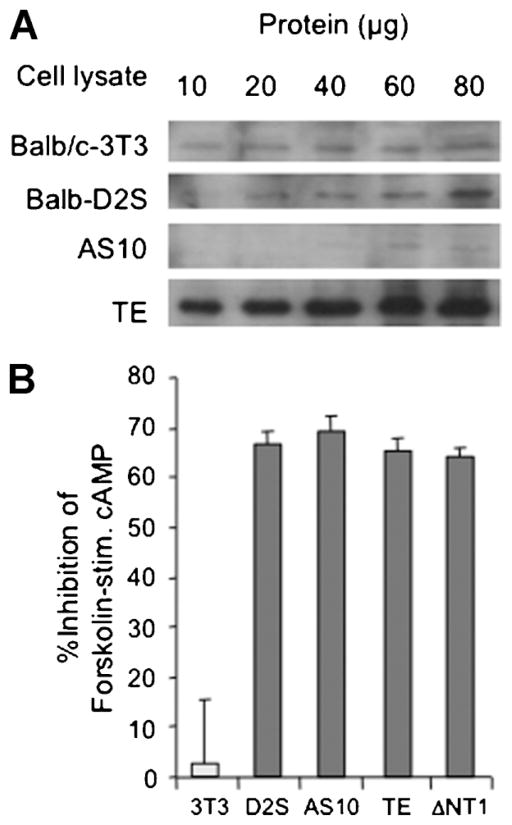
The function of the TNFAIP8–Gαi interaction was addressed using Balb-D2S cells stably transfected with sense (TE clone), ΔNT or antisense (AS6, AS10 clones) TNFAIP8 constructs. Clones were screened by RT-PCR for TNFAIP8 RNA, and positive clones examined by Western blot analysis, using rabbit polyclonal TNFAIP8 antibody (Fig. 4A). Compared to Balb/c-3T3 or parental Balb-D2S cells, the clone over-expressed TNFAIP8, while the AS-10 clone had nearly undetectable levels. The level of D2S receptor binding was reduced in most clones compared to wild-type Balb-D2S cells (Table 1), hence data from clones with similar levels of D2S receptors are compared.
Fig. 4.
D2S-induced inhibition of cAMP is unaffected in TNFAIP8 antisense or over-expressing cell lines. A: TNFAIP8 protein in antisense/over-expressing Balb-D2S clones. Positive clones of Balb-D2S cells stably transfected with sense or antisense TNFAIP8 constructs were subjected to Western blot analysis using anti-TNFAIP8. The indicated amounts of input cell lysate from Balb/c-3T3, Balb-D2S, antisense TNFAIP8 clone AS10, and sense TNFAIP8 clone TE are shown. B: D2S-induced Gαi-dependent inhibition of forskolin-stimulated cAMP. Balb/c-3T3 (3T3), Balb-D2S (D2S), and derivative sense (TE), antisense (AS10), and N-terminal truncated (ΔNT1) TNFAIP8 clones are shown. Cells were challenged with 10 μM forskolin in the absence or presence of D2S receptor agonist apomorphine (1 μM), and cAMP levels measured by radioimmunoassay. Data from at least three independent experiments are expressed as % inhibition by apomorphine of forskolin-induced cAMP level.
TABLE 1.
D2 receptor binding levels in balb/c-3T3 clones
| Clone | D2 Receptor (fmol/mg) |
|---|---|
| Balb/c 3T3 | 4 ± 2 |
| Balb-D2S | 115 ± 9 |
| TE | 33 ± 7 |
| TJ | 32 ± 8 |
| TB | 64 ± 10 |
| T4 | 16 ± 3 |
| ΔNT-3 | 23 ± 7 |
| ΔNT-I | 108 ± 10 |
| ΔNT-A | 50 ± 10 |
| ASD | 56 ± 9 |
| AS10 | 42 ± 17 |
| AS6 | 24 ± 11 |
Binding assays performed on membrane preparations from the indicated cell lines. Shown is the specific [3H]-spiperone binding (fmol/mg protein). The data are expressed as mean ± SEM (n = 5–9).
The role of the TNFAIP8–Gαi interaction in G-protein coupling was addressed by examining D2S-induced inhibition of forskolin-stimulated cAMP formation (Fig. 4B), a pathway mediated by Gαi2 and Gαi3 in Balb-D2S cells (Ghahremani et al., 2000). Treatment with D2 agonist APO inhibited forskolin-induced cAMP by 65% in Balb-D2S cells and derivative TNFAIP8 clones, while D2-negative Balb/c-3T3 cells lacked response to APO. Thus, modifying TNFAIP8 levels did not affect D2S signaling via Gαi2/3 proteins to inhibit AC activity.
TNFAIP8 mediates D2S/Gαi-induced transformation
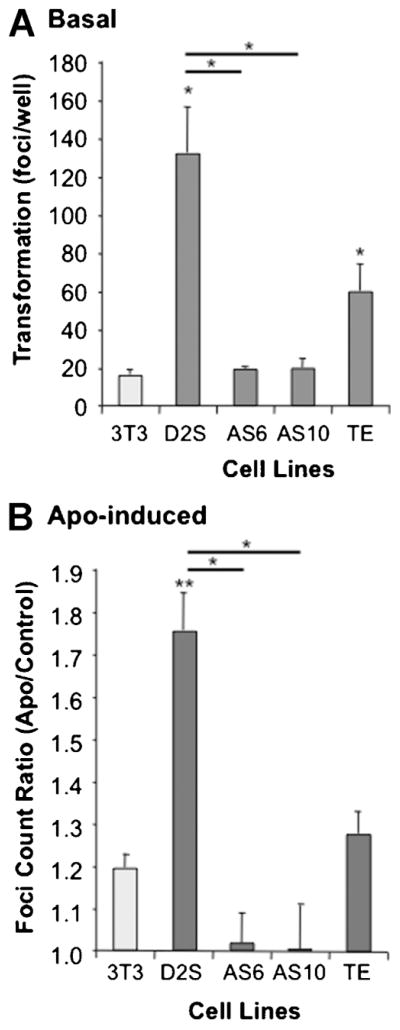
In order to address the role of TNFAIP8 in D2S-induced cell transformation, basal and APO-induced colony formation in soft agar was measured over 3 weeks (Fig. 5). Compared to non-transfected Balb/c-3T3 cells, basal foci formation was 3- to 6-fold higher in Balb-D2S and TE cells (Fig. 5A), suggesting that the D2S receptor has constitutive activity in this assay. Consistent with constitutive D2S signaling, we have previously observed that PTX, which blocks Gαi signaling, also reduces basal foci formation in Balb-D2S cells (Ghahremani et al., 2000). Similarly, spontaneous foci formation was completely blocked in AS10 and AS6 cells, suggesting that depletion of TNFAIP8 prevents spontaneous transformation of the D2S-expressing cells. In addition, APO-induced foci formation was completely blocked in AS6 and AS10 cells compared to Balb-D2S cells (Fig. 5B). In these experiments, the TNFAIP8-over-expressing TE cells showed a reduced response to APO compared to Balb-D2S cells, which may reflect the lower D2S receptor levels. These results suggest that endogenous TNFAIP8 is required for D2S receptor-induced transformation of Balb-D2S cells.
Fig. 5.
Basal and D2S-induced transformation of Balb-D2S cells requires TNFAIP8. Balb/c-3T3, Balb-D2S or derivative TNFAIP8 sense (TE) or antisense (AS6, AS10) clones were plated in soft agar and cultured for 3 weeks in the absence (Basal) or presence of 1 μM apomorphine (APO), and stained, images captured and the number of foci/well of transformed cells counted using image analysis software. Data from triplicate wells are presented as mean ±SE of at least three independent experiments/cell line. A: Basal transformation. *P < 0.05 versus 3T3 or D2S (as indicated) by one-way ANOVA with Dunnett’s post-test. B: APO-induced transformation. APO-induced transformation was measured as—fold basal transformation. **P < 0.01, *P < 0.05 versus 3T3 or D2S (as indicated) by one-way ANOVA and Bonferroni test.
TNFAIP8 mediates D2S signaling to inhibit TNFα-induced cell death
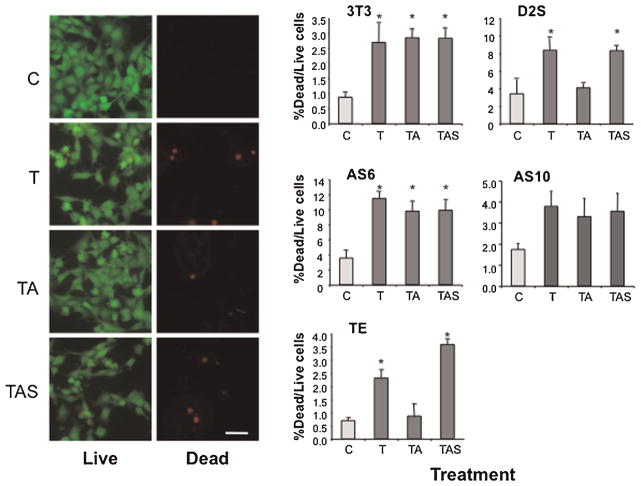
TNFα signals to apoptosis cascades to eliminate pro-oncogenic cells and prevent transformation (Sethi et al., 2008). Since TNFAIP8 has been implicated as an inhibitor of basal TNFα-induced apoptosis (Kumar et al., 2000), we hypothesized that D2S signaling via TNFAIP8 may inhibit TNFα-induced cell death, possibly contributing to D2S-induced transformation. Cells were treated or not with TNFαin the presence of CHX to inhibit survival pathways, and cell death was assayed as the ratio of dead/live cells (Fig. 6). Interestingly, the basal level of cell death was greater in Balb-D2S cells (4%) compared to Balb/c-3T3 cells (1%), and this was reduced in the TE cells (<1%), consistent with the anti-apoptotic activity of TNFAIP8. In Balb/c-3T3, Balb-D2S, and TNFAIP8 clones, TNFα significantly increased cell death by 2- to 3-fold. This effect was blocked by APO in Balb-D2S and TE but not Balb/c-3T3 cells and reversed by D2 antagonist SPP, indicating that D2S receptors inhibit TNFα-induced cell death. Importantly, in AS6 and AS10 cells depleted of TNFAIP8, TNFα-induced cell death was not blocked by APO, indicating that TNFAIP8 is required for D2S-induced inhibition of TNFα-induced cell death.
Fig. 6.
D2S-induced inhibition of TNFα-induced cell death requires TNFAIP8. Serum-starved(20 h,0.5%FBS)Balb/c-3T3(3T3),Balb-D2S(D2S), or TNFAIP8 clones (antisense AS6, AS10; or sense TE) were untreated (control, C) or treated with TNFα (5 ng/ml) alone (T), TNFα and 0.3 μM apomorphine (TA), or TNFα, apomorphine and 1 μM D2 antagonist spiperone (TAS), in the presence of CHX (10 μg/ml) for 3 h. Cell death was measured using the Live/Dead assay. Left panels representative photomicrographs of calcein (Live, fluorescein filter) or ethidium (Dead, rhodamine filter) stained Balb-D2S cells with treatment indicated; 10× magnification; scale bar, 25 μm. Right panels data from at least three independent experiments are presented as mean ±SE of the % of dead cells compared to live cells. *P < 0.05 versus control by one-way ANOVA with Dunnett’s post-test.
TNFα triggers cell death by caspase-dependent and -independent mechanisms (Thorburn, 2004). We addressed the effect of TNFα on caspase activation and whether this pathway is regulated by D2S-TNFAIP8 signaling. To measure caspase-active cells, cells were treated without or with TNFα, APO, or SPP, labeled with an irreversible fluorescein-conjugated caspase-3/7 inhibitor (Ekert et al., 1999) and fluorescent caspase-active cells counted (Fig. 7). The basal level of caspase-active cells was reduced in Balb-D2S and TE (1–2%) compared to Balb/c-3T3, AS6, or AS10 cells (>5%), suggesting that reduced apoptotic signaling in D2S-expressing cells is reversed upon depletion of TNFAIP8 consistent with the anti-apoptotic role of TNFAIP8. In Balb/c-3T3, Balb-D2S, and TNFAIP8 clones, TNFα significantly increased caspase-active cells except in AS10 cells (data not shown), suggesting that TNFα-induced cell death is caspase3/7-independent in AS10 cells. TNFα-induced caspase activation was blocked by D2S activation in Balb-D2S, TE, and AS6 cells, but not in Balb/c-3T3 cells and was reversed by SPP, indicating that D2S receptors inhibit TNFα-induced caspase-3/7 activation. However, the D2S response in the TNFAIP8-depleted AS6 cells indicates that D2S-inhibition of caspase3/7 activation may be independent of TNFAIP8. Taken together these data demonstrate that the D2S receptor inhibits TNFα-induced caspase activation and cell death, but that TNFAIP8 may mediate D2S inhibition of TNFα-mediated cell death by caspase3/7-independent pathways.
Fig. 7.
D2S signaling inhibits TNFα-induced caspase3/7 activity independent of TNFAIP8. Serum-starved (16 h, 0.5% FBS) Balb/c-3T3 (3T3), Balb-D2S (D2S), or TNFAIP8 clones (antisense AS6 or sense TE) were untreated (control, C) or treated with TNFα(5 ng/ml) alone (T), TNFα and 0.3 μM apomorphine (TA), or TNFα, apomorphine and 1 μM D2 TAS for 2.5 h, followed by addition of CaspaTag reagent for 1.5 h to stain cells with activated caspase3/7. Top: representative photomicrograph of TNFα-treated Balb-D2S cells in phase contrast or fluorescence to identify CaspaTag-stained cells; 20T magnification; scale bar, 25 μm. Below, data from ≥3 independent experiments are presented as mean ± SE of the % caspase-positive cells in triplicate wells. *P < 0.05 compared to control or as indicated by one-way ANOVA with Bonferroni post-test.
Discussion
TNFAIP8: a novel Gαi effector
Previously, we found that the D2S receptor coupled via Gαi3 to stimulate transformation of Balb/c-3T3 cells, independent of D2S-induced MAPK activation or cell proliferation (Ghahremani et al., 2000). This suggested that a new Gαi3-mediated pathway was involved in transformation. Here, we report the identification by two-hybrid screening of TNFAIP8 as a novel Gαi effector that couples the D2S receptor to cell transformation. These results were verified by yeast mating, in vitro pull-down and co-immunoprecipitation studies, as well as BRET studies in living cells, which show that TNFAIP8 directly and preferentially interacts with activated Gαi2 and Gαi3. However, in cells, both non-activated and activated Gαi3 interacted with TNFAIP8 suggesting that a preformed complex between the two proteins may exist, consistent with coupling between Gαi proteins and TNFAIP8.
Deletion analysis showed that the TNFAIP8 DED and extreme C-terminal domains were necessary for TNFAIP8–Gαi protein interactions. Recently, the crystal structure of a member of the TNFAIP8 family (TIPE2) has revealed that the central DED-like domain differs from a classical DED domain present in cFLIP or caspase 8 (Zhang et al., 2009). The TNFAIP DED domain extends to the C-terminal region, and is folded to bring the N- and C-termini of this DED-like domain in close proximity creating a hydrophobic cavity, which may be a site of protein interactions. Our findings suggest that the predicted proximity of TNFAIP8 DED and C-terminal regions could form the binding pocket for Gα proteins.
Previous studies have found that over-expression of TNFAIP8 enhances oncogenic activity in transformed cell lines and increases their tumor growth and metastasis in vivo (Kumar et al., 2000, 2004). By contrast, in non-transformed Balb-D2S cells TNFAIP8 over-expression did not enhance basal or APO-induced foci formation, indicating that TNFAIP8 alone is not oncogenic. Similarly, over-expression of TNFAIP8 did not affect inhibition of cAMP (Gαi-mediated). However, TNFAIP8 was required for D2S-mediated transformation, since upon depletion of TNFAIP8 basal and APO-induced transformation was completely blocked. The Gαi-TNFAIP8 pathway appears selective for D2S-induced transformation since antisense depletion of TNFAIP8 did not block other signaling pathways of the D2S receptor, including D2S-induced inhibition of cAMP. These results suggest that TNFAIP8 is selective for coupling to transformation, perhaps by forming a complex with Gαi proteins via direct interaction. While these findings indicate the role of TNFAIP8 in D2S signaling to transformation, the mechanisms involved remain unclear.
Gαi-TNFAIP8 signaling to cell death and transformation
To begin to elucidate possible mechanisms of TNFAIP8 in D2S receptor action, we addressed the role of TNFAIP8 in TNFα signaling. Previous studies have shown that TNFAIP8 inhibits TNFα-induced activation of caspase 8 and TNFα-induced apoptosis (You et al., 2001), while depletion of TNFAIP8 enhances cell death (Zhang et al., 2004, 2006). Our data indicate that TNFAIP8 over-expression failed to inhibit TNFα-induced caspase3/7 activation and cell death in the TE clone, while antisense to TNFAIP8 did not appear to enhance these TNFα actions. The ability of TNFAIP8 to inhibit these TNFα pathways in non-transformed Balb-D2S cells required activation of Gαi signaling by D2S receptors, suggesting that upon transformation Gαi or other TNFAIP8 pathways are constitutively activated to prevent cell death. Upon activation of D2S receptor signaling, TNFα-induced cell death was completely blocked. This action required TNFAIP8, since it was completely reversed upon depletion of TNFAIP8 in AS6 and AS10 cells. By contrast, TNFα-induced cell death was not inhibited in the AS6 clone, suggesting that alternate caspase-independent pathways may be recruited by D2S-TNFAIP8 coupling to induce cell death. Recently, glucocorticoid-induced apoptosis was shown to require TNFAIP8 (Woodward et al., 2010), and the possibility that Gi-TNFAIP8 interaction may prevent this signaling remains to be addressed. Structural differences between the TNFAIP8-related DED domain and classical DED domains may allow recruitment of additional mechanisms. For example, TNFAIP8 was found to interact with PTP4A2/PRL2 (Ewing et al., 2007), a family of tyrosine phosphatases implicated in liver regeneration, cell proliferation, and transformation (Stephens et al., 2005). Taken together, these data support a novel D2S-mediated cell survival signaling pathway that is dependent on TNFAIP8 expression which also recruits both caspase-dependent and caspase-independent mechanisms to prevent death receptor-induced cell death (Debatin and Krammer, 2004; Thorburn, 2004). For example, cell death by loss of cell adhesion (anoikis) triggers both extrinsic and intrinsic death pathways via integrin-dependent signaling that is consistent with the proposed interaction between TNFAIP8 and talin, a modulator of integrin signaling (Zhang et al., 2006).
Enhanced Gαi-TNFAIP8-mediated survival of pre-oncogenic cells may promote increased formation of foci of transformed cells (Debatin and Krammer, 2004). In these studies, several lines of evidence suggested the presence of constitutive activity of the D2S receptor in Balb-D2S cells, leading to increased transformation. The basal level of foci formation was increased in Balb-D2S compared to Balb/c-3T3 cells, and depletion of TNFAIP8 also reduced spontaneous transformation in D2S receptor expressing cells. It may be that over the 3-week time course of foci formation, components of the serum or cells were sufficient to activate the D2S receptor to contribute to the basal transformation rate. These findings could have important implications for regulation of oncogenesis in general. TNFAIP8 is over-expressed in human breast and renal cell carcinomas compared to normal tissue (Kumar et al., 2004) and leads to the up-regulation of genes involved in metastasis and tissue remodeling, leading to more aggressive and invasive tumors in nude mice (Kumar et al., 2004). Gαi-mediated signaling to activate this pathway may promote tumor progression in a variety of tissues. G-proteins have been implicated in tumor formation and appear to recruit multiple pathways to both enhance cell survival/proliferation and to reduce cell death (Dorsam and Gutkind, 2007). For example, the Gαi-coupled lysophosphatidic acid (LPA) receptor has been implicated in LPA-induced oncogenesis (Jonkers and Moolenaar, 2009). Increased LPA signaling to G-proteins by MMTV-driven transgenic over-expression of LPA receptor or autotaxin (which synthesizes LPA) induces breast cancer initiation, growth, invasion, and metastasis (Liu et al., 2009), consistent with the actions of TNFAIP8 over-expression (Kumar et al., 2004; Zhang et al., 2006). By blocking the Gαi-TNFAIP8 pathway, it may be possible to selectively enhance elimination of damaged pre-oncogenic cells to reduce tumor formation or progression. Blockers of Gαi-TNFAIP8 signaling would also be predicted to enhance response to death receptor activating compounds to treat established tumors (Ashkenazi, 2008).
Acknowledgments
Contract grant sponsor: National Cancer Institute of Canada.
Contract grant sponsor: Canadian Institutes of Health Research.
Contract grant sponsor: National Science and Engineering Research Council.
Contract grant sponsor: National Science and Engineering Research Council Canada Studentship.
Contract grant sponsor: Ontario Graduate Scholarship.
Contract grant sponsor: Heart and Stroke Foundation Centre for Stroke Recovery.
We thank Drs. Michel Bouvier (Univ. de Montréal) and Eric Accili (Univ. of British Columbia) for the Gαi1 and HCN constructs used for BRET assay. This study was supported by National Cancer Institute of Canada, Canadian Institutes of Health Research to P.R.A., National Science and Engineering Research Council, Canada Studentship to B.L. and Ontario Graduate Scholarship to H.N.
Literature Cited
- Albert PR. Heterologous expression of G protein-linked receptors in pituitary and fibroblast cell lines [Review] Vit Horm. 1994;48:59–109. doi: 10.1016/s0083-6729(08)60496-3. [DOI] [PubMed] [Google Scholar]
- Albert PR. G protein preferences for dopamine D2 inhibition of prolactin secretion and DNA synthesis in GH(4) pituitary cells. Mol Endocrinol. 2002;16:1903–1911. doi: 10.1210/me.2001-0329. [DOI] [PubMed] [Google Scholar]
- Albert PR, Robillard L. G protein specificity. Traffic direction required. Cell Signal. 2002;14:407–418. doi: 10.1016/s0898-6568(01)00259-5. [DOI] [PubMed] [Google Scholar]
- Albert PR, Ghahremani MH, Morris SJ. Mechanisms of dopaminergic regulation of prolactin secretion. In: Neve KA, Neve RL, editors. The dopamine receptors. Totowa, NJ: Humana Press Inc; 1997. pp. 359–381. [Google Scholar]
- Albert PR, Sajedi N, Lemonde S, Ghahremani MH. Constitutive G(i2)-dependent activation of adenylyl cyclase type II by the 5-HT1A receptor. Inhibition by anxiolytic partial agonists. J Biol Chem. 1999;274:35469–35474. doi: 10.1074/jbc.274.50.35469. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A. Targeting the extrinsic apoptosis pathway in cancer. Cytokine Growth Factor Rev. 2008;19:325–331. doi: 10.1016/j.cytogfr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Banihashemi B, Albert PR. Dopamine-D2S receptor inhibition of calcium influx, adenylyl cyclase, and mitogen-activated protein kinase in pituitary cells: Distinct Galpha and Gbetagamma requirements. Mol Endocrinol. 2002;16:2393–2404. doi: 10.1210/me.2001-0220. [DOI] [PubMed] [Google Scholar]
- Bockaert J, Claeysen S, Becamel C, Pinloche S, Dumuis A. G protein-coupled receptors: Dominant players in cell–cell communication. Int Rev Cytol. 2002;212:63–132. doi: 10.1016/s0074-7696(01)12004-8. [DOI] [PubMed] [Google Scholar]
- Dave RH, Saengsawang W, Yu JZ, Donati R, Rasenick MM. Heterotrimeric G-proteins interact directly with cytoskeletal components to modify microtubule-dependent cellular processes. Neurosignals. 2009;17:100–108. doi: 10.1159/000186693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatin KM, Krammer PH. Death receptors in chemotherapy and cancer. Oncogene. 2004;23:2950–2966. doi: 10.1038/sj.onc.1207558. [DOI] [PubMed] [Google Scholar]
- Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- Ekert PG, Silke J, Vaux DL. Caspase inhibitors. Cell Death Differ. 1999;6:1081–1086. doi: 10.1038/sj.cdd.4400594. [DOI] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O’Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, Vasilescu J, Abu-Farha M, Lambert JP, Duewel HS, Stewart II, Kuehl B, Hogue K, Colwill K, Gladwish K, Muskat B, Kinach R, Adams SL, Moran MF, Morin GB, Topaloglou T, Figeys D. Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin L-G, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Gales C, Van Durm JJ, Schaak S, Pontier S, Percherancier Y, Audet M, Paris H, Bouvier M. Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nat Struct Mol Biol. 2006;13:778–786. doi: 10.1038/nsmb1134. [DOI] [PubMed] [Google Scholar]
- Ghahremani MH, Forget C, Albert PR. Distinct roles for Galpha(i)2 and Gbetagamma in signaling to DNA synthesis and Galpha(i)3 in cellular transformation by dopamine D2S receptor activation in BALB/c 3T3 cells. Mol Cell Biol. 2000;20:1497–1506. doi: 10.1128/mcb.20.5.1497-1506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group INEW. Cellular and molecular mechanisms of cell transformation and standardization of transformation assays of established cell lines for the prediction of carcinogenic chemicals: Overview and recommended protocols. Cancer Res. 1985;45:2395–2399. [Google Scholar]
- Hamdan FF, Percherancier Y, Breton B, Bouvier M. Monitoring protein–protein interactions in living cells by bioluminescence resonance energy transfer (BRET) Curr Protoc Neurosci. 2006;Chapter 5(Unit 5):23. doi: 10.1002/0471142301.ns0523s34. [DOI] [PubMed] [Google Scholar]
- Horrevoets AJ, Fontijn RD, van Zonneveld AJ, de Vries CJ, ten Cate JW, Pannekoek H. Vascular endothelial genes that are responsive to tumor necrosis factor-alpha in vitro are expressed in atherosclerotic lesions, including inhibitor of apoptosis protein-1, stannin, and two novel genes. Blood. 1999;93:3418–3431. [PubMed] [Google Scholar]
- Jonkers J, Moolenaar WH. Mammary tumorigenesis through LPA receptor signaling. Cancer Cell. 2009;15:457–459. doi: 10.1016/j.ccr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Kumar D, Whiteside TL, Kasid U. Identification of a novel tumor necrosis factor-alpha-inducible gene, SCC-S2, containing the consensus sequence of a death effector domain of fas-associated death domain-like interleukin-1beta-converting enzyme-inhibitory protein. J Biol Chem. 2000;275:2973–2978. doi: 10.1074/jbc.275.4.2973. [DOI] [PubMed] [Google Scholar]
- Kumar D, Gokhale P, Broustas C, Chakravarty D, Ahmad I, Kasid U. Expression of SCC-S2, an antiapoptotic molecule, correlates with enhanced proliferation and tumorigenicity of MDA-MB 435 cells. Oncogene. 2004;23:612–616. doi: 10.1038/sj.onc.1207123. [DOI] [PubMed] [Google Scholar]
- Liu S, Umezu-Goto M, Murph M, Lu Y, Liu W, Zhang F, Yu S, Stephens LC, Cui X, Murrow G, Coombes K, Muller W, Hung MC, Perou CM, Lee AV, Fang X, Mills GB. Expression of autotaxin and lysophosphatidic acid receptors increases mammary tumorigenesis, invasion, and metastases. Cancer Cell. 2009;15:539–550. doi: 10.1016/j.ccr.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima H, Ohno K, Tanaka-Azuma Y, Nakano S, Yamada T. Identification of tumor promotion marker genes for predicting tumor promoting potential of chemicals in BALB/c 3T3 cells. Toxicol In Vitro. 2009;23:148–157. doi: 10.1016/j.tiv.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Mao H, Zhao Q, Daigle M, Ghahremani MH, Chidiac P, Albert PR. RGS17/RGSZ2, a novel regulator of Gi/o, Gz, and Gq signaling. J Biol Chem. 2004;279:26314–26322. doi: 10.1074/jbc.M401800200. [DOI] [PubMed] [Google Scholar]
- Miura D, Kobayashi M, Kakiuchi S, Kasahara Y, Kondo S. Enhancement of transformed foci and induction of prostaglandins in Balb/c 3T3 cells by palytoxin: In vitro model reproduces carcinogenic responses in animal models regarding the inhibitory effect of indomethacin and reversal of indomethacin’s effect by exogenous prostaglandins. Toxicol Sci. 2006;89:154–163. doi: 10.1093/toxsci/kfi342. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Cho G, Wen B, Insel PA. Identification and cDNA cloning of a novel human mosaic protein, LGN, based on interaction with G alpha i2. Gene. 1996;181:39–43. doi: 10.1016/s0378-1119(96)00456-8. [DOI] [PubMed] [Google Scholar]
- Nafisi H, Banihashemi B, Daigle M, Albert PR. GAP1(IP4BP)/RASA3 mediates Galphai-induced inhibition of mitogen-activated protein kinase. J Biol Chem. 2008;283:35908–35917. doi: 10.1074/jbc.M803622200. [DOI] [PubMed] [Google Scholar]
- Sethi G, Sung B, Aggarwal BB. TNF: A master switch for inflammation to cancer. Front Biosci. 2008;13:5094–5107. doi: 10.2741/3066. [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens BJ, Han H, Gokhale V, Von Hoff DD. PRL phosphatases as potential molecular targets in cancer. Mol Cancer Ther. 2005;4:1653–1661. doi: 10.1158/1535-7163.MCT-05-0248. [DOI] [PubMed] [Google Scholar]
- Takesono A, Cismowski MJ, Ribas C, Bernard M, Chung P, Hazard S, III, Duzic E, Lanier SM. Receptor-independent activators of heterotrimeric G-protein signaling pathways. J Biol Chem. 1999;274:33202–33205. doi: 10.1074/jbc.274.47.33202. [DOI] [PubMed] [Google Scholar]
- Thorburn A. Death receptor-induced cell killing. Cell Signal. 2004;16:139–144. doi: 10.1016/j.cellsig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Whitaker GM, Accili EA. Using bioluminescence resonance energy transfer to measure ion channel assembly. Methods Mol Biol. 2008;491:189–197. doi: 10.1007/978-1-59745-526-8_15. [DOI] [PubMed] [Google Scholar]
- Wise A, Jupe SC, Rees S. The identification of ligands at orphan G-protein coupled receptors. Annu Rev Pharmacol Toxicol. 2004;44:43–66. doi: 10.1146/annurev.pharmtox.44.101802.121419. [DOI] [PubMed] [Google Scholar]
- Woodward MJ, de Boer J, Heidorn S, Hubank M, Kioussis D, Williams O, Brady HJ. Tnfaip8 is an essential gene for the regulation of glucocorticoid-mediated apoptosis of thymocytes. Cell Death Differ. 2010;17:316–323. doi: 10.1038/cdd.2009.125. [DOI] [PubMed] [Google Scholar]
- You Z, Ouyang H, Lopatin D, Polver PJ, Wang CY. Nuclear factor-kappa B-inducible death effector domain-containing protein suppresses tumor necrosis factor-mediated apoptosis by inhibiting caspase-8 activity. J Biol Chem. 2001;276:26398–26404. doi: 10.1074/jbc.M102464200. [DOI] [PubMed] [Google Scholar]
- Zhang HG, Hyde K, Page GP, Brand JP, Zhou J, Yu S, Allison DB, Hsu HC, Mountz JD. Novel tumor necrosis factor alpha-regulated genes in rheumatoid arthritis. Arthritis Rheum. 2004;50:420–431. doi: 10.1002/art.20037. [DOI] [PubMed] [Google Scholar]
- Zhang C, Chakravarty D, Sakabe I, Mewani RR, Boudreau HE, Kumar D, Ahmad I, Kasid UN. Role of SCC-S2 in experimental metastasis and modulation of VEGFR-2, MMP-1, and MMP-9 expression. Mol Ther. 2006;13:947–955. doi: 10.1016/j.ymthe.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang J, Fan C, Li H, Sun H, Gong S, Chen YH, Shi Y. Crystal structure of TIPE2 provides insights into immune homeostasis. Nat Struct Mol Biol. 2009;16:89–90. doi: 10.1038/nsmb.1522. [DOI] [PubMed] [Google Scholar]