Abstract
Serotonin (5-hydroxytryptamine, 5-HT) neurotransmission is negatively regulated by 5-HT1A autoreceptors on raphe neurons, and is implicated in mood disorders. Pet-1/FEV is an ETS transcription factor expressed exclusively in serotonergic neurons and is essential for serotonergic differentiation, although its regulation of 5-HT receptors has not yet been studied. Here, we show by electrophoretic mobility shift assay that recombinant human Pet-1/FEV binds directly to multiple Pet-1 elements of the human 5-HT1A receptor promoter to enhance its transcriptional activity. In luciferase reporter assays, mutational analysis indicated that while several sites contribute, the Pet-1 site at −1406 bp had the greatest effect on 5-HT1A promoter activity. To address the effect of Pet-1 on 5-HT1A receptor regulation in vivo, we compared the expression of 5-HT1A receptor RNA and protein in Pet-1 null and wild-type littermate mice. In the raphe nuclei of Pet-1−/− mice tryptophan hydroxylase 2 (TPH2) RNA, and 5-HT and TPH immunostaining were greatly reduced, indicating a deficit in 5-HT production. Raphe 5-HT1A RNA and protein levels were also reduced in Pet-1-deficient mice, consistent with an absence of Pet-1-mediated transcriptional enhancement of 5-HT1A autoreceptors in serotonergic neurons. Interestingly, 5-HT1A receptor expression was up-regulated in the hippocampus, but down-regulated in the striatum and cortex. These data indicate that, in addition to transcriptional regulation by Pet-1 in raphe neurons, 5-HT1A receptor expression is regulated indirectly by alterations in 5-HT neurotransmission in a region-specific manner that together may contribute to the aggressive/anxiety phenotype observed in Pet-1 null mice.
Keywords: depression, development, ETS transcription factor, serotonin, transcription, transgenic
The serotonin (5-hydroxytryptamine, 5-HT) system, which originates from neurons of the midbrain raphe nuclei and projects widely throughout the brain (Dahlstrom and Fuxe 1964), has been implicated in behavioural disorders, including major depression, anxiety and aggression (Mann 1999; Albert and Lemonde 2004; Gordon and Hen 2004; Lesch 2005; Akimova et al. 2009). Gene knockout mouse models have demonstrated contrasting roles for 5-HT1A and 5-HT1B receptors in anxiety and aggressive behaviours, with 5-HT1B null mice having an aggressive phenotype and 5-HT1A null mice having an anxious phenotype (Zhuang et al. 1999; Lesch 2005). Tissue-specific, conditional rescue of the 5-HT1A receptor in the 5-HT1A−/− background and pharmacological studies have identified a critical timeframe of forebrain 5-HT1A receptor expression during embryonic and early postnatal development to establish a normal anxiety phenotype in the adult (Gross et al. 2002; Lo Iacono and Gross 2008). In addition, the 5-HT1A receptor is expressed pre-synaptically on serotonergic neurons where it functions as a negative regulator of neuronal activity and differentiation (Sotelo et al. 1990; Rumajogee et al. 2004). Thus, the developmental regulation of the 5-HT1A receptor is a critical determinant of the activity of the serotonin system, as well as the behavioural phenotype in the adult.
In response to elevation of 5-HT using serotonin-selective reuptake blockers, the pre-synaptic 5-HT1A autoreceptor appears to preferentially desensitize compared with post-synaptic 5-HT1A receptors (Le Poul et al. 2000), and this correlates with disinhibition of raphe neurons and clinical improvement of depression in patients (Pineyro and Blier 1999). Similarly, in 5-HT transporter (5-HTT) null mice, a preferential down-regulation of 5-HT1A RNA and protein was observed in the dorsal raphe nucleus compared with post-synaptic sites (Fabre et al. 2000; Li et al. 2000). These data suggest that adaptive changes in 5-HT1A receptor expression are implicated in depression and response to antidepressants.
Several transcription factors have been implicated in the development of the serotonin system, including Pet-1/FEV [pheochromocytoma 12 ETS (E26 transformation-specific) factor/Fifth Ewing Variant], an ETS-domain factor that is exclusively localized in 5-HT neurons (Maurer et al. 2004; Iyo et al. 2005). Pet-1 is expressed approximately 1 day before 5-HT (E11 in the mouse) (Hendricks et al. 1999). Pet-1 null mice (Hendricks et al. 2003) have a severe deficit in the serotonin system, with a 70–80% reduction in 5-HT levels, and display an anxious-aggressive phenotype that is reminiscent of the combined phenotypes of 5-HT1A and 5-HT1B knockout mice (Zhuang et al. 1999; Lesch 2005). These mice further display reduced expression of a number of Pet-1 target serotonergic genes that contain consensus Pet-1 elements (Hendricks et al. 1999), including the serotonin transporter (5-HTT) and tryptophan hydroxylase (TPH); however, the expression of raphe-specific TPH2 or specific 5-HT receptors in these mice has not been examined.
The presence of a Pet-1 consensus DNA element identified in the 5-HT1A receptor promoter suggests its potential role in regulation of pre-synaptic 5-HT1A receptor expression (Hendricks et al. 1999). Pet-1 was shown to enhance transcription of a reporter containing four copies of the 5-HT1A Pet-1 element, but the role of Pet-1 in the regulation of 5-HT1A gene transcription has not been addressed. In this study, we examined the role of Pet-1 (i) as a direct regulator of the 5-HT1A receptor gene and (ii) in the postnatal expression of 5-HT1A receptors in vivo. Our results demonstrate that Pet-1 acts by direct and indirect mechanisms to regulate the expression of pre- and post-synaptic 5-HT1A receptors in vivo, which could contribute to the mature affective phenotype of Pet-1-null mice.
Materials and methods
Cell culture
Human SK-N-AS neuroblastoma cells (from Dr Michael Bannon, Wayne State University, Detroit, MI, USA), human SK-N-SH neuroblastoma cells (from Dr Lakshmi Devi, New York University; Fricker et al. 2005) and human embryonic kidney cells (HEK293 cells) were cultured on polystyrene plates (Fisher, Ottawa, ON, Canada) in Dulbecco’s Modified Eagle’s Medium (Wisent, St. Bruno, QC, Canada), supplemented with 8% v/v heat inactivated foetal bovine serum at 37°C in 5% CO2. Human retinoblastoma Y79 cells were cultured in suspension in RPMI-1640 medium supplemented with 1.5 g/L NaHCO3, 10 mM HEPES, 1 mM sodium pyruvate, 20% foetal bovine serum and 4.5 g/L glucose (Wisent).
Nuclear fractionation
Nuclear extracts from HEK293 cells were obtained as described previously (Czesak et al. 2006). Successful nuclear fractionation was validated by Western blotting using monoclonal mouse antibodies raised against Histone H1 (1 : 2500; Upstate, Lake Placid, NY, USA) as a nuclear marker and against c-raf (1 : 1000; BD Biosciences, Mississauga, ON, Canada) as a cytosolic marker.
Plasmid constructs
Human Pet-1 (FEV) was PCR-amplified from human SK-N-AS cells and cloned into the EcoRV sites of pcDNA3.1 and the Kpn1 and EcoRV sites of pTriEx-4 and verified by DNA sequence analysis. The human 5-HT1A promoter pGL3-Basic luciferase reporter constructs were described previously (Lemonde et al. 2004). The −1517 wt 5-HT1A construct was mutated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, Cedar Creek, TX, USA) at the core GGAA nucleotides of the Pet-1 consensus motif using complementary primers: 111–101: 5′-ggtctctgcattccctAGctccgaaacttcccaggagaagg; 136–126: 5′-cgccgaagcagta agaactAGctgcttgggt ctctgc; 449–439: 5′-ggagagagggaaggaagCTaat agggagaggagggtc; 532–522: 5′-ggaagagg gagactgaaTCggaaggcagg tggggag; 931–921: 5′ccattcaggctccctatgctAGcttttctacatctcctattgc; 1406–1396: 5′-catatgcaaaatattCGcatccctgaattgactagccacaaagc.
Transfection
HEK293 cells were split into 12-well plates 16 h prior to transfection with luciferase constructs and pCMV-β-gal as control for transfection efficiency. Transfection was done using Lipofectamine Plus (Invitrogen, Burlington, ON, Canada) and 0.5 μg DNA/well at a ratio of 1 : 1 : 1 DNA : lipofectamine : plus for 3 h in serum-free OptiMEM (Invitrogen). After 48 h, plates were rinsed with phosphate-buffered saline (PBS) and lysed with 200 μL 1× reporter lysis buffer at 4°C (Promega, Madison, WI, USA). Luciferase and β-galactosidase activity were assessed using L-MaxII and SpectroMax M2 (Molecular Devices, Woodbridge, ON, Canada), respectively.
Protein expression
Pet-1 : pTriEx-4 was transformed into BL21 (DE3) Escherichia coli and grown at 37°C to OD600 of 0.6, and induced with addition of 1 mM isopropyl-β-D-thioglactopyranoside (Wisent) for 3.5 h. Cells were harvested by centrifugation and Pet-1 protein was isolated under denaturing conditions following the QIAexpressionist protocol (Qiagen, Mississauga, ON, Canada) and using Ni-nitrilotriacetic acid beads to select for His-tagged proteins. Fractions with the highest expression were combined and dialysed against DNA binding buffer (20 mM HEPES, 0.2 mM EDTA, 0.2 mM EGTA, 100 mM KCl, 5% glycerol, pH 7.9) containing 2 mM dithiothreitol. Protein was quantified using the BCA Protein Assay Kit (Pierce, Rockford, IL, USA).
Electrophoretic mobility shift assay (EMSA)
Oligonucleotides corresponding to known 5-HT1A Pet-1 binding elements (5-HT1A −137), were annealed and end-labelled with [α-32P]deoxy-CTP using 2.5 U Klenow (New England Biolabs, Pickering, ON, Canada) and purified using a Sephadex G-50 column (GE HealthCare, Baie D’Urfe, QC, Canada). Bacterially expressed, purified human Pet-1 was pre-incubated with or without annealed competitor probes: polyomavirus enhancer activator 3 (PEA3) wt (5′-gggatccaggaagtga), PEA3 mut (5′-gggatccatcaagtga), E2F (5′-tatagtgtactctactattctgctc), 5-HT1A −111/101 (5′-ggtgcattcccttcctccgaaac), 5-HT1A −136/126 (ggagtaagaacttcctgcttggg), 5-HT1A −450/440 (5′-ggggaaggaaggaaatagggaga), 5-HT1A −532/522 (5′-ggaaataaagggaagtgaggagg), 5-HT1A −931/921 (5′-ggtccctatgcttccttttctca), 5-HT1A −1406/1396 (5′-gggcaaaatatttccatccctga), 5-HTT −1154/-1144 (5′-gggaacgataggaagtagaagac), 5-HTT −1172/-1162 (5′-gggaccgaaaggaaatagcagtg), some of which have been described previously (Hendricks et al. 1999; Rogaeva and Albert 2007). Binding was done in 25-μL reaction containing DNA binding buffer, 250 ng of herring sperm DNA (Roche, Laval, QC, Canada) for 30 min at 22°C, and 32P-labelled 5-HT1A −137 probe (50 000 cpm/reaction) was added and incubated 20 min, followed by electrophoresis on a 5% native polyacrylamide gel at 4°C, vacuum-drying and exposure to film. Band intensity was quantified in Adobe Photoshop (Adobe Systems, San Jose, CA, USA), normalizing to the specific Pet-1 + 32P-136/126 band (in %) to combine multiple experiments and to calculate SEM.
Animals
The University of Ottawa Animal Care Committee approved all animal care procedures, in accordance with requirements of the Canadian Council on Animal Care. Pet-1 null (B6.129-Pet1Tm Eds/SJL) mice were generously provided by Dr Evan S. Deneris, Case Western Reserve University, Cleveland, OH, Canada (Hendricks et al. 2003) and were genotyped by PCR for Pet-1 using the following primers: (forward, wild-type) 5′-gcgacttggggggtcattatcac, (reverse) 5′-gcctgatgttcaaggaagacctcgg, (forward knockout) 5′-cggtggatgtggaatgtgtgcg with products of (wild-type) 209 bp or 361 bp (knockout). Equal proportions of male and female adult knockout mice or wild-type littermates (8–10 weeks) were killed using sodium pentobarbital (0.01 mL/g body wt, intraperitoneal; Somnitol; MTC Pharmaceuticals, Cambridge, ON, Canada), brains removed, dissected and extracted; dorsal raphe region was dissected as described (Czesak et al. 2007). For immunofluorescence, mice were perfused transcardially with 25 mL PBS followed by 50 mL 4% paraformaldehyde containing 0.2% picric acid in PBS, pH 6.9. Brains were removed and post-fixed for 90 min and then placed in 10% sucrose containing 0.01% sodium azide for storage at 4°C. Brains were rapidly frozen with dry ice and cryostat sections were cut at 10 μm and thaw mounted onto chrome alum gelatin-coated glass slides. Coronal brain slices were obtained according to the Mouse Brain Atlas (Paxinos and Franklin 2001) using the following coordinates: rostral raphe, which includes dorsal and median areas (Bregma −4.2 to −4.96 mm), hippocampus (Bregma 1.46 to −2.30 mm), and prefrontal cortex (Bregma 2.96-2.80 mm). Sections were allowed to air dry and then rinsed in PBS prior to incubation with antibodies.
Immunofluorescence
Slides were incubated overnight using the following antibodies: 1 : 50 rabbit anti-5-HT1A (custom made primary antibody designed to the i2 loop of the 5-HT1A receptor sequence; Cedarlane, Hornby, ON, Canada); 1 : 100 sheep anti-TPH antibody (Chemicon International, Temecula, CA, USA); 1 : 100 rat anti-5-HT antibody (Chemicon) or rabbit anti-5-HT (1 : 1000; Protos Biotech Corp, New York, NY, USA); 1 : 50 mouse anti-NeuN antibody (Chem-icon); and 1 : 500 rabbit anti-glial acidic fibrillary protein antibody (DAKO Inc., Mississauga, ON, Canada), in PBS containing 0.3% Triton-X100 and incubated under gentle vibration for 3 h at 22°C (Jacobsen and Staines 2004). For the 5-HT1A primary antibody control, pre-immune rabbit serum 1 : 5000 (Cedarlane). Secondary antibody (goat anti-rabbit Alexa Fluor 488, 1 : 200, Alexa Fluor 488 donkey anti-rabbit IgG; Alexa Fluor 594 goat anti-rat IgG; anti-sheep Alexa Fluor 488, or Alexa Fluor 594 rabbit anti-mouse; Molecular Probes, Eugene, OR, USA) in PBS was incubated at 37°C for 45 min. Sections were examined using an Axiophot II microscope (Carl Zeiss Canada, Toronto, ON). Images were acquired using Northern Eclipse imaging software (EMPIX, Toronto, ON, Canada) and a Retiga Qimage CCD camera (QIMAGING, Burnaby, BC, Canada). Blind counts of positive-labelled cells were performed on images taken at 20× magnification using Adobe Photoshop CS3. A constant area and minimum-threshold of 15% brightness (compared with background) was defined for each image counted.
In situ hybridization
Plasmid containing a 417-bp sequence of the mouse 5-HT1A receptor cDNA from Dr Alexandre Bonnin (Bonnin et al. 2006) was linearized and transcribed to incorporate digoxygenin-UTP (Roche Diagnostics) to generate sense or anti-sense riboprobes using SP6 or T7 polymerases, respectively (Jacobsen et al. 2008). Tissue sections were incubated overnight with riboprobes (1 : 1000) in hybridization buffer (150 mM NaCl, 15 mM sodium citrate, pH 7.0 (SSC), 50% deionized formamide, 10% dextran sulphate, 1 mg/mL rRNA, 1× Denhardt’s solution). Slides were washed three times for 30 min each, followed by two washes, 30 min each with 1× MABT (100 mM maleic acid, 150 mM NaCl, pH 7.5, 0.1% Tween-20). Slides were then blocked for 1 h at 22°C in blocking reagent and followed by an overnight incubation with 1 : 1000 anti-digoxygenin antibody (Roche). The following day slides were washed five times for 20 min with MABT, equilibrated with pre-staining buffer (100 mM NaCl, 50 mM MgCl2, 100 mM Tris pH 9.5, 0.1% Tween- 20) three times for 10 min at 22°C, developed using staining solution (100 mM NaCl, 50 mM MgCl2, 100 mM Tris pH 9.5, 0.1% Tween-20, 10% polyvinyl alcohol) containing 3.5 μL/mL of 5-bromo-4-chloro-3-indolyl-phosphate (Roche) and 4.5 μL/mL of 4-nitro blue tetrazolium chloride (Roche) for 2–24 h at 22°C and terminated by rinsing in PBS. Sections were examined using a Zeiss Axiophot II light microscope at 10× magnification. Image data were acquired using Northern Eclipse imaging software and an Axiovert S100 Zeiss microscope (EMPIX).
RNA isolation
Total RNA was extracted by trituration in 800 μL TRIzol (Invitrogen). For RT-PCR, 800 μL TRIzol (Invitrogen) was added directly to cells pre-rinsed in PBS. Chloroform (160 μL) was added and centrifuged for 25 min at 4°C and RNA precipitated using isopropanol. RNA was treated with 1 μL TURBO DNase (Ambion, Austin, TX, USA) to remove genomic DNA in 50 μL, for 30 min at 37°C, terminated using 5 μL DNase Inactivation Reagent and converted to cDNA using the Cells-to-cDNA™ II kit (Ambion). Reverse Transcriptase was omitted in -RT control.
RT-qPCR
Real-time PCR of cDNA prepared as described above was done using a Rotor-Gene RG-3000 (Corbett research, Sydney, Australia) and with TaqMan Gene Expression Assay using fluorophore labelled probes designed to discriminate between genomic DNA and cDNA (Applied Biosystems, Foster City, CA, USA): GAPDH (Mm99999915_g1), Htr1a (Mm00434106_s1) and TPH2 (Mm00557717_m1). A standard was included for comparison with standard curves, with GAPDH as an internal control; 1.5 μL of cDNA was added to 0.5 μL 20× specific probe, 5 μL 2× TaqMan Universal PCR Master Mix and 3 μL H2O, to make up a final volume 10 μL. For each probe, no-template controls and -RT controls demonstrated no amplification. The cycling programme was: 95°C, 10 min; 40 cycles of 95°C, 15 s; 60°C, 45 s. Data were analysed using Rotor Gene v6.0 software. Data were analysed by two-tailed, paired Students t-test, where *p < 0.05, **p < 0.005, ***p < 0.0005.
Western blot
Tissue was homogenized on ice in 50 μL of IP Buffer (25 mM Tris pH 7.4, 150 mM NaCl, 1 mM CaCl2, 1% Triton X-100) containing phenylmethylsulfonyl fluoride (Sigma, Oakville, ON, Canada), Protease Inhibitors (Roche, Basel, Switzerland) and dithiothreitol (Fisher). Western blot analysis was performed (Banihashemi and Albert 2002) using (1 : 4000) anti-5-HT1A (Millipore, Mississauga, ON, Canada), and developed using BM Chemiluminescence Blotting Substrate (Roche). The membranes were re-probed with anti-β-actin (1 : 1000) to control for loading. Band intensity of four independent experiments was quantified using Adobe Photoshop (Adobe Systems).
Statistical analyses
One-way ANOVA was performed for luciferase experiments using the Newman-Keuls Multiple Comparison post hoc test. Comparison between +/+ and −/− samples was by two-tailed unpaired t-test.
Results
Pet-1 regulates 5-HT1A receptor transcription through multiple sites
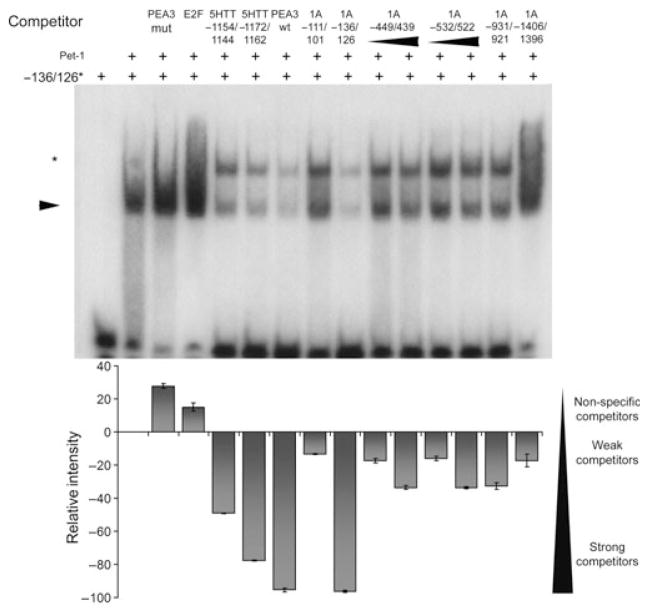
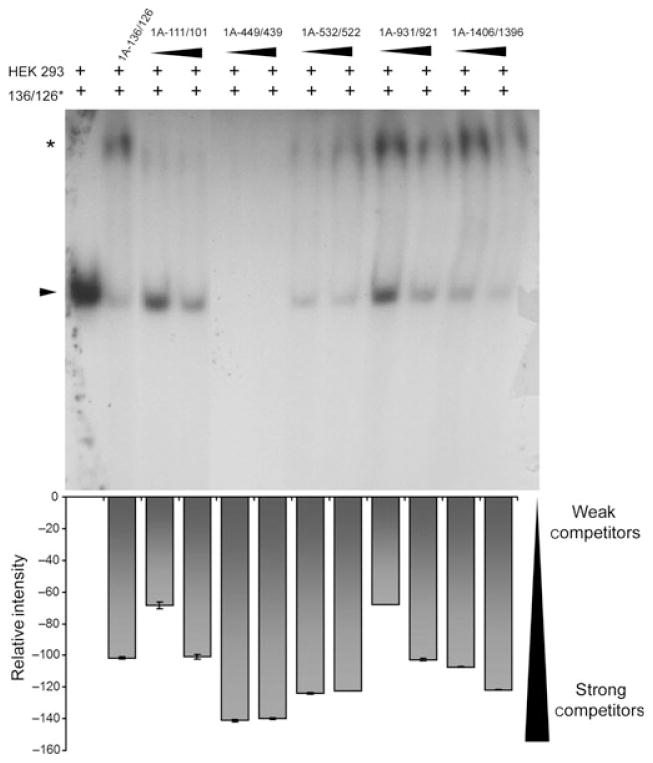
Based on the consensus Pet-1 binding element (Hendricks et al. 1999), we identified six putative Pet-1 elements within the first 1.5 Kb of the human 5-HT1A promoter (Table 1), several of which were highly conserved between human, rat and mouse 5-HT1A promoters (Table 2). To address Pet-1 binding to these elements, EMSA was done using recombinant human Pet-1 incubated with radio-labelled human 5-HT1A element 1A-136/126 and competing with excess unlabelled primers (Fig. 1). A major Pet-1/DNA complex (arrow) was identified that was competed by primers designed against the related PEA3 element, 5-HTT Pet-1 elements or unlabelled 1A −136/126 primers, but not by non-specific competitors (PEA3 mut or E2F). The other putative 5-HT1A Pet-1 elements all competed, but more weakly than the 1A −136/126 site. In the presence of excess competitor primers, a low mobility species (Fig. 1, asterisk) was observed that may represent a low affinity dimeric Pet-1 complex, as observed for other ETS proteins (Lee et al. 2005; Green et al. 2010). We next isolated nuclear extract from HEK293 cells, which express endogenous Pet-1 (Fig. S1), for use in EMSA experiments (Fig. 2). In the presence of labelled 1A-136/126 and HEK293 nuclear extract, a single band was observed (arrowhead). This experiment revealed a strong competition by all five putative Pet-1 elements, with 1A-449/439 and 1A-533/522 competing better than the 1A-136/126 element. Again, a low mobility complex was observed with excess competitors (Fig. 2, asterisk, and data not shown). Together, these data indicate that like recombinant Pet-1, a Pet-1-like nuclear component binds to multiple Pet-1 response elements in the 5-HT1A promoter, suggesting a role for Pet-1 in the regulation of 5-HT1A receptor transcription.
Table 1.
Multiple Pet-1 elements in serotonergic genes
| Gene | Site | Sequence |
|---|---|---|
| h5-HTT | −1154/1144 | GATAGGAAGTA |
| h5-HTT | −1172/1162 | GAAAGGAAATA |
| m5-HTT | −2424/2414 | CCCAGGAAATG |
| m5-HTT | −2024/2014 | GGGAGGAAATG |
| hTPH1 | −1154/1144 | ATACGGAAATT |
| mTPH1 | −790/780 | TACAGGATATA |
| h-AADC | Intron | TTCAGGAAATT |
| h5-HT1A | −136/126 R | AGCAGGAAGTT |
| h5-HT1A | −111/101 R | CGGAGGAAGGG |
| h5-HT1A | −449/439 F | GGAAGGAAATA |
| h5-HT1A | −532/522 F | GAAGGGAAAGT |
| h5-HT1A | −931/921 R | AAAAGGAAGCA |
| h5-HT1A | −1406/1396 R | GGATGGAAATA |
| PEA3 | TCCAGGAAGTG | |
| Consensus | RRMAGGAARTR |
Alignment of consensus Pet-1 elements previously identified (Hendricks et al. 1999), as well as putative additional elements in the 5-HT1A promoter identified using the Transcription Element Search Software (TESS) (Schug 2008), including their respective positions with respect to the start ATG and presence on the coding (F) or non-coding (R) strand. 5-HTT, serotonin transporter; AADC, amino acid decarboxylase; TPH1, tryptophan hydroxylase-1; PEA3, polyomavirus enhancer activator 3. R=G or A, M=A or C. Highlighted is the position of the minimal GGAA Pet-1 binding core.
Table 2.
Multiple conserved Pet-1 elements in the 5-HT1A promoter
| Species | Site (human) | Sequence |
|---|---|---|
| Human | −111/101 R | CGGAGGAAGGG |
| Rat | CAGAGGGAGGG | |
| Mouse | CGAAGGGAGGG | |
| Human | −136/126 R | AGCAGGAAGTT |
| Rat | AGCGGGAAGTT | |
| Mouse | AGCGGGAAGTT | |
| Human | −449/439 F | GGAAGGAAATA |
| Rat | AGAAGCAACCA | |
| Mouse | CGAAGGAACTA | |
| Human | −532/522 F | GAAGGGAAAGT |
| Rat | GAAGGGAAGGT | |
| Mouse | GGAAGGAAGGT | |
| Human | −931/921 R | AAAAGGAAGCA |
| Rat | TAAACAGAGCC | |
| Mouse | TAAATAGAGCC | |
| Human | −1406/1396 R | GGATGGAAATA |
| Rat | GGAAAGAAATC | |
| Mouse | GGAAAGAAATT |
Comparison of putative Pet-1 elements in the promoter of the 5-HT1A receptor. Position of the human element with respect to the start ATG is indicated as well as presence on the coding (F) or non-coding (R) strand.
Fig. 1.
Human Pet-1 directly binds multiple Pet-1 elements in the human 5-HT1A promoter. Electrophoretic mobility shift assay (EMSA) experiment showing Pet-1 binding to the previously identified Pet-1 element in the 5-HT1A promoter (−136/126, arrow) and subsequent competition by known specific probes (5-HTT −1154/1144, 5-HTT −1172/1162, PEA3 wt, 1A −136/126) but not of known non-specific probes (PEA3 mut, E2F). Putative 5-HT1A Pet-1 elements (1A −111/101, −450/440, −532/522, −931/921 and −1406/1396) were also able to compete for Pet-1 binding, although to a smaller extent than the 5-HT1A −136/126 itself. In all cases, 200× molar excess competitor was used, and where a concentration range was tested, the lower concentration of competitor was 100× molar excess. Intensity of the specific Pet-1 band (arrow) is quantified below each lane and expressed as intensity relative to the baseline Pet-1 interaction with the radiolabelled 1A −136/126 element. (*) A low mobility complex was observed in the presence of excess specific competitors. Error bars represent SEM for band intensity quantified for four independent experiments.
Fig. 2.
Specific binding of human embryonic kidney cells (HEK293) nuclear protein and 5-HT1A promoter Pet-1 elements. Electrophoretic mobility shift assay (EMSA) experiment showing complex formed between nuclear protein isolated from HEK293 cells and previously identified Pet-1 element in the 5-HT1A promoter (−136/126, arrow) and competition by putative 5-HT1A promoter Pet-1 elements (1A −111/101, −450/440, −532/522, −931/921 and −1406/1396). In all cases, a concentration range of 100× to 200× molar excess competitor was used, with 200× excess unlabelled 1A-136/126. Intensity of the specific Pet-1 band (arrow) is quantified below each lane and expressed as intensity relative to the baseline Pet-1 interaction with the radiolabelled 1A −136/126 element. (*) A weak low mobility complex was observed in the presence of excess specific, but not non-specific, competitors. Error bars represent SEM of band intensity quantified for four independent experiments.
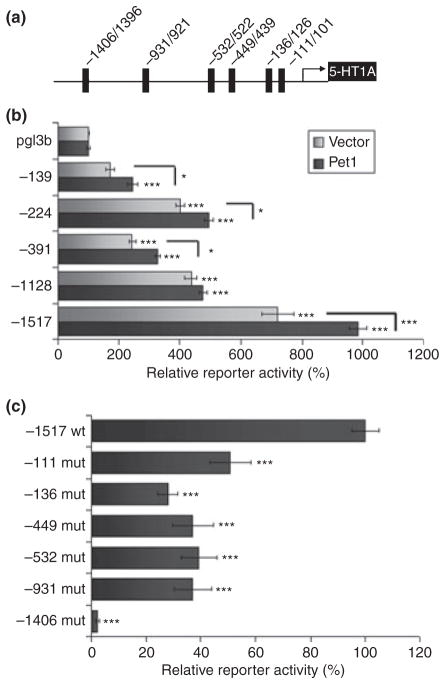
We screened for a Pet-1-deficient cell line for use in expression studies, but Pet-1 RNA was detected in several human cell lines, including HEK293 cells, retinoblastoma Y79, neuroblastoma SK-N-AS and SK-N-SH cells (Fig. S1, data not shown). The presence of Pet-1 mRNA suggests that endogenous Pet-1 protein is expressed in these cells and may contribute to basal transcription of 5-HT1A reporter constructs. To determine the role of Pet-1 sites of the 5-HT1A receptor gene, HEK293 cells were co-transfected with human Pet-1 expression plasmid or vector, and a series of human 5-HT1A promoter-luciferase reporter constructs (Fig. 3a) and luciferase activity was measured (Fig. 3b). All constructs except the −139 construct displayed significantly increased luciferase activity relative to the pGL3-Basic vector. The shorter constructs (−139, −224 and −391), containing Pet-1 binding elements 1A-111/101 and 1A-136/126, displayed modest but significant activation by Pet-1. In contrast to the −1128 construct which showed negligible activation, the greatest activation was obtained with the longest 5-HT1A promoter construct (−1517), which contains an additional Pet-1 element at −1406 bp. The low level of activation by transfected Pet-1 may reflect elevated basal activity of these constructs because of the presence of endogenous Pet-1 expression in these cells (Fig. S1). These data suggest a role for several Pet-1 response elements in Pet-1-mediated activation of the 5-HT1A promoter.
Fig. 3.
Human Pet-1 activates transcription through multiple Pet-1 elements in the human 5-HT1A promoter. (a) Map showing position of Pet-1 elements in the 5-HT1A promoter. (b) Transcriptional activity of Pet-1 on 5-HT1A promoter constructs containing increasing lengths of promoter sequence in human embryonic kidney cells (HEK293 cells). Pet-1 activation is strongest for the longest construct that contains all of the putative Pet-1 elements (−1517). Luciferase activity was normalized to β-galactosidase activity and expressed as percent of pGL3B vector (100%). (c) Co-transfection of Pet-1 with wild-type and mutated −1517 bp 5-HT1A promoter reporter constructs. Pet-1-mediated transcriptional activation of the 5-HT1A promoter construct is significantly reduced when any of the putative Pet-1 binding elements is mutated. Luciferase/β-galactosidase activity ratio is expressed as percent of activity of the non-mutated −1517 construct. Error bars represent SEM. ***p < 0.0005, **p < 0.005, *p < 0.05; N = 4.
To address the role of specific Pet-1 elements to enhance 5-HT1A transcription, the GGAA core sequence of each putative Pet-1 element (Table 2) was mutated in the −1517 5-HT1A promoter construct and co-transfected with the Pet-1 construct in HEK293 cells (Fig. 3c). Each mutant construct displayed substantially reduced reporter activity relative to the non-mutated 5-HT1A promoter construct, indicating that each Pet-1 element contributes to Pet-1 mediated 5-HT1A activation. Strikingly, mutation of the −1406 bp Pet-1 site almost completely abolished transcriptional activity of the −1517 5-HT1A promoter construct, suggesting its importance for both endogenous and transfected Pet-1 action in 5-HT1A transcription.
Pet-1 is critical for 5-HT1A receptor and TPH2 expression in vivo
We addressed whether Pet-1 regulates the expression of 5-HT1A receptors in vivo, using Pet-1 null mice as a model system. Pet-1 null mice display a 70–80% reduction in serotonin levels as well as a disruption in serotonergic gene expression (Hendricks et al. 2003). The level of 5-HT1A receptor RNA in the raphe region was assessed by in situ hybridization and was greatly reduced in the dorsal raphe nucleus of Pet-1 knockout compared with wild-type mice (Fig. 4a), consistent with the loss Pet-1 enhancer activity. The level of TPH and 5-HT1A receptor protein in these mice was examined by immunofluorescent staining (Fig. 4b). Compared with wild-type mice, Pet-1 null mice displayed strongly reduced levels of TPH and 5-HT1A immunoreactivity in the dorsal raphe region. Pet-1 knockout mice also displayed significant reductions in 5-HT staining (data not shown) and 5-HT1A-immunoreactive cells in the dorsal and median raphe nuclei, although this reduction was greater in the rostral than caudal portion (Fig. 4c). The smaller reduction in 5-HT1A-positive cells in the dorsal raphe nucleus (Fig. 4c) may represent the presence of 5-HT1A receptors in non-serotonergic cells that become up-regulated because of low 5-HT levels. In contrast, in the hippocampal dentate gyrus and prefrontal cortex, 5-HT1A receptor immunostaining and cell counts were similar in wild-type and Pet-1 knockout brains (Fig. S2). In contrast, there was no change in a glial marker (glial fibrillary acidic protein, Fig. S3), but a 25% reduction in NeuN-positive cells was observed in Pet-1−/− dorsal raphe nucleus (Fig. S4), which may reflect a partial loss of 5-HT neurons. Thus, the loss of Pet-1 leads to a reduction in 5-HT1A receptor expression in the raphe nuclei.
Fig. 4.
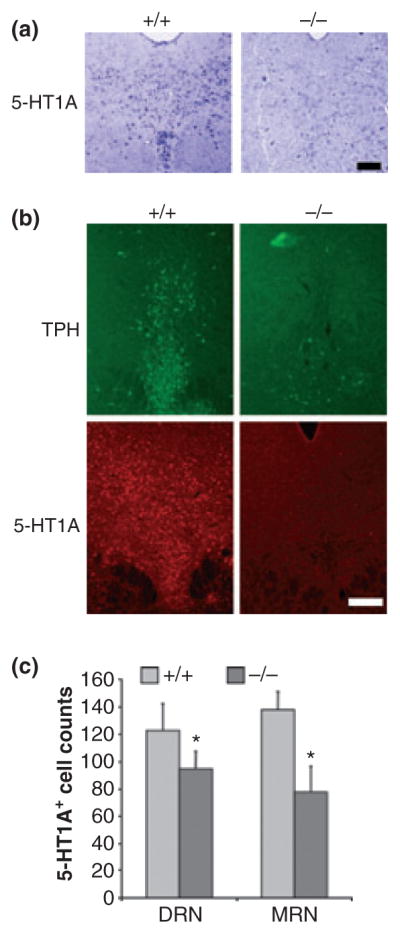
Reduced 5-HT1A mRNA and protein expression in Pet-1−/− raphe nuclei. (a) In situ hybridization showing 5-HT1A mRNA anti-sense probe staining in the dorsal raphe of wild-type (+/+) and Pet-1 knockout (−/−) mice; scale bar = 50 μm. (b) Immunofluorescent labelling for tryptophan hydroxylase (TPH) or 5-HT1A receptor proteins in sections of DRN from the same wild-type or knockout mice. Representative images are shown at 10× magnification with a scale bar = 100 μm; n = 3 mice/group. (c) Quantification of 5-HT1A-immu-nopositive cells. Cell counts are presented from the dorsal (DR) and median raphe (MRN) of wild-type (+/+) and Pet-1 knockout (−/−) mice; n = 3 mice/group. Statistical significance was measured using two-tailed unpaired t-test, where *p < 0.05.
To quantify changes in 5-HT1A receptor expression, brain tissue was analysed using RT-qPCR (Fig. 5a and b) or Western blot analysis (Fig. 5c). In Pet-1−/− cortex, 5-HT1A RNA levels were reduced compared with wild-type controls; while a reduction in 5-HT1A protein was detected in striatum. In contrast, in Pet-1−/− hippocampus, 5-HT1A protein but not mRNA was elevated (Fig. 5c). The discrepancy between the changes observed in cortex and hippocampus by Western blot compared with 5-HT1A immunofluorescence (Fig. 4) could be owing to analysis of whole tissues using RT-qPCR and Western blot, or the small magnitude of changes and the non-quantitative nature of staining. In the dorsal raphe region (Fig. 4c), TPH2 RNA levels were nearly abolished, consistent with the observed loss of TPH protein in Pet-1−/− raphe (Fig. 4b; Hendricks et al. 2003). Similarly, raphe 5-HT1A RNA levels were reduced by 40%, consistent with but less striking than the reduction observed by in situ hybridization (Fig. 4a). Up-regulation of 5-HT1A receptors in non-serotonergic neurons in the midbrain could account for the smaller effects observed when whole midbrain tissue was analysed by Western blot or RT-qPCR. Taken together, these data suggest that while Pet-1 may act as a transcriptional enhancer of 5-HT1A receptor activity in the raphe serotonergic neurons, additional compensatory mechanisms that could involve the serotonergic deficit in Pet-1−/− mice and lead to region-specific alterations in post-synaptic 5-HT1A receptor expression.
Fig. 5.
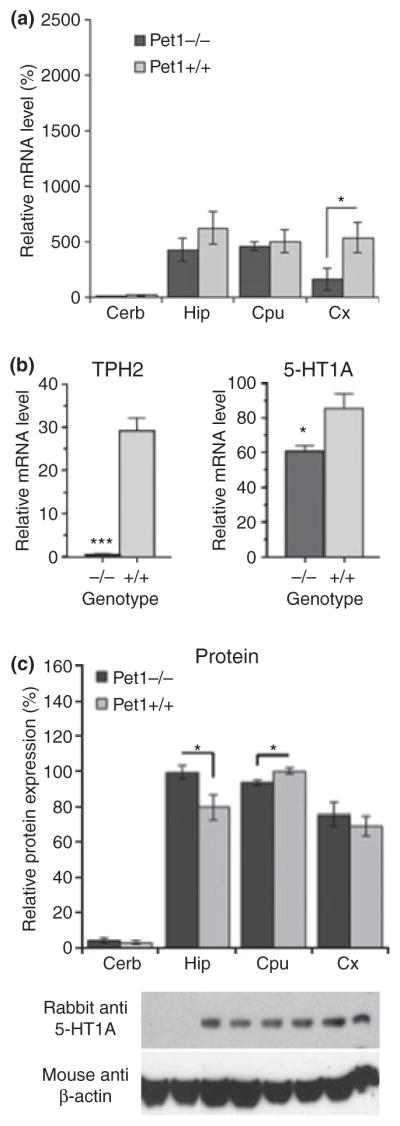
Pet-1 knockout mice display altered 5-HT1A receptor expression. Levels of 5-HT1A mRNA (a) or both 5-HT1A and tryptophan hydroxylase 2 (TPH2) RNA (b), and 5-HT1A protein (c) were measured in indicated brain regions by Q-RT-PCR and Western blot, respectively. Regions examined included: 5-HT1A-negative cerebellum (Cerb) as a negative control; hippocampus (Hip), striatum (CPu, including caudate, putamen and globus pallidus), and cortex (Cx); and the dorsal raphe region (DR). (a) 5-HT1A mRNA was reduced in the cortex of Pet-1 null mice. GAPDH was used as an internal control; N = 6. (b) TPH2 and 5-HT1A RNA levels were quantified in dorsal raphe tissue. GAPDH was used as an internal control; N = 4. (c) 5-HT1A protein is elevated in hippocampus (HIP), and reduced in the striatum. Band intensity of four independent experiments was quantified. β-Actin was used as a loading control. Error bars represent SEM. ***p < 0.001, *p < 0.05.
Discussion
Transcriptional regulation of the 5-HT1A receptor gene by Pet-1
Our data illustrate a role for Pet-1 as an enhancer of 5-HT1A transcription that binds to multiple Pet-1 elements in the 5-HT1A promoter to enhance its transcription (Figs 1–3). In EMSA experiments performed with recombinant human Pet-1, the 5-HT1A −136/126 element displayed the greatest binding efficacy (Fig. 1); however, in HEK293 nuclear extracts, all of the Pet-1 elements displayed similar binding efficacy (Fig. 2). While HEK293 cells express Pet-1, other ETS transcription factors or co-regulators may be present in this cell line to influence the binding efficiency. The enhancement provided by co-transfection of Pet-1 with 5-HT1A promoter constructs (Fig. 3) suggests a direct role for Pet-1 in transcriptional regulation of the 5-HT1A receptor. The reduced promoter activity upon mutation of any one of the Pet-1 elements suggests that they all contribute to transcriptional regulation of the 5-HT1A receptor. The nearly complete loss of transcriptional activity upon mutation of the −1406 Pet-1 site indicates its predominant role in basal and Pet-1 mediated transcription of the 5-HT1A promoter. Since Pet-1 is expressed in the brain exclusively in 5-HT neurons (Hendricks et al. 1999), these data suggest that Pet-1 is crucial for the basal expression of the 5-HT1A autoreceptor expression. These findings demonstrate for the first time the activity of human Pet-1 to enhance transcription of a human gene target and to validate the role of Pet-1 in transcriptional regulation of the human 5-HT1A receptor promoter. The human Pet-1 homologue (FEV) is exclusively expressed in serotonergic neurons of the raphe nuclei (Maurer et al. 2004; Iyo et al. 2005), hence Pet-1 is likely to regulate pre-synaptic 5-HT1A receptor expression in humans.
Regulation of TPH2 and 5-HT1A receptor expression by Pet-1 in vivo
To address the impact of Pet-1 on 5-HT1A receptor expression in vivo, we compared the 5-HT1A expression in Pet-1 knockout and wild-type brains. The Pet-1 null mice displayed a marked reduction in 5-HT immunoreactive cell bodies in all raphe nuclei examined (Fig., data not shown), reduced dorsal raphe 5-HT levels, and almost undetectable levels of TPH2 RNA (Fig. 5b). This result provides evidence suggesting that the TPH2 gene specifically is regulated by Pet-1, although the presence and identification of functional Pet-1 sites on the TPH2 gene remains to be elucidated. Previous studies have shown that the proximal TPH2 promoter is strongly repressed by RE-1 silencer of transcription/neural restrictive silencing factor, and activated by a cell-specific and calcium-dependent mechanism (Lenicov et al. 2007; Patel et al. 2007). Additionally, Lmx1b appears to coordinate with Pet-1 to regulate TPH2 expression (Zhao et al. 2006). The presence of detectable 5-HT levels in Pet-1−/− mice, despite the loss of TPH2 RNA, suggests that sufficient serotonin re-uptake activity is present to capture serotonin derived from pineal gland or serotonin entry via circumventricular organs.
The loss of Pet-1 reduced 5-HT1A RNA and protein levels in the dorsal and median raphe nuclei (Fig. 4), and this was confirmed by quantification of 5-HT1A mRNA from the dorsal raphe region (Fig. 5b), which contains the majority of the serotonergic raphe nuclei (Dahlstrom and Fuxe 1964). Since 5-HT-positive cells express Pet-1, the reduction in detectable 5-HT- and 5-HT1A-positive cells raphe nuclei indicates that 5-HT1A autoreceptor expression in vivo is strongly regulated by Pet-1. The remaining 5-HT1A receptor expression in the Pet-1-deficient raphe region may be because of compensatory up-regulation in response to low 5-HT levels. In contrast, in 5-HTT knockout mice, which display a persistent increase in synaptic 5-HT (Schmitt et al. 2007), 5-HT1A receptor binding sites and mRNA levels were significantly decreased in the dorsal raphe (Fabre et al. 2000; Li et al. 2000). Consistent with this, the 5-HTT Long Polymorphic Repeat short allele, which reduces 5-HTT expression, has been associated with a reduction in 5-HT1A receptor levels in humans (David et al. 2005). Despite the complete loss of Pet-1, a fraction of 5-HT neurons are still able to differentiate (Hendricks et al. 2003). Thus, while Pet-1 appears required for 5-HT1A autoreceptor expression, compensatory mechanisms that permit serotonergic differentiation observed in a fraction of cells may account for low levels of 5-HT1A receptors observed in the Pet-1−/− mice.
Consistent with our findings, a substantial reduction in 5-HT1A receptor mRNA levels was recently reported in the dorsal raphe nuclei of mice in which Pet-1 gene was disrupted at the onset of 5-HT differentiation (E12.5) (Liu et al. 2010). Importantly, whole cell recordings of Pet-1 promoter-driven yellow fluorescent protein-labelled raphe neurons demonstrate that 5-HT1A autoreceptor function is almost completely impaired in these conditional Pet-1-null mice. These results clearly indicate a major desensitization of functional 5-HT1A autoreceptors, consistent with the strong reduction in 5-HT1A RNA that we observe. Hence, the residual 5-HT1A protein in Pet-1−/− midbrain is likely because of up-regulation of 5-HT1A expression in non-serotonergic cells.
In target regions for 5-HT neurons, which receive a substantially reduced input of functional 5-HT projections in Pet-1 null mice (Hendricks et al. 2003), several alterations in 5-HT1A RNA and protein were observed. These changes were not obvious in immunofluorescence, where 5-HT1A-positive cell counts in dentate gyrus were not altered. Western blot measured an increase in whole hippocampal 5-HT1A protein, suggesting that 5-HT1A protein/cell could be increased, which would not be detectable by immunofluorescence. Lesion of 5-HT projections to the hippocampus using the 5-HT neurotoxin, 5,7-dihydroxytryptamine, results in increased levels of 5-HT1A protein in the hippocampus (Patel et al. 1996), and thus the lack of 5-HT in the Pet-1 knockout mice may also result in up-regulation of these post-synaptic 5-HT1A receptors. In agreement with this, acute treatment with 5-HT1A antagonist WAY100,635 was also shown to up-regulate 5-HT1A receptor number in cortex and hippocampus (Abbas et al. 2007). The reduction in 5-HT1A RNA levels in the cortex agrees with pharmacological studies demonstrating that reduced 5-HT signalling delays expression of cortical 5-HT type-1 receptors (Whitaker-Azmitia et al. 1996; Lauder et al. 2000). The increase of 5-HT1A receptor expression in the hippocampus versus a decrease in striatum of Pet-1 knockout mice, suggests that reduced serotonergic signalling may alter receptor expression in a site-specific manner.
Adaptive regulation of 5-HT1A receptor expression and behaviour in Pet-1 null mice
Pet-1 null mice display a heightened anxiety/aggressivity phenotype (Hendricks et al. 2003). Interestingly, 5-HT1A receptor knockout studies have demonstrated that early developmental expression of the forebrain 5-HT1A receptor is critical for establishing normal anxiety-like behaviour (Gross et al. 2002; Lo Iacono and Gross 2008). The alterations in hippocampal and cortical 5-HT1A receptors observed in Pet-1−/− mice may contribute to the anxiety phenotype of these mice. On the other hand, 5-HT1B receptor knockout mice display an aggressive phenotype with reduced anxiety (Saudou et al. 1994; Zhuang et al. 1999). In addition to 5-HT1A receptors, several other 5-HT receptors (5-HT1B, 5-HT2A, 5-HT2C, 5-HT4 and others) have been implicated in affective disorders and in the therapeutic actions of antidepressants (Schechter et al. 2005; Weisstaub et al. 2006; Lucas et al. 2007), and may be altered in Pet-1−/− mice. In stress-sensitive monkeys, FEV (Pet-1) is down-regulated and can be restored by antidepressant treatment, suggesting a role for Pet-1 expression in depression-related phenotypes in primates (Lima et al. 2009). It is as of yet unclear whether there is compensation by any other neurotransmitter system in Pet-1 null mice. Our preliminary data indicate an increase in tyrosine hydroxylase-positive projections (largely noradrenergic) in certain brainstem regions of Pet-1 knockout mice, such as the dorsal raphe (data not shown). The implications of this finding have yet to be elucidated, but suggest that additional adaptive changes may contribute to the behavioural phenotype of the Pet-1 null mice.
Taken together, the above data demonstrate that Pet-1 can activate 5-HT1A receptor expression through multiple Pet-1 response elements in the 5-HT1A promoter, and indicate that Pet-1 is critical for 5-HT1A autoreceptor expression in the raphe nuclei. The Pet-1 null mice illustrate the critical role of Pet-1 in the development of serotonin neurons, and their subsequent role as a differentiation signal for target cells. The Pet-1-dependent cascade that regulates 5-HT neuron differentiation and 5-HT1A autoreceptor expression, also indirectly affects 5-HT1A heteroreceptor expression in 5-HT-innervated target regions, such as cortex or hippocampus. The combination of altered pre- and post-synaptic receptor expression has inextricable implications for the role of this gene in the development of affective behaviours.
Supplementary Material
Acknowledgments
We thank Dr Evan Deneris, Case Western Reserve University, Cleveland OH, for providing the Pet-1 null mice, and for helpful advice and comments on the manuscript; and Heather Kooiman for helpful assistance. The Heart and Stroke Foundation Centre for Stroke Recovery supported some of the equipment used for these studies. KXJ was supported by a Natural Sciences and Engineering Research Council (NSERC) Canada Graduate Scholarship. MC was supported by a Canada Graduate Scholarship from Canadian Institutes of Health Research (CIHR) and Ontario Mental Health Foundation; MD was supported by Summer Studentships from NSERC. This work was funded by a grant from the CIHR to PRA.
Abbreviations used
- 5-HT
5-hydroxytryptamine
- 5-HTT
5-hydroxy-tryptamine transporter
- EMSA
electrophoretic mobility shift assay
- HEK293 cells
human embryonic kidney cells
- PBS
phosphate-buffered saline
- PEA3
polyomavirus enhancer activator 3
- Pet-1/FEV
pheochromocytoma 12 ETS (E26 transformation-specific) factor/Fifth Ewing Variant
- TPH2
tryptophan hydroxylase 2
Footnotes
The authors declare no conflicts of interest.
Additional supporting information may be found in the online version of this article:
Figure S1. Pet-1 mRNA is expressed in multiple cell lines.
Figure S2. 5-HT1A protein expression in prefrontal cortex and hippocampus.
Figure S3. GFAP expression in DRN.
Figure S4. NeuN expression in raphe nuclei.
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Abbas SY, Nogueira MI, Azmitia EC. Antagonist-induced increase in 5-HT1A-receptor expression in adult rat hippocampus and cortex. Synapse. 2007;61:531–539. doi: 10.1002/syn.20399. [DOI] [PubMed] [Google Scholar]
- Akimova E, Lanzenberger R, Kasper S. The serotonin-1A receptor in anxiety disorders. Biol Psychiatry. 2009;66:627–635. doi: 10.1016/j.biopsych.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Albert PR, Lemonde S. 5-HT1A receptors, gene repression, and depression: guilt by association. Neuroscientist. 2004;10:575–593. doi: 10.1177/1073858404267382. [DOI] [PubMed] [Google Scholar]
- Banihashemi B, Albert PR. Dopamine-D2S receptor inhibition of calcium influx, adenylyl cyclase, and mitogen-activated protein kinase in pituitary cells: distinct Galpha and Gbetagamma requirements. Mol Endocrinol. 2002;16:2393–2404. doi: 10.1210/me.2001-0220. [DOI] [PubMed] [Google Scholar]
- Bonnin A, Peng W, Hewlett W, Levitt P. Expression mapping of 5-HT1 serotonin receptor subtypes during fetal and early postnatal mouse forebrain development. Neuroscience. 2006;141:781–794. doi: 10.1016/j.neuroscience.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Czesak M, Lemonde S, Peterson EA, Rogaeva A, Albert PR. Cell-specific repressor or enhancer activities of Deaf-1 at a serotonin 1A receptor gene polymorphism. J Neurosci. 2006;26:1864–1871. doi: 10.1523/JNEUROSCI.2643-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czesak M, Burns AM, Lenicov FR, Albert PR. Characterization of rat rostral raphe primary cultures: multiplex quantification of serotonergic markers. J Neurosci Methods. 2007;164:59–67. doi: 10.1016/j.jneumeth.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Dahlstrom A, Fuxe K. Localization of monoamines in the lower brain stem. Experientia. 1964;20:398–399. doi: 10.1007/BF02147990. [DOI] [PubMed] [Google Scholar]
- David SP, Murthy NV, Rabiner EA, Munafo MR, Johnstone EC, Jacob R, Walton RT, Grasby PM. A functional genetic variation of the serotonin (5-HT) transporter affects 5-HT1A receptor binding in humans. J Neurosci. 2005;25:2586–2590. doi: 10.1523/JNEUROSCI.3769-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre V, Beaufour C, Evrard A, Rioux A, Hanoun N, Lesch KP, Murphy DL, Lanfumey L, Hamon M, Martres MP. Altered expression and functions of serotonin 5-HT1A and 5-HT1B receptors in knock-out mice lacking the 5-HT transporter. Eur J Neurosci. 2000;12:2299–2310. doi: 10.1046/j.1460-9568.2000.00126.x. [DOI] [PubMed] [Google Scholar]
- Fricker AD, Rios C, Devi LA, Gomes I. Serotonin receptor activation leads to neurite outgrowth and neuronal survival. Brain Res Mol Brain Res. 2005;138:228–235. doi: 10.1016/j.molbrainres.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Gordon JA, Hen R. The serotonergic system and anxiety. Neuromol Med. 2004;5:27–40. doi: 10.1385/NMM:5:1:027. [DOI] [PubMed] [Google Scholar]
- Green SM, Coyne HJ, 3rd, McIntosh LP, Graves BJ. DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association. J Biol Chem. 2010;285:18496–18504. doi: 10.1074/jbc.M109.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C, Zhuang X, Stark K, Ramboz S, Oosting R, Kirby L, Santarelli L, Beck S, Hen R. Serotonin1A receptor acts during development to establish normal anxiety-like behaviour in the adult. Nature. 2002;416:396–400. doi: 10.1038/416396a. [DOI] [PubMed] [Google Scholar]
- Hendricks T, Francis N, Fyodorov D, Deneris ES. The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J Neurosci. 1999;19:10348–10356. doi: 10.1523/JNEUROSCI.19-23-10348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–247. doi: 10.1016/s0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Iyo AH, Porter B, Deneris ES, Austin MC. Regional distribution and cellular localization of the ETS-domain transcription factor, FEV, mRNA in the human postmortem brain. Synapse. 2005;57:223–228. doi: 10.1002/syn.20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen KX, Staines WA. Vibration enhancement of slide-mounted immunofluorescence staining. J Neurosci Methods. 2004;137:71–77. doi: 10.1016/j.jneumeth.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Jacobsen KX, Vanderluit J, Slack RS, Albert PR. HES1 regulates 5-HT1A receptor gene transcription at a functional polymorphism: essential role in developmental expression. Mol Cell Neurosci. 2008;38:349–358. doi: 10.1016/j.mcn.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Lauder JM, Liu J, Grayson DR. In utero exposure to serotonergic drugs alters neonatal expression of 5-HT(1A) receptor transcripts: a quantitative RT-PCR study. Int J Dev Neurosci. 2000;18:171–176. doi: 10.1016/s0736-5748(99)00085-4. [DOI] [PubMed] [Google Scholar]
- Le Poul E, Boni C, Hanoun N, Laporte AM, Laaris N, Chauveau J, Hamon M, Lanfumey L. Differential adaptation of brain 5-HT1A and 5-HT1B receptors and 5-HT transporter in rats treated chronically with fluoxetine. Neuropharmacology. 2000;39:110–122. doi: 10.1016/s0028-3908(99)00088-x. [DOI] [PubMed] [Google Scholar]
- Lee GM, Donaldson LW, Pufall MA, Kang HS, Pot I, Graves BJ, McIntosh LP. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J Biol Chem. 2005;280:7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- Lemonde S, Rogaeva A, Albert PR. Cell type-dependent recruitment of trichostatin A-sensitive repression of the human 5-HT1A receptor gene. J Neurochem. 2004;88:857–868. doi: 10.1046/j.1471-4159.2003.02223.x. [DOI] [PubMed] [Google Scholar]
- Lenicov FR, Lemonde S, Czesak M, Mosher TM, Albert PR. Cell-type specific induction of tryptophan hydroxylase-2 transcription by calcium mobilization. J Neurochem. 2007;103:2047–2057. doi: 10.1111/j.1471-4159.2007.04903.x. [DOI] [PubMed] [Google Scholar]
- Lesch KP. Serotonergic gene inactivation in mice: models for anxiety and aggression? Novartis Found Symp. 2005;268:111–140. discussion 140–116, 167–170. [PubMed] [Google Scholar]
- Li Q, Wichems C, Heils A, Lesch KP, Murphy DL. Reduction in the density and expression, but not G-protein coupling, of serotonin receptors (5-HT1A) in 5-HT transporter knockout mice: gender and brain region differences. J Neurosci. 2000;20:7888–7895. doi: 10.1523/JNEUROSCI.20-21-07888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima FB, Centeno ML, Costa ME, Reddy AP, Cameron JL, Bethea CL. Stress sensitive female macaques have decreased fifth Ewing variant (Fev) and serotonin-related gene expression that is not reversed by citalopram. Neuroscience. 2009;164:676–691. doi: 10.1016/j.neuroscience.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Maejima T, Wyler SC, Casadesus G, Herlitze S, Deneris ES. Pet-1 is required across different stages of life to regulate serotonergic function. Nat Neurosci. 2010;13:1190–1198. doi: 10.1038/nn.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Iacono L, Gross C. Alpha-Ca2+/calmodulin-dependent protein kinase II contributes to the developmental programming of anxiety in serotonin receptor 1A knock-out mice. J Neurosci. 2008;28:6250–6257. doi: 10.1523/JNEUROSCI.5219-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas G, Rymar VV, Du J, et al. Serotonin(4) (5-HT(4)) receptor agonists are putative antidepressants with a rapid onset of action. Neuron. 2007;55:712–725. doi: 10.1016/j.neuron.2007.07.041. [DOI] [PubMed] [Google Scholar]
- Mann JJ. Role of the serotonergic system in the pathogenesis of major depression and suicidal behavior. Neuropsychopharmacology. 1999;21:99S–105S. doi: 10.1016/S0893-133X(99)00040-8. [DOI] [PubMed] [Google Scholar]
- Maurer P, Rorive S, de Kerchove d’Exaerde A, Schiffmann SN, Salmon I, de Launoit Y. The Ets transcription factor Fev is specifically expressed in the human central serotonergic neurons. Neurosci Lett. 2004;357:215–218. doi: 10.1016/j.neulet.2003.12.086. [DOI] [PubMed] [Google Scholar]
- Patel TD, Azmitia EC, Zhou FC. Increased 5-HT1A receptor immunoreactivity in the rat hippocampus following 5,7-dihydroxytryptamine lesions in the cingulum bundle and fimbria-fornix. Behav Brain Res. 1996;73:319–323. doi: 10.1016/0166-4328(96)00122-2. [DOI] [PubMed] [Google Scholar]
- Patel PD, Bochar DA, Turner DL, Meng F, Mueller HM, Pontrello CG. Regulation of tryptophan hydroxylase-2 gene expression by a bipartite RE-1 silencer of transcription/neuron restrictive silencing factor (REST/NRSF) binding motif. J Biol Chem. 2007;282:26717–26724. doi: 10.1074/jbc.M705120200. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. 2. Academic Press; San Diego, CA: 2001. [Google Scholar]
- Pineyro G, Blier P. Autoregulation of serotonin neurons: role in antidepressant drug action. Pharmacol Rev. 1999;51:533–591. [PubMed] [Google Scholar]
- Rogaeva A, Albert PR. The mental retardation gene CC2D1A/Freud-1 encodes a long isoform that binds conserved DNA elements to repress gene transcription. Eur J Neurosci. 2007;26:965–974. doi: 10.1111/j.1460-9568.2007.05727.x. [DOI] [PubMed] [Google Scholar]
- Rumajogee P, Verge D, Hanoun N, Brisorgueil MJ, Hen R, Lesch KP, Hamon M, Miquel MC. Adaptation of the serotoninergic neuronal phenotype in the absence of 5-HT autoreceptors or the 5-HT transporter: involvement of BDNF and cAMP. Eur J Neurosci. 2004;19:937–944. doi: 10.1111/j.0953-816x.2004.03194.x. [DOI] [PubMed] [Google Scholar]
- Saudou F, Amara DA, Dierich A, LeMeur M, Ramboz S, Segu L, Buhot MC, Hen R. Enhanced aggressive behavior in mice lacking 5-HT1B receptor. Science. 1994;265:1875–1878. doi: 10.1126/science.8091214. [DOI] [PubMed] [Google Scholar]
- Schechter LE, Ring RH, Beyer CE, Hughes ZA, Khawaja X, Malberg JE, Rosenzweig-Lipson S. Innovative approaches for the development of antidepressant drugs: current and future strategies. NeuroRx. 2005;2:590–611. doi: 10.1602/neurorx.2.4.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A, Benninghoff J, Moessner R, et al. Adult neurogenesis in serotonin transporter deficient mice. J Neural Transm. 2007;114:1107–1119. doi: 10.1007/s00702-007-0724-6. [DOI] [PubMed] [Google Scholar]
- Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. In: Baxevanis AD, editor. Current Protocols in Bioinformatics. Unit 2.6. Chapter 21. J. Wiley and Sons; Somerset, NJ, USA: 2008. pp. 1–15. [DOI] [PubMed] [Google Scholar]
- Sotelo C, Cholley B, El Mestikawy S, Gozlan H, Hamon M. Direct immunohistochemical evidence of the existence of 5-HT1A autoreceptors on serotoninergic neurons in the midbrain raphe nuclei. Eur J Neurosci. 1990;2:1144–1154. doi: 10.1111/j.1460-9568.1990.tb00026.x. [DOI] [PubMed] [Google Scholar]
- Weisstaub NV, Zhou M, Lira A, et al. Cortical 5-HT2A receptor signaling modulates anxiety-like behaviors in mice. Science. 2006;313:536–540. doi: 10.1126/science.1123432. [DOI] [PubMed] [Google Scholar]
- Whitaker-Azmitia PM, Druse M, Walker P, Lauder JM. Serotonin as a developmental signal. Behav Brain Res. 1996;73:19–29. doi: 10.1016/0166-4328(96)00071-x. [DOI] [PubMed] [Google Scholar]
- Zhao ZQ, Scott M, Chiechio S, Wang JS, Renner KJ, Gereau RW, 3rd, Johnson RL, Deneris ES, Chen ZF. Lmx1b is required for maintenance of central serotonergic neurons and mice lacking central serotonergic system exhibit normal locomotor activity. J Neurosci. 2006;26:12781–12788. doi: 10.1523/JNEUROSCI.4143-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Gross C, Santarelli L, Compan V, Trillat AC, Hen R. Altered emotional states in knockout mice lacking 5-HT1A or 5-HT1B receptors. Neuropsychopharmacology. 1999;21:S52–S60. doi: 10.1016/S0893-133X(99)00047-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.