Abstract
During oogenesis and early embryonic development in Drosophila, translation of proteins from maternally deposited mRNAs is tightly controlled. We and others have previously shown that translational regulatory proteins that function during oogenesis also have essential roles in the nervous system. Here we examine the role of Cup in neuromuscular system development. Maternal Cup controls translation of localized mRNAs encoding the Oskar and Nanos proteins and binds to the general translation initiation factor eIF4E. In this paper, we show that zygotic Cup protein is localized to presynaptic terminals at larval neuromuscular junctions (NMJs). cup mutant NMJs have strong phenotypes characterized by the presence of small clustered boutons called satellite boutons. They also exhibit an increase in the frequency of spontaneous glutamate release events (mEPSPs). Reduction of eIF4E expression synergizes with partial loss of Cup expression to produce satellite bouton phenotypes. The presence of satellite boutons is often associated with increases in retrograde bone morphogenetic protein (BMP) signaling, and we show that synaptic BMP signaling is elevated in cup mutants. cup genetically interacts with four genes (EndoA, WASp, Dap160, and Synj) encoding proteins involved in endocytosis that are also neuronal modulators of the BMP pathway. Endophilin protein, encoded by the EndoA gene, is downregulated in a cup mutant. Our results are consistent with a model in which Cup and eIF4E work together to ensure efficient localization and translation of endocytosis proteins in motor neurons and control the strength of the retrograde BMP signal.
Keywords: translational repression, mRNA localization, synapse, transmitter release
Introduction
Most key developmental regulatory proteins are regulated by multiple mechanisms. These include transcriptional regulation, mRNA localization, translational control, protein localization, and protein lifetime. In Drosophila, many proteins involved in oogenesis and early development are translated from maternal mRNAs. Because these mRNAs are deposited in the egg, their translation must be precisely regulated in order to allow production of their protein products at the appropriate times and places. Most genes encoding maternally expressed regulators are also zygotically transcribed later in development, and some of these genes function during nervous system development. The translational control mechanisms used to regulate expression and localization of these maternal gene products are also operative in the nervous system. However, translational regulatory circuits in neurons can have different organizations from those that operate during early development.
Pumilio (Pum) and Nanos (Nos) are key maternal translational regulators. They form a complex that binds to the 3′ UTR of hunchback (hb) mRNA and represses its translation in the posterior part of the embryo (Sonoda and Wharton, 1999). We and others have shown that zygotic Pum and Nos are also required for neural development and function (Baines, 2005; Menon et al., 2009; Menon et al., 2004; Muraro et al., 2008; Ye et al., 2004). Pum, Nos, and the general translational initiation factor eIF4E are components of a regulatory circuit in the neuromuscular system that controls postsynaptic translation of glutamate receptor (GluR) mRNAs. In postsynaptic muscles, Pum binds to the 3′ UTRs of the eIF4E(Menon et al., 2004), GluRIIA, and nos mRNAs (Menon et al., 2009) and represses their translation. Postsynaptic Nos represses expression of the alternate GluR subunit, GluRIIB, by an unknown mechanism that is not dependent on Pum (Menon et al., 2009). In neurons, the Pum/Nos complex binds to and represses translation of paralytic (para) mRNA, which encodes a voltage-gated sodium channel (Muraro et al., 2008). Pum and Nos are also required for normal development of neuromuscular junction (NMJ) presynaptic terminals (Menon et al., 2009; Menon et al., 2004), and they regulate branching of the dendrites of peripheral sensory neurons (Ye et al., 2004).
Since Pum and Nos function in the nervous system, we wished to investigate molecules that interact with these translational regulators during oogenesis or early embryonic development and define their roles at the larval NMJ. In this paper, we examine the zygotic functions of Cup, which is a maternal regulator of nos mRNA translation in oocytes. Cup also binds to eIF4E (Nakamura et al., 2004; Nelson et al., 2004; Wilhelm et al., 2003; Zappavigna et al., 2004), and eIF4E expression is controlled by Pum in the neuromuscular system (Menon et al., 2004). Thus, we were interested in determining whether Cup is also important for neuromuscular system development.
Cup is encoded by the female-sterile gene fs(2)cup, which is required for production of functional oocytes (Keyes and Spradling, 1997; Schupbach and Wieschaus, 1991). Cup’s major role during oogenesis is to regulate translation of mRNAs that are localized to specific domains within the oocyte. oskar (osk) mRNA is localized to the posterior pole of the oocyte and is required for the establishment of the germ line and for posterior patterning (Ephrussi et al., 1991). Cup is required for osk mRNA localization, and it represses translation of osk mRNA until it reaches its final location. Translation of osk mRNA is prematurely derepressed in cup mutants, resulting in the expression of Osk protein at the wrong pole of the oocyte (Wilhelm et al., 2003). Cup itself does not bind to mRNAs, but engages with osk mRNA by forming a complex with Bruno, a sequence-specific RNA-binding protein (Nakamura et al., 2004). Cup also represses translation of nos mRNA that is not localized at the posterior pole of the embryo. It engages with nos mRNA through its interactions with Smaug, another sequence-specific RNA-binding protein (Nelson et al., 2004). Cup was also identified as a binding partner of Nos in a yeast two-hybrid screen (Verrotti and Wharton, 2000).
Cup represses translation through a variety of mechanisms. One proposed mechanism is to obstruct the formation of the elongation initiation factor 4F (eIF4F) complex. The eIF4F complex includes an RNA helicase, eIF4A, a scaffolding protein, eIF4G, and the cap-binding protein, eIF4E. eIF4E is the target for translational repressors known as eIF4E-binding proteins (4E-BPs). By competing with eIF4G for binding to eIF4E, 4E-BPs inhibit the recruitment of the 43S preinitiation complex and block translation (Wilhelm and Smibert, 2005). Cup is a 4E-BP, and contains two eIF4E-binding motifs located within its N-terminal domain (Nakamura et al., 2004; Nelson et al., 2004; Wilhelm et al., 2003; Zappavigna et al., 2004). In addition to binding each other directly in vitro and in cell culture assays, eIF4E and cup genetically interact to regulate ovary development. Cup is also required for localizing eIF4E to the posterior pole in developing oocytes (Zappavigna et al., 2004).
Since Nos and Cup interact and function together in the germline, and Cup regulates nos mRNA translation, we anticipated that if zygotically expressed Cup has a function at the NMJ, zygotic cup mutants might have phenotypes that resembled either loss-of-function (LOF) or gain-of-function (GOF) nos phenotypes (Menon et al., 2009). In the present study, we show that Cup is indeed expressed by motor neurons and localized to NMJ presynaptic terminals. However, we found that the cup zygotic phenotype is quite distinct from those of nos LOF or GOF mutants. There was also no obvious change in Nos protein levels in cup mutants.
Given these results, we decided to perform a broader analysis of the functions of Cup and eIF4E at the NMJ. We find that cup mutant NMJs contain many small, clustered boutons. These are known as satellites (reviewed by (Menon et al., 2013; O’Connor-Giles and Ganetzky, 2008; Oh and Robinson, 2012). The frequency of spontaneous glutamate release events (minis) is altered in cup mutants. cup and eIF4E genetically interact, as in the ovary (Zappavigna et al., 2004), and the nature of these interactions suggest that Cup is a positive regulator of eIF4E function in motor neurons. We also show that, like other genes for which mutations produce satellite bouton phenotypes, Cup is a negative regulator of the retrograde Bone Morphogenetic Protein (BMP) signaling pathway, in which Glass Bottom Boat (Gbb), a BMP ligand secreted by the muscle, activates BMP receptors on the presynaptic terminal and thereby increases expression of genes required for synaptic growth (Marques and Zhang, 2006). Cup appears to indirectly affect BMP signaling via positive regulation of endocytic proteins that are neuronal modulators of the pathway.
Our data are consistent with a model in which Cup and eIF4E facilitate localization and translation of endocytic proteins at the proper time(s) and place(s). Cup is known as a translational repressor and competes with eIF4G for binding to eIF4E (Nelson et al., 2004). However, Cup’s binding to eIF4E is not necessarily a repressive mechanism. Based on studies in cultured cells, it was recently proposed that eIF4E binding to Cup tethers eIF4E to mRNAs destined for specific locations, so that translation can begin promptly when Cup dissociates from the mRNA complex (Igreja and Izaurralde, 2011). In this way, Cup can both repress translation of mRNAs via a non-eIF4E-dependent mechanism, and positively control translation by facilitating rapid assembly of the eIF4F complex when repression is relieved. We suggest that the second mechanism operates in motor neurons. It is consistent with our findings that Cup is a positive regulator of direct and indirect modulators of the BMP signaling pathway and works in concert with eIF4E to facilitate normal development and function of NMJ presynaptic terminals.
Results
Cup protein localizes to NMJ presynaptic terminals
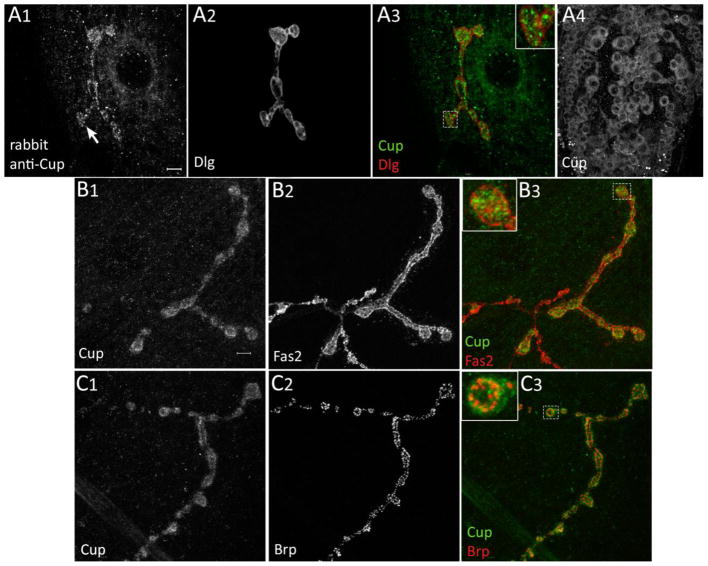
To evaluate Cup’s functions during NMJ development, we first determined whether Cup is expressed in the nervous system. Third-instar larvae were dissected and stained with antibodies against Cup and visualized by confocal microscopy. Three polyclonal antibodies made against different regions of the protein by different research groups were tested for immunoreactivity. Rabbit and mouse anti-Cup antibodies were made against amino acids 595-975 of Cup, while the rat anti-Cup antibody was generated against amino acids 201-499 (Keyes and Spradling, 1997; Nakamura et al., 2004). We found that all three antibodies stained the larval NMJ (Figs. 1 and S1). The postsynaptic marker Discs-Large (Dlg) was used to outline the boutons (Fig. 1A2). Rabbit (Fig. 1A1) and rat and mouse (Figure S1) Cup antibodies all showed punctate Cup staining within the presynaptic bouton and to a lesser extent on the postsynaptic muscle membrane. In addition to staining at the NMJ, Cup is also found in the cytoplasm of neuronal cell bodies in the ventral nerve cord (VNC)(Figs. 1A4 and S1). These cells were identified as neurons using antibodies against horseradish peroxidase (HRP), which selectively stain the surfaces of insect neurons and are commonly used as neuronal markers.
Fig. 1.
Cup protein is localized to NMJ presynaptic terminals. (A1-A4) A wild type third instar larva stained with rabbit anti-Cup and mouse anti-Dlg antibodies. (A1) Cup is found in punctate structures in NMJ presynaptic terminal and is also expressed at much lower levels in muscles. (A2) Anti-Dlg staining of the same NMJ as in A1. (A3) Merged images of A1 and A2 with the inset showing a zoomed in view of the highlighted bouton to display the Cup puncta (green). (A4) Cup also localizes to cell bodies in the ventral nerve cord. (B1–B3) Colocalization of Fas2 and Cup at the muscle 4 NMJ in a wild type larva. (B1) Anti-Cup staining, (B2) Anti-Fas2 staining, and (B3) overlap of Cup (green) and Fas2 (red) from B1 and B2, respectively. Inset is a zoomed-in view of the highlighted bouton to show Cup puncta within the presynaptic terminal. (C1-C3) Colocalization of Cup and Brp at the muscle 4 NMJ in a wild type larva. (C1) Anti-Cup staining, (C2) Anti-Brp staining, and (C3) overlap of Cup (green) and Brp (red) staining from C1 and C2, respectively. Inset is a magnified view of the highlighted bouton. Scale bar: 5μm.
Presynaptic localization of Cup at the NMJ was confirmed by double staining with antibodies for the synaptic markers Fasciclin 2 (Fas2), a cell adhesion molecule in the periactive zone, and Bruchpilot (Brp), an active zone protein (Figs. 1B and C, respectively). Anti-Fas2 stains in a network-like pattern within the presynaptic bouton, and also stains postsynaptically around the bouton (Fig. 1B2). Brp shows punctate staining marking active zones (Fig. 1C2). Cup puncta inside presynaptic boutons show partial overlap with both Fas2 and Brp (insets in 1B3 and 1C3). Cup is enriched in the type 1b boutons and localizes faintly at type 1s boutons (asterisk in Fig. S1). Anti-Cup does not stain the axon trunks.
To confirm that the presynaptic staining we observed is due to Cup protein, we next examined Cup immunoreactivity at NMJs in cup mutants and in larvae where Cup was knocked down using transgenic RNAi. The cup alleles used in this study are cupΔ212 (Nakamura et al., 2004), a P-element imprecise excision line that produces an N-terminally truncated Cup protein, two ethyl methane sulfonate (EMS)-induced alleles (cup20 and cup8), and two deficiencies that remove cup, Df(2L)BSC7 (26D10-27C1) and Df(2L)BSC187 (26F3-27A1). The two EMS-induced mutations are not protein nulls, and their sequences are not known (Keyes and Spradling, 1997). During oogenesis, cup20 and cup8 have stronger defects in egg chamber development than does cupΔ212 (Nakamura et al., 2004).
We observed a decrease in Cup protein levels at the NMJ relative to wild-type, as visualized with the rabbit antibody, in cup20/Df(2L)BSC7 transheterozygotes (Fig. S2). Wild-type and cup NMJs exhibited similar staining intensities with anti-HRP (Fig. S2). The residual Cup antibody immunoreactivity seen in the mutant is likely due to expression of mutant protein from the cup20 allele, since cup20 is not a protein null (Keyes and Spradling, 1997). Complete elimination of Cup protein at NMJs was not observed with any cup allelic combination. We also knocked down Cup presynaptically by expressing cup RNAi using a pan-neuronal driver, C155-Gal4. NMJs in cup RNAi larvae recapitulated the decreased staining seen in cup mutants relative to wild-type (Fig. S2).
Loss of Cup function causes alterations in NMJ morphology
To evaluate cup phenotypes, we stained larvae bearing transheterozygous combinations of the various cup alleles. Third-instar larvae from controls and cup mutants were dissected and stained with anti-HRP, and NMJs were analyzed by confocal microscopy. Our previous studies of Nos showed that it regulates NMJ growth. In nos mutants, there was an increase in bouton number (Menon et al., 2009), and neuronal Nos overexpression decreased the number of boutons. In cup transheterozygotes, however, there was no statistically significant change in the total number of full-size NMJ boutons (data not shown). Thus, although loss of maternal Cup produces patterning defects due to Nos mislocalization and Cup was identified as a Nos interactor using the yeast two-hybrid assay, loss of zygotic Cup does not recapitulate nos LOF or GOF phenotypes in the nervous system.
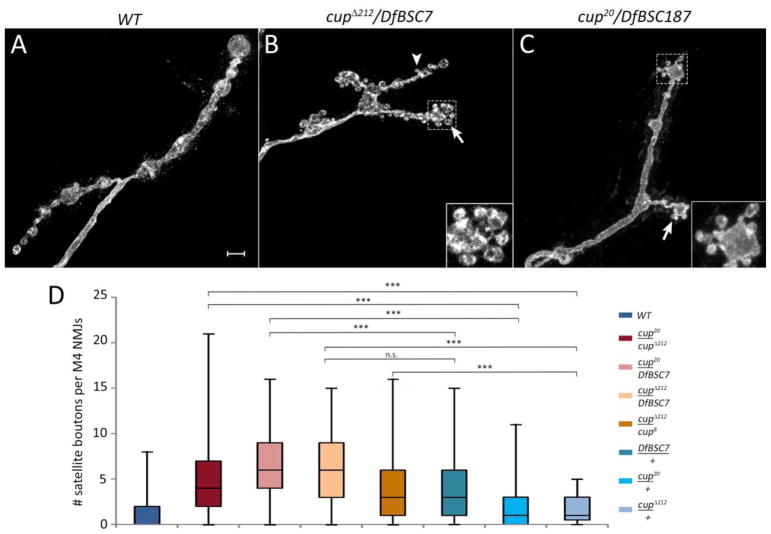
cup mutants, however, displayed a strong NMJ phenotype affecting bouton organization. Wild-type muscle 4 NMJs show a simple pattern of type 1b boutons connected to each other in a linear chain (Fig. 2A). In cup transheterozygotes, there are multiple small boutons budding from the parent type 1b boutons. These are rarely seen in control animals. Clustered small boutons were most commonly observed at the terminal parent bouton (arrows and insets, Fig. 2B, C). Small boutons were also seen emerging from regions connecting two boutons, or from a branching bouton, although this was less frequent (Fig. 2B, arrowhead). These small, clustered boutons have been observed in many other studies, and are known as satellite boutons.
Fig. 2.
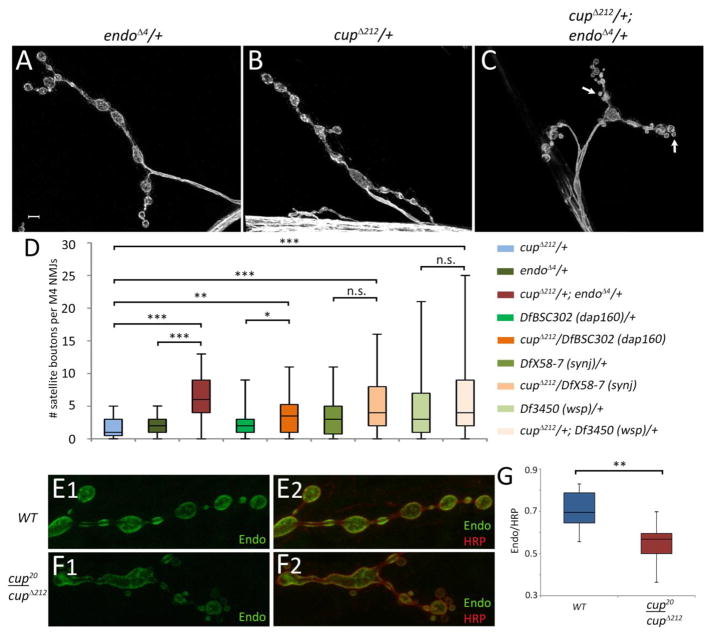
Loss of Cup function produces NMJ satellite bouton phenotypes. Muscle 4 NMJs in wild type larvae (A), cupΔ212/DfBSC7 (B), and cup20/DfBSC187 (C). Arrows in (B) and (C) highlight satellite boutons sprouting from terminal parent boutons. Arrowhead in (B) shows a satellite bouton on the axon trunk. Insets in (B) and (C) are blowups of the outline areas. (D) Box plots showing number of satellite boutons per muscle 4 NMJs in different genotypes. Wild type (WT; n=157), cup20/cupΔ212 (n=228); cup20/DfBSC7 (n=124); cupΔ212/DfBSC7 (n=46); cupΔ212/cup8 (n=55); DfBSC7/+ (n=80); cup20/+ (n=45); cup 212/+ (n=43). 25th to 75th percentiles (boxes), medians (line in boxes) and ranges (whiskers, 1.5 times the interquartile range extended from both ends of the box) are shown for each genotype. Significance of satellite bouton numbers as determined by the Kolmogorov-Smirnov (K-S) non-parametric test is shown with asterisks. *** p<0.001, ** p<0.01, * p<0.05, n.s. p>0.05. Scale bar: 5μm.
The number of satellite boutons per muscle 4 NMJ from segments A2-A5 was quantitated in different mutant combinations and controls (Fig. 2D). Due to the non-normal distribution of the satellite bouton numbers, the non-parametric Kolmogorov-Smirnov (K-S) test was used to calculate the D statistic and evaluate the statistical significance (see Experimental Methods). Box plots were used to represent the data and significance is indicated above each genotype. Satellite bouton numbers in cup mutants were significantly higher than in wild type (WT) or heterozygous controls. The number of satellite boutons in cup20/cupΔ212 was 4-fold higher than in the corresponding heterozygous controls (K-S test, cup20/+ D=0.34, p<0.001; cupΔ212/+ D=0.40, p<0.001; Fig. 2D). The differences in satellite bouton numbers between mutants and controls were highly statistically significant for other transheterozygous combinations as well, including cup20/DfBSC7 and cupΔ212/cup8 (compared to heterozygous controls (cup20/+ D=0.59, p<0.00; DfBSC7/+ D=0.31, p<0.001; cupΔ212/+ D=0.42, p<0.001). However, satellite bouton numbers in cupΔ212/DfBSC7 did not differ significantly from those in DfBSC7/+ (D=0.22, p=0.09). All heterozygous controls, especially the DfBSC7/+, showed an increase in satellite bouton numbers compared to WT (cup20/+ D=0.22, p>0.05; cupΔ212/+ D=0.25, p<0.05; DfBSC7/+ D=0.46, p<0.001) suggesting that satellite bouton formation is very sensitive to Cup levels.
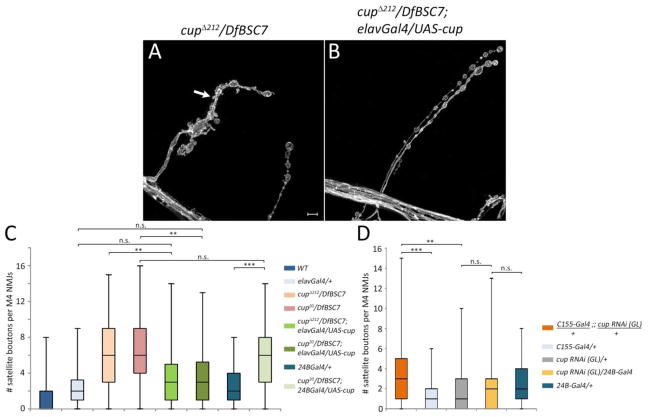
Cup staining at NMJs suggests that it functions in the motor neurons, since it is localized predominantly presynaptically, although there are a few puncta within the postsynaptic muscles (Fig. 1). To evaluate whether Cup function is required pre- or postsynaptically to facilitate normal synaptic growth, we expressed a cup cDNA in cup mutant backgrounds using 24B-Gal4 (postsynaptic) and elav-Gal4 (panneuronally). Expression of Cup postsynaptically in a cup20/DfBSC7 background did not rescue the satellite bouton phenotype (Figure 3C; cup20/DfBSC7 D=0.08, p>0.05). However, when Cup was expressed panneuronally in both cupΔ212/DfBSC7 and cup20/DfBSC7 backgrounds, this produced significant rescue of satellite bouton phenotypes (cupΔ212/DfBSC7 D=0.33, p<.01; cup20/DfBSC7 D=.32, p<0.001, as compared to their corresponding rescue genotypes). When the presynaptic rescues were compared to elav-Gal4/+ controls, there was no significant difference, suggesting a complete rescue of the mutant phenotype (cupΔ212/DfBSC7; elavGal4/UAS-cup D=0.19, p>0.05; cup20/DfBSC7; elavGal4/UAS-cup D=0.21, p>0.05, as compared to elav-Gal4/+). To further confirm the presynaptic function of cup, we used RNAi to knock down Cup expression pre- and postsynaptically (Fig. 3D). Knockdown of Cup presynaptically resulted in a 3-fold increase in satellite bouton number compared to the cup RNAi control (D=0.24, p<0.01), whereas postsynaptic Cup, cup RNAi (GL)/24B-Gal4, knockdown did not produce a phenotype (cup RNAi (GL)/+ D=0.05, p>0.05; 24B-Gal4/+ D=0.17, p>0.05; Fig. 3D). This provides further evidence that Cup functions in the presynaptic neuron rather than in the postsynaptic muscle.
Fig. 3.
Cup is required in motor neurons. (A) A cupΔ212/DfBSC7 mutant shows an elevated number of satellite boutons (arrow). (B) Expression of a Cup transgene in neurons in a cupΔ212/DfBSC7 mutant background rescues the satellite bouton phenotype. (C) Box plots showing number of satellite boutons per muscle 4 NMJs in control, mutant, and rescued genotypes. Wild type (WT; n=157), elav-Gal4 (n=80), cupΔ212/DfBSC7 (n=46); cup20/DfBSC7 (n=124); cupΔ212/DfBSC7; elav-Gal4/UAS-cup (n=56); cup20/DfBSC7 (n=124); cup20/DfBSC7; elav-Gal4/UAS-cup (n=48), 24B-Gal4 (n=80), cup20/DfBSC7; 24B-Gal4/UAS-cup (n=68). 25th to 75th percentiles (boxes), medians (line in boxes) and ranges (whiskers, 1.5 times the interquartile range extended from both ends of the box) are shown for each genotype. (D) Box plots showing number of satellite boutons per muscle 4 NMJs in panneuronal expression of cup RNAi (genotype C155-Gal4/+;; cup RNAi (GL)/+), muscle expression of cup RNAi (genotype cup RNAi (GL)/24B-Gal4 cup) and their corresponding controls. C155-Gal4/+;;cup RNAi (GL)/+ (n=94), C155-Gal4/+ (n=56), cup RNAi (GL)/+ (n=95), cup RNAi (GL)/24B-Gal4 (n=88), 24B-Gal4/+ (n=80). Significance of satellite bouton numbers as determined by the K-S test is shown with asterisks. *** p<0.001, ** p<0.01, * p<0.05, n.s. p>0.05. Scale bar: 5μm.
Electrophysiological analysis of cup mutants shows an increase in spontaneous release frequency
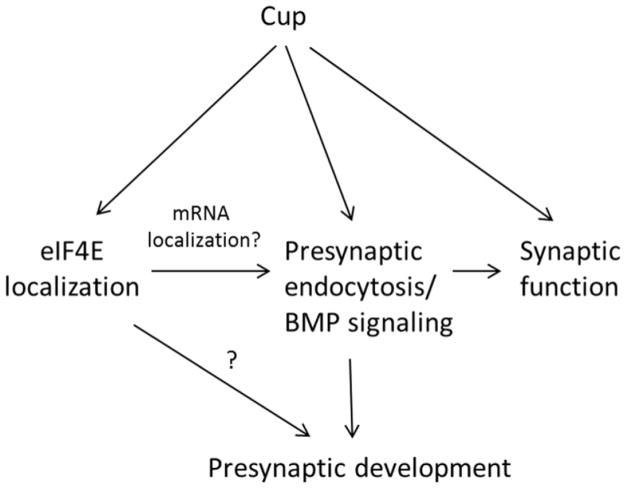
To determine whether Cup is important for synaptic function, we recorded evoked and spontaneous activity from muscle 6/7 NMJs of cup mutants and controls (Fig. 4). The K-S test for non-parametric distributions was used to compare the mutant genotypes to their respective controls (see Experimental Methods). None of the cup transheterozygote combinations revealed a change in evoked excitatory postsynaptic potential (EPSP) amplitudes (Figure 4B). Representative EPSP traces are shown in Figure 4A. The resting membrane potentials were also similar among all the genotypes and controls (data not shown).
Fig. 4.
Cup mutants have an elevated frequency of spontaneous glutamate release events. (A) Representative EPSPs in wild type, cup20/cupΔ212, and cup 20/DfBSC7. (B) Box plots showing EPSP amplitudes in wild type, various cup alleles, and eIF4E knockdown larvae. Wild type (WT; n=112), cup20/cupΔ212 (n=77); cup20/DfBSC7 (n=106); cupΔ212/DfBSC7 (n=85); DfBSC7/+ (n=59); cup20/+ (n=60); cupΔ212/+ (n=46); C155-Gal4/+; eIF4E RNAi (GD)/+ (n=91); eIF4E RNAi (GD)/+ (n=77). 25th to 75th percentiles (boxes), medians (line in boxes) and ranges (whiskers, 1.5 times the interquartile range extended from both ends of the box) are shown for each genotype. No statistically significant differences were observed. (C) Representative traces of miniature EPSPs (mEPSPs) in wild type, cup20/cup 212, and cup20/DfBSC7. (D) Box plots showing the mEPSP frequencies in the various cup mutants and eIF4E RNAi (TRiP) lines. Significance of mEPSP frequencies as determined by the K-S test is shown with asterisks. *** p<0.001, ** p<0.01, * p<0.05, n.s. p>0.05. Wild type (WT; n=89), cup20/cupΔ212 (n=72); cup20/DfBSC7 (n=97); cupΔ212/DfBSC7 (n=71); DfBSC7/+ (n=58); cup20/+ (n=61); cupΔ212/+ (n=45); C155-Gal4; eIF4E RNAi (GD)/+ (n=76); eIF4E RNAi (GD)/+ (n=73). (E) Box plots showing mEPSP amplitudes in wild type, various cup mutants, and eIF4E knockdown larvae. No statistically significant differences as determined by the K-S test were observed. The number of samples were the same as in panel D.
Miniature (mini) EPSPs (mEPSPs) are due to spontaneous release of single vesicles of neurotransmitter. Analysis of mEPSP frequency showed an approximate 1.5-fold increase over heterozygous controls for all cup mutant combinations (Fig. 4D). All values were statistically significant (p<0.001) when analyzed by the K-S test (cup20/cupΔ212 vs. cup20/+ D=0.41, cupΔ212/+ D=0.38; cup20/DfBSC7 vs. cup20/+ D=0.34, DfBSC7/+ D=0.45; cupΔ212/DfBSC7 vs. cupΔ212/+ D=0.32, DfBSC7/+ D=0.46). Representative mEPSP traces are shown in Figure 4C for wild type and two cup transheterozygotes. These results are consistent with a role for cup in motor neurons, since changes in frequencies of spontaneous release events are normally associated with presynaptic function. mEPSP amplitudes did not show significant changes for any of the genotypes tested (Figs. 4E).
Cup interacts with eIF4E to regulate synaptic growth at the larval NMJ
eIF4E is part of the eIF4F translation initiation complex. Cup has been shown to directly interact with eIF4E, and there are two eIF4E-binding sites in Cup (Nakamura et al., 2004; Nelson et al., 2004; Zappavigna et al., 2004). There is a single eIF4E gene in Drosophila, which is expressed in all cells and is required for cell viability. There are also 5 eIF4E-related genes, all of which are primarily expressed in the testis. Their functions are unknown. None of them are detectably expressed in the larval CNS (see Flybase for details). Maternal cup and eIF4E interact genetically, and these interactions suggest that they function collaboratively in the developing oocyte (Zappavigna et al., 2004).
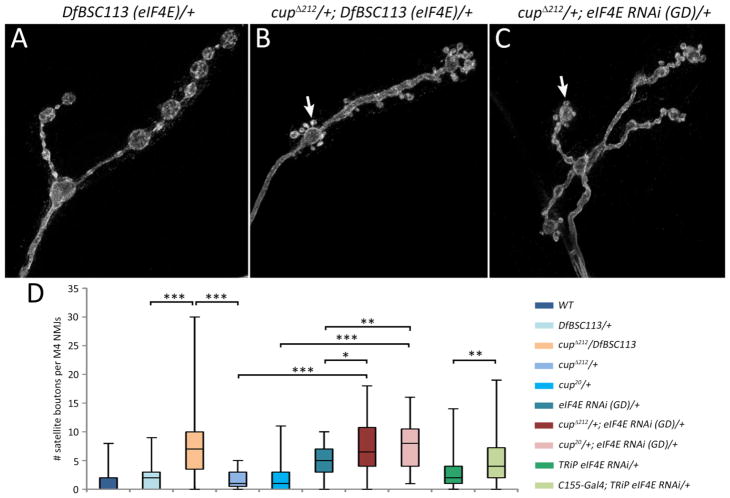
To determine if Cup-eIF4E interactions are required for NMJ development, we carried out a genetic interaction test and quantitated satellite bouton formation in cup/+, eIF4E/+ transheterozygotes (Fig. 5). We used cupΔ212, which eliminates the primary eIF4E binding site, 4E-BM1 (Nakamura et al., 2004), and a deficiency, DfBSC113, that removes eIF4E and thus represents a null mutation. NMJs of cupΔ212/+; DfBSC113 (eIF4E)/+ transheterozygotes showed a 3.5-fold increase in satellite boutons compared to DfBSC113 (eIF4E)/+ controls (D=0.54, p<0.001; Fig. 5D).
Fig. 5.
cup genetically interacts with eIF4E. (A) DfBSC113 (eIF4E)/+ heterozygotes have normal NMJs. (B) A transheterozygous cupΔ212/+; DfBSC113 (eIF4E)/+ mutant has numerous satellite boutons. (C) RNAi knockdown of eIF4E in a cupΔ212/+ background also shows the satellite bouton phenotype. (D) Box plots showing number of satellite boutons per muscle 4 NMJs in various genotypes. Partial knockdown of both cup and eIF4E together greatly increases the number of satellite boutons. Wild type (WT; n=157), DfBSC113 (eIF4E)/+ n=48; cupΔ212/+ (n=43); cupΔ212/+; DfBSC113 (eIF4E)/+ (n=55); cup20/+ (n=45); eIF4E RNAi (GD)/+ (n=31); cupΔ212/+; eIF4E RNAi (GD)/+ (n=30); cup20/+; eIF4E RNAi (GD)/+ (n=39); eIF4E RNAi (TRiP)/+ (n=72); C155-Gal4; eIF4E RNAi (TRiP) (n=64). 25th to 75th percentiles (boxes), medians (line in boxes) and ranges (whiskers, 1.5 times the interquartile range extended from both ends of the box) are shown for each genotype. Significance of satellite bouton numbers as determined by the K-S test is shown with asterisks. *** p<0.001, ** p<0.01, * p<0.05, n.s. p>0.05. Scale bar: 5μm.
We used two eIF4E RNAi lines (GD from VDRC collection, Vienna, and TRiP from the TRiP RNAi collection, Harvard) to confirm the role of eIF4E, since DfBSC113 also removes other genes. Because eIF4E is required for cell viability, strong knockdown of eIF4E causes pleotropic phenotypes and early lethality. Many UAS transgenes, however, have leaky, low level expression even without a Gal4 driver. The GD and TRiP eIF4E RNAi lines both showed satellite boutons, although to different extents, in the absence of any driver (Fig 5D). We used this low level knockdown of eIF4E to our advantage and examined if satellite bouton numbers changed when the eIF4E RNAi (GD) line was combined with two heterozygous cup alleles. Indeed, satellite bouton numbers increased (eIF4e RNAi (GD)/+ vs. cupΔ212/+; eIF4E RNAi (GD)/+ D=.34, p<0.05 and eIF4e RNAi (GD)/+ vs. cup20/+; eIF4E RNAi (GD)/+ D=0.44, p<0.001; Fig. 5D).
The specificity of the cup/eIF4E interaction led us to more closely investigate eIF4E itself. Even though leaky eIF4E RNAi expression showed a slight increase in the number of satellite boutons, we wanted to further reduce eIF4E levels in neurons to see if the phenotype would become stronger. The neuronal driver C155-Gal4 was crossed to the eIF4E RNAi (TRiP) line, because this line showed less leaky expression as compared to the GD RNAi line. C155-driven eIF4E RNAi did not cause larval lethality or obvious larval motility phenotypes. We observed that there was a 2-fold increase in the number of satellite boutons when eIF4E RNAi was expressed presynaptically, as compared to the non-driven RNAi line (C155-Gal4; eIF4E RNAi (TRiP) vs. eIF4E RNAi (TRiP)/+ D=0.33, p<0.001; Fig. 5D).
BMP signaling is upregulated in cup mutants
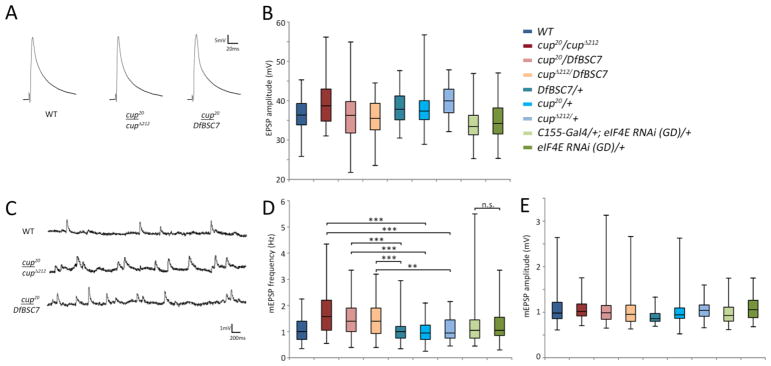
Most mutations that produce satellite boutons have been found to upregulate the retrograde BMP signaling pathway. To examine whether cup mutations also do this, we employed two tests that are standard in the field (reviewed by (O’Connor-Giles and Ganetzky, 2008)). BMP signaling acts through Mothers against decapentaplegic (Mad). When Mad is phosphorylated, it translocates to the nucleus where it serves as a transcription factor. To evaluate if BMP signaling is involved in the cup satellite bouton phenotype, we examined synaptic phosphorylated Mad (pMad), which is a marker of pathway activation at presynaptic terminals (Aberle et al., 2002; Marques et al., 2002). We stained wild-type and cup mutant larvae with antibody against pMad. We found that pMad immmunoreactivity was increased in cupΔ212/DfBSC7 as compared to control NMJs (Figs. 6A1, B1). The levels of anti-HRP reactivity were unchanged between mutant and control animals (Figs. 6A2, B2).
Fig. 6.
BMP signaling is elevated in cup mutants. (A1–A3) Wild type larva, muscle 4 NMJ. (A) pMad staining at the NMJ is almost undetectable (NMJ outline is not visible with anti-pMad in A1). (A2) Anti-HRP staining of neuronal membranes. (A3) Overlay of pMad and anti-HRP staining from A1 and A2. (B1–B3) A cupΔ212/DfBSC7 mutant. (B1) pMad staining is greatly increased compared to (A1); the NMJ outline is clearly visible. (B2) Anti-HRP staining. (B3) Overlap of pMad and anti-HRP staining. Anti-HRP was normalized between A2 and B2 in order to accurately compare pMad intensities. Inset, pMad puncta within satellite boutons. (C) Box plots showing number of satellite boutons per muscle 4 NMJs in cupΔ212/+, Dadj1E4/+, and cupΔ212/+; Dadj1E4/+ transheterozygotes. Dadj1E4/+ (n=55); cupΔ212/+; Dadj1E4/+ (n=69); cupΔ212/+ (n=43). 25th to 75th percentiles (boxes), medians (line in boxes) and ranges (whiskers, 1.5 times the interquartile range extended from both ends of the box) are shown for each genotype. Significance of satellite bouton numbers as determined by the K-S test is shown with asterisks. *** p<0.001, ** p<0.01, * p<0.05, n.s. p>0.05.
The increase in pMad levels in cup mutants suggests that Cup and BMP signaling may function together to regulate satellite bouton formation. We investigated this further using genetic interaction analysis. We made transheterozygotes for Daughters against decapentaplegic (Dad), a negative regulator of BMP signaling (Sweeney and Davis, 2002), and cup. We reasoned that if BMP signaling is involved in Cup’s function, then the allelic combination of cup and dad should have a greater number of satellite boutons, as compared to single heterozygote controls. This was in fact observed: satellite boutons are increased by ~5-fold in cupΔ212/+; Dad1jE4/+ as compared to heterozygotes, cupΔ212/+ (D=0.50, p<0.001) or Dad1jE4/+ (D=0.56, p<0.001; Fig. 6C).
The endocytic protein Endophilin may be a Cup target
During oogenesis, Cup is required for localization and translational repression of osk mRNA prior to when it reaches its final posterior location in the oocyte (Nakamura et al., 2004). Cup is also required for efficient translation of posteriorly localized osk mRNA (Ottone et al., 2012). In order to identify potential targets in the neuromuscular system, we reasoned that partial loss of Cup might result in abnormal expression of proteins encoded by target mRNAs. As an initial step towards identifying targets of Cup in the nervous system, we searched the literature on satellite boutons to see which genes produced strong satellite bouton phenotypes when mutated. Some of these genes encode proteins that are involved in endocytosis and/or in the organization of the actin cytoskeleton in the presynaptic terminal (Dickman et al., 2006; Koh et al., 2004; Marie et al., 2004; Rodal et al., 2011). We tested four of these (Endophilin A (EndoA; also referred to as Endo), Synaptojanin (Synj), Dap160, and Wiskott-Aldrich Syndrome protein (WASp)) for genetic interactions with cup by quantitating satellite boutons in heteroallelic combinations involving cup and each of the potential target genes. The most statistically significant interaction was with Endo, which showed approximately a 3-fold increase when combined with cup as compared to endoΔ4/+ (D=0.60, p<0.001) or cupΔ212/+ (D=0.64, p<0.001) alone (Fig. 7D). cupΔ212/+,DfBSC302 (dap160)/+ transheterozygotes also had a statistically significant increase in satellite bouton numbers compared to cupΔ212/+ (D=0.36, p<0.01) and DfBSC302 (dap160)/+ (D=0.29, p<0.05). Transheterozygotes of cupΔ212 and synj or WASp did not show any statistically different number of satellite boutons when compared to their heterozygous controls.
Fig. 7.
Genetic interactions between cup and genes encoding modulators of synaptic BMP signaling. endo is another name for EndoA, and wsp is another name for WASp. (A) endoΔ4/+ muscle 4 NMJs look similar to wild type (see Figure 3). (B) cupΔ212/+also shows no phenotype. (C) A cupΔ212/+; endoΔ4/+ transheterozygote has excess satellite boutons (arrows). (D) Box plots showing number of satellite boutons per muscle 4 NMJs in cup and heterozygotes of candidate genes and cupΔ212/+, candidate gene/+ transheterozgotes. cupΔ212/+; endoΔ4/+ (n=40); cupΔ212/DfBSC302 (Dap160)/+ (n=40); cupΔ212/DfX58-7 (Synj) (n=69); cupΔ212/+; Df3450 (wsp)/+ (n=53); endoAΔ4/+ (n=40); DfBSC302 (Dap160)/+ (n=96); DfX58-7 (Synj)/+ (n=48); Df3450 (wsp) /+ (n=125); cupΔ212/+ (n=43). 25th to 75th percentiles (boxes), medians (line in boxes) and ranges (whiskers, 1.5 times the interquartile range extended from both ends of the box) are shown for each genotype. (E, F) Endophilin expression at the NMJ in wild-type (E) and in a cup mutant (F). Endophilin staining (green) is brighter in wild-type (E1) than in cup20/cupΔ212 (F1). Endophilin staining was normalized to anti-HRP staining intensity (red in (E2, F2). (G) Box plots showing that Endophilin staining intensity differs significantly between wild-type and mutant NMJs. WT (n=14); cup20/cupΔ212 (n=18). Significance of all data sets as determined by the K-S test is shown with asterisks. *** p<0.001, ** p<0.01, * p<0.05, n.s. p>0.05. Scale bar: 5μm.
Since EndoA and cup showed the strongest interaction, and a good antibody against EndoA was available (Verstreken et al., 2002), we next investigated if EndoA protein levels were altered in cup mutants. Indeed, EndoA NMJ staining in cup20/cupΔ212 is decreased when compared to wild type (Figs. 7E1, F1). This reduction in EndoA immunoreactivity at the NMJ was statistically significant (D=0.72, p<0.01, Fig. 7G).
Discussion
Maternal translational regulators that function during oogenesis and early embryo development, including Pum, Nos, FMRP (dFmr1), Staufen, and Brat, are also zygotically expressed at the larval NMJ and have functions in the neuromuscular system (Gardiol and St Johnston, 2014; Menon et al., 2009; Menon et al., 2004; Shi et al., 2013; Zhang et al., 2001). Here we show that Cup, another of these maternal regulators, controls larval NMJ structure and function. Cup is localized to the cytoplasm of CNS neurons and in punctate structures within presynaptic NMJ terminals (Fig. 1). cup mutants have strong NMJ phenotypes characterized by the accumulation of small satellite boutons that bud from parent boutons or axonal segments (Fig. 2). Satellite bouton phenotypes are also observed when cup is knocked down with cup RNAi in presynaptic neurons, but not in postsynaptic muscles (Fig. 3D). Moreover, the satellite boutons phenotype in cup mutants is only rescued by presynaptic, and not by postsynaptic, expression of Cup (Fig. 3). cup mutant NMJs also display an increase in the frequency of spontaneous release events (minis), but have no change in evoked responses (Fig. 4).
Cup interacts physically and genetically with eIF4E, the cap-binding protein, which is required for translation in all cells (Nakamura et al., 2004; Nelson et al., 2004; Wilhelm et al., 2003; Zappavigna et al., 2004). We tested whether the Cup-eIF4E interaction is important for NMJ development by examining the phenotypes of transheterozygotes lacking one copy of each gene. We also reduced eIF4E expression in neurons using RNAi in a cup heterozygous background. In both cases, the resulting larvae with reductions in Cup and eIF4E had strong satellite bouton phenotypes. Neuronal knockdown of eIF4E alone also produced the phenotype (Fig. 5).
The presence of satellite boutons is often associated with elevation of retrograde BMP signaling from the muscle to the presynaptic terminal. To evaluate whether cup behaves genetically as a regulator of the BMP pathway, we examined transheterozygotes missing one wild-type copy of cup and one copy of Dad, a negative regulator of BMP signaling. cup/+, Dad/+ transheterozygotes displayed strong satellite bouton phenotypes. NMJs in cup mutants also showed elevated levels of synaptic pMad, a marker for activation of BMP signaling (Fig. 6).
A number of genes for which mutations produce satellite boutons encode proteins involved in endocytosis and the organization of the presynaptic actin cytoskeleton. It is thought that endocytic mutants have satellite bouton phenotypes because they have excessive levels of BMP receptors on the plasma membrane of presynaptic terminals (O’Connor-Giles and Ganetzky, 2008). We examined whether genes encoding endocytic/cytoskeletal proteins might be Cup targets by making transheterozgyotes between cup mutations and mutations in four of these genes: EndoA, Synj, Dap160, and WASp. The strongest interaction was with the EndoA gene, which encodes an Endophilin protein (Fig. 7).
During oogenesis, Cup regulates translation of localized osk and nos mRNAs. Cup represses translation of osk mRNA until it arrives at its appropriate posterior location within the oocyte (Wilhelm et al., 2003). nos mRNA is concentrated at the posterior pole of the oocyte, but is also present in the remainder of the oocyte cytoplasm. However, unlocalized nos mRNA is not translated. In the absence of Cup, Osk and Nos proteins are translated from unlocalized mRNAs and are present in the anterior regions of the oocyte or embryo, resulting in pattern formation defects (Nakamura et al., 2004; Nelson et al., 2004; Verrotti and Wharton, 2000; Wilhelm et al., 2003). Cup does not bind to mRNA directly, but is recruited to osk and nos mRNAs by sequence-specific RNA-binding proteins (Nakamura et al., 2004; Nelson et al., 2004).
Cup is a component of a macromolecular complex that includes eIF4E and a variety of proteins involved in mRNA dynamics (Nakamura et al., 2004; Wilhelm et al., 2003). Cup binds to eIF4E through two binding motifs, denoted as 4E-BM1 and 4E-BM2. 4E-BM1 is required for eIF4E binding, and it interacts with the same parts of eIF4E as eIF4G and 4E-BPs. 4E-BM2 contributes to binding but is not required, and interacts with a different part of eIF4E (Kinkelin et al., 2012). Because Cup, like other 4E-BPs, can compete with eIF4G for binding to eIF4E and thereby prevent formation of the eIF4F complex, it was thought that this was the mechanism by which Cup represses translation of osk and nos mRNAs. However, cup and eIF4E genetically interact during oogenesis in a manner that suggests that eIF4E function is reduced, not increased, in a cup mutant (Zappavigna et al., 2004). Consistent with this, eIF4E is depleted from the posterior pole in cup mutants (Wilhelm et al., 2003; Zappavigna et al., 2004). Also, even correctly localized osk mRNA may be translated less efficiently in cup mutants than in wild-type, because Osk staining at the posterior pole is decreased in the mutants (Nakamura et al., 2004; Wilhelm et al., 2003).
Igreja and Izaurralde (2011) showed that eIF4E binding is not required for translational repression by Cup. Rather, Cup promotes mRNA deadenylation and represses translation via eIF4E-independent mechanisms. Based on these findings, these authors proposed that Cup’s binding to eIF4E brings eIF4E to mRNAs, so that translation can begin as soon as Cup repression is relieved, and thereby increases the efficiency of translation of localized mRNAs. This is consistent with our findings in the neuromuscular system, where we show that eIF4E function is reduced when one copy of the cup gene is mutant (Fig. 5). Furthermore, cup interacts genetically with potential target genes in a manner that suggests that it is a positive rather than a negative regulator of function (Fig. 7).
What is Cup’s function in motor neurons? Our genetic data suggest that one function of Cup is to positively regulate endocytic proteins that are modulators of retrograde BMP signaling. Consistent with this, there is a small but statistically significant decrease in Endophilin protein levels at the NMJ in cup mutants (Fig. 7). We suggest that the cup satellite bouton phenotype is the consequence of simultaneous downregulation of expression of a number of BMP modulators, and that the amount of any single modulator protein is not dramatically altered in cup mutants. Like Endophilin, correctly localized Osk is reduced but not eliminated in cup mutants (Nakamura et al., 2004; Wilhelm et al., 2003), so the increase in translation efficiency that Igreja and Izaurralde (2011) propose is contributed by Cup-eIF4E interactions may be a relatively subtle effect.
Since Cup’s function during oogenesis is to regulate translation of localized mRNAs, this raises the question of whether Cup’s mRNA targets in motor neurons are also subcellularly localized, and, if so, to which compartment(s). Cup localizes to punctate structures in the NMJ presynaptic terminal. An epitope-tagged version of eIF4E expressed in motor neurons is also detected in puncta at presynaptic terminals (data not shown). This overexpression experiment, however, does not prove that endogenous eIF4E is normally localized at the NMJ. It is not possible to use anti-eIF4E staining to evaluate whether endogenous eIF4E is in presynaptic terminals, since eIF4E is present at very high levels in the underlying muscle fibers. Localization of epitope-tagged eIF4E was not visibly altered in a cup mutant.
Zucker and colleagues (Beaumont et al., 2001) showed that local translation occurs within presynaptic NMJ terminals in crayfish, so it is possible that this also could occur in Drosophila. We and others have also observed that a variety of translational regulators, including Pum, Nos, Staufen, FMRP, and Brat, localize to presynaptic terminals. If translation of localized mRNA does occur in presynaptic terminals, it might to controlled by Cup, in a similar manner to Cup’s selective regulation of translation of mRNAs localized to the posterior pole of the oocyte. However, we have no evidence for this model. An alternative, and perhaps more likely, model is that mRNAs controlled by Cup are in a compartment in the motor neuron cell body that is dedicated to production of proteins that are destined for the presynaptic terminal (Fig. 8).
Fig. 8.
Summary diagram of Cup’s actions at the NMJ. Based on its functions in ovaries and on the data presented here, we suggest that Cup affects eIF4E localization in neurons and thereby regulates localization and translation of mRNAs encoding endocytic proteins that modulate BMP signaling and thereby affect presynaptic terminal development. Cup may also have other effects on these mRNAs that do not involve eIF4E. Cup and the endocytic proteins also regulate mEPSP frequency.
Experimental Methods
Drosophila Stocks
“Wild type” controls were w1118 crossed to Canton S. cup8 and cup20 alleles were provided by J. Wilhelm (University of California, San Diego, CA) (Keyes and Spradling, 1997), and cup 212 was obtained from A. Nakamura (Institute of Molecular Embryology and Genetics, Kumamato University, Japan). EndoA 4, Df(Dap160), Synj, WASp, dad1jE4, DfBSC7(cup), DfBSC113(eIF4E), and DfBSC187(cup) were obtained from the Drosophila Stock Center (Bloomington, IN). The Gal4 lines used were C155 (Elav)-Gal4 and elav-Gal4 (on first and third chromosome, respectively) for neuronal expression and 24B-Gal4 for muscle expression (3rd chromosome). The cup RNAi (GL) line was obtained from the Harvard TRiP collection (Boston, MA). The eIF4E RNAi (GD) line was obtained from the Vienna Drosophila RNAi Center (Dietzl et al., 2007) and the eIF4E RNAi (TRiP) line was obtained from the Harvard TRiP collection (Boston, MA). Both of the eIF4E RNAi lines have leaky expression and the non-driven lines were used to cross to cup alleles in order to perform the genetic interaction tests. The control and experimental crosses for eIF4E RNAi were done at room temperature since elevated temperature seemed to restrict satellite bouton growth.
The UAS-cup transgenic line was constructed with UAS-cup cDNA clone kindly provided by C. Smibert (University of Toronto, Ontario, Canada). The DNA was sent to Rainbow Transgenic Flies (Camarillo, CA) for injection. We used a transgenic line with UAS-Cup on the third chromosome.
Antibodies
The primary antibodies used in this study are below. The rabbit and mouse Cup antibodies were obtained from Akira Nakamura (Institute of Molecular Embryology and Genetics, Kumamato University, Japan) and used at a dilution of 1:2000 and 1:1000, respectively (Nakamura et al, 2004). Rat anti-Cup was obtained from Allan Spradling (Carnegie Institution for Science, Baltimore, MD). Guinea pig anti-EndoA was obtaIned from Hugo Bellen and was used at 1:200. The Discs-Large (4F3; 1:100) and Bruchpilot (nc82; 1:100) antibodies were obtained from the Developmental Studies Hybridoma Bank (University of Iowa, IA). Mouse anti-Fas2 was used at 1:3 (Van Vactor 1993). The rabbit pMad antiserum was kindly provided by Carl-Henrik Heldin (Ludwig Institute for Cancer Research, Uppsala, Sweden) and P. ten Dijke (Leiden University, Leiden, The Netherlands) and used at a dilution of 1:100. Tetramethylrhodamine isothiocyanate-horseradish peroxidase (HRP; Jackson ImmunoResearch Laboratories) was used at 1:50. The secondary antibodies, Alexa-Fluor 488 anti-mouse, anti-rabbit, and anti-rat were used at 1:500 and Alexa-Fluor 568 anti-mouse was used at 1:500 also.
Immunohistochemistry
Third instar larvae were dissected, stained, and processed as described previously (Menon et al., 2009). For EndoA and p-Mad staining experiments, 4% paraformaldehyde was used instead of Bouin’s fix (Sigma-Aldrich, St. Louis, MO) and incubated for 30 min. Larvae were then incubated with the appropriate primary and secondary antibodies and imaged on a Zeiss LSM 510 confocal microscope.
In larval preps where EndoA staining intensity was compared between cup mutants and controls, the HRP signal was used for normalization. Average intensity projections were created from z-stacks collected on Zeiss LSM510 and processed in Image J (NIH, Bethesda, MD). NMJs were outlined to create a region of interest and corresponding red and green channel intensities were measured. The green/red ratio was then calculated and used to compare the genotypes.
Electrophysiology
Third instar larvae were dissected as described in (Carrillo et al., 2010). The dissection was performed in HL3.1 solution containing (in mM): 70 NaCl, 5 KCl, 4 MgCl2, 12.5 CaCl2, 10 NaHCO3, 5 trehalose, 115 sucrose, and 5 HEPES, pH 7.2. For recording, HL3.1 solution was also used with 1mM CaCl2. Excitatory postsynaptic potentials (EPSPs) were recorded using 20–30 MΩ microelectrodes filled with 3M KCL. Evoked EPSPs were recorded from ventral longitudinal muscles by using a 2–4 μm suction electrode to stimulate nerve branches. Recordings were obtained from muscles 6 and 7 in segments 3–5. The average EPSP amplitude was calculated from 10 sequential EPSPs per muscle at a frequency of 0.5 Hz. Miniature EPSPs were recorded for 20 seconds and the number of events and amplitudes were analyzed using Synaptosoft (by J. Lee; Synaptosoft). Both evoked and spontaneous EPSPs were recorded using an Axoclamp-2B amplifier (Axon Instruments, Foster City, CA). The K-S test was used to determine significance.
Statistics
We used the Kolmogorov-Smirnov (K-S) test to evaluate the statistical significance of all data sets. The KS test is a non-parametric test that calculates a D statistic, which is the maximum difference between the empirical distribution functions of two data sets. The p value significance is as follows: *** p<0.001, ** p<0.01, * p<0.05, n.s. p>0.05.
Supplementary Material
Highlights.
The maternal translational regulator Cup is localized to puncta in larval NMJs
cup mutants have increased numbers of satellite boutons
Cup interacts physically and genetically with translation initiation factor eIF4E
Cup positively regulates endocytic proteins that modulate retrograde BMP signaling
Acknowledgments
We thank Jim Wilhelm (UC San Diego) for helpful discussions, Craig Smibert (University of Toronto, Toronto, Canada) for the UAS-Cup cDNA, Akira Nakamura (Riken Center for Developmental Biology, Japan), Jim Wilhelm, Allan Spradling (Carnegie Institution for Science, Maryland), Hugo Bellen (Baylor University), the Developmental Studies Hybridoma Bank, and the Drosophila Stock Center for stocks and antibodies. We thank Elena Armand for technical assistance and the Biological Imaging facility (CalTech) for access to Zeiss LSM 510 Confocal. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic RNAi fly stocks. This work was supported by an NIH RO1 grant to K.Z., NS62821.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberle H, Haghighi AP, Fetter RD, McCabe BD, Magalhaes TR, Goodman CS. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002;33:545–558. doi: 10.1016/s0896-6273(02)00589-5. [DOI] [PubMed] [Google Scholar]
- Baines RA. Neuronal homeostasis through translational control. Molecular neurobiology. 2005;32:113–121. doi: 10.1385/MN:32:2:113. [DOI] [PubMed] [Google Scholar]
- Beaumont V, Zhong N, Fletcher R, Froemke RC, Zucker RS. Phosphorylation and local presynaptic protein synthesis in calcium- and calcineurin-dependent induction of crayfish long-term facilitation. Neuron. 2001;32:489–501. doi: 10.1016/s0896-6273(01)00483-4. [DOI] [PubMed] [Google Scholar]
- Carrillo RA, Olsen DP, Yoon KS, Keshishian H. Presynaptic activity and CaMKII modulate retrograde semaphorin signaling and synaptic refinement. Neuron. 2010;68:32–44. doi: 10.1016/j.neuron.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman DK, Lu Z, Meinertzhagen IA, Schwarz TL. Altered synaptic development and active zone spacing in endocytosis mutants. Curr Biol. 2006;16:591–598. doi: 10.1016/j.cub.2006.02.058. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Gardiol A, St Johnston D. Staufen targets coracle mRNA to Drosophila neuromuscular junctions and regulates GluRIIA synaptic accumulation and bouton number. Developmental biology. 2014;392:153–167. doi: 10.1016/j.ydbio.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igreja C, Izaurralde E. CUP promotes deadenylation and inhibits decapping of mRNA targets. Genes & development. 2011;25:1955–1967. doi: 10.1101/gad.17136311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes LN, Spradling AC. The Drosophila gene fs(2)cup interacts with otu to define a cytoplasmic pathway required for the structure and function of germ-line chromosomes. Development. 1997;124:1419–1431. doi: 10.1242/dev.124.7.1419. [DOI] [PubMed] [Google Scholar]
- Kinkelin K, Veith K, Grunwald M, Bono F. Crystal structure of a minimal eIF4E-Cup complex reveals a general mechanism of eIF4E regulation in translational repression. Rna. 2012;18:1624–1634. doi: 10.1261/rna.033639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh TW, Verstreken P, Bellen HJ. Dap160/intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron. 2004;43:193–205. doi: 10.1016/j.neuron.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Marie B, Sweeney ST, Poskanzer KE, Roos J, Kelly RB, Davis GW. Dap160/intersectin scaffolds the periactive zone to achieve high-fidelity endocytosis and normal synaptic growth. Neuron. 2004;43:207–219. doi: 10.1016/j.neuron.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Marques G, Bao H, Haerry TE, Shimell MJ, Duchek P, Zhang B, O’Connor MB. The Drosophila BMP type II receptor Wishful Thinking regulates neuromuscular synapse morphology and function. Neuron. 2002;33:529–543. doi: 10.1016/s0896-6273(02)00595-0. [DOI] [PubMed] [Google Scholar]
- Marques G, Zhang B. Retrograde signaling that regulates synaptic development and function at the Drosophila neuromuscular junction. International review of neurobiology. 2006;75:267–285. doi: 10.1016/S0074-7742(06)75012-7. [DOI] [PubMed] [Google Scholar]
- Menon KP, Andrews S, Murthy M, Gavis ER, Zinn K. The translational repressors Nanos and Pumilio have divergent effects on presynaptic terminal growth and postsynaptic glutamate receptor subunit composition. J Neurosci. 2009;29:5558–5572. doi: 10.1523/JNEUROSCI.0520-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KP, Carrillo RA, Zinn K. Development and plasticity of the Drosophila larval neuromuscular junction. Wiley interdisciplinary reviews Developmental biology. 2013;2:647–670. doi: 10.1002/wdev.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon KP, Sanyal S, Habara Y, Sanchez R, Wharton RP, Ramaswami M, Zinn K. The translational repressor Pumilio regulates presynaptic morphology and controls postsynaptic accumulation of translation factor eIF-4E. Neuron. 2004;44:663–676. doi: 10.1016/j.neuron.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Muraro NI, Weston AJ, Gerber AP, Luschnig S, Moffat KG, Baines RA. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J Neurosci. 2008;28:2099–2109. doi: 10.1523/JNEUROSCI.5092-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Nelson MR, Leidal AM, Smibert CA. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. Embo J. 2004;23:150–159. doi: 10.1038/sj.emboj.7600026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor-Giles KM, Ganetzky B. Satellite signaling at synapses. Fly. 2008;2:259–261. doi: 10.4161/fly.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor-Giles KM, Ho LL, Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58:507–518. doi: 10.1016/j.neuron.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Robinson I. Barfly: sculpting membranes at the Drosophila neuromuscular junction. Developmental neurobiology. 2012;72:33–56. doi: 10.1002/dneu.20923. [DOI] [PubMed] [Google Scholar]
- Ottone C, Gigliotti S, Giangrande A, Graziani F, Verrotti di Pianella A. The translational repressor Cup is required for germ cell development in Drosophila. Journal of cell science. 2012;125:3114–3123. doi: 10.1242/jcs.095208. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Blunk AD, Akbergenova Y, Jorquera RA, Buhl LK, Littleton JT. A presynaptic endosomal trafficking pathway controls synaptic growth signaling. The Journal of cell biology. 2011;193:201–217. doi: 10.1083/jcb.201009052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Chen Y, Gan G, Wang D, Ren J, Wang Q, Xu Z, Xie W, Zhang YQ. Brain tumor regulates neuromuscular synapse growth and endocytosis in Drosophila by suppressing mad expression. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:12352–12363. doi: 10.1523/JNEUROSCI.0386-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13:2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney ST, Davis GW. Unrestricted synaptic growth in spinster-a late endosomal protein implicated in TGF-beta-mediated synaptic growth regulation. Neuron. 2002;36:403–416. doi: 10.1016/s0896-6273(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Verrotti AC, Wharton RP. Nanos interacts with cup in the female germline of Drosophila. Development. 2000;127:5225–5232. doi: 10.1242/dev.127.23.5225. [DOI] [PubMed] [Google Scholar]
- Verstreken P, Kjaerulff O, Lloyd TE, Atkinson R, Zhou Y, Meinertzhagen IA, Bellen HJ. Endophilin mutations block clathrin-mediated endocytosis but not neurotransmitter release. Cell. 2002;109:101–112. doi: 10.1016/s0092-8674(02)00688-8. [DOI] [PubMed] [Google Scholar]
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. The Journal of cell biology. 2003;163:1197–1204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Smibert CA. Mechanisms of translational regulation in Drosophila. Biology of the cell / under the auspices of the European Cell Biology Organization. 2005;97:235–252. doi: 10.1042/BC20040097. [DOI] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- Zappavigna V, Piccioni F, Villaescusa JC, Verrotti AC. Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14800–14805. doi: 10.1073/pnas.0406451101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.