Abstract
Genome-wide association studies have identified twenty loci associated with late-onset Alzheimer disease (LOAD). We examined each of the twenty loci, specifically the ±50kb region surrounding the most strongly associated variant, for changes in gene(s) transcription specific to LOAD. Post-mortem human brain samples were examined for expression, methylation, and splicing differences. LOAD specific differences were detected by comparing LOAD to normal and “disease” controls. Eight loci, prominently ABCA7, contain LOAD specific differences. Significant changes in the CELF1 and ZCWPW1 loci occurred in genes not located nearest the associated variant, suggesting that these genes should be investigated further as LOAD candidates.
Keywords: Late-Onset Alzheimer Disease, genome-wide association study, transcription, Differential Expression, RNA-Seq, microarray, DNA methylation, Splicing, Dementia with Lewy bodies
Introduction
Late-Onset Alzheimer disease (LOAD) is a neurodegenerative disease that affects individuals 60 years or older. LOAD heritability estimates of up to 70% indicates that there is a considerable genetic component to the disease[1]. In 1993, APOE was the first gene to be unequivocally established as a susceptibility gene for LOAD [2, 3]. Recently, genome-wide association studies (GWAS) have identified an additional twenty loci significantly associated with LOAD that fall within or near the ABCA7, BIN1, CASS4, CD2AP, CD33, CELF1, CLU, CR1, EPHA1, FERMT2, HLA, INP55D, MEF2C, MS4A6A, NME8, PICALM, PTK2B, SLC2A4, SORL1, and ZCWPW1 genes [4-9].
In an effort to understand how these variants influence LOAD etiology, several studies have attempted to elucidate how these loci contribute to LOAD by examining transcription and splicing of the genes nearest the GWAS variants with the strongest association. To date, increased CD33 molecule (CD33) expression has been shown to be associated with Alzheimer disease (AD) and to inhibit microglial uptake of amyloid beta [10, 11]. Alternate isoform expression of Clusterin (CLU) results in an increase of CLU protein secretion, an effect observed in AD [12]. Increased copy number variants located within the complement component (3b/4b) receptor 1 (Knops blood group) (CR1) gene are significantly associated with LOAD [13]. Lastly, sortilin-related receptor, L (DLR class) A repeats containing (SORL1) harbors an intronic polymorphism associated with decreased expression in LOAD [14, 15]. In total, transcriptional alterations have been identified in the ABCA7, BIN1, CD33, CLU, CR1, and SORL1 loci[10]. While the majority of studies examined the gene nearest the strongest associated variant, it is important to note that all significant GWAS variants fall outside of known exons and that some areas of strong association contain multiple genes. Fourteen of the twenty strongly associated variants lie within intronic regions and six variants fall completely outside of known gene boundaries.
In this study, we wanted to examine all the genes located with a 100kb region surrounding each of the most strongly LOAD associated variants to examine gene transcription for abnormalities and potentially identify the gene(s) that may play a role in LOAD etiology. To do this, we examined each LOAD loci for changes in gene expression, methylation, and splicing specific to LOAD by performing RNA sequencing (RNA-Seq) on a total of ten cases and ten cognitively normal controls. Changes in gene expression and splicing were examined within the twenty loci. DNA methylation, a known regulator of expression, was examined in eight LOAD and eight cognitively normal controls in the same samples used for RNA-Seq. To determine if alterations were LOAD specific, or were secondary effects of neurodegeneration, alterations in expression, methylation, and splicing observed in LOAD were also compared to a “disease control”, Dementia with Lewy bodies (DLB). Patients with DLB exhibit similar phenotypes to LOAD; however, their pathological attributes differ substantially. This characteristic allowed us to use the disease control to potentially filter out the differences observed in LOAD from those due to DLB neurodegeneration and enabled us to hopefully identify processes specifically contributing to LOAD. We confirmed expression differences using quantitative Real Time PCR (qRT-PCR) and compared findings to a previous microarray study. This study revealed a total of eight loci with significant changes in expression, methylation, and splicing in seventeen genes throughout the loci specifically altered in LOAD. These findings may provide mechanistic insights into the role these loci play in LOAD.
Materials and Methods
Tissue samples
RNA transcription was investigated using tissue samples isolated from the temporal pole from a total of thirty brain samples. Ten samples were collected from each of the following three groups: subjects with late-onset Alzheimer disease (LOAD), neurologically normal controls, and disease controls, subjects with dementia with Lewy bodies (DLB). Samples were extracted from the temporal pole (Brodmann area 38) of age-matched Caucasian males (Table 1). The mean (SD) ages were LOAD: 77.4 (±5.7) years; DLB: 79.1 (±5.6) years; cognitively normal controls: 74.6 (±7.8) years. Samples were frozen and stored at −80C.
Table 1.
Sample Information.
| Original Data Set | Group | Sex (M/F) | Age | PMI | Autolysis | Brain Weight |
|---|---|---|---|---|---|---|
| LOAD | 10/0 | 76.5(±1.8) | -- | 6.5(±1.0) | 1261.2(±42.2) | |
| Control | 10/0 | 79.1(±1.9) | -- | 14.9(±2.5) | 1413.0(±50.1) | |
| DLB | 10/0 | 74.6(±2.6) | -- | 7.3(±0.8) | 1231.0(±37.3) | |
| GEO (GSE15222) | ||||||
| LOAD | 88/88 | 83.6(±0.5) | 7.1(±0.3) | -- | -- | |
| Control | 105/85 | 81.1(±0.7) | 9.8(±0.7) | -- | -- | |
The original data set consists of the three cohorts. All samples from the original data set are from the temporal pole; Brodmann's Area 38. The second data set (GES15222) consists of the replication data set and is from the pre-frontal cortex. CAU: Caucasian.
All cases underwent a standardized neuropathological assessment with evaluation of gross and microscopic findings and quantitative analysis of Alzheimer's type pathology.
LOAD cases were selected according to dementia status, staged for LOAD pathology according to Braak (III, IV), and were positive for Aβ and PHF-Tau in two brain areas (Brodmann areas 9 and 39)[1]. Semi quantitative grading of Lewy body pathology and assignment of Lewy body type were determined according to the Third CDLB recommendations [2, 3]. Cases of DLB were selected based on the distribution of Lewy bodies and the severity of Alzheimer-type pathology.
Normal control samples were confirmed to be cognitively normal, and died from underlying causes of death unrelated to neurological disease. Autopsies and neuropathologic diagnoses were performed in accordance with published guidelines by a consultant neuropathologist.
RNA and DNA isolation
RNA was isolated from the frozen tissue samples using the QIAGEN Qiashredder and TRIzol reagents (Invitrogen). RNA was extracted using the miRNeasy mini kit (Invitrogen) and each sample was treated with an on-column DNase treatment (Invitrogen). RNA was dissolved in DNase/RNase-free water (Invitrogen) and concentration was determined using the Qubit florometer with the Qubit™ RNA kit. The quality of the RNA was determined using the Bioanalyzer 2100 (Agilent). All RNA samples were stored at −80C.
Genomic DNA was isolated from 24 of the 30 frozen tissue samples (N=8 LOAD patients, N=8 cognitively normal controls, and N=8 DLB patients). Isolation was carried out using the QIAamp® DNA Mini and Blood Mini Handbook (Qiagen) in accordance with the manufacturer's specifications. DNA concentration was determined using the Qubit fluorometer with the Qubit™ DNA Broad Range kit. DNA integrity was assessed using gel electrophoresis.
RNA-Seq library preparation
RNA-Seq libraries were prepared from 10ug of total RNA isolated from each sample. The RNA integrity number (RIN) was determined using the Bioanalyzer 2100 and all samples had a RIN number ≥6. Ribosomal RNA was depleted using the RiboMinus™ Eukaryote Kit for RNA-Seq (Invitrogen). Depletion was confirmed using the Agilent RNA Nano chip and Bioanalyzer 2100. RNA concentrations were determined using the Qubit™ RNA kit. Approximately 600 ng of ribosomal depleted RNA was utilized for library preparation using Script-Seq (Epicentre®) along with the Phusion® Polymerase enzyme (Kappa Biosystems). Library completion was confirmed using the DNA High Sensitivity Kit on the Bioanalzyer 2100 and concentration was determined using the Library Quantification Kit-Illumina (Kappa Biosystems).
Microarray Validation Analysis
The microarray data set was downloaded from the publicly available GEO database (GEO accession number GSE15522)[8]. All subsequent data manipulations and analyses were done using the Limma R Bioconductor package [9]. Initially, four AD samples were removed as they were determined to be outliers. We adjusted for confounders to account for age, gender, and sex variables on expression. To do this, we used a robust linear regression model for covariate corrections as rlm
| (1) |
and the residuals were used for differential gene expression. For each gene, differential expression between two conditions was assessed by a two-tailed t-test, using a threshold P-value of 0.05. All P-values were corrected using the Benjamini and Hochberg False Discovery Rate (FDR) [10].
DNA methylation library preparation
Bisulfite conversion of 500ng of genomic DNA was achieved with the EZ DNA Methylation Gold kit (Zymo Research). DNA samples were prepared according to the Illumina© Infinium protocol and run on the Illumina© Infinium HumanMethylation450 bead chip.
RNA-Seq analysis
Libraries were sequenced on the Illumina HiSeq2000 with an average of ~50 million 100bp paired-end reads being sequenced per library. Reads were aligned using GSNAP software and only unique reads were used in analyses [4]. To ensure the greatest percentage of aligned reads, bar codes were clipped off the sequencing read prior to alignment. Strandedness was assigned based on the Script-seq library protocol.
To examine expression and splicing differences, reads were assembled to transcripts from the Gencode v15 database to generate count data for each transcript using SAMtools [5]. To reduce noise, only transcripts with a count above five were used in subsequent analysis. Transcriptional differences were determined using the DESeq2 software [6]. Splicing differences were resolved using the DEXSeq v1.12.0 software [4]. To accurately assess splicing, only genes of four or more exons were examined, which resulted in a total of 17,076 genes evaluated for splicing differences. The Kolmogorov–Smirnov test was used to examine differences in exon distribution. Both of these analysis, as well as downstream analysis, were carried out using R software, version 3.1.0 (http://www.r-project.org).
DNA methylation analysis
DNA methylation was performed using the IMA (Illumina Methylation Analyzer) R package available from bioconductor [7]. All default parameters were used except that the β-value, which is the ratio of methylation probes to both un-methylated and methylated probes, was quantile normalized. The CpG sites and CpG islands were then assessed for differential methylation using a generalized linear model.
Quantitative RT-PCR
Quantitative rtPCR was used to validate RNA-Seq findings. RNA from all three groups was reverse transcribed with SuperScript®III (Invitrogen). 5-50ng of cDNA was used per reverse transcription PCR (rt-PCR) reaction. Q-PCR was performed using isoform specific Taqman® Assays. The housekeeping genes, ACTB, GAPDH, and GUSB were used for normalization. All of the housekeeping genes utilized for this study were located at least 55 Mb from the closet AD loci. Fold differential expression between LOAD cases and both controls were calculated using the delta-delta Ct method (LightCycler®480 Software, Version 1.5).
Results
Expression in the twenty loci
Using RNA-Seq, we examined expression in post mortem human brain tissue from 10 LOAD cases and 10 normal controls (Table 1). A total of 100 genes are located throughout the 100kb surrounding the twenty loci, 87 with detectable expression (Table 2). The gene located closest to the most strongly associated variant has, by convention, been used to refer to each locus (Supplemental Table 1). Comparison of the 87 genes between LOAD and normal controls revealed nine genes that were differentially expressed in LOAD after correcting for multiple testing using false discovery rate (FDR < 0.1). These genes were CNN2 within the ABCA7 locus, CR1 and a lincRNA within the CR1 locus, MS4A14, MS4A7, MS46E within the MS4A6A locus, TRIM35 within the PTK2B locus, and C7orf61 and TSC22D4 within the ZCWPW1 locus (Table 2).
Table 2.
All genes located within ±100kb of the twenty LOAD associated variants.
| Locus | Variant Position | RefSeq Gene Name | LOAD | CON | DLB | LOAD vs. CON | LOAD vs. DLB | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Avg. Expression | Avg. Expression | Avg. Expression | log2FC | P-value | adj.p | log2FC | P-value | adj.p | |||
| ABCA7 | chr19:1063443 | ABCA7 | 69.74 | 60.04 | 58.06 | 0.209 | 0.486 | 0.867 | 0.197 | 0.514 | 0.756 |
| ABCA7 | chr19:1063443 | AC011558 | 4.21 | 0.97 | 1.42 | −0.346 | 0.342 | 0.792 | −0.176 | 0.626 | 0.828 |
| ABCA7 | chr19:1063443 | CNN2 | 20.63 | 7.1 | 12.52 | 1.137 | 0.002 | 0.076 | 1.169 | 0.002 | 0.036 |
| ABCA7 | chr19:1063443 | GPX4 | 7.26 | 7.67 | 8.76 | −0.621 | 0.024 | 0.278 | −0.665 | 0.016 | 0.126 |
| ABCA7 | chr19:1063443 | GRIN3B | 10.08 | 9.44 | 6.9 | 0.365 | 0.342 | 0.792 | 0.277 | 0.475 | 0.863 |
| ABCA7 | chr19:1063443 | HMHA1 | 16.14 | 20.09 | 19.74 | 0.120 | 0.679 | 0.928 | 0.127 | 0.663 | 0.850 |
| ABCA7 | chr19:1063443 | POLR2E | 131.13 | 197.75 | 127.07 | −0.484 | 0.125 | 0.565 | −0.521 | 0.099 | 0.336 |
| ABCA7 | chr19:1063443 | TMEM259 | 207.63 | 213.95 | 160.49 | −0.007 | 0.984 | 0.996 | −0.059 | 0.866 | 0.948 |
| ABCA7 | chr19:1063443 | SBNO2 | 117.21 | 71.87 | 115.35 | 0.626 | 0.025 | 0.283 | 0.658 | 0.019 | 0.138 |
| BIN1 | chr2:127892810 | BIN1 | 1465.96 | 1236.07 | 1135.52 | 0.227 | 0.429 | 0.841 | 0.207 | 0.474 | 0.728 |
| BIN1 | chr2:127892810 | CYP27C1 | 10.08 | 9.44 | 6.9 | 0.086 | 0.816 | 0.956 | 0.024 | 0.948 | 0.982 |
| BIN1 | chr2:127892810 | RP11-521O16.1 | NA | NA | NA | Not Expressed | |||||
| CASS4 | chr20:55018260 | AL121914.1 | NA | NA | NA | Not Expressed | |||||
| CASS4 | chr20:55018260 | CASS4 | 83.11 | 67.66 | 110.58 | 0.288 | 0.214 | 0.687 | 0.320 | 0.168 | 0.438 |
| CASS4 | chr20:55018260 | CSTF1 | 97.44 | 91.23 | 101.49 | 0.064 | 0.705 | 0.937 | 0.066 | 0.694 | 0.865 |
| CASS4 | chr20:55018260 | FAM209A+RTFDC1 | 554.33 | 810.41 | 504.93 | −0.506 | 0.018 | 0.239 | −0.525 | 0.014 | 0.116 |
| CASS4 | chr20:55018260 | GCNT7 | 328.78 | 417.78 | 324.59 | −0.327 | 0.061 | 0.421 | −0.333 | 0.056 | 0.249 |
| CD2AP | chr6:47487762 | AL355353.1 | NA | NA | NA | Not Expressed | |||||
| CD2AP | chr6:47487762 | CD2AP | 535.25 | 468.8 | 482.4 | 0.181 | 0.294 | 0.757 | 0.179 | 0.299 | 0.585 |
| CD2AP | chr6:47487762 | GPR111 + GPR115 | 1.52 | 0.45 | 6.13 | 0.911 | 0.018 | 0.238 | 0.920 | 0.018 | 0.133 |
| CD33 | chr19:51727962 | C19orf75 | NA | NA | NA | Not Expressed | |||||
| CD33 | chr19:51727962 | CD33 | 20.13 | 12.78 | 23.95 | 0.507 | 0.115 | 0.548 | 0.577 | 0.075 | 0.292 |
| CD33 | chr19:51727962 | CTD-3187F8.11+CTD-3187F8.12 | 1.52 | 0.45 | 6.13 | −0.014 | 0.941 | 0.987 | −0.013 | 0.948 | 0.982 |
| CD33 | chr19:51727962 | CTD-3187F8.14 | 7.26 | 7.67 | 8.76 | −0.095 | 0.795 | 0.954 | −0.061 | 0.869 | 0.949 |
| CD33 | chr19:51727962 | ETFB+CLDN2+CTD-26166J11.11 | 135.27 | 233.4 | 131.1 | −0.632 | 0.054 | 0.401 | −0.684 | 0.038 | 0.201 |
| CD33 | chr19:51727962 | IGLON5 | 554.33 | 810.41 | 504.93 | −0.018 | 0.950 | 0.989 | −0.002 | 0.994 | 0.998 |
| CD33 | chr19:51727962 | SIGLEC22P | NA | NA | NA | Not Expressed | |||||
| CD33 | chr19:51727962 | SIGLEC9 | 33.88 | 24.02 | 43.24 | 0.410 | 0.187 | 0.654 | 0.475 | 0.129 | 0.387 |
| CD33 | chr19:51727962 | SIGLECL1 | 3.96 | 0.74 | 0.2 | 1.219 | 0.006 | 0.134 | 1.114 | 0.014 | 0.500 |
| CD33 | chr19:51727962 | VSIG10L | 17.74 | 13.95 | 26.74 | 0.288 | 0.342 | 0.792 | 0.369 | 0.225 | 0.511 |
| CELF1 | chr11:47557871 | AC093673.5 | 10.14 | 10.33 | 19.24 | −0.130 | 0.697 | 0.934 | −0.012 | 0.972 | 0.990 |
| CELF1 | chr11:47557871 | C1QTNF4 | 25.19 | 31.2 | 29.99 | −0.249 | 0.468 | 0.861 | −0.241 | 0.485 | 0.736 |
| CELF1 | chr11:47557871 | CELF1 | 2128.61 | 2502.02 | 2329.1 | −0.229 | 0.020 | 0.251 | −0.229 | 0.020 | 0.140 |
| CELF1 | chr11:47557871 | FAM180B | 16.14 | 20.09 | 19.74 | −0.203 | 0.552 | 0.891 | −0.181 | 0.597 | 0.810 |
| CELF1 | chr11:47557871 | KBTBD4 | 328.78 | 417.78 | 324.59 | −0.251 | 0.221 | 0.694 | −0.270 | 0.188 | 0.465 |
| CELF1 | chr11:47557871 | KBTBD4 | 43.89 | 60.17 | 38.86 | −0.351 | 0.224 | 0.697 | −0.387 | 0.181 | 0.455 |
| CELF1 | chr11:47557871 | MTCH2 | 332.59 | 377.02 | 306.78 | −0.172 | 0.326 | 0.781 | −0.178 | 0.308 | 0.593 |
| CELF1 | chr11:47557871 | NDUFS3+PTPMT1 | 307.51 | 382.2 | 298.87 | −0.297 | 0.082 | 0.479 | −0.303 | 0.076 | 0.294 |
| CELF1 | chr11:47557871 | RAPSN | 7.53 | 5.79 | 5.96 | 0.264 | 0.527 | 0.883 | 0.250 | 0.552 | 0.890 |
| CELF1 | chr11:47557871 | RN7SL652P | NA | NA | NA | Not Expressed | |||||
| CELF1 | chr11:47557871 | RP11-750H9.7 | NA | NA | NA | Not Expressed | |||||
| CLU | chr8:27467686 | C19orf6 | 207.63 | 213.95 | 160.49 | −0.007 | 0.984 | 0.996 | −0.059 | 0.866 | 0.948 |
| CLU | chr8:27467686 | CLU | 11795.04 | 11132.05 | 11399.36 | 0.080 | 0.662 | 0.924 | 0.080 | 0.663 | 0.850 |
| CLU | chr8:27467686 | SCARA3 | 548.69 | 391.78 | 530.12 | 0.441 | 0.075 | 0.462 | 0.455 | 0.067 | 0.276 |
| CR1 | chr1:207692049 | AL691452 | 0.1407136 | 0 | 0.2097543 | 0.060 | 0.834 | 0.960 | 0.258 | 0.393 | 0.999 |
| CR1 | chr1:207692049 | CR1 | 105.09 | 24.07 | 50.04 | 1.650 | 7.31E-06 | 0.003 | 1.677 | 6.44E-06 | 0.002 |
| CR1 | chr1:207692049 | CR1L+CD46P1 | 37.09 | 23.37 | 43.46 | 0.580 | 0.031 | 0.311 | 0.627 | 0.020 | 0.142 |
| CR1 | chr1:207692049 | CR2 | 6.88 | 2.85 | 9.67 | Not Expressed | |||||
| CR1 | chr1:207692049 | RP11-78B10.2 | 49.5 | 9.47 | 26.03 | 1.674 | 2.44E-05 | 0.006 | 1.759 | 1.27E-05 | 0.002 |
| EPHA1 | chr7:143110762 | CLCN1 | 11.11 | 13.4 | 16.96 | −0.282 | 0.366 | 0.806 | 0.217 | 0.485 | 0.736 |
| EPHA1 | chr7:143110762 | EPHA1 | 29.99 | 26 | 26.5 | 0.229 | 0.455 | 0.856 | 0.227 | 0.462 | 0.720 |
| EPHA1 | chr7:143110762 | EPHA1-AS1 | 28.95 | 36.18 | 27.2 | −0.237 | 0.381 | 0.814 | −0.256 | 0.344 | 0.628 |
| EPHA1 | chr7:143110762 | TAS2R41 | NA | NA | NA | Not Expressed | |||||
| EPHA1 | chr7:143110762 | TAS2R60 | 0 | 0.155 | 0.408 | −0.280 | 0.362 | 0.803 | 0.000 | 0.999 | 1.000 |
| EPHA1 | chr7:143110762 | ZYX | 232.27 | 231.44 | 248.32 | 0.006 | 0.983 | 0.996 | 0.014 | 0.958 | 0.985 |
| FERMT2 | chr14:53400629 | FERMT2 | 1027.22 | 899.73 | 943.86 | 0.182 | 0.322 | 0.779 | 0.181 | 0.325 | 0.610 |
| HLA | chr6:32578530 | HLA-DQA1 | 338.02 | 334.7 | 343.81 | 0.402 | 0.230 | 0.704 | 0.366 | 0.278 | 0.566 |
| HLA | chr6:32578530 | HLA-DQB1-AS1 | 29.99 | 26 | 26.5 | 0.663 | 0.095 | 0.509 | 0.701 | 0.081 | 0.303 |
| HLA | chr6:32578530 | HLA-DQB1+BPG254F23.6 | 28.95 | 36.18 | 27.2 | 0.031 | 0.936 | 0.986 | 0.053 | 0.892 | 0.959 |
| HLA | chr6:32578530 | HLA-DRB1 | 135.27 | 233.4 | 131.1 | 0.248 | 0.437 | 0.847 | 0.241 | 0.452 | 0.713 |
| INPP5D | chr2:234068476 | ATG16L1 | 654.5 | 762.19 | 659.43 | 0.154 | 0.743 | 0.946 | −0.228 | 0.635 | 0.990 |
| INPP5D | chr2:234068476 | INPP5D | 1027.22 | 899.73 | 943.86 | 0.296 | 0.203 | 0.673 | 0.301 | 0.197 | 0.476 |
| MEF2C | chr5:88223420 | MEF2C | 5828.35 | 7852.58 | 6726.79 | −0.410 | 0.026 | 0.285 | −0.411 | 0.026 | 0.162 |
| MS4A6E | chr11:59923508 | AP001257 | NA | NA | NA | Not Expressed | |||||
| MS4A6E | chr11:59923508 | MS4A14 + MS4A7 + MS4A6E | 387.37 | 147.35 | 379.33 | 1.105 | 0.001 | 0.052 | 1.199 | 0.000 | 0.015 |
| MS4A6E | chr11:59923508 | MS4A2 | NA | NA | NA | Not Expressed | |||||
| MS4A6E | chr11:59923508 | MS4A3 | NA | NA | NA | Not Expressed | |||||
| MS4A6E | chr11:59923508 | MS4A4A | 74.82 | 45.71 | 58.75 | 0.390 | 0.156 | 0.407 | 0.555 | 0.152 | 0.421 |
| MS4A6E | chr11:59923508 | MS4A4E | 41.41 | 18.94 | 32.98 | 0.749 | 0.075 | 0.462 | 0.814 | 0.056 | 0.249 |
| MS4A6E | chr11:59923508 | MS4A6A | 116.54 | 65.41 | 140.01 | 0.599 | 0.103 | 0.525 | 0.707 | 0.056 | 0.248 |
| NME8 | chr7:37841534 | NME8+GPR141+EPDR1 | 679.45 | 652.88 | 577.82 | 0.054 | 0.769 | 0.949 | 0.047 | 0.799 | 0.918 |
| PICALM | chr11:85867875 | EED | NA | NA | NA | Not Expressed | |||||
| PICALM | chr11:85867875 | PICALM | 3970.26 | 2907.2 | 3646.87 | 0.423 | 0.031 | 0.309 | 0.428 | 0.029 | 0.175 |
| PKT2B | chr8:27195121 | PTK2B | 2251.03 | 3099.49 | 3213.15 | −0.438 | 0.044 | 0.363 | −0.425 | 0.051 | 0.237 |
| PKT2B | chr8:27195121 | TRIM35 | 136.21 | 192.66 | 134.92 | −0.482 | 0.009 | 0.168 | −0.491 | 0.008 | 0.083 |
| SLC24A4 | chr14:92926952 | SLC24A4 | 934.57 | 1278.8 | 1191.77 | −0.429 | 0.040 | 0.347 | −0.424 | 0.042 | 0.213 |
| SORL1 | chr11:121435587 | SORL1 | 5264.46 | 6457.45 | 5391.24 | −0.288 | 0.022 | 0.264 | −0.290 | 0.021 | 0.145 |
| ZCWPW1 | chr7:100004446 | C7orf61+TSC22D4 | 337.4 | 182.56 | 210.02 | 0.812 | 0.003 | 0.085 | 0.797 | 0.003 | 0.051 |
| ZCWPW1 | chr7:100004446 | CTB-161A2.3+PILRB+PILRA+CTB-161A2.4 | 1078.42 | 1256.73 | 1079.03 | −0.202 | 0.370 | 0.807 | −0.208 | 0.356 | 0.639 |
| ZCWPW1 | chr7:100004446 | MEPCE | 127.05 | 157.81 | 155.73 | −0.272 | 0.373 | 0.810 | −0.258 | 0.399 | 0.672 |
| ZCWPW1 | chr7:100004446 | PPP1R35 | 7.72 | 8.15 | 8.06 | 0.023 | 0.951 | 0.989 | 0.028 | 0.942 | 0.980 |
| ZCWPW1 | chr7:100004446 | ZCWPW1 | 164.06 | 181.56 | 188.61 | −0.125 | 0.586 | 0.901 | −0.118 | 0.608 | 0.818 |
A total of 82 genes were examined for differences in expression, splicing, and methylation in LOAD when compared to normal and disease controls. Of these 82, expression was only detected in 66 genes. Genes that overlap are denoted as gene1+gene2. Differences in expression between the cohorts was determined using DEseq. Differential gene expression was examined between LOAD vs. normal controls (LOADvsCON) and LOAD vs. the disease controls (LOADvsDLB). The direction of expression is given by the log2fold-change (log2FC) with the exppression fold-change being relative either the normal control, under the LOAD vs. CON, or the disease control, under the LOAD vs. DLB.. THe adjusted p-value (adj.p) was determined using FDR. Genes with an asterisk (*) are significant (FDR<0.1).
To determine whether the differential expression of the nine genes is LOAD-specific and not a secondary result of the neurodegenerative process, we compared expression of these nine genes in LOAD to that observed in DLB samples from the temporal pole, which we term “disease controls.” Expression of C7orf61, CNN2, CR1, MS4A14, MS4A7, MS46E and a lincRNA (RP11-78B10.2) within the CR1 locus, were statistically different in LOAD when compared to both normal and disease controls within our RNA-Seq data
Verifying RNA-Seq results
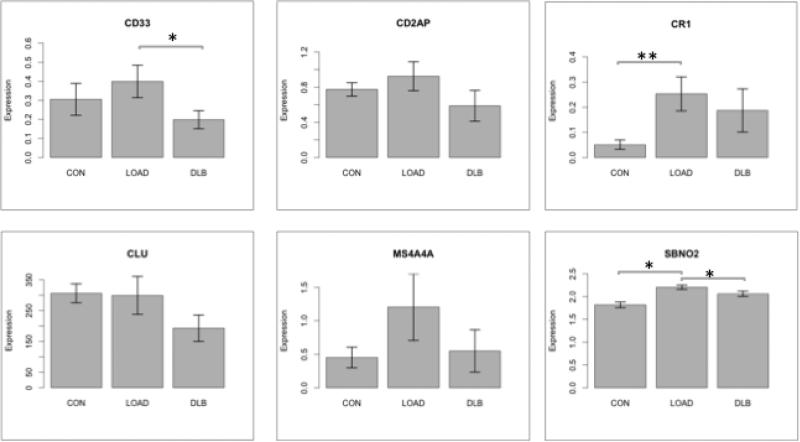
To confirm the RNA-Seq findings, six genes were validated using qRT-PCR. The direction of differential expression of all six genes was consistent with that observed using the RNA-Seq data (Figure 1). The qRT-PCR results of one gene, CR1, had a similar direction of expression; however, it varied in significance. While CR1 differed significantly between LOAD and both normal and DLB controls in the RNA-Seq data, CR1 only differed significantly between LOAD and normal controls in the qRT-PCR results (Figure 1). There was a similar increase in CR1 in LOAD when compared to DLB, indicating that CR1 is differentially expressed in LOAD; however, these findings may be due to the increased sensitivity of RNA-Seq. In summary, qRT-PCR results verify our RNA-Seq findings.
Figure 1. qPCR results verify RNA-Seq findings.
We validated RNA-Seq results by performing qPCR on several genes within the 20 loci. Ten samples were used for each cohort. Differential expression observed in the RNA-Seq results correlated with the qPCR results. Genes expression differences were determined using the Wilcox-Rank Sum test (*:p<0.05; **:p<0.005). CON: Normal controls; LOAD: Late-onset Alzheimer disease; DLB: Dementia with Lewy Bodies.
Replicating RNA-Seq results
To examine if our transcriptional changes overlapped with previous findings, we evaluated publicly available microarray data (GSE15222) from autopsied prefrontal cortical tissue of 163 LOAD cases and 196 controls[16]. Expression values were normalized and corrected for sex, age, and post mortem interval (Table 1). These values were then tested for differential expression between LOAD and controls.
In the microarray data set, only 52 of the 87 genes had detectable expression. Thirty of these genes were significant for differential expression (FDR < 0.05). Although this transcriptome analysis was performed in another brain region, we found that five of the nine genes with altered expression in LOAD were similarly altered in LOAD samples in the prefrontal cortex (Supplemental Table 2). The lincRNA (RP11-78B10.2) within the CR1 locus, MS4A14, and C7orf61 were undetectable. While TRIM35 and MS4A6A were not differentially expressed, their direction of expression was similar to the genes’ expression in our original data set. These findings replicate most of the expression changes observed in our study as well as suggest that the differential expression in LOAD can be observed in at least 2 different brain regions.
Splicing in LOAD
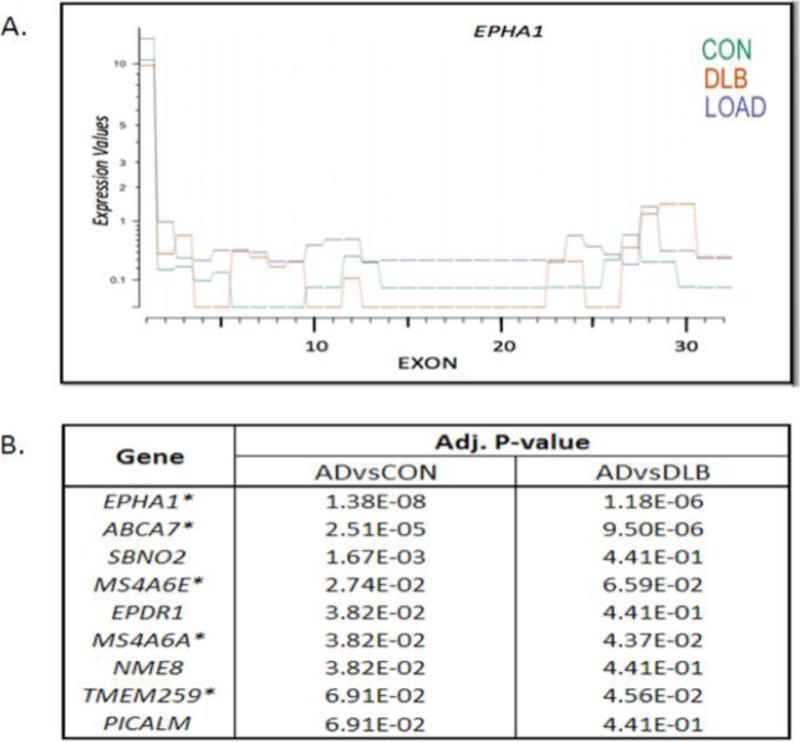
Splicing differences can give rise to proteins that vary in structure and as a result can differentially influence cellular processes [17]. We examined splicing in two different ways: significant changes in the use of individual exons and differences in exon distribution across the gene. Expression differences of individual exons within the 20 loci were observed in LOAD; however, no individual exon was significant after FDR (data not shown). While no individual exon was significantly altered in LOAD, differences in overall exon distribution would reveal if several exons throughout the gene were altered in LOAD, which would be suggestive of isoform expression differences (Figure 2A). At this time, no established method exists to test isoform expression using count data; therefore, we examined this by using the Kolmogorov-Smirnov test to determine if the normalized exon values in LOAD differed from controls. This test showed that ABCA7, TMEM259, EPDR1, EPHA1, MS4A6A, MS4A6E, NME8, PICALM, and SBNO2 have differential exon distributions when comparing LOAD to normal controls (FDR < 0.1) (Figure 2B).
Figure 2. Five genes have LOAD specific differences in overall exon distribution.
(A) EPHA1 has the biggest difference in overall exon distribution in LOAD when compared to both controls (test: Kolmogorov–Smirnov test, p-value <0.001). The exons are represented on the x-axis and the pre-normalized expression level is on the y-axis. LOAD is purple, normal controls (CON) is green, and DLB is orange (A). The nine genes with overall exon distribution are listed the nine genes in Fig2B. The first comparison is between LOAD and normal controls (ADvsCON). All nine genes with differential exon distribution between LOAD and normal controls were then compared to DLB (ADvsDLB). Genes with an asterisk indicate those that were differentially spliced in LOAD when compared to both normal and disease controls.
Using the disease controls, we conducted the same analysis to examine differential exon distribution between LOAD and DLB to identify LOAD specific splicing changes. Of the nine genes found to differ between LOAD and normal controls, five genes ABCA7, TMEM259, EPHA1, MS4A6A, and MS4A6E had differing exon distribution when compared LOAD to disease controls (Figure 2B). These findings suggest that differential splicing in multiple exons across these genes are specific to LOAD, at least when compared to DLB.
Overall DNA methylation is drastically altered in LOAD associated loci
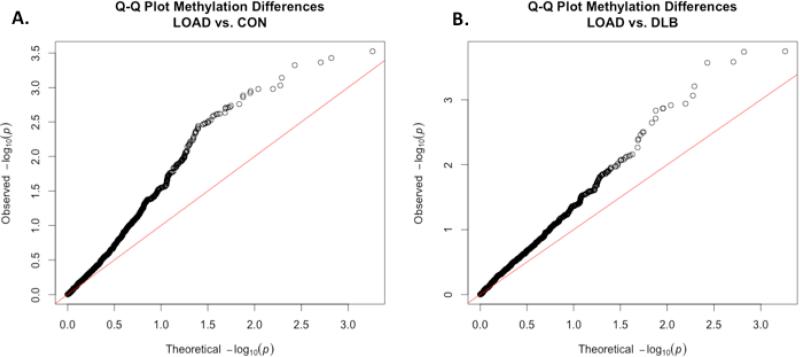
DNA methylation is a known regulator of transcription and can affect transcription of multiple genes; thus, we examined each locus for significant disruptions of methylation[18]. A comprehensive analysis of DNA methylation was performed using the Illumina 450K methylation bead array in eight of the same LOAD cases, eight DLB samples, and eight normal controls.
After quality control, a total of 1,324 CpG sites were identified within the twenty loci. A Q-Q plot of the p-values generated from the comparisons of LOAD to normal controls revealed widespread methylation differences across the loci (Figure 3A). Because q-q plots are generally performed on genotype data, we wanted to make sure this deviation in p-values was due to DNA methylation and not an artifact of the approach. Examination of DNA methylation when comparing samples within the same cohort (e.g. 4 LOAD samples vs. 4 LOAD samples) revealed that the number of CpG sites that differed were in accordance with the expected number, the number of tests * 0.05. Thus, the differences observed when comparing LOAD to CON and DLB samples above are likely to be real (Supplemental Figure 1).
Figure 3. Methylation differences in 20 gene loci.
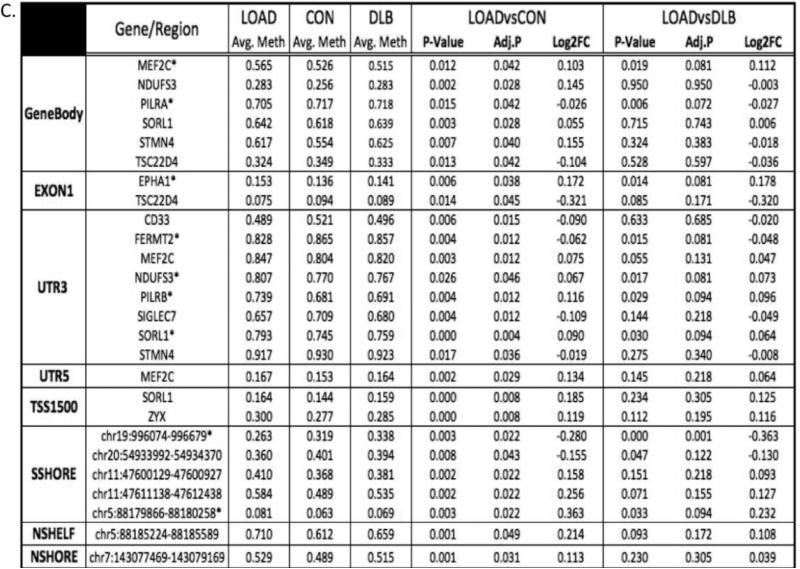
1324 CpG sites were detected within the 20 loci. Comparison of these sites between LOAD and normal controls (A) and LOAD and DLB (B) revealed that a significant proportion of CpG sites within the loci have p-values that are lower than expected under the null., suggesting there is wide-spread changes in DNA methylation within the loci. Figure 3C displays the CpG sites that are differentially methylated in LOAD when compared to normal controls. LOAD specific changes in methylation were identified after comparing these same sites between LOAD and DLB. LOG2FC is the fold-change in DNA methylation of LOAD relative either the normal control, under the LOAD vs. CON, or the disease control, under the LOAD vs. DLB. A linear model was used to determine differences in methylation (P-value). The adjusted p-value (ADJ.P) was determined using FDR. Genes with an asterisk (*) are significant in LOAD when compared to both normal and disease controls (FDR<0.1).
Because DNA methylation located in the promoter region is known to regulate gene expression, we examined correlation of promoter-based methylation with gene expression across the 20 loci. On average, correlation between DNA methylation located in the promoter regions and gene expression in LOAD (r =−0.013) across the 20 loci was slightly altered (student's t-test; p-value= 0.06) when compared to the correlation observed in normal controls (r = −0.151). However, the correlation between DNA-methylation and gene expression in DLB (r = -0.021) was similar to that observed in LOAD (student's t-test; p= 0.59).
In total, we identified ten loci with differences in DNA methylation within LOAD: ABCA7, CASS4, CELF1, CD33, EPHA1, FERMT2, MEF2C, PTK2B, SORL1, and ZCWPW1. Closer examination revealed CpGs within twelve genes and five CpG islands within these loci to have significant differential methylation in LOAD after FDR between LOAD and normal controls. Interestingly, eight of the twelve genes with CpG differences were located within the 3’UTR (Figure 3C), a phenomenon known to regulate transcription elongation and stabilize splicing [19].
We then compared the DNA methylation differences observed above in LOAD to disease controls to determine whether the changes were specific to LOAD. DNA methylation differed across the twenty loci when comparing LOAD to the disease controls (Figure 3B). Of the twelve loci with differential methylation between LOAD and normal controls, six loci remained differentially methylated when compared to disease controls (Figure 3C). Six of the genes in these loci were differentially methylated in LOAD (FDR<0.1) when compared to DLB: MEF2C, NDUFS3, FERMT2, PILRA, PILRB, and SORL1. The biggest methylation difference was observed in the CpG island shore (chr19:1045074-1045679) located in a non-coding region within the ABCA7 locus. The CpG shore was hypomethylated in LOAD compared to both normal and disease controls (FDR<0.05).
Discussion
Recent GWAS have shed light on variants associated with LOAD. While APOE has known coding variants directly associated with LOAD, most of the twenty closely associated variants identified in the recent large-scale GWAS studies fall outside coding regions [9]. The absence of known direct effects on the genes makes it difficult to interpret how these variants influence the disease state. It is currently unknown whether the GWAS associated variants directly contribute to LOAD or are in linkage disequilibrium with the disease causing variant(s). It has been hypothesized that GWAS variants located outside of coding regions may play regulatory roles, such as altering expression, DNA methylation, or splicing [20]. The recent ENCODE project, for example, found that up to 54% of non-coding variants in previous GWAS studies overlap regulatory regions [21-23]. To understand which genes and processes are contributing to LOAD, this study examined expression, methylation, or splicing across the 20 associated loci for changes specific to LOAD.
The ABCA7 locus, the locus with the largest effect size after APOE [9], had the greatest amount of differential expression, methylation, and splicing changes (Figure 4). Interestingly, the gene with the strongest change in expression within this locus CNN2 (Calponin 2), is flanked by two genes with differences in exon distribution: ABCA7 (ATP-binding cassette member 7) and TMEM259 (transmembrane protein 259). In addition to the increased expression of CNN2, overall expression in the ABCA7 locus is increased in LOAD. Because methylation is often inversely correlated with expression, the decreased methylation of the CpG island shore (chr19: 1045074-1045679) located 49kb downstream from the (rs3764650) ABCA7 variant suggests that there may be a correlation to the increased expression observed across the loci. Previous findings suggest that the three genes with LOAD specific differences are involved in similar processes: ABCA7 and TMEM259 are thought to be involved in host-defense mechanisms [24]. Interestingly, ABCA7 is involved in the host-defense system through phagocytosis, a process that necessitates cellular structure reorganization of actin [25, 26] and CNN2 functions in the structural organization of actin filaments [27]. Together, these findings suggest that major disruption in regulatory processes is occurring within the ABCA7 locus and that several genes within this locus could be contributing to LOAD etiology.
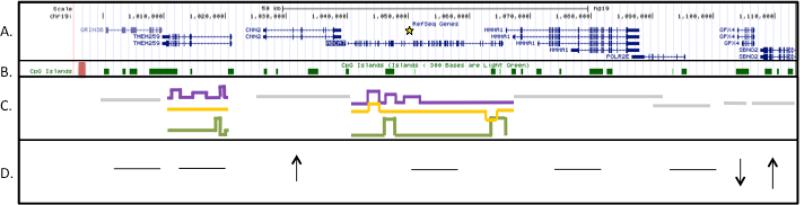
Figure 4. Illustration of expression, methylation, and splicing differences across the ABCA7 locus.
Panel A displays the eight genes located within the ABCA7 locus and the LOAD associated SNP (rs115550680) is marked by the yellow star (A). In panel B, a CpG Island upstream of ABCA7 has LOAD specific hypomethylation and is displayed as a red rectangle (B). Panel C displays the LOAD specific splicing differences identified in TMEM259 and ABCA7. Genes with splicing differences are in color(C). LOAD (purple); disease control (yellow); CON (green); Genes with no difference (grey). LOAD specific expression differences of three genes are observed in this locus (D): CNN2 is significantly increased (adj.p<0.05), a moderate decrease of GPX4 (p-value<0.05), and the moderate increase of SBNO2 expression (p-value<0.05) are illustrated in panel D. An arrow located underneath the gene indicates the direction of LOAD specific expression of the gene.
Similar to the ABCA7 locus, the MS4A6A locus contains several genes that are involved in immune-related processes and have LOAD specific changes. All of the genes within the MS4A6A loci belong to the membrane-spanning 4-domain family. Expression of MS4A14, MS4A7, and MS4A6E is increased in LOAD. In addition, splicing of MS4A6A and MS4A6E is specifically altered in LOAD. Interestingly, the strongest variant within this locus (rs983392) lies in between MS4A6A and the other genes, suggesting that several genes within this locus are contributing to LOAD.
LOAD specific changes are only observed within one gene in the CELF1 locus. The NDUFS3, NADH-ubiquinone oxidoreductase fe-s protein 3 gene has a LOAD specific increase in methylation within the 3’UTR. Moreover, NDUFS3 expression is moderately decreased (p<0.05) in LOAD. Interestingly, NADH-ubiquinone oxidoreductase fe-s protein 3 (NDUFS3) is a subunit of the mitochondrial complex 1 that is disrupted within the brains of patients with Down syndrome and patients with LOAD [28, 29].
Similar to the CELF1 locus, the ZCWPW1 locus does not have differential expression of the ZCWPW1 (zinc finger, CW type with PWWP domain 1) gene itself. Methylation differences specific to LOAD are observed in the PILRA and PILRB genes within this locus; increased methylation was observed in a the CpG sites located within the gene body of PILRA and a CpG sites located on the 3’UTR of PILRB has decreased methylation. No significant expression differences of PILRA or PILRB were observed in LOAD; however, two genes of unknown function, TSC22D4 (TSC22 domain family protein 4) and C7ORF61, are significantly increased in LOAD. Interestingly, paired immunoglobulin-like type 2 receptor, alpha (PILRA) and beta (PILRB) help regulate cell-to-cell signaling, a process significantly decreased in LOAD [30, 31].
Notable differences were observed within five loci. In the EPHA1 loci, overall exon distribution of EPHA1 (EPH receptor A1) and methylation within the 1st exon differed in LOAD when compared to both controls. Methylation differences were observed in the FERMT2 and SORL1 loci. Methylation within the 3’UTR region was significantly increased in SORL1 (Sortilin-related receptor) and decreased in FERMT2 (Fermitin family homolog) in LOAD when compared to both controls. In addition to SORL1 having increased methylation, the expression of SORL1 was decreased, as seen in previous studies [14, 15]. LOAD specific differences in the MEF2C locus consist of decreased methylation within the gene body of ME2FC (Myocyte enhancer factor 2C) and a shore off a CpG island downstream of MEF2C. Interestingly, MEF2C gene expression was moderately decreased in LOAD. The CR1 locus contains two genes with significant expression after FDR. Increased expression of the first gene, CR1 (complement component (3b/4b) receptor 1), in LOAD has been previously observed [32]; however, our utilization of a disease control allows us to show this increase is LOAD specific. The second gene with increased expression in the CR1 locus is an ncRNA (RP11-78B10.2) of unknown function. Interestingly, this ncRNA overlaps a region that is thought to be a repeat region of CR1 that is expanded in LOAD [32]. Taken together, LOAD specific changes in these five indicate that loci the genes located closest to the variant are likely the LOAD contributing gene
The expression and splicing differences found are likely to be those genes with the strongest effect on expression and splicing given our stringent requirement for at least five mappable reads. While we observed DNA methylation differences across the 20 loci in LOAD, there was only a slight difference in promoter based methylation and expression. Our findings replicated previously observed expression changes of CR1 and SORL1; however, expression differences of other genes found to differ in LOAD samples did not (e.g. ABCA7, BIN1, CD33, and CLU). Because we observe similar changes of these four genes in the microarray dataset, it is thought that the lack of differences may be due to sample size and that with an increase of sample size, it might be possible to identify additional LOAD specific differences.
The utilization of Dementia with Lewy bodies (DLB) as a “disease control” allowed us to distinguish between the changes within the loci that were LOAD specific from those that resulted from the neurodegenerative process. DLB is a neurodegenerative disease and its hallmark pathological feature is the accumulation of Lewy bodies within the brain. Some of this accumulation occurs in the same areas of the brain affected by αβ plaques, including the temporal lobe [33]. While the pathology of DLB and LOAD differs, the two diseases share many similarities and phenotypic characteristics[34].
In addition to LOAD and DLB sharing many characteristics, they also shared disruptions in some of the same genes across the 20 loci. Interestingly, six of the CpG sites located nearest the variant were similarly methylated in both LOAD and DLB samples. These six consisted of CpG sites near CELF1, EPHA1, CD33, CASS4, PTK2B, ZCWPW1 in the CELF1, EPHA1, CD33, CASS4, PTK2B, ZCWPW1 loci, respectively. Similar changes in splicing were also observed in SBNO2, MS4A7A, NME8, and PICALM of the ABCA7 MS4A6E, NME8, and PICALM locus, respectively. The similarities between LOAD and DLB tend to suggest that certain genes within these loci may play a role in the overall neurodegenerative process.
We were able to identify expression, methylation, and splicing of genes within eighteen of the twenty loci. Of these eighteen loci, eight had differences in expression, methylation, and/or splicing between LOAD and normal controls. By using a disease control, we were able to distinguish between expression and methylation changes due to LOAD verses the more general neurodegenerative process and pinpoint the changes specifically altered in LOAD. Interestingly, ABCA7, CELF1, MS4A6A, and ZCWPW1 loci had LOAD specific changes in expression, methylation, and splicing in genes that were not located closest to the variant. These findings suggest that genes in addition to, or in place of, the gene located closest to the variant may also be contributing to LOAD. This seems most evident in the ABCA7 and MS4A6A loci. In addition, the slight increase in methylation observed across all 20 loci, suggest the possibility that GWAS studies may have identified a combination of genes that contribute to LOAD. Characterization of these changes through the 20 loci has given insight into the disrupted processes that are occurring throughout the loci and as a result may be directly contributing to LOAD.
Supplementary Material
Highlights.
Examined expression and DNA methylation differences in the twenty AD GWAS loci
Identified LOAD specific changes in eight of the twenty AD GWAS loci
LOAD specific changes in five genes and one CpG island identified within the ABCA7 locus
LOAD specific differences observed in the CR1, EPHA1, FERMT2, MEF2C, and SORL1 loci
Acknowledgments
We would like to thank Krista John-Williams, Ryan Gentry, and Simone Clark for technical support. We would also like to acknowledge the Brain Endowment Bank at the University of Miami for the tissue samples. We are grateful to Dr. Holly Cukier for critically reading the manuscript. This work was supported by the NIH aging grant 1R01AG027944 and the Alzheimer's association grant IRG-09-1333827.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All authors report no conflicts of interest.
References
- 1.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Archives of general psychiatry. 2006;63(2):168–74. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 2.Corder E, Saunders A, Strittmatter W, Schmechel D, Gaskell P, Small G, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 3.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. JAMA: the journal of the American Medical Association. 1997;278(16):1349–56. [PubMed] [Google Scholar]
- 4.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nature genetics. 2009;41(10):1088–93. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nature genetics. 2011;43(5):436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nature genetics. 2011;43(5):429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jun G, Naj AC, Beecham GW, Wang L-S, Buros J, Gallins PJ, et al. Meta-analysis confirms CR1, CLU, and PICALM as Alzheimer disease risk loci and reveals interactions with APOE genotypes. Archives of neurology. 2010 doi: 10.1001/archneurol.2010.201. archneurol. 2010.201 v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert J-C, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nature genetics. 2009;41(10):1094–9. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 9.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45(12):1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer's disease brains. PloS one. 2012;7(11):e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, et al. Alzheimer's Disease Risk Gene< i> CD33</i> Inhibits Microglial Uptake of Amyloid Beta. Neuron. 2013 doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ling I-F, Bhongsatiern J, Simpson JF, Fardo DW, Estus S. Genetics of Clusterin Isoform Expression and Alzheimer's Disease Risk. PloS one. 2012;7(4):e33923. doi: 10.1371/journal.pone.0033923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brouwers N, Van Cauwenberghe C, Engelborghs S, Lambert J, Bettens K, Le Bastard N, et al. Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Molecular psychiatry. 2011;17(2):223–33. doi: 10.1038/mp.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy JJ, Saith S, Linnertz C, Burke JR, Hulette CM, Welsh-Bohmer KA, et al. The Alzheimer's associated 5′ region of the< i> SORL1</i> gene< i> cis</i> regulates< i> SORL1</i> transcripts expression. Neurobiology of aging. 2012;33(7):1485. e1–. e8. doi: 10.1016/j.neurobiolaging.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, et al. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. Journal of Biological Chemistry. 2007;282(45):32956–64. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- 16.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, et al. A survey of genetic human cortical gene expression. Nature genetics. 2007;39(12):1494–9. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 17.Thomas-Chollier M, Watson LC, Cooper SB, Pufall MA, Liu JS, Borzym K, et al. A naturally occuring insertion of a single amino acid rewires transcriptional regulation by glucocorticoid receptor isoforms. Proceedings of the National Academy of Sciences. 2013;110(44):17826–31. doi: 10.1073/pnas.1316235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aran D, Sabato S, Hellman A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013;14(3):R21. doi: 10.1186/gb-2013-14-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi JK, Bae JB, Lyu J, Kim TY, Kim YJ. Nucleosome deposition and DNA methylation at coding region boundaries. Genome Biol. 2009;10(9):R89. doi: 10.1186/gb-2009-10-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol. 2012;30(11):1095–106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Degner JF, Pai AA, Pique-Regi R, Veyrieras J-B, Gaffney DJ, Pickrell JK, et al. DNase [thinsp] I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482(7385):390–4. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464(7289):768–72. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M. Linking disease associations with regulatory information in the human genome. Genome research. 2012;22(9):1748–59. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Wang L, Berman M, Kong YY, Dorf ME. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35(3):426–40. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka N, Abe-Dohmae S, Iwamoto N, Yokoyama S. Roles of ATP-binding cassette transporter A7 in cholesterol homeostasis and host defense system. J Atheroscler Thromb. 2011;18(4):274–81. doi: 10.5551/jat.6726. [DOI] [PubMed] [Google Scholar]
- 26.May RC, Machesky LM. Phagocytosis and the actin cytoskeleton. J Cell Sci. 2001;114(Pt 6):1061–77. doi: 10.1242/jcs.114.6.1061. [DOI] [PubMed] [Google Scholar]
- 27.Hossain MM, Crish JF, Eckert RL, Lin JJ, Jin JP. h2-Calponin is regulated by mechanical tension and modifies the function of actin cytoskeleton. J Biol Chem. 2005;280(51):42442–53. doi: 10.1074/jbc.M509952200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leuner K, Schulz K, Schutt T, Pantel J, Prvulovic D, Rhein V, et al. Peripheral mitochondrial dysfunction in Alzheimer's disease: focus on lymphocytes. Mol Neurobiol. 2012;46(1):194–204. doi: 10.1007/s12035-012-8300-y. [DOI] [PubMed] [Google Scholar]
- 29.Kim SH, Fountoulakis M, Dierssen M, Lubec G. Decreased protein levels of complex I 30-kDa subunit in fetal Down syndrome brains. J Neural Transm Suppl. 2001;(61):109–16. doi: 10.1007/978-3-7091-6262-0_9. [DOI] [PubMed] [Google Scholar]
- 30.Mousseau DD, Banville D, L'Abbe D, Bouchard P, Shen SH. PILRalpha, a novel immunoreceptor tyrosine-based inhibitory motif-bearing protein, recruits SHP-1 upon tyrosine phosphorylation and is paired with the truncated counterpart PILRbeta. J Biol Chem. 2000;275(6):4467–74. doi: 10.1074/jbc.275.6.4467. [DOI] [PubMed] [Google Scholar]
- 31.Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004;101(7):2173–8. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brouwers N, Van Cauwenberghe C, Engelborghs S, Lambert JC, Bettens K, Le Bastard N, et al. Alzheimer risk associated with a copy number variation in the complement receptor 1 increasing C3b/C4b binding sites. Mol Psychiatry. 2012;17(2):223–33. doi: 10.1038/mp.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iseki E. Dementia with Lewy bodies: reclassification of pathological subtypes and boundary with Parkinson's disease or Alzheimer's disease. Neuropathology. 2004;24(1):72–8. doi: 10.1111/j.1440-1789.2003.00530.x. [DOI] [PubMed] [Google Scholar]
- 34.Gomez-Tortosa E, Irizarry MC, Gomez-Isla T, Hyman BT. Clinical and neuropathological correlates of dementia with Lewy bodies. Ann N Y Acad Sci. 2000;920:9–15. doi: 10.1111/j.1749-6632.2000.tb06899.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.