Abstract
Enhancers control the timing, location and expression levels of their target genes. Nucleotide variation in enhancers has been shown to lead to numerous phenotypes, including human disease. While putative enhancer sequences and nucleotide variation within them can now be detected in a rapid manner using various genomic technologies, the understanding of the functional consequences of these variants still remains largely unknown. Massively parallel reporter assays (MPRAs) can overcome this hurdle by providing the ability to test thousands of sequences and nucleotide variants within them for enhancer activity en masse. Here, we describe this technology and specifically focus on how it is being used to obtain an increased understanding of enhancer regulatory code and grammar.
Keywords: Enhancer, Massively parallel reporter assays, Transcriptional regulation
1. Introduction
Enhancers regulate the location, timing and levels of gene transcription. Enhancers can be located in non-coding sequences and also in coding exons [1, 2] and can regulate their target gene/s both in cis and in trans (on another chromosome) [3, 4]. Enhancers were also shown to be transcribed, termed as enhancer RNA (eRNA), and it is thought that their transcribed sequence might contribute to their function [5–7]. Nucleotide variation in enhancers has been shown to lead to a multitude of phenotypes, including morphological differences between species [8] and human disease [9]. For example, the majority of disease-associated genome wide association studies (GWAS) hits fall in non-coding regions of the human genome [10, 11].
Enhancers themselves are thought to be regulated by the binding of transcription factors, which are expressed in a cell-type specific manner, to particular DNA motifs within their sequence. Functional transcription factor binding sites (TFBS) tend to be clustered and conserved among species [12–15]. In addition, transcription factors recruit histone acetyltransferase (HAT), such as EP300 and CBP to enhancers. These proteins are thought to affect the chromatin environment by acetylating lysine residues in core histone tails. Amongst these marks, histone H3 acetyl Lys27 (H3K27ac) was shown to correlate with active enhancers [16–18]. H3K4me1 was shown to correlate with both active and poised enhancers [16–18]. Transcription factor binding and histone modifications can be used to identify potential enhancers in a genome-wide manner through chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq). Enhancers that associate with active marks and transcription factors are located in open chromatin [17, 19]. This trait can also be used to identify enhancers via technologies such as DNase-seq, FAIRE-seq, and ATAC-seq [20–23]. While these and other genomic technologies can efficiently identify potential enhancer sequences in a genome-wide manner, they are primarily descriptive and the sequences they detect are not necessarily functional enhancers.
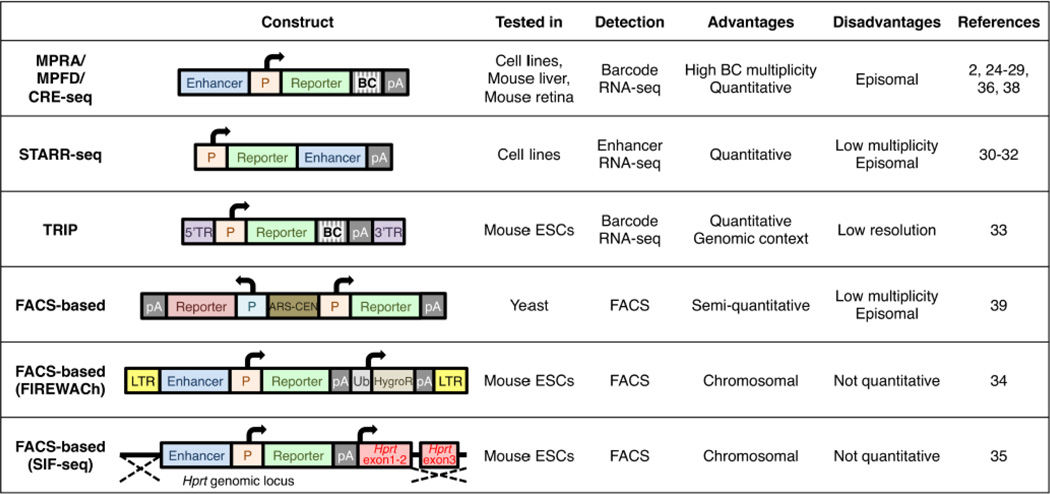
To validate enhancer function, an experimental assay needs to be carried out. Enhancers are generally characterized by a reporter assay that links a candidate enhancer sequence to a minimal promoter (a promoter that is not sufficient to drive reporter expression without a functional enhancer) and a reporter gene (GFP, LacZ, luciferase or others). The reporter vectors are introduced into cell lines or organisms, and the reporter gene expression is examined. If the candidate sequence acts as an enhancer, it should activate the minimal promoter and the reporter gene expression in the tissue/cell type of interest. However, in this ‘classic’ method, enhancer activity is examined in a ‘one by one’ manner, and is thus low-throughput and time-consuming. This is especially limiting with the current genomic revolution that is producing in a rapid manner genome-wide enhancer prediction datasets and human whole-genome sequences with thousands of potential phenotype causing nucleotide variants whose function needs to be evaluated. Recently developed high-throughput technologies that include massively parallel reporter assays (MPRA [2, 24, 25]), MPFD [26], CRE-seq [27–29], STARR-seq [30–32], TRIP [33], FIREWACh [34] and SIFseq [35], can enable us to overcome this obstacle (Fig. 1). In this review, we will focus on MPRA, MPFD and CRE-seq that share a basic methodology. These three methods will be mentioned as “MPRA” in this review.
Fig. 1.
Comparison of plasmid construct, assay model, strategy of reporter detection and advantage and disadvantages of different MPRA technologies [2, 24–36, 38, 39]. ARS-CEN, yeast autonomously replicating sequence and centromere; BC, barcode; HygroR, hygromycin-resistance gene; LTR, long terminal repeat; P, promoter; pA, polyadenylation signal; TR, terminal repeat, Ub, ubiquitin promoter.
2. Massively parallel reporter assays
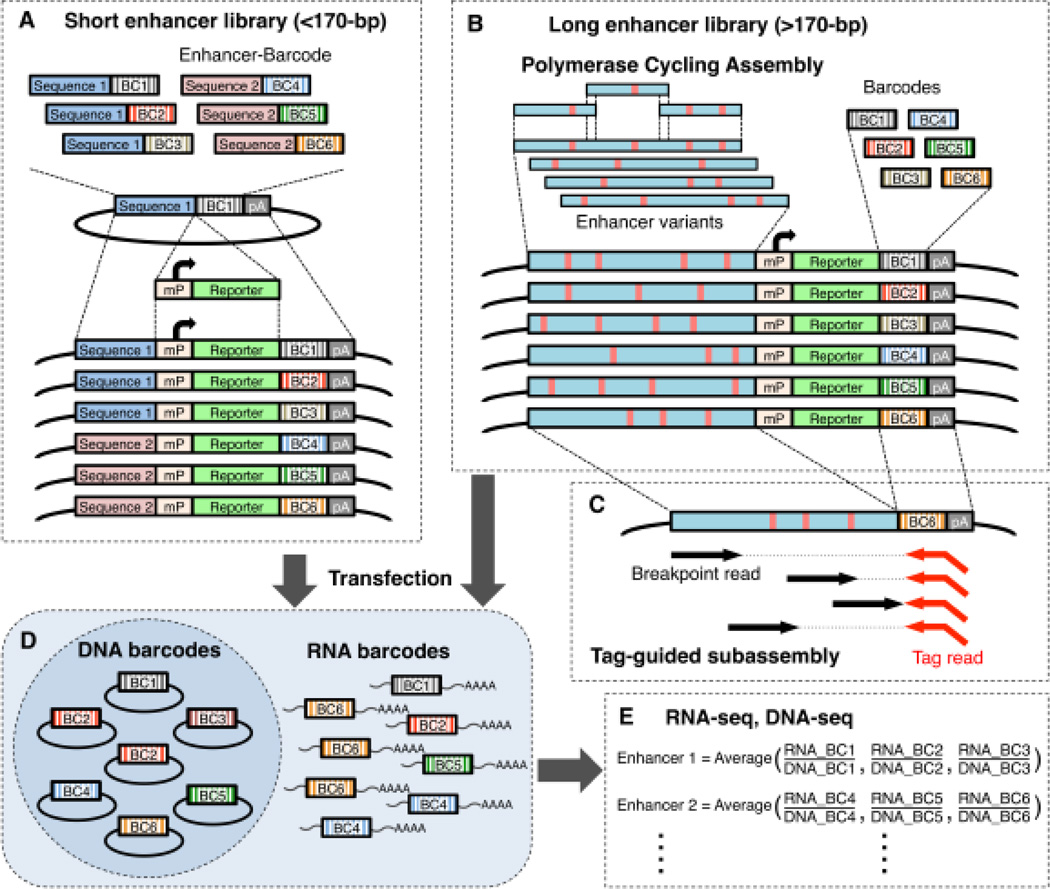
MPRA is a high-throughput technology that enables the analysis of transcriptional activities of thousands of regulatory elements in a single experiment (Fig. 2A-E). The principal of this technology was first developed by Patwardhan et al. in 2009 for promoter assays [36]. In this study, saturation mutagenesis of three bacteriophage promoters (T3, T7, and SP6) was carried out in vitro. All possible point mutations and small indels of these promoters were assayed for their function by synthesizing in parallel, on a programmable microarray, the promoters along with a transcriptional start site that was followed by a specific 20-bp of ‘barcode’ sequence. Thus, if the sequence is a promoter it will lead to the transcription of the barcode sequence. Each mutant variant was coupled to six distinct barcodes to provide a quantitative measurement for the activity of each variant, and assisted in reducing barcode-dependent bias, such as a contingent gain of transcriptional activity in the barcode sequence. Following in vitro transcription of the promoter-barcode library, transcribed barcodes were then sequenced via RNA-seq to serve as a single readout for the differential activity of distinct promoter variants. This assay was also used to test three mammalian core promoters (CMV, HBB, and S100A4) in HeLa nuclear extract, and successfully detected mutations and deletions that caused a significant drop in efficiency of transcription within essential regions of these promoters, such as the TATA box and the initiator element.
Fig. 2.
Overview of the massively parallel reporter assay. (A) To generate a short enhancer MPRA library, candidate enhancer sequences and barcode sequences are synthesized on a programmable microarray and cloned into a plasmid vector. Minimal promoter (mP) and EGFP reporter gene are inserted between the enhancer and barcode. (B) Polymerase cycling assembly is used to generate a long enhancer library. Long enhancer variants (red vertical lines) and barcodes are separately inserted upstream of mP or within the 3’ UTR of EGFP gene, respectively. (C) To associate enhancer and barcodes, the downstream end of the enhancer and upstream end of the barcode are digested and re-ligated to bring them adjacent to one another, followed by sequencing with tag-guided subassembly. (D) The MPRA library is introduced into cell lines or tissues. Transcribed barcode RNAs are extracted from the cells. (E) The relative transcriptional activities of distinct enhancers are measured by sequencing and counting their corresponding barcode RNAs. The counts of transcribed RNA barcodes are normalized by DNA barcode counts that are from the library. BC, barcode; mP, minimal promoter; pA, polyadenylation signal.
3. Massively parallel enhancer saturation mutagenesis
Similar to the promoter mutagenesis experiments, MPRA technology was next used to analyze via saturation mutagenesis the function of three liver enhancers [ALDOB (hg19: chr9: 104195570-104195828), ECR11 (hg19: chr2: 169939082-169939701) and LTV1 (mm9: chr7: 29161443-29161744)] [26]. A library of >100,000 enhancer variants was synthesized and cloned upstream of a minimal promoter and a luciferase reporter gene. A 20-bp barcode was pre-cloned in the 3’ UTR of the reporter gene, resulting in each enhancer haplotype being randomly coupled to a distinct barcode. The library was then introduced into the mouse liver via hydrodynamic tail vein injection [37], livers were dissected after 24 hours and RNA-seq was carried out to determine the enhancer function of the various haplotypes. Transcribed barcode counts were normalized by DNA barcode counts from the plasmid library. These assays found that most enhancer mutations have modest effects on enhancer activity, and that a TFBS was often but not always concordant with their function, emphasizing the importance of direct experimental characterization.
A similar assay by another group was also used to analyze the effect of all possible mutations or deletions of two inducible enhancers [synthetic cAMP-regulated enhancer (CRE) and the enhancer of the IFNB gene (hg19: chr9: 21077976-21078062)] on transcriptional activity in HEK293T cells [25]. They identified mutations and indels that alter the enhancer activity within CREB binding sites in the CRE enhancer, as well as ATF-2/c-Jun, IRFs, and NF-κB binding sites in the IFNB enhancer. Furthermore, they trained quantitative sequence-activity models (QSAMs) using the MPRA data, and demonstrated that these QSAMs can be combined to compare the activity of the enhancers in their induced and uninduced states.
Following these assays, additional MPRA saturation mutagenesis assays have been published. Kwasnieski et al focused on the RhoCRE3 enhancer (mm9: chr6: 115881830-115881881), which regulates mouse Rho gene expression in rod photoreceptors, and examined the effects of single nucleotide variants on transcription in the mouse retina [28]. They found that the majority (86%) of single nucleotide substitutions showed significant effects on enhancer activity. Changes of the activities were explained not only by mutations within putative TFBS but also by complex phenomena, including transcription factor competition and TFBS turnover during evolution, as they found gain-of-function mutations that re-create CRX binding sites that are conserved among mammals but disrupted in rodents.
In another study, the enhancer activity of three coding exons that also function as enhancers, [SORL1 exon 17 (hg19: chr11: 121424477-121425074), TRAF3IP2 exon 2 (hg19: chr6: 111912400-111913095), PPARG exon 6 (hg19: chr11: chr3: 12447098-12447677)], termed eExons, was dissected in mouse liver and HeLa cells [2]. In these assay, both synonymous and non-synonymous mutations showed similar effects on enhancer activity and many of the deleterious mutation clusters overlapped known liver-associated TFBS, demonstrating that mutation in eExons could lead to multiple phenotypes by disrupting both the protein sequence and enhancer activity. Interestingly, the mutation profile of these eExons was different in HeLa cells when compared to mouse liver, demonstrating that sequences that function as enhancers in multiple tissues could have different operating profiles in each tissue. This would suggest that mutations in these enhancers could cause different phenotypes depending on their location in the enhancer.
4. Decoding using synthesized enhancers/promoters
MPRA can also be used in an opposing manner to saturation mutagenesis, testing for example thousands of synthetic sequences for their enhancer activity, thereby obtaining a better understanding of the ‘enhancer code’. In one such experiment, 4,970 synthetic regulatory element sequences (SRESs), which harbor different combinations of 12 liver-associated TFBS was tested [38]. This experiment assayed the effect of TFBS number, spacing, combination and order on enhancer function. In terms of number, it was observed that for some TFBS, having more copies increases enhancer activity, but not for all. Sequences having different TFBS (heterotypic) were stronger than sequences that had the same TFBS (homotypic). For pairs of two different TFBS, some combinations were favored than others, while one combination (HNF1A and XBP1) actually interfered with one another. As for order and spacing, flexibility was observed, supporting a ‘billboard model’, in which heterotypic TFBS clusters constitute a flexible mechanism for fine-tuning robust gene expression.
A similar experiment was also carried out in yeast. Sharon et al. used fluorescence-based high-throughput reporter assays [39]. In their method, a mixed barcoded oligonucleotide pool was inserted upstream of a core promoter followed by the yellow fluorescent protein (YFP) gene to generate a library, and transformed yeast were sorted using fluorescence activated cell sorting (FACS) based on YFP intensity to obtain transcriptionally active sequences. They analyzed 6,500 synthesized sequences, and found that TFBS number, location, and affinity were important for the activity, while orientation was flexible in most cases. They also found a ~10-bp periodicity of TFBS location for the transcriptional activity.
5. Massively parallel testing of thousands of enhancer candidates
MPRA has also been utilized to gain an increased understanding of the enhancer code by examining the enhancer activity of thousands of different candidate enhancer sequences. In one such study, 2,104 ENCODE-predicted enhancers and 3,314 enhancer variants that contain targeted motif disruptions were studied in HepG2 and K562 cells [24]. This study found that disruption of TFBS of selected transcription activators (HNF1, HNF4 and FOXA in HepG2; GATA and NFE2L2 in K562) led to reduced activity in the matched cell-type (i.e. disruption of HNF1 motif caused reduced expression in HepG2, but no effect in K562), while disruption of repressor motifs (GFI1 in HepG2 and ZFP161 in K562) showed an effect only in the unmatched cell type (i.e. disruption of GFI1 motif caused misexpression in K562, but no effect in HepG2). This finding suggested an ‘enhancer activator and repressor model’, in which the cell-type specificity of enhancers is maintained by the combined action of activators that are expressed and bind in the matched cell type, and repressors that are expressed and bind in the unmatched cell type. It also showed that evolutionary conservation, nucleosome exclusion, binding of other factors, and strength of the motif all correlated with enhancer activity.
In another study, 2,100 randomly chosen regulatory elements, which were predicted as enhancer, weak enhancer, or repressed in K562 and/or H1 human embryonic stem cells based on histone modification data from the ENCODE Project were tested [27]. They showed that only ~26% of the ENCODE enhancer predictions portrayed gene regulatory activity. They also showed that the weak enhancer predictions led to stronger expression levels than did enhancer predictions. This discrepancy between predictions was partially explained by their finding that lower levels of H3K27ac and H3K36me3 (a histone mark of transcribed genes) were associated with higher enhancer activity in their assay. These results highlight the need to carry out these functional assays so as to better improve these predictions. In another MPRA experiment, 1,298 Crx-bound regions were compared to 3,035 control sequences in explanted mouse retina. They demonstrated that Crx-bound regions with high GC contents drove expression, whereas unbound sequences with low GC contents did not, even when liberated from their larger genomic context, suggesting the importance of local sequence features for transcriptional activation rather than genomic context [29]. Combined, these reports demonstrate the ability of the MPRA technology to functionally test thousands of different enhancer predictions, obtain a better understanding of their code and grammar and refine these predictions.
6. MPRA caveats
While MPRAs are becoming extremely efficient in obtaining a better understanding of regulatory elements in a high-throughput manner, they have several caveats. Many MPRAs take advantage of oligonucleotide arrays to synthesize their assayed sequence. In these arrays, the current maximum length of the synthesized sequences is usually 200-bp. This limitation is problematic, since enhancers could be longer in length and thus they would miss out on potentially important sequences. In the saturation mutagenesis experiment, one way around that was to use the polymerase cycling assembly, in which overlapping oligonucleotides (~90-bp) that contain a programmed level of degeneracy were synthesized and assembled to construct long enhancer variants that contained multiple substitutions (Fig. 2B). Using this technique longer enhancers could be assayed (>600-bp) [2, 26]. Substitutions in the long enhancer variants were determined by tag-guided subassembly, which enabled full-length and high-accuracy sequencing of individual enhancer variants in association with their downstream barcodes [40] (Fig. 2C). In this method, a long fragment library is converted to a population of nested sub-libraries, and a tag sequence guides grouping of short reads derived from the same long fragment, enabling localized assembly of long fragments. However, it is not currently feasible to test thousands of variants in thousands of long enhancers (>200bp) using this method. DNA synthesis technologies that can allow the generation of longer sequences at a cost efficient manner could overcome this problem.
MPRA results include a certain level of false positives and false negatives because of technical problems including synthesis, sequencing, and enhancer-barcode association errors. It is thus important to carry out multiple replicate experiments, include positive and negative control sequences in MPRA libraries, and compare the level of reporter gene expression to that of controls. Once such example is the use of scrambled controls as done by White et al who compared the activity of 1,298 enhancer candidates to 3,035 scrambled DNA controls [29]. The scrambled DNA sequences produced distinct levels of reporter expressions that can overlap with the activity of many functional sequences. From this they concluded that enhancer function couldn’t be assessed solely by applying a threshold level of activity [29]. In addition, to further confirm MPRA results, individual validation of a few selected enhancers should also be carried out. Patwardhan et al for example, tested six enhancer variants chosen from their MPRA result for their activities individually in the mouse liver and showed high correlation of the individual data and MPRA [26]. An alternative method to MPRA is STARR-seq, in which candidate enhancer sequences are subcloned 3’ to a reporter gene, and if functional enhancers activate the transcription of themselves, thus DNA synthesis and enhancer-barcode association is not required (Fig. 1). However, because each enhancer induces only one readout (the enhancer itself), unlike other MPRAs that use multiple barcodes per enhancer, STARR-seq does not provide high multiplicity of enhancer activity if coverage is low for a specific sequence and can be biased by enhancer RNA stability.
Most of the MPRAs are carried out in an episomal manner and thus may not acquire chromatin marks or be influenced by other intrinsic factors. The development of MPRA methods that allow for genomic integration such as lentivirus libraries or embryonic stem cell (ESC) recombination, which were previously employed in similar MPRA studies, such as FIREWACh [34] or SIF-seq [35] (Fig. 1), can allow for enhancer assays in a chromosomal context. TRIP, a high-throughput enhancer trap technology, allows enhancer analysis in genomic context [33]. However, these studies could not provide specific quantitative measurements.
Since enhancers tend to be tissue-specific, it is important to try and carry out MPRA for these sequences in a homogenous cell population, if possible. Applying sorting technologies, such as FACS, microfluidics, and Transcribed Ribosome Affinity Purification followed by sequencing (TRAP-seq) [41], can allow MPRA to be utilized in heterogenic cell populations or complex tissues. Also, although Patwardhan et al successfully used hydrodynamic tail vein injection to introduce an MPRA library into the mouse liver [26], this technique cannot deliver these libraries into other tissues at ample concentrations that could allow MPRA measurements.
Another caveat of the MPRA and also the ‘standard’ one-by-one enhancer assays is that query sequences are removed from their genomic context to construct an artificially designed reporter plasmid. Therefore, the effect of enhancer-promoter distance, looping and the chromosomal environment on transcriptional activity is not taken into account in the MPRA. In addition, most MPRA use a standard minimal promoter that is not the actual promoter that is being targeted by the candidate enhancer. Ultimately, to characterize the functional effects of enhancer mutations, they need to be assayed in their endogenous setting. One potential tool that could be used to edit enhancer sequence in the genomic context is saturation genome editing via CRISPR/Cas9 technology. This technology was recently used to test the effect of every possible mutation in exons of two different genes, BRCA1 and DBR1, within HEK293T and Hap1 cells, respectively [42].
7. Future directions
Enhancer identification assays, such as ChIP-seq, DNase-seq, ATAC-seq, ChIA-PET and others, are only able to highlight whether a certain sequence could be an enhancer. MPRA compensates for these descriptive assays by experimentally characterizing functional regulatory elements in a high-throughput scale, and provides a powerful resource to decode functional regions and nucleotide variants in the human genome and other genomes. This technology can be applied to verify the functionality of annotated regulatory regions, such as TFBS, methylated or unmethylated DNA regions, open chromatin regions, and transposon-derived enhancers. Other than promoters or enhancers, MPRAs can also be adopted to analyze the function of silencers or insulators, of which molecular mechanisms are not well known. Furthermore, translational regulatory elements, including miRNAs, can be also be used in assays that are similar to MPRA. For example, Kosuri et al analyzed the function of ribosome binding sites in E. coli, using FACS-based high-throughput assay [43] and Zhao et al assayed the function of 3’ UTR on RNA stability in human cell lines using ‘fast-UTR’ [44].
MPRAs have the potential to assist numerous biological disciplines such as development, evolution, and disease. Enhancers are thought to be responsible for dynamic expression of key developmental genes. Many developmental enhancers have been identified [45, 46], however, the gene regulatory network that govern embryonic development is yet to be fully characterized. MPRAs can be a useful tool to facilitate genome-wide discovery of developmental enhancers. Morphological differences between species have been shown to be caused also by changes in gene regulatory elements during evolution [47–49]. MPRA could essentially be used to rapidly characterize the impact of nucleotide changes that cause gain, loss, or any enhancer activity changes that could lead to these phenotypic differences. As for human disease, with the rapid identification of nucleotide variation in whole genome datasets, there is an unmet need for the functional characterization of these variants. MPRA could address this need for testing for example all GWAS variants of a certain phenotype, all expression quantitative trait loci (eQTLs) of a certain tissue or even all nucleotide variants in a certain individual.
Highlights.
We describe massively parallel reporter assays.
We describe how these assays are used to decipher the regulatory code and grammar.
We describe caveats and future directions of massively parallel reporter assays.
Acknowledgments
This work was supported in part by the National Human Genome Research Institute grant number 1R01HG006768 and the National Cancer Institute grant number 1R01HG008123.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Fumitaka Inoue, Email: fumitaka.inoue@ucsf.edu.
Nadav Ahituv, Email: nadav.ahituv@ucsf.edu.
References
- 1.Birnbaum RY, Clowney EJ, Agamy O, Kim MJ, Zhao J, Yamanaka T, Pappalardo Z, Clarke SL, Wenger AM, Nguyen L, Gurrieri F, Everman DB, Schwartz CE, Birk OS, Bejerano G, Lomvardas S, Ahituv N. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 2012 doi: 10.1101/gr.133546.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnbaum RY, Patwardhan RP, Kim MJ, Findlay GM, Martin B, Zhao J, Bell RJ, Smith RP, Ku AA, Shendure J, Ahituv N. Systematic dissection of coding exons at single nucleotide resolution supports an additional role in cell-specific transcriptional regulation. PLoS Genet. 2014;10:e1004592. doi: 10.1371/journal.pgen.1004592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 4.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jorgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Muller F, Forrest AR, Carninci P, Rehli M, Sandelin A. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam MT, Li W, Rosenfeld MG, Glass CK. Enhancer RNAs and regulated transcriptional programs. Trends in biochemical sciences. 2014;39:170–182. doi: 10.1016/j.tibs.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahituv N. Gene Regulatory Sequences and Human Disease. New York: Springer; 2012. p. x. 283 pages. [Google Scholar]
- 10.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010;20:565–577. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy S, Hannenhalli S, Workman C. Enrichment of regulatory signals in conserved non-coding genomic sequence. Bioinformatics. 2001;17:871–877. doi: 10.1093/bioinformatics/17.10.871. [DOI] [PubMed] [Google Scholar]
- 14.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 15.Pennacchio LA, Loots GG, Nobrega MA, Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 20.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, Liu Z, London D, McDaniell RM, Shibata Y, Showers KA, Simon JM, Vales T, Wang T, Winter D, Zhang Z, Clarke ND, Birney E, Iyer VR, Crawford GE, Lieb JD, Furey TS. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kheradpour P, Ernst J, Melnikov A, Rogov P, Wang L, Zhang X, Alston J, Mikkelsen TS, Kellis M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23:800–811. doi: 10.1101/gr.144899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Jr, Kinney JB, Kellis M, Lander ES, Mikkelsen TS. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nature biotechnology. 2012;30:271–277. doi: 10.1038/nbt.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, Lee C, Andrie JM, Lee SI, Cooper GM, Ahituv N, Pennacchio LA, Shendure J. Massively parallel functional dissection of mammalian enhancers in vivo. Nature biotechnology. 2012;30:265–270. doi: 10.1038/nbt.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwasnieski JC, Fiore C, Chaudhari HG, Cohen BA. High-throughput functional testing of ENCODE segmentation predictions. Genome Res. 2014;24:1595–1602. doi: 10.1101/gr.173518.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwasnieski JC, Mogno I, Myers CA, Corbo JC, Cohen BA. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc Natl Acad Sci U S A. 2012;109:19498–19503. doi: 10.1073/pnas.1210678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White MA, Myers CA, Corbo JC, Cohen BA. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc Natl Acad Sci U S A. 2013;110:11952–11957. doi: 10.1073/pnas.1307449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold CD, Gerlach D, Spies D, Matts JA, Sytnikova YA, Pagani M, Lau NC, Stark A. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat Genet. 2014;46:685–692. doi: 10.1038/ng.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 32.Shlyueva D, Stelzer C, Gerlach D, Yanez-Cuna JO, Rath M, Boryn LM, Arnold CD, Stark A. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol Cell. 2014;54:180–192. doi: 10.1016/j.molcel.2014.02.026. [DOI] [PubMed] [Google Scholar]
- 33.Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LF, van Lohuizen M, van Steensel B. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell. 2013;154:914–927. doi: 10.1016/j.cell.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 34.Murtha M, Tokcaer-Keskin Z, Tang Z, Strino F, Chen X, Wang Y, Xi X, Basilico C, Brown S, Bonneau R, Kluger Y, Dailey L. FIREWACh: high-throughput functional detection of transcriptional regulatory modules in mammalian cells. Nature methods. 2014;11:559–565. doi: 10.1038/nmeth.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickel DE, Zhu Y, Nord AS, Wylie JN, Akiyama JA, Afzal V, Plajzer-Frick I, Kirkpatrick A, Gottgens B, Bruneau BG, Visel A, Pennacchio LA. Function-based identification of mammalian enhancers using site-specific integration. Nature methods. 2014;11:566–571. doi: 10.1038/nmeth.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nature biotechnology. 2009;27:1173–1175. doi: 10.1038/nbt.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim MJ, Ahituv N. The hydrodynamic tail vein assay as a tool for the study of liver promoters and enhancers. Methods Mol Biol. 2013;1015:279–289. doi: 10.1007/978-1-62703-435-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, Ovcharenko I, Ahituv N. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet. 2013;45:1021–1028. doi: 10.1038/ng.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharon E, Kalma Y, Sharp A, Raveh-Sadka T, Levo M, Zeevi D, Keren L, Yakhini Z, Weinberger A, Segal E. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nature biotechnology. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hiatt JB, Patwardhan RP, Turner EH, Lee C, Shendure J. Parallel, tag-directed assembly of locally derived short sequence reads. Nature methods. 2010;7:119–122. doi: 10.1038/nmeth.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hupe M, Li MX, Gertow Gillner K, Adams RH, Stenman JM. Evaluation of TRAP-sequencing technology with a versatile conditional mouse model. Nucleic Acids Res. 2014;42:e14. doi: 10.1093/nar/gkt995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kosuri S, Goodman DB, Cambray G, Mutalik VK, Gao Y, Arkin AP, Endy D, Church GM. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci U S A. 2013;110:14024–14029. doi: 10.1073/pnas.1301301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao W, Pollack JL, Blagev DP, Zaitlen N, McManus MT, Erle DJ. Massively parallel functional annotation of 3' untranslated regions. Nature biotechnology. 2014;32:387–391. doi: 10.1038/nbt.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, Hansen DV, Nord AS, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, Kaplan T, Kriegstein AR, Rubin EM, Ovcharenko I, Pennacchio LA, Rubenstein JL. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 48.Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2012;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- 49.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]