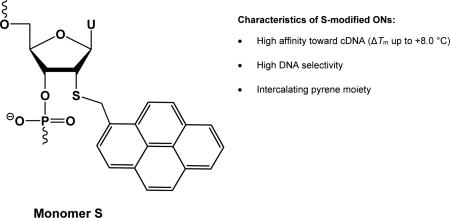
Abstract
Pyrene-functionalized oligonucleotides are intensively explored for applications in materials science and diagnostics. Here, we describe a short synthetic route to 2′-S-(pyren-1-yl)methyl-2′-thiouridine monomer S, its incorporation into oligodeoxyribonucleotides (ONs), and biophysical characterization thereof. Pseudorotational analysis reveals that the furanose ring of this monomer has a slight preference for South-type conformations. ONs modified with monomer S display high cDNA affinity but decreased binding specificity. Hybridization is associated with bathochromic shifts of pyrene absorption bands and quenching of pyrene fluorescence consistent with an intercalative binding mode of the pyrene moiety. Monomer S was also evaluated as a building block for mixed-sequence recognition of double-stranded DNA via the Invader strategy. However, probes with +1 interstrand arrangements of monomer S were found to be less efficient than Invader probes based on 2′-O-(pyren-1-yl)methyluridine or 2′-N-(pyren-1-yl)methyl-2′-N-methyl-2′-aminouridine.
Graphical Abstract
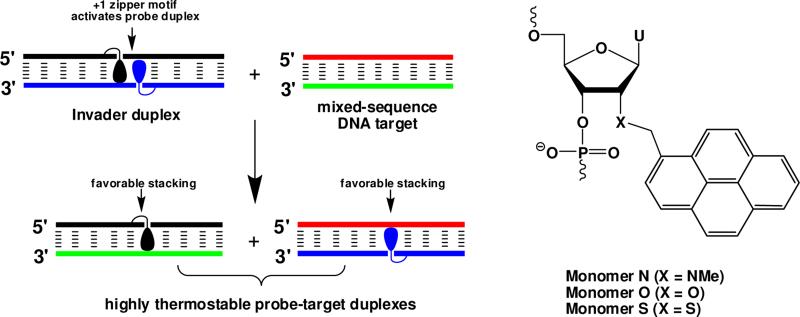
Development of pyrene-functionalized oligonucleotides is an area that continues to attract considerable interest due to the prospect of tools for a range of applications in materials science and diagnostics,1 including generation of self-assembled helical pyrene arrays2 and the development of probes for detection of complementary DNA/RNA3 (cDNA/cRNA) and single nucleotide polymorphisms (SNPs).4 As part of our growing interest in pyrene-functionalized oligonucleotides, we recently introduced an unique approach for recognition of double-stranded DNA (dsDNA),5-7 which is based on double-stranded oligonucleotide probes that are energetically activated through modification with +1 interstrand zipper8 arrangements of pyrenefunctionalized nucleotides such as 2'-O-(pyren-1-yl)methyl RNA or 2′-N-(pyren-1-yl)methyl-2′-N-methyl-2′-amino DNA monomers (Figure 1). This particular motif forces the two pyrene moieties to intercalate into the same region of the probe, leading to local perturbation and duplex destabilization as the ‘nearest neighbor exclusion principle’ is violated.9 In contrast, each of the two probe strands form very stable duplexes with cDNA as the intercalating pyrene moieties are engaged in efficient π-stacking with neighboring base-pairs. This generates a thermodynamic gradient, which, unlike most other hybridization-based strategies,10 allows for recognition of mixed-sequence dsDNA target regions at physiologically relevant conditions.7,11
Figure 1.
Recognition of dsDNA via the Invader strategy and structures of studied monomers. Droplets denote the intercalating pyrene moiety.
Our earlier efforts at optimizing the dsDNA-recognition efficiency of these so-called Invader probes have focused on varying: the number and relative position of the key activating monomers, the nature of the nucleobase and intercalator, and the length of the linker and the orientation between the intercalator and sugar skeleton.6,12-15 In the present work, we set out to study the influence of the 2′-heteroatom of the pyrene-functionalized nucleotide monomer on the dsDNA-recognition characteristics of Invader probes. We hypothesized that the lower electronegativity of the sulfur atom of 2′-S-(pyren-1-yl)methyl-2′-thiouridine monomer S would weaken the gauche effect between O4′ and the 2′-substituent, and thus increase the population of C2′-endo (South-type) furanose conformations.16 This, in turn, was expected to result in more favorable conditions for pyrene intercalation, leading to higher cDNA affinity relative to ONs modified with current-generation Invader building blocks 2′-O-(pyren-1-yl)methyluridine monomer O17 or 2′-N-(pyren-1-yl)methyl-2′-N-methyl-2′-aminouridine monomer N17 (Figure 1).
Here we describe i) a short synthetic route to 2′-S-(pyren-1-yl)methyl-2′-thiouridine phosphoramidite 4 and its incorporation in ONs, and results from ii) coupling constant analyses, which provide insights into conformational preferences of monomer S, iii) thermal denaturation experiments and thermodynamic parameter analysis, iv) UV-Vis absorption and fluorescence experiments, and v) dsDNA-recognition experiments, all discussed in relation to ONs and Invader probes based on O and N monomers.
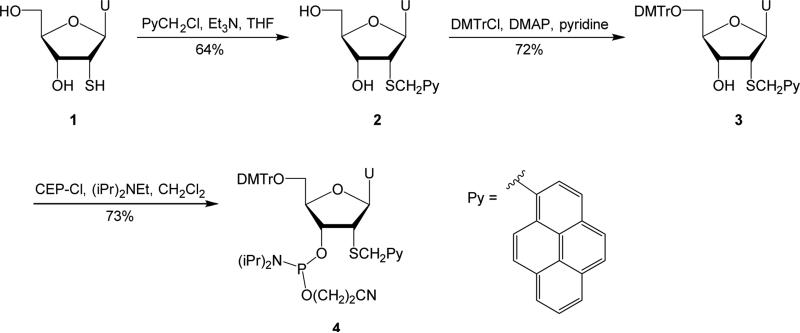
2′-Deoxy-2′-thiouridine 1, which was used as the starting material for the synthesis of phosphoramidite 4 (Scheme 1), was prepared from uridine in ~50% yield over three steps as described in the literature.18 Nucleoside 1 was then alkylated at the S2′-position using 1-pyrenylmethyl chloride under mildly basic conditions,19 to afford nucleoside 2 in 64% yield. Similar yields were obtained when 1-pyrenylmethyl bromide was used as the alkylating agent (results not shown). Standard O5′-DMT protection afforded nucleoside 3 in 72% yield, which was treated with 2-cyanoethyl-N,N-diisopropylchlorophosphoramidite (CEP-Cl) and Hünig's base to give target phosphoramidite 4 in 73% yield.
Scheme 1.
Synthesis of target nucleoside 4. U = uracil-1-yl; DMTr = 4,4′-dimethoxytrityl; DMAP = 4-dimethylaminopyridine; CEP-Cl = 2-cyanoethyl-N,N-diisopropylchlorophosphoramidite.
A coupling constant analysis was performed to determine if the lower electronegativity of the 2′-sulfur of monomer S induces a greater proportion of South type furanose conformations relative to monomers N and O. Thus, 3JHH scalar coupling constants for the endocyclic sugar protons of nucleoside 3 were used as input in a Matlab-based pseudorotational analysis program,20 which facilitates determination of pseudorotation phase angles (P) and puckering amplitudes (ϕm) for five-membered ring systems, by solving modified Karplus-Diez-Donders equations (Table S1).20-22 From this analysis, nucleoside 3 is predicted to have a slight preference for South conformations (P = 143°, ϕm = 38°, %S = 61%), while the corresponding nucleoside of monomer O is predicted to be in two more equally populated conformations, i.e., a North conformation (P = 11°, ϕm = 38°, %N = 51%) and a South conformation (P = 130°, ϕm = 33°; %S = 49%). Interestingly, the corresponding nucleoside of monomer N is predicted to exclusively adopt South type conformations (main conformer P = 145°, ϕm = 38°, 61% frequency; secondary conformer P = 122°, ϕm = 27°), presumably due to additional steric interactions in North conformations between the 2′-N-methyl group and the 3′-oxygen. However, it is important to appreciate that stereoelectronic effects on the nucleoside level may not necessarily fully translate to the oligonucleotide or duplex level. For example, crystal structures of A- and B-type DNA duplexes modified with 2'-S-methyl-uridines show that the modified residues adopt RNA-like C3'-endo puckers, demonstrating that replacement of the electronegative 2′-oxygen by a sulfur, does not fundamentally alter the conformational preference of the sugar in the oligonucleotide context,23 even though these monomers were predicted to adopt DNA-like C2′-endo puckers.24 Ultimately, high-resolution X-ray or solution NMR structures of N-, O-, and S-modified duplexes will be necessary to fully understand the structural underpinnings of the observed trends in thermal denaturation temperatures (Tm) (vide infra).
Phosphoramidite 4 was used in automated solid phase DNA synthesis to incorporate monomer S into ONs using extended hand-coupling times (15 min) and 4,5-dicyanoimidazole as an activator, resulting in stepwise coupling yields of >95%. The identity and purity of the modified ONs was established through MALDI-MS (Table S2) and ion-pair reverse phase HPLC (>90% purity).
Thermal denaturation temperatures of duplexes between S-modified ONs and cDNA/cRNA were determined from thermal denaturation curves recorded in medium salt phosphate buffer and were compared relative to unmodified duplexes, as well as, N- or O-modified duplexes. Duplexes between S-modified ONs and cDNA are considerably more stable than the corresponding unmodified reference duplex (ΔTm between ±0.0 and +8.0 °C, Table 1), while duplexes with cRNA are less stable (ΔTm between -10.0 to -2.0 °C, Table 1). ONs in which monomer S is flanked by 3′-purines form particularly stable duplexes (compare ΔTm's for B2- and B4- series, Table 1), which, together with the observed DNA selectivity (Table S3), are typical observations for ONs modified with intercalating pyrene moieties.12-15,25 Surprisingly, S-modified ONs display lower cDNA/cRNA affinity than N- and O-modified ONs (Tm's lower by 1.5-7.0 °C vs cDNA, and 4.0-6.0 °C vs cRNA, Table 1), most likely due to steric interference of the larger 2′-sulfur atom and/or perturbation of hydration layers at the brim of the minor groove.
Table 1.
ΔTm's of duplexes between B1-B6 and cDNA/cRNA.a
| ΔTm [°C] |
||||||||
|---|---|---|---|---|---|---|---|---|
| + cDNA |
+ cRNA |
|||||||
| ON | Sequence | B = | S | Ob | Nc | S | Ob | Nc |
| B1 | 5′-GBG ATA TGC | +2.0 | +5.0 | +5.0 | −7.0 | −2.0 | −2.0 | |
| B2 | 5′-GTG ABA TGC | +8.0 | +12.5 | +15.0 | −2.0 | +4.0 | +3.0 | |
| B3 | 5′-GTG ATA BGC | +5.0 | +8.0 | +9.0 | −4.5 | ±0.0 | −0.5 | |
| B4 | 3′-CAC BAT ACG | ±0.0 | +3.5 | +1.5 | −10.0 | −4.5 | −6.5 | |
| B5 | 3′-CAC TAB ACG | +8.0 | +11.5 | +15.0 | −2.0 | +2.5 | +3.0 | |
| B6 | 3′-CAC BAB ACG | +8.0 | +14.0 | +14.0 | −10.5 | −1.0 | −3.0 | |
ΔTm = change in Tm relative to reference duplexes D1:D4 (Tm = 29.5 °C), D1:R4 (Tm = 27.5 °C) or R1:D4 (Tm = 27.5 °C), where D1: 5′-GTG ATA TGC, D4: 3′-CAC TAT ACG, R1: 5′-GUG AUA UGC and R4: 3′-CAC UAU ACG. Tm's were determined as the maximum of the first derivative of melting curves (A260 vs T) recorded in medium salt buffer ([Na+] = 110 mM, [Cl−] = 100 mM, pH 7.0 (NaH2PO4/Na2HPO4)), using 1.0 μM of each strand. Tm's are averages of at least two measurements within 1.0 °C. A = adenin-9-yl DNA monomer, C = cytosin-1-yl DNA monomer, G = guanin-9-yl DNA monomer and T = thymin-1-yl DNA monomer. For structures of monomers S, O, and N, see Figure 1.
From reference 17.
From reference 15.
The binding specificity of centrally modified ONs (B2-series) was studied using DNA strands that have mismatched nucleotides opposite to the pyrene-functionalized monomer (Table 2). As it is frequently observed with intercalator-modified ONs,14,17,26 mismatched DNA targets are less efficiently discriminated than with unmodified reference strands. While S2 and O2 display similar binding fidelity, significantly better discrimination is observed with N2. Additional specificity data are presented and discussed in the supporting information (Tables S4 and S5).
Table 2.
Discrimination of mismatched DNA targets by S2/O2/N2 and reference strands.a
| DNA: 3′-CAC TBT ACG |
||||||
|---|---|---|---|---|---|---|
| Tm [°C] | ΔTm [°C] | |||||
| ON | Sequence | B = | A | C | G | T |
| D1 | 5′-GTG ATA TGC | 29.5 | −16.5 | −9.5 | −17.0 | |
| S2 | 5′-GTG ASA TGC | 37.5 | −15.0 | −3.0 | −7.0 | |
| O2b | 5′-GTG AOA TGC | 42.0 | −13.0 | −5.0 | −6.5 | |
| N2b | 5 -GTG ANA TGC | 44.5 | −23.0 | −3.5 | −13.0 | |
For conditions of thermal denaturation experiments, see Table 1. Tm's of fully matched duplexes are shown in bold. ΔTm = change in Tm relative to fully matched duplex
From reference 17.
UV-vis absorption and steady-state fluorescence emission spectra of S-modified ONs, in the presence or absence of cDNA/cRNA targets, were recorded to further ascertain the binding mode of the pyrene moiety, as intercalation is known to induce bathochromic shifts of pyrene absorption bands due to ground-state electronic interactions with nucleobases,27 and nucleobase-mediated quenching of pyrene fluorescence.28 Indeed, hybridization of S-modified ONs with cDNA and cRNA results in bathochromic shifts of the pyrene absorption maxima (Table 3 and Figure S2), although the shifts are smaller than with O- or N-modified ONs, suggesting weaker interactions with nucleobases and less pronounced intercalation.
Table 3.
Absorption maxima in the 300-400 nm region for S/O/N-modified ONs and the corresponding duplexes with cDNA/cRNA.a
|
λmax [Δλmax] /nm |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B = | S | Ob | Nb | ||||||||
| ON | Sequence | SSP | +cDNA | +cRNA | SSP | +cDNA | +cRNA | SSP | +cDNA | +cRNA | |
| B1 | 5′-GBG ATA TGC | 355 | 354 [±0] | 354 [−1] | 350 | 353 [+3] | 352 [+2] | 349 | 353 [+4] | 351 [+2] | |
| B2 | 5′-GTG ABA TGC | 353 | 354 [+1] | 357 [+4] | 348 | 353 [+5] | 352 [+4] | 348 | 353 [+5] | 351 [+3] | |
| B3 | 5′-GTG ATA BGC | 353 | 354 [+1] | 356 [+3] | 350 | 353 [+3] | 352 [+2] | 349 | 353 [+4] | 354 [+5] | |
| B4 | 3′-CAC BAT ACG | 353 | 357 [+4] | 356 [+3] | 350 | 352 [+2] | 352 [+2] | 349 | 354 [+5] | 349 [±0] | |
| B5 | 3′-CAC TAB ACG | 352 | 354 [+2] | 356 [+4] | 349 | 352 [+2] | 352 [+3] | 348 | 354 [+6] | 352 [+4] | |
| B6 | 3′-CAC BAB ACG | 353 | 357 [+4] | 355 [+2] | --- | --- | --- | 348 | 352 [+4] | 347 [−1] | |
Measurements were performed at 5 °C (monomers O and N) or 10 °C (monomer S) using a spectrophotometer and quartz optical cells with 1.0 cm path lengths. Buffer conditions are as in thermal denaturation experiments.
From references 15 and 17.
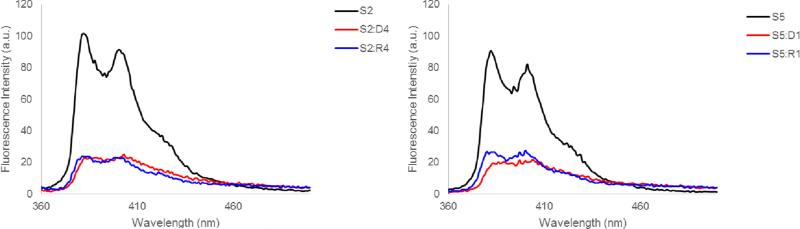
Steady-state fluorescence emission spectra of S-modified ONs and the corresponding duplexes with cDNA/cRNA feature two vibronic bands at λem = 383±1 nm and 401±2 nm, as well as a small shoulder at ~420 nm. As expected for intercalating pyrene moieties, the fluorescence intensity decreases upon hybridization with DNA/RNA targets (Figure 2).
Figure 2.
Steady-state fluorescence emission spectra of representative S-modified ONs and the corresponding duplexes with cDNA/cRNA. Spectra were recorded at T = 10 °C using λex = 350 nm. Each strand was used at 1.0 μM concentration in Tm buffer.
DNA duplexes with different interstrand arrangements of S monomers were studied to determine their suitability for recognition of mixed-sequence dsDNA targets following the Invader strategy. Consistent with our previous observations with other Invader chemistries,6,7,12,14,15 duplexes with +1 interstrand zippers of S monomers are less stable than probes with other zipper arrangements (compare Tm's for S2:S5 relative to other probe duplexes, Table 4). The energetically activated nature of S2:S5 was verified through analysis of thermodynamic parameters, which were obtained from denaturation curves.29 Thus, formation of S2:S5 is considerably less favorable than formation of reference duplexes or probe duplexes with other S-zippers (compare ΔΔG293 values in third ΔG293 column, Table 4). The energetic activation of S2:S5 is enthalpic in origin (ΔΔH = +11 kJ/mol, Table S6), most likely as the nearest neighbor exclusion principle is violated, leading to perturbation of local base pairs. The activated nature of S2:S5 is even more evident when estimating the binding energy for recognition of isosequential dsDNA targets as (dsDNA) where ONA:ONB is a duplex with an interstrand zipper arrangement of monomers. Probes that are activated for dsDNA recognition via the process depicted in Figure 1, display strongly negative values since the products of the recognition process (i.e., probe-target duplexes) are more stable than the reactants (i.e., double-stranded probes and target duplexes). Indeed, much lower values are observed for S2:S5 than for other probe duplexes ( trend: S2:S5<<S2:S4≤S1:S2<S1:S5, Table 4). However, contrary to our initial expectations, S2:S5 is less activated for dsDNA recognition than O2:O5 or N2:N5 , in large part because the probe-target duplexes are significantly less stable (ΔG293 for S2:cDNA and S5:cDNA = −10 kJ/mol, Table 4, compared to ΔG293 for O2:cDNA, O5:cDNA, N2:cDNA and N5:cDNA = −14, −12, −20 and −19 kJ/mol, respectively14,15).
Table 4.
Biophysical properties of S-modified duplexes.a
| ΔG293[ΔΔG293] (kJ/mol) |
||||||||
|---|---|---|---|---|---|---|---|---|
| ON |
ZP |
Sequence |
Tm (°C) |
upper ON vs cDNA |
lower ON vs cDNA |
probe duplex |
(kJ/mol) |
λmax (nm) |
| S1 | +4 | 5′-GSG ATA TGC | 40.5 | −49±0 [−4] | −55±1 [−10] | −59±1 [−14] | ±0 | 353 |
| S5 | 3′-CAC TAS ACG | |||||||
| S1 | +2 | 5′-GSG ATA TGC | 30.5 | −49±0 [−4] | −48±0 [−3] | −46±0 [−1] | −6 | 353 |
| S4 | 3′-CAC SAT ACG | |||||||
| S2 | +1 | 5′-GTG ASA TGC | 25.5 | −55±1 [−10] | −55±1 [−10] | −44±1 [+1] | −21 | 351 |
| S5 | 3′-CAC TAS ACG | |||||||
| S2 | −1 | 5′-GTG ASA TGC | 33.5 | −55±1 [−10] | −48±0 [−3] | −50±0 [−5] | −8 | 355 |
| S4 | 3′-CAC SAT ACG | |||||||
ZP = zipper. For conditions of thermal denaturation and absorption experiments, see Table 1 and Table 3, respectively. ΔΔG293 is measured relative to ΔG293 for D1:D4 = −45 kJ/mol. (ONA:ONB) = ΔG293 (ONA:cDNA) + ΔG293 (cDNA:ONB) - ΔG293 (ONA:ONB) - ΔG293 (dsDNA). “±” denotes standard deviation. For UV/Vis absorption spectra of double-stranded probes, see Figure S4.
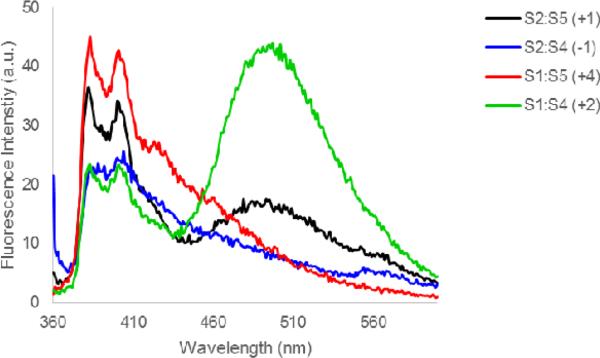
Another characteristic of DNA duplexes with +1 interstrand arrangements of intercalator-modified nucleotide monomers,13-15 which is shared by S2:S5, is the blue-shifted pyrene absorption, which is indicative of reduced pyrene-nucleobase interactions due to locally perturbed duplex geometries (compare λmax for S2:S5 with λmax for other probe duplexes, Table 4, or probe-target duplexes, Table 3). Moreover, steady-state fluorescence emission spectra of S2:S5 (and of +2 zipper probe S1:S4) exhibit prominent and unstructured emission at λem ~490 nm, which is consistent with pyrene-pyrene excimers.30 Probe duplexes with other zipper arrangements do not display prominent emission at λem ~490 nm (Figure 3). Based on our previously published molecular modeling structures of O2:O5,14 we speculate that the two pyrene moieties of S2:S5 co-stack inside the duplex core leading to excimer formation, while the excimer emission of S1:S4 is due to pyrene stacking in the major groove as suggested for other probes with +2 zipper arrangements of intercalator-modified nucleotides.31
Figure 3.
Steady-state fluorescence emission spectra of DNA duplexes with different interstrand monomer arrangements of S. For experimental conditions, see Figure 2.
Our previous studies have shown that efficient dsDNA recognition via the Invader strategy requires probes that are strongly energetically activated ().6,13-15 We therefore selected to evaluate the dsDNA-recognition efficiency of S2:S5 using a 3′-digoxigenin (DIG) labeled DNA hairpin (DH) as a model dsDNA target, which is comprised of a 9-mer double-stranded mixed sequence stem linked by a T10 loop (Figure 4). However, incubation of DH1 with S2:S5 in a HEPES buffer for 12-16 hours at ambient temperatures did not result in formation of slower-migrating recognition complexes on non-denaturing PAGE gels even at 500-fold molar probe excess (Figure 4). This contrasts the observations with O2:O514 and N2L:N515, which result in ~50% dsDNA recognition when used at ~20-fold molar excess, but is consistent with the comparatively low dsDNA-targeting potential of S2:S5 as judged by the values. A similar outcome was obtained when DH1 was annealed in the presence of S2:S5 followed by room temperature incubation (Figure S5), indicating that the recognition complex is not stable at these experimental conditions. Hence, the results suggest that Invader probes based on 2'-O-(pyren-1-yl)methyluridine monomer O or 2′-amino-2′-deoxy-2′-N-(pyren-1-ylmethyl)-2′-N-methyl-uridine monomer N, but not 2′-thio-2′-deoxy-2′-S-(pyren-1-ylmethyl)uridine monomer S, are suitable for dsDNA recognition via the Invader strategy.
Figure 4.
Attempted recognition of model dsDNA target DH1 using Invader S2:S5. (a) Illustration of recognition process. Sequence of DH1: 5′-GTGATATGC-(T10)-GCTTATCACDIG-3′. (b) Representative electrophoretogram upon incubation of DH1 with 1-500 fold molar excess of S2:S5. Experimental conditions for electrophoretic mobility shift assay: separately preannealed targets (34.4 nM) and S2:S5 (variable molar excess) were incubated at ambient temperature for 12-16 h in 1X HEPES buffer (50 mM HEPES, 100 mM NaCl, 5 mM MgCl2, 10% sucrose, 1.4 mM spermine tetrahydrochloride, pH 7.2) and then resolved on 16% nondenaturing PAGE (performed at 70 V, 2.5 h, ~4 °C) using 0.5x TBE as a running buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA); DIG: digoxigenin.
In conclusion, a short, high yielding synthetic route to 2′-thio-2′-deoxy-2′-S-(pyren-1-yl)methyluridine has been developed. Pseudorotational analysis indicates that the furanose ring predominantly predominantly adopts a South-type conformation. ONs modified with these building blocks display prominent cDNA affinity, but less so than corresponding ONs modified with 2'-O-(pyren-1-yl)methyluridine or 2′-N-(pyren-1-yl)methyl-2′-N-methyl-2′-aminouridine. Several observations strongly suggest that the pyrene moiety of the title compound in intercalating into nucleic acid duplexes, including prominent DNA selectivity, decreased thermodynamic mismatch discrimination, and bathochromic shifts of pyrene absorption maxima and quenching of fluorescence upon hybridization with cDNA. Although double-stranded probes with +1 interstrand zipper arrangements of 2′-thio-2′-deoxy-2′-S-(pyren-1-yl)methyluridines are activated for recognition of mixed-sequence dsDNA following the Invader strategy, these probes were not able to recognize a DNA hairpin model target. Nonetheless, 2′-thio-2′-deoxy-2′-S-(pyren-1-yl)methyluridine are an interesting addition to the toolbox of affinity-enhancing building blocks for use in oligonucleotide chemistry.
Supplementary Material
Acknowledgements
This study was supported by Award Number GM088697 from the National Institute of General Medical Sciences, National Institutes of Health. We thank Dr. Alex Blumenfeld (Dept. Chemistry, Univ. Idaho) and Dr. Lee Deobald (EBI Murdock Mass Spectrometry Center, Univ. Idaho) for assistance with NMR and mass spectrometric analysis, and Prof. Carolyn Bohach (Food Science, Univ. Idaho) for access to gel documentation stations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data (all experimental protocols, NMR spectra and additional biophysical characterization data) associated with this article can be found, in the online version, at XXX.
References and Notes
- 1.Østergaard ME, Hrdlicka P. J. Chem. Soc. Rev. 2011;40:5771. doi: 10.1039/c1cs15014f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a Varghese R, Wagenknecht HA. Chem. Commun. 2009:2615. doi: 10.1039/b821728a. [DOI] [PubMed] [Google Scholar]; b Malinovskii VL, Wenger D, Haner R. Chem. Soc. Rev. 2010;39:410. doi: 10.1039/b910030j. [DOI] [PubMed] [Google Scholar]; c Teo YN, Kool ET. Chem. Rev. 2012;112:4221. doi: 10.1021/cr100351g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a Yamana K, Zako H, Asazuma K, Iwase R, Nakano H, Murakami A. Angew. Chem. Int. Ed. 2001;40:1104. doi: 10.1002/1521-3773(20010316)40:6<1104::aid-anie11040>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]; b Østergaard ME, Cheguru P, Papasani MR, Hill RA, Hrdlicka PJ. J. Am. Chem. Soc. 2010;132:14221. doi: 10.1021/ja1057295. [DOI] [PubMed] [Google Scholar]
- 4.a Okamoto A, Kanatani K, Saito I. J. Am. Chem. Soc. 2004;126:4820. doi: 10.1021/ja039625y. [DOI] [PubMed] [Google Scholar]; b Østergaard ME, Kumar P, Baral B, Guenther DC, Anderson BA, Ytreberg FM, Deobald L, Paszczynski AJ, Sharma PK, Hrdlicka P. J. Chem. Eur. J. 2011;17:3157. doi: 10.1002/chem.201002109. [DOI] [PubMed] [Google Scholar]; c Karmakar S, Hrdlicka P. J. Chem. Sci. 2013;4:3447. doi: 10.1039/C3SC50726B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hrdlicka PJ, Kumar TS, Wengel J. Chem. Commun. 2005:4279. doi: 10.1039/b506986f. [DOI] [PubMed] [Google Scholar]
- 6.Sau SP, Kumar TS, Hrdlicka PJ. Org. Biomol. Chem. 2010;8:2028. doi: 10.1039/b923465a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Didion BA, Karmakar S, Guenther DC, Sau SP, Verstegen JP, Hrdlicka PJ. ChemBioChem. 2013;4:3447. doi: 10.1002/cbic.201300414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.See supplementary data for definition of zipper nomenclature.
- 9.Intercalators bind with a maximum loading of one ligand per two base-pairs due to limitations in local helix expandability and/or to avoid disruption of highly stable stacking interactions between nucleobases and the first bound intercalator: Tsai C, Jain SC, Sobell HM. J. Mol. Biol. 1977;114:301. doi: 10.1016/0022-2836(77)90252-2. Williams LD, Egli M, Gao Q, Rich A. Structure and Function. In: Sarma RH, Sarma MH, editors. Nucleic Acids. Vol. 1. Adenine press; 1992. pp. 107–125. Persil O, Hud NV. Trends Biotechnol. 2007;25:433. doi: 10.1016/j.tibtech.2007.08.003.
- 10.a Rogers FA, Lloyd JA, Glazer PM. Curr. Med. Chem.: Anti-Cancer Agents. 2005;5:319. doi: 10.2174/1568011054222300. [DOI] [PubMed] [Google Scholar]; b Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. Nucleic Acids Res. 2008;36:5123. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Nielsen PE. Chem. Biodiv. 2010;7:786. doi: 10.1002/cbdv.201000005. [DOI] [PubMed] [Google Scholar]; d Lohse J, Dahl O, Nielsen PE. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11804. doi: 10.1073/pnas.96.21.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Ishizuka T, Yoshida J, Yamamoto Y, Sumaoka J, Tedeschi T, Corradini R, Sforza S, Komiyama M. Nucleic Acids Res. 2008;36:1464. doi: 10.1093/nar/gkm1154. [DOI] [PMC free article] [PubMed] [Google Scholar]; f Bahal R, Sahu B, Rapireddy S, Lee C-M;, Ly DH. ChemBioChem. 2012;13:56. doi: 10.1002/cbic.201100646. [DOI] [PMC free article] [PubMed] [Google Scholar]; g Bentin T, Larsen HJ, Nielsen PE. Biochemistry. 2003;42:13987. doi: 10.1021/bi0351918. [DOI] [PubMed] [Google Scholar]; h Kaihatsu K, Shah RH, Zhao X, Corey D. Biochemistry. 2003;42:13996. doi: 10.1021/bi035194k. [DOI] [PubMed] [Google Scholar]; i Moreno PMD, Geny S, Pabon YV, Bergquist H, Zaghloul EM, Rocha CSJ, Oprea II, Bestas B, El-Andaloussi S, Jørgensen PT, Pedersen EB, Lundin KE, Zain R, Wengel J, Smith CIE. Nucleic Acids Res. 2013;41:3257. doi: 10.1093/nar/gkt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denn B, Karmakar S, Guenther DC, Hrdlicka P. J. Chem. Commun. 2013;49:9851. doi: 10.1039/c3cc45705b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sau SP, Madsen AS, Podbevsek P, Andersen NK, Kumar TS, Andersen S, Rathje RL, Anderson BA, Guenther DC, Karmakar S, Kumar P, Plavec J, Wengel J, Hrdlicka PJ. J. Org. Chem. 2013;78:9560. doi: 10.1021/jo4015936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karmakar S, Guenther DC, Hrdlicka PJ. J. Org. Chem. 2013;78:12040. doi: 10.1021/jo402085v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karmakar S, Madsen AS, Guenther DC, Gibbons BC, Hrdlicka P. J. Org. Biomol. Chem. 2014;12:7758. doi: 10.1039/c4ob01183j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson BA, Onley JJ, Hrdlicka PJ. J. Org. Chem. 2015;80:5395. doi: 10.1021/acs.joc.5b00742. [DOI] [PubMed] [Google Scholar]
- 16.Imazawa M, Ueda T, Ukita T. Chem. Pharm. Bull. 1975;23:604. [Google Scholar]
- 17.Karmakar S, Anderson BA, Rathje RL, Andersen S, Jensen T, Nielsen P, Hrdlicka PJ. J. Org. Chem. 2011;76:7119. doi: 10.1021/jo201095p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a Divakar KJ, Mottah A, Reese CB, Sanghvi YS. J. Chem. Soc. Perkin Trans. 1990;1:969. [Google Scholar]; b Divakar KJ, Reese CB. J. Chem. Soc. Perkin Trans. 1. 1982:1625. [Google Scholar]
- 19.Ozaki H, Momiyama S, Yokotsuka K, Sawai H. Tetrahedron Lett. 2001;42:677. [Google Scholar]
- 20.Hendrickx PMS, Martins JC. Chem. Cent. J. 2008;2:20. doi: 10.1186/1752-153X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donders LA, de Leeuw FAAM, Altona C. Magn. Res. Chem. 1989;27:556. [Google Scholar]
- 22.a Diez E, Esteban A, Guilleme J, Bermejo F. J. Mol. Struct. 1981;70:61. [Google Scholar]; b Diez E, Esteban A, Bermejo F, Altona C, de Leeuw FAAM. J. Mol. Struct. 1984;125:49. [Google Scholar]; c de Leeuw FAAM, van Kampen P, Altona C, Diez E, Esteban A. J. Mol. Struct. 1984;125:67. [Google Scholar]
- 23.Pallan PS, Prakash TP, Li F, Eoff RL, Manoharan M, Egli M. Chem. Commun. 2009:2017. doi: 10.1039/b822781k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venkateswarlu D, Lind KE, Mohan V, Manoharan M, Ferguson DM. Nucleic Acids Res. 1999;27:2189. doi: 10.1093/nar/27.10.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura M, Fukunaga Y, Sasa K, Ohtoshi Y, Kanaori K, Hayashi H, Nakano H, Yamana K. Nucleic Acids Res. 2005;33:5887. doi: 10.1093/nar/gki889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.a Kumar TS, Madsen AS, Østergaard ME, Sau SP, Wengel J, Hrdlicka PJ. J. Org. Chem. 2009;74:1070. doi: 10.1021/jo802037v. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Korshun VA, Stetsenko DA, Gait MJ. J. Chem. Soc., Perkin Trans. 2002;1:1092. [Google Scholar]; c Dohno C, Saito I. ChemBioChem. 2005;6:1075. doi: 10.1002/cbic.200400325. [DOI] [PubMed] [Google Scholar]
- 27.Asanuma H, Fujii T, Kato T, Kashida HJ. Photochem. Photobiol. C. 2012;13:124. [Google Scholar]
- 28.a Dougherty G, Pilbrow JR. Int. J. Biochem. 1984;16:1179. doi: 10.1016/0020-711x(84)90215-5. [DOI] [PubMed] [Google Scholar]; b Manoharan M, Tivel KL, Zhao M, Nafisi K, Netzel TL. J. Phys. Chem. 1995;99:17461. [Google Scholar]; c Wilson JN, Cho Y, Tan S, Cuppoletti A, Kool ET. ChemBioChem. 2008;9:279. doi: 10.1002/cbic.200700381. [DOI] [PubMed] [Google Scholar]
- 29.Mergny JL, Lacroix L. Oligonucleotides. 2003;13:515. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 30.Winnik FM. Chem. Rev. 1993;93:587. [Google Scholar]
- 31.a Dioubankova NN, Malakhov AD, Stetsenko DA, Gait MJ, Volynsky PE, Efremov RG, Korshun VA. ChemBioChem. 2003;4:841. doi: 10.1002/cbic.200300678. [DOI] [PubMed] [Google Scholar]; b Astakhova IV, Malakhov AD, Stepanova IA, Ustinov AV, Bondarev SL, Paramonov AS, Korshun VA. Bioconj. Chem. 2007;18:1972. doi: 10.1021/bc700280h. [DOI] [PubMed] [Google Scholar]; c Astakhova IV, Ustinov AV, Korshun VA, Wengel J. Bioconj. Chem. 2011;22:533. doi: 10.1021/bc1005027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.