Abstract
The nociceptin/orphanin FQ (NOP) receptor, the fourth member of the opioid receptor family, is involved in many processes common to the opioid receptors including pain and drug abuse. To better characterize receptor location and trafficking, knock-in mice were created by inserting the gene encoding enhanced green fluorescent protein (eGFP) into the NOP receptor gene (Oprl1) and producing mice expressing a functional NOP-eGFP C-terminal fusion in place of the native NOP receptor. The NOP-eGFP receptor was present in brain of homozygous knock-in animals in concentrations somewhat higher than in wild-type mice and was functional when tested for stimulation of [35S]GTPγS binding in vitro and in patch-clamp electrophysiology in dorsal root ganglia (DRG) neurons and hippocampal slices. Inhibition of morphine analgesia was equivalent when tested in knock-in and wild-type mice. Imaging revealed detailed neuroanatomy in brain, spinal cord, and DRG and was generally consistent with in vitro autoradiographic imaging of receptor location. Multicolor immunohistochemistry identified cells coexpressing various spinal cord and DRG cellular markers, as well as coexpression with μ-opioid receptors in DRG and brain regions. Both in tissue slices and primary cultures, the NOP-eGFP receptors appear throughout the cell body and in processes. These knock-in mice have NOP receptors that function both in vitro and in vivo and appear to be an exceptional tool to study receptor neuroanatomy and correlate with NOP receptor function.
SIGNIFICANCE STATEMENT The NOP receptor, the fourth member of the opioid receptor family, is involved in pain, drug abuse, and a number of other CNS processes. The regional and cellular distribution has been difficult to determine due to lack of validated antibodies for immunohistochemical analysis. To provide a new tool for the investigation of receptor localization, we have produced knock-in mice with a fluorescent-tagged NOP receptor in place of the native NOP receptor. These knock-in mice have NOP receptors that function both in vitro and in vivo and have provided a detailed characterization of NOP receptors in brain, spinal cord, and DRG neurons. They appear to be an exceptional tool to study receptor neuroanatomy and correlate with NOP receptor function.
Keywords: eGFP, GPCR, histochemistry, knock-in, N/OFQ, NOP receptor
Introduction
The nociceptin/orphanin FQ (NOP) receptor (previously called ORL1) is the fourth member of the opioid receptor family (Cox et al., 2015). Although this G-protein-coupled receptor (GPCR) is distinguished from the other members of the receptor family (μ, δ, and κ) by its low affinity for opioid peptides and most high affinity opiate ligands, it has homology to the other family members equivalent to the homology they have for each other (Mogil and Pasternak, 2001). The NOP receptor is found in many brain regions, spinal cord, and dorsal root ganglia (DRG), as well as many peripheral organs (Neal et al., 1999; Mollereau and Mouledous, 2000). Although the endogenous ligand for NOP receptors, nociceptin/orphanin FQ (now called N/OFQ) was originally found to block opiate analgesia when administered intracerebroventricularly (Meunier et al., 1995; Reinscheid et al., 1995), it was subsequently determined to have antinociceptive properties when administered intrathecally (Xu et al., 1996; Jhamandas et al., 1998), suggesting a close but complicated interaction with opiate receptors. N/OFQ also has a large number of CNS and peripheral actions, influencing memory, feeding, stress/anxiety, drug reward, and renal and cardiovascular activity. Furthermore, NOP receptors have demonstrated significant plasticity, with mRNA and receptor levels modified by many factors including chronic pain and chronic opiate treatment (Darland et al., 1998; Ueda et al., 2000; Lambert, 2008).
The determination of the precise location of NOP receptors in the brain has been difficult due to the lack of a suitable antibody for immunohistochemical characterization, a common problem for GPCRs. In situ hybridization has been used to identify NOP receptor mRNA containing cell bodies, and in vitro autoradiographic determinations of [3H]N/OFQ binding sites have been published (Ikeda et al., 1998; Neal et al., 1999). However, these methods lack the resolution and sensitivity of fluorescently tagged antibodies. One method to examine the location and trafficking of GPCRs has recently been developed with the production of knock-in mice with a fluorescent tag covalently attached to the C-terminal of the receptor. Such knock-in mice have been produced with enhanced green fluorescent protein (eGFP) fused to δ-opioid receptors (Scherrer et al., 2006) and mCherry fused to μ-opioid receptors (Scherrer et al., 2006; Erbs et al., 2015). δ-eGFP mice have been very useful for the determination of the location of δ-opiate receptors and for the identification of DRG and primary afferent neurons that are involved in the antinociceptive and anti-allodynic actions of δ receptor agonists (Scherrer et al., 2006; Scherrer et al., 2009; Bardoni et al., 2014). Very recently the crossing of the δ-eGFP mice with the μ-mCherry mice allowed an accurate mapping of μ and δ receptor colocalization (Gardon et al., 2014; Erbs et al., 2015), a particularly controversial topic.
To study the location, trafficking and plasticity of NOP receptors, we have knocked eGFP into the NOP receptor gene (Oprl1) and produced mice expressing a functional NOP-eGFP C-terminal fusion in place of the native NOP receptor. These mice have NOP receptors that function both in vitro and in vivo and prove to be an exceptional tool to study receptor neuroanatomy.
Materials and Methods
Antibodies.
We used the following antibodies: anti-GFP Abcam (rabbit; 1:1000 for DRG neurons, primary cultures, and spinal cord, and 1:1500 for the brain; and chicken, 1:2500 for double-labeling with rabbit anti-μ antibody); sheep anti-CGRP (calcitonin gene related peptide), Abcam (1:1500); rabbit anti-PKCγ, Santa Cruz Biotechnology (1:1500); mouse anti-NF200: Sigma-Aldrich (1:20,000); and rabbit monoclonal anti-μ-opioid receptor, UMB3; Abcam (1:200). For the isolectin B-4 (IB4) binding cells, biotinylated IB4 (Sigma-Aldrich, 1:500) and streptavidin-conjugated to AlexaFluor555 (1:2000, Life Technologies) were used. All of the secondary antibodies conjugated to fluorophore were obtained from either Life Technologies or Jackson ImmunoResearch Laboratories.
Drugs.
SR16835 was synthesized at Astraea Therapeutics and suspended in 2% dimethyl sulfoxide and 0.5% aqueous hydroxypropylcellulose. Morphine sulfate, N/OFQ and SB612111 were provided by the National Institute of Drug Abuse Drug Supply Program. SR16835 (0, 10, and 30 mg/kg) and morphine (free base, 3 mg/kg) were subcutaneously injected in a volume of 5 ml/kg.
Animals
Generation of Oprl1-eGFP knock-in mice
A targeting construct was produced whereby the Oprl1 gene was modified so that a floxed neomycin resistant gene was inserted before exon 4 and the stop codon in exon 5 was replaced by a Gly-Ser-Ile-Ala-Thr-eGFP encoding cDNA followed by a stop codon. This was subsequently transfected into ES cells. A positive ES clone, where homologous recombination had properly occurred, was electroporated with a Cre-expressing plasmid to excise the neomycin gene and microinjected into C57BL6J blastocysts. Chimeric mice were crossed with C57BL6J mice to obtain F1 heterozygous progenies. Heterozygous animals were intercrossed to generate Oprl1-eGFP mice that were fertile and developed normally.
Male and female Oprl1-eGFP-homozygous (NOPeGFP/eGFP), heterozygous (NOP+/eGFP) or their wild-type (NOP+/+) littermates weighing 20–25 g were used. Animals were group-housed under standard laboratory conditions and kept on a 12 h day/night cycle (lights on at 7:00 A.M.). Animals were handled three times before the experiment. Mice were maintained in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All methods used were preapproved by the Institutional Animal Care and Use Committee at the Torrey Pines Institute for Molecular Studies (Port St Lucie, FL).
In vitro pharmacology
[3H]N/OFQ binding to NOP receptors in mouse brain membranes was conducted as described previously (Adapa and Toll, 1997). Briefly, brains both male and female, from each genotype, were homogenized in 50 mm Tris HCl, pH 7.7, centrifuged twice at 15,000 rpm and resuspended in the Tris buffer containing 1 mg/ml BSA, at a concentration of 160 μg of protein per milliliter. Binding was conducted in triplicate in a 96 well format in 1.0 ml volumes and containing [3H]N/OFQ in concentrations ranging from 0.2 to 5.3 nm, with or without 1 μm N/OFQ to determine nonspecific binding. Samples were filtered after 1 h using a Tomtec cell harvester and counted in a Wallac β plate reader. Kd and Bmax values were determined using Graphpad, Prism. Stimulation of [35S]GTPγS binding was conducted basically as described by Scherrer et al. (2006), based on the original method of Traynor and Nahorski (1995). The same membrane preparation was used as described above, except the final pellet was suspended in Buffer A (100 mm NaCl, 10 mm MgCl2) containing 30 μm GDP at a concentration of 15 μg of protein per milliliter. Samples were filtered, counted, and analyzed as described above.
NOP-eGFP mouse genotyping.
Total DNA was isolated from the mouse tail using DNeasy Kit (Qiagen). The NOP primers were 5′-CCCTGCACCGGGAGATGCA-3′ (forward) and 5′-GACAGAGGCCATGGAGGCC-3′ (reverse), which were used to amplify a 319 bp NOP DNA fragment. The NOP-eGFP primers were 5′-CCCTGCACCGGGAGATGCA-3′ (forward) and 5′-GCGGACTGGGTGCTCAGGTA-3′ (reverse), to amplify a 733 bp NOP-eGFP transgenic DNA fragment. PCR was performed at an annealing temperature of 60°C using the GoTaq FlexiDNA kit (Promega).
Real-time reverse transcription PCR (RT-PCR).
Total RNA was isolated from NOP+/+, NOP+/eGFP, and NOPeGFP/eGFP mice (n = 4 of each genotype) using RNAeasy Mini kit (Qiagen). To measure mRNA levels, real-time quantitative RT-PCR was performed with the iCycler IQ5 multicolor Real-Time PCR detection system (Bio-Rad Laboratories) using QuantiFast Probe PCR Kit (Qiagen). Gene-specific primers and dual-labeled probes for NOP/NOP-eGFP and GAPDH were designed using the NCBI primer-BLAST software and synthesized by Qiagen). The following real-time RT-PCR protocol was used for all genes: cDNA was synthesized using QuantiTect Reverse transcription kit (Qiagen) and cDNA corresponding to 50 ng total RNA was used as a DNA template for real-time PCRs: 95°C for 3 min to activate the HotStart enzyme followed by 45 cycles of amplification and quantification (10 s at 95°C; 30 s at 60°C) each with a single FAM fluorescence measurement. GAPDH was used as a housekeeping gene and relative NOP mRNA levels were calculated by subtracting mean GAPDH Ct values from NOP Ct values using the 2−ΔΔCt method (Wu et al., 2010). Primers and probes for real-time PCR were as follows: GAPDH_F581, 5′-GTGGAAGGGCTCATGACCAC-3′; GAPDH_R698, 5′-ATGCAGGGATGATGTTCTGG-3′; GAPDH_Probe, 5′-[6∼FAM]AGCTGTGGCGTGATGGCCGT[Tamra∼Q]-3′; NOPf432, 5′-TGGGGAACTGCCTCGTCATGT-3′; NOPr590, 5′-TCCCAAATGGCCAGAAGCCCA-3′; NOP_Probe, 5′-[6∼FAM]AATCTGGCACTGGCTGATACCCTGG[Tamra∼Q]-3′.
Immunohistochemistry
Six- to 8-week-old male NOP-eGFP (NOPeGFP/eGFP) C57BL/6J mice were transcardially perfused in 4% paraformaldehyde (PFA) in PBS. The brains, DRG (L2–L6) and spinal cord (Lumbar cord) were dissected from the mice and cryoprotected in 30% sucrose in PBS. Tissues were then frozen in O.C.T. (Sakura Finetek, INC., Torrence, CA). Tissue sections (30 μm for the brains; 40 μm for spinal cord; and 10 μm for DRG) were prepared by using a cryostat (Leica Biosystems, Buffalo Grove, IL) and blocked with PBS containing 5% normal donkey serum and 0.3% Triton X-100 for 1 h at room temperature. The sections were then incubated with primary antibodies, indicated in each figure, at 4°C, overnight. For the chicken anti-GFP antibody, the incubation was performed at 37°C for 2 h. After extensive wash with PBS containing 1% normal donkey serum and 0.3% Triton X-100, sections were incubated with appropriate secondary antibody conjugated to AlexaFluor for 2 h at room temperature. In certain experiments, sections from DRG and spinal cord were incubated with a biotinylated IB4 (1:500) followed by incubation with streptavidin-conjugated to AlexaFluor 555 (1:2000). Images were collected under either a DV Elite fluorescent microscope (GE Healthcare) and SoftWoRx software (GE Healthcare), or a Leica TCS SP5II confocal microscope and LAS AF Lite software (Leica Microsystems). ImageJ (National Institute of Health) was also used to measure the size of DRG neurons.
Primary cultures
Hippocampi were dissected from mouse pups (P0) and digested by papain (Brewer, 1997). Cells were plated on poly-d-lysine-coated glass coverslips and cultured in B27/neurobasal A medium (Life Technologies) containing 0.5 mm glutamine, 5 ng/ml basic fibroblast growth factor (Life Technologies). Fully matured primary neurons (10 d in culture) were used for the internalization experiments. Cells were treated with 1 μm N/OFQ for the time periods indicated in Figure 3. Cells were then washed with PBS three times and fixed in 4% PFA for 15 min at room temperature and blocked with PBS containing 5% normal donkey serum and 0.3% Triton X-100 for 1 h at room temperature. Cells were then incubated with rabbit anti-GFP antibody. After extensive washing with PBS containing 1% normal donkey serum and 0.3% Triton X-100, cells were incubated with donkey anti-rabbit IgG AlexaFluor 488. Images were collected and analyzed by the same procedure as described above. ImageJ was also used for the quantification of the fluorescent intensity in the cells.
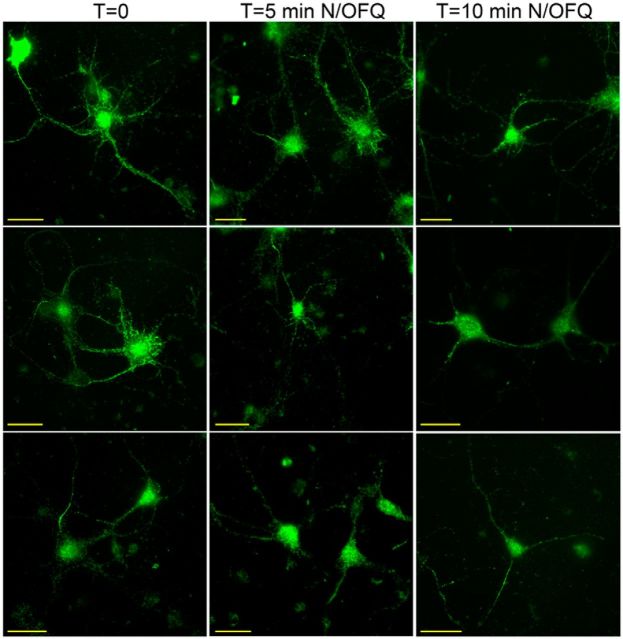
Figure 3.
N/OFQ activates NOP-eGFP receptors in primary neuron cells. Hippocampal primary neurons (10 d in vitro) isolated from NOP-eGFP mice were treated with 1 μm N/OFQ for the indicated time periods to examine the changes in the subcellular localization of NOP-eGFP. NOP-eGFP receptors were visualized with an anti-GFP antibody and secondary antibody conjugated to AlexaFluor488. Scale bar, 25 μm. The 5 min time point appears to have more punctate processes in some cells.
Tail withdrawal assay
Assessment of thermal nociception using the tail-flick assay.
Nociception was assessed by the tail-flick assay with a thermal stimulator (Ugo Basile, Stoelting) that uses radiant heat. During testing, the focused beam of light was applied to the lower half of the animal's tail, and tail-flick latency was recorded. A 15 s cutoff time was scheduled to avoid tissue damage. Baseline values for tail-flick latency were determined before drug administration in each animal. After baseline measures were taken, NOP-eGFP-homozygous (N = 30) and wild-type (N = 30) mice received injection of SR16835 0, 10, or 30 mg/kg, and tail-flick latency was assessed in each animal 30 min later. Immediately after the second pain assessment, morphine was administered to all mice (T = 0), and tail-flick latency measured 30 (T = 30) and 60 min (T = 60) following morphine administration. Tail-flick latency is reported as percentage of maximum possible effect (%MPE) that was quantified by the following formula: %MPE = 100 × [(test latency − baseline latency)/(15 − baseline latency)]. A score of 100% was assigned if the animal did not respond before the 15 s cutoff.
Electrophysiology
Animals were handled in accordance with institutionally approved protocols and the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Mice were deeply anesthetized with halothane and decapitated. The brain was quickly removed and placed into ice-cold artificial CSF (ACSF) continuously bubbled with 5% CO2/95% O2. The ACSF was composed as 124 mm NaCl, 2.5 mm KCl, 1.2 mm KH2PO4, 2.4 mm CaCl2, 1.3 mm MgSO4, 26 mm NaHCO3, and 10 mm glucose, pH 7.4. Hippocampal slices were prepared using a tissue chopper (Stoelting, 400 μm) or vibratome (Leica VT100S, 260 μm). Slices were incubated at room temperature in ACSF, bubbled with 5% CO2/95% O2 continuously for at least 1 h before being transferred to the recording chamber. Extracellular recording was made in a submerged mode at room temperature (Harvard Apparatus). Data were collected through an Axopatch-2B amplifier or Multiclamp 700B (Axon CNS, Molecular Devices) with program pClamp 10.4 (Molecular Devices). Slices were continuously perfused with oxygenated ACSF at a flow rate of 2 ml/min with peristaltic pump or through reservoirs by gravity feeding.
Field potential (EPSP) was recorded using a glass microelectrode filled with ACSF (resistance: 1–3 MΩ) or 2 m NaCl internal solution composed as 140 mm KGlu, 1 mm K-EGTA, 0.1 mm CaCl2, 2 mm MgSO4, 10 mm HEPES. Approximately 10 min of stable baseline was recorded before drug application. Biphasic current pulses (0.2 ms duration for 1 phase, 0.4 ms in total) were delivered in 10 s intervals through a concentric bipolar stimulating electrode (FHC) placed in the middle of stratum radiatum to stimulate the Schaffer collateral fibers. Stimulation pulses were given through an isolator (ISO-Flex, AMPI), which was driven by computer-generated pulses with pClamp10.4 program. To record field EPSP in CA1, the recording electrodes were placed in the middle of stratum radiatum ∼100 μm apart from the stimulating electrode (lateral or inner side) in parallel to the cell body layer. Recording electrodes were fabricated with borosilicate capillary glass tubing (outer diameter: 1.5 mm, inner diameter: 0.86 mm;Warner Instruments) using a Flaming Brown microelectrode puller (model P-87). No obvious synaptic depression or facilitation was observed with this frequency stimulation. Slices were recorded within 8 h after dissection. The potassium gluconate internal solution worked well and apparently facilitated the stability of baseline when the recording electrodes had relatively long shoulder and sharper tip.
Signals were digitized at 20 kHz and not filtered. Input/output curves were obtained for each slice using stimulus intensity from threshold (usually 0.02 mA) to a maximum of 0.4 mA. Test pulse intensities were adjusted to evoke EPSP ∼35% of maximal response. The slope of EPSP was measured from the initial phase of negative wave. The inhibition ratio was calculated as percentage of the value after application of drugs in comparison with the value of control (before drug application). Each data point was measured as the average of three consecutive traces. Data were analyzed with Pclamp 10.4, Microsoft Excel, StatView 5.0., and presented as mean ± SEM. One-way ANOVA was used for multiple group comparison.
Preparation of DRG neurons.
Mouse DRG neurons were prepared from 1- to 2-month-old NOP-eGFP mice. Briefly, the spine was taken out and split into two halves from the middle line after sacrificing the mice by decapitation. Lumber DRGs were collected into modified Kreb's solution (130 mm NaCl, 10 mm HEPES-Na, 5 mm KCl, 1 mm CaCl2, 10 mm glucose, 2 mm MgCl2, pH adjusted to 7.35 with 1N HCl) in a 1.5 ml tube. For digestion, the DRGs were removed into 0.5 ml of Hank's balanced salt solution (HBSS) with 1 mg/ml collagenase and 0.5 mg/ml trypsin added. The DRGs were minced with a fine scissors and incubated at 35°C for 50 min. After removing the HBSS solution, the DRGs were dispersed into modified Kreb's solution and triturated with fire polished glass pipettes until no clump was visible. Finally, the cells were dispersed onto poly-l-ornithine-coated (Sigma-Aldrich) coverslips and maintained in a modified Kreb's solution with streptomycin sulfate (0.2 mm), penicillin G sodium (0.3 mm), and gentamycin (0.1 mm) at 21°C.
Whole-cell voltage-clamp recordings were performed on large size (diameter, 30–40 μm) mouse DRG neurons after acutely dissociation. All experiments were performed at room temperature (∼21°C). Whole-cell patch-clamp recordings were made using a MultiClamp 700B amplifier and analyzed offline with pCLAMP10.4 software (Axon CNS, Molecular Devices). For current-clamp recording, the external solution was a modified Kreb's solution specified above. The internal solution was composed of 65 mm KCl, 80 mm KF, 5 mm KOH, 10 mm EGTA, 2 mm MgATP (pH 7.35–7.4 adjusted with KOH, and the osmolality verified as 295 mOsm/kg). Recording electrodes were pulled by P-87 puller (Sutter Instrument). The tip of resistance was 3–4 MΩ in bath and the series resistance was <10 MΩ after whole-cell configuration. N/OFQ (1 μm) was applied through a local perfusion system with the opening of the tube located ∼150 μm from the cell.
Statistical Analyses.
Baseline values for tail-flick latency before drug administration were analyzed by one-way ANOVA using mouse genotype as a between factor. The same approach was used for qPCR data. Thermal nociception data were analyzed by using three-way ANOVA with mouse genotype and drug treatment (SR16835) as between-subject factors and time course of morphine effect (0, 30, and 60 min) as a within-subject factor. The level of significance was set at p ≤ 0.05. Student Newman–Keuls post hoc tests were used where appropriate.
Results
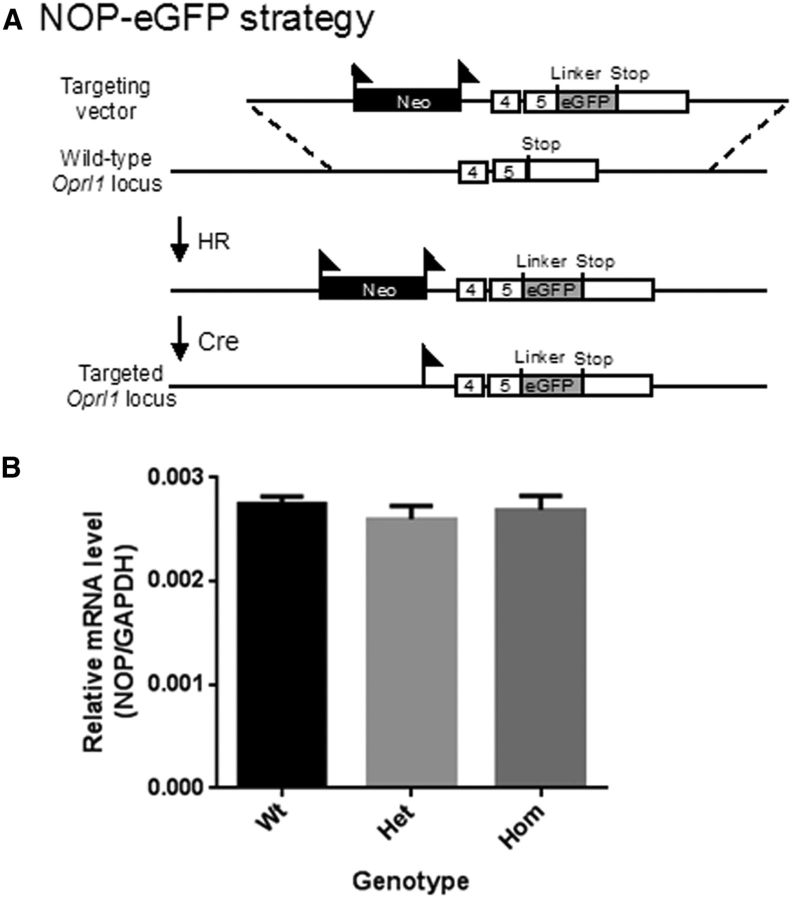
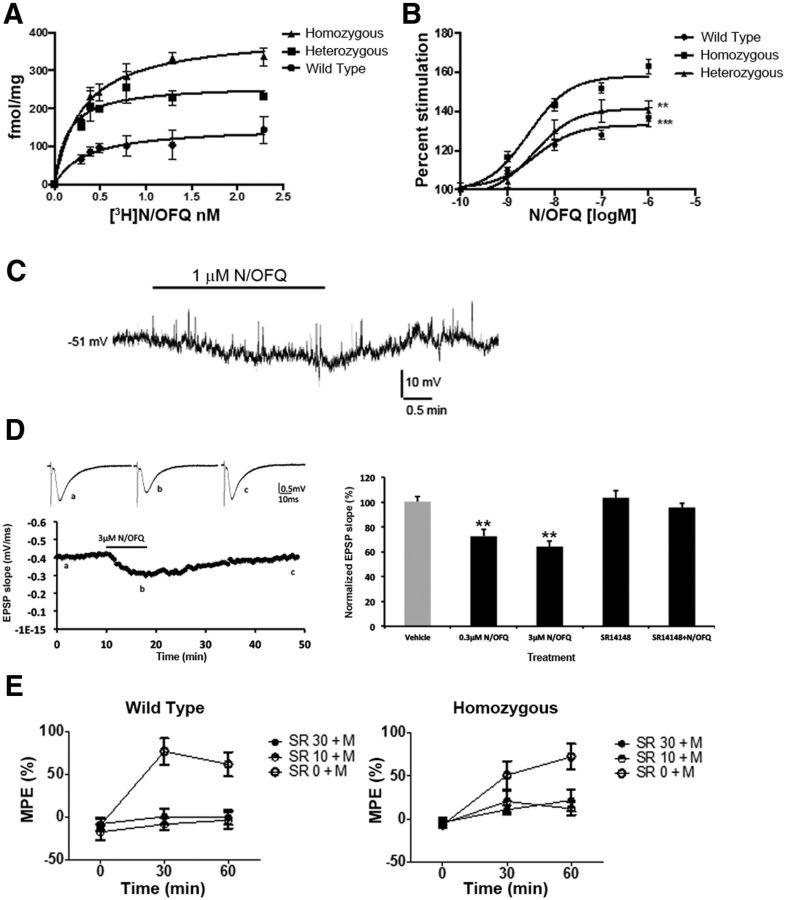
We used homologous recombination to introduce the eGFP cDNA into exon 5 of the Oprl1 mouse gene, in frame and 5′ from the stop codon (Fig. 1A), in a manner similar to that described for the δ and μ receptors (Scherrer et al., 2006; Erbs et al., 2015). Quantitative RT-PCR indicated that the genomic modification did not affect transcription, as there was no significant difference between mRNA levels of NOP receptor in the NOP+/+, NOP+/eGFP, and NOPeGFP/eGFP mice. Conversely, receptor binding studies using [3H]N/OFQ on brain membranes derived from the NOP+/+, NOP+/eGFP, and NOPeGFP/eGFP mice indicated that there was a progressive increase in receptor number in wild-type, heterozygous, and homozygous knock-in mice, respectively. Although there is no significant difference in Kd in the three genotypes (0.22 ± 0.1, 0.21 ± 0.1, 0.31 ± 0.08 nm for NOP+/+, NOP+/eGFP, and NOPeGFP/eGFP mice, respectively; n = 3 mice of each genotype), the presence of the NOP-eGFP fusion protein increases Bmax (144 ± 17.6, 282 ± 22, 404 ± 21.4 fmol/mg protein), suggesting that the eGFP improves receptor translation or perhaps stability of the fusion protein, Figure 2A. The affinity of the antagonist SB612111 and the small molecule NOP receptor agonist SR16835 were also similar when determined using the three genotypes, indicating no change in the binding properties of the NOP-eGFP receptor (Table 1). [35S]GTPγS binding studies conducted in membranes derived from NOP+/+, NOP+/eGFP, and NOPeGFP/eGFP mice indicated that the homozygous knock-in mice more efficiently transduced a signal with higher N/OFQ induced [35S]GTPγS binding than the other two genotypes, without a change in potency (EC50 5.8 ± 2.1, 3.4 ± 0.3, 5.3 ± 2.1 nm for NOP+/+, NOP+/eGFP, and NOPeGFP/eGFP mice, respectively; n = 3 mice of each genotype), consistent with the increase in receptor number Figure 2B.
Figure 1.
Knock-in construct, genotyping scheme and NOP-eGFP mRNA levels in NOP+/+, NOP+/eGFP, and NOPeGFP/eGFP mice. A, Targeting strategy. Oprl1 exons, eGFP cDNA, and the floxed (triangles) neomycin cassette are displayed as empty, gray, and black boxes, respectively. Homologous recombination (HR) was followed by Cre recombinase treatment (Cre) in ES cells. B, mRNA levels were determined by performing RT-PCR using whole brains, as described in Materials and Methods. N = 4 mice of each genotype. Error bars denote SD.
Figure 2.
In vitro and in vivo activity of N/OFQ at NOP receptors in NOP+/+, NOP+/eGFP, and NOPeGFP/eGFP mice. [3H]N/OFQ binding and [35S]GTPγS binding to brain membranes, electrophysiological recordings, and the radiant heat tail-flick assay were performed as described in Materials and Methods. In vitro experiments shown in A and B are from single experiments conducted in quadruplicate ([3H]N/OFQ binding) or triplicate ([35S]GTPγS binding), with error bars denoting SD among the replicates. Each experiment was repeated at least 2 additional times with similar results. C, Electrophysiological response to N/OFQ in a DRG neuron with current-clamp recording. Application of 1 μm N/OFQ reversibly hyperpolarized membrane potential. D, The effect of N/OFQ on field EPSP in slices of hippocampus. Application of N/OFQ (0.3 μm, 3 μm) reversibly inhibited the field EPSP slopes. Left, Top, The representative traces of field EPSP before, during and after application of 3 μm of N/OFQ. Left, Bottom, The time course of the inhibitory effect of N/OFQ. The bar indicated the period of drug application. Right, The summary results of N/OFQ and antagonist, SR14148. Application of vehicle (0.01% of DMSO, n = 4) or SR14148 alone (n = 3) had no significant effect on field EPSP. E, Nociceptive response to thermal stimulation in the tail-flick test in NOP+/+ (left) and NOPeGFP/eGFP (right) mice following the combined administration of SR16835 (0, 10, and 30 mg/kg) and morphine (M; 3 mg/kg). Tail-flick latency was determined immediately before morphine administration (T = 0) that followed SR16835 treatment. Tail flick response was then repeated 30 (T = 30) and 60 min (T = 60) following morphine treatment. Data are mean %MPE ± SEM. ***p < 0.001 difference from T = 0; ##p < 0.01, ###p < 0.001 difference from SR 0 + morphine treatment.
Table 1.
Binding of selected ligands to brain membranes derived from NOP+/+, NOP+/eGFP, and NOPeGFP/eGFP mice
| NOP+/+ |
NOP+/eGFP |
NOPeGFP/eGFP |
||||
|---|---|---|---|---|---|---|
| Ki nm | Hill | Ki nm | Hill | Ki nm | Hill | |
| N/OFQ | 0.17 ± 0.05 | 1.1 ± 0.01 | 0.24 ± 0.1 | 1.07 ± 0.06 | 0.30 ± 0.04 | 1.1 ± 0.06 |
| SB612111 | 4.81 ± 0.68 | 0.85 ± 0.02 | 4.63 ± 0.9 | 0.80 ± 0.04 | 7.27 ± 1.2 | 0.8 ± 0.01 |
| SR16835 | 51.3 ± 17 | 0.97 ± 0.03 | 37.1 ± 1.1 | 0.81 ± 0.06 | 50.8 ± 3.0 | 0.72 ± 0.01 |
Binding to brain membranes from mice of each genotype, as well as data analysis were conducted as described in Materials and Methods. Data presented are from two experiments each conducted in triplicate. Values shown are average ± SD.
Patch-clamp experiments on DRG neurons demonstrated that N/OFQ (1 μm) induced a hyperpolarizing response in DRG neurons (Fig. 2C), as has been demonstrated in DRG neurons and other neuronal cells prepared from wild-type mice (Xie et al., 2008; Murali et al., 2012). Patch-clamp electrophysiology was also conducted on hippocampal slices. Application of 0.3 or 3 μm N/OFQ reversibly inhibited the field EPSP recorded in CA1 by stimulating Schaffer collateral fibers (one-way ANOVA, F(4,18) = 10.25, p < 0.001). The inhibitory effect of N/OFQ was revealed by paired t test before and after drug application (p < 0.001). The effect of 3 μm N/OFQ (64.1 ± 4.6%, n = 6) is slightly larger than that of 0.3 μm N/OFQ (72.3 ± 5.9%, n = 7) but the difference was not significant, implicating that the effect was close to the maximum. The inhibitory effect of 0.3 μm N/OFQ was completely blocked by 10 μm antagonist, SR14148 (n = 3; one-way ANOVA, F(4,18) = 10.25, p < 0.001). There was slight and insignificant enhancement of field EPSP by applying 10 μm of SR14148, suggesting a potential endogenous tonic effect of N/OFQ in hippocampal slices. The effects of N/OFQ were tested in two setups of recording systems with consistent results. Therefore, data were pooled for statistical analysis.
NOP-eGFP mice were tested for antinociceptive activity to determine whether the modified receptor functioned in vivo. Systemic administration of NOP receptor agonists block morphine antinociception, and therefore NOP+/+ and NOPeGFP/eGFP mice were tested to determine whether systemic administration of the NOP receptor agonist SR16835 could block morphine-induced antinociception Figure 2D. Baseline values for tail-flick latency determined before drug administration were 4.3 ± 0.1 s for the homozygous line and 4.6 ± 0.2 s for the wild types. As expected, ANOVA revealed no changes in basal values of thermal nociception between the two lines (F(1,58) = 1.8, NS). When analyzing nociceptive response following administration of SR16835 and morphine, overall ANOVA revealed a main effect of both SR16835 (F(2,54) = 29.8, p < 0.001) and morphine (F(2,108) = 58.1, p < 0.001) treatments. These effects were not accompanied by a main effect of genotype (F(1,58) = 3.3, NS), SR16835 treatment × genotype interaction (F(2,58) = 2.0, NS) and time course of morphine treatment × genotype interaction (F(2,108) = 1.4, NS), indicating that the homozygous and the wild-type lines showed similar sensitivity to drug effects. In this experiment, genotype × SR16835 treatment × morphine treatment interaction barely reached significance (F(4,108) = 2.8, p = 0.05). On post hoc analysis a robust analgesic response to morphine at 30 and 60 min time points was observed in both mouse lines (p < 0.001 at T = 30 and T = 60 in both lines). This response was fully prevented by both SR16835 doses examined in the wild-type mice at T = 30 (p < 0.001), as well as T = 60 (p < 0.001). Both SR16835 doses significantly blocked morphine-induced analgesia at T = 60 (p < 0.01 for the dose of 30 mg/kg, p < 0.001 for the dose of 10 mg/kg) but not at T = 30 in the homozygous NOPeGFP/eGFP mice.
Subcellular localization of NOP receptors
To investigate the biological function within cells, we examined the subcellular localization and trafficking of NOP-eGFP receptors using hippocampal primary neurons prepared from the NOPeGFP/eGFP mice. As seen in Figure 3, NOP-eGFP fluorescence appears generally throughout the cells, rather than mostly along the plasma membrane, although there are some cells in which plasma membrane fluorescence is evident. This subcellular localization of NOP receptors is unlike the results using NOP receptor-transfected continuous cell lines, in which NOP receptors distribute on the plasma membrane rather than intracellularly (Spampinato et al., 2002; Corbani et al., 2004). This is also in contrast to δ-eGFP receptors in knock-in mice, in which most cells appear to have a large portion of plasma membrane fluorescence (Scherrer et al., 2006), but similar to the recently derived μ-mCherry mice, in which receptor fluorescence also appears throughout the cells (Erbs et al., 2015). Because the fluorescent receptors are not mostly on the plasma membrane in the NOP-eGFP containing cells, visualization of actual “internalization” into the cytoplasm is not clearly evident. When primary neurons were treated with 1 μm N/OFQ, some cells and cell processes appear to obtain a more punctate appearance (Fig. 3). This process was rapid, taking place within 5 min. However, this was neither consistent to all cells nor amenable to quantification. Therefore, the ubiquitous nature of the NOP receptors in the primary hippocampal cultures makes the observation of trafficking difficult and correlating visualized receptor trafficking with some agonist function is not possible at this time.
Anatomical profiling of NOP receptor distribution in the NOP-eGFP mice
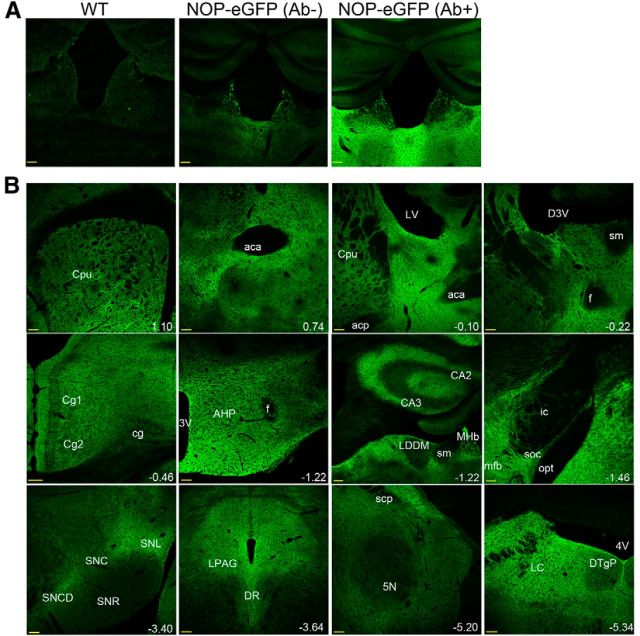
Previous literature has reported that NOP receptors are distributed among many different regions of the brain (Neal et al., 1999). Taking advantage of NOP-eGFP knock-in mice, we performed immunostaining with tissues, using anti-eGFP antibody, to better understand the locations of NOP receptors. Initial experiments demonstrated that anti-eGFP staining was specific to NOP-eGFP expression and not present in wild-type mice (Fig. 4A). We also found that the level of the eGFP fluorescence was greatly reduced in the NOP-eGFP brain, compared with the sections immunostained with an anti-GFP antibody. Based on these results, we conducted all of the immunostaining with anti-GFP antibodies to enhance the NOP-eGFP signal intensity (Fig. 4A).
Figure 4.
NOP-eGFP receptor expression in the brain. A, Evaluation of the specific NOP-eGFP receptor expression in the brain derived from the wild-type (WT) and NOP-eGFP mice. Brain sections from NOP-eGFP mice were incubated with (Ab+) or without (Ab−) an anti-GFP antibody. B, NOP-eGFP receptor distribution was observed in a wide range of brain region. The position of all sections is given relative to bregma (mm); the numbers highlighted in white. Scale bars, 100 μm.
NOP-eGFP immunoreactivities were detected throughout the mouse nervous system including the brains, spinal cord, and DRG neurons (Figs. 4–7) derived from NOP-eGFP mice. In the brain, NOP-eGFP receptors were distributed widely through: the nucleus accumbens (NAc; bregma, 0.74 mm), bed nucleus of the stria terminalis (bregma, −0.10 mm), cingulate cortex (bregma, −0.46 mm), amygdala, hippocampus (bregma, −1.22 mm), medial habenula (MHb; bregma, −1.22 mm), interpeduncular nucleus (IPN), periaqueductal gray (PAG; bregma, −3.64 mm), raphe nucleus (bregma, −3.64 mm), and ventral tegmental area (Figs. 4B, 7). These highly NOP-expressing regions are important for pain related biological actions as well as drug abuse and reward. In addition to the brain regions described above, NOP-eGFP expression is present in a part of the hippocampus, striatum, hypothalamus, and substantia nigra, which are essential to other known biological actions of NOP receptor, such as metabolic systems, locomotor activity, and mood regulation (Fig. 4B; for review, see Witkin et al., 2014). The distribution of NOP receptors is in general agreement with the previously reported brain structures provided from the radioligand binding study using rat brains (Neal et al., 1999). One significant difference is in the caudate-putamen. Neal et al. (1999) found this region to be devoid of NOP receptors, though they did detect a small amount of NOP receptor mRNA. We find NOP-eGFP receptors to be sparse, but not absent from this brain region. This result is consistent with studies that demonstrated that NOP receptor activation blocks DA release and other parameters of dopamine neurotransmission in striatal slices and synaptosomes (Flau et al., 2002; Olianas et al., 2008).
Figure 5.
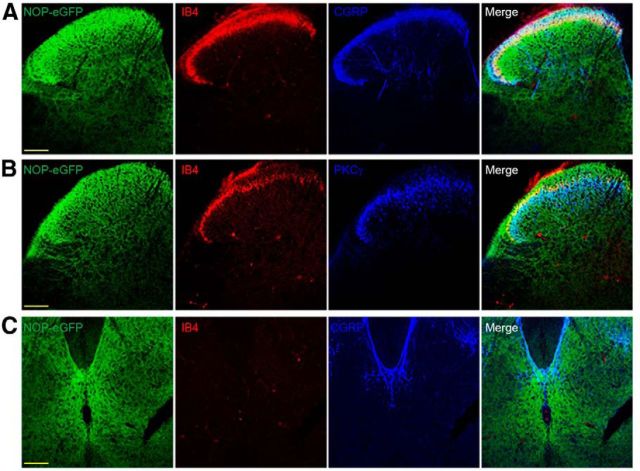
NOP-eGFP receptors are highly distributed in laminae I–III and X. Tissue sections from the spinal cord were incubated with anti-GFP, and -CGRP (laminae I and IIo; A) or -PKCγ (ventral border of lamina Iii; B). Tissues were also treated with biotinylated IB4 (dorsal border of lamina IIi) and streptavidin-conjugated to AlexaFluor 555 (A, B). C, Immunostaining of lamina X. Immunoreactivity of NOP-eGFP was observed in laminae I–III, where CGRP-immunoreactive terminals, PKCγ interneurons and IB4-positive interneurons are located. Scale bars, 100 μm.
Figure 6.
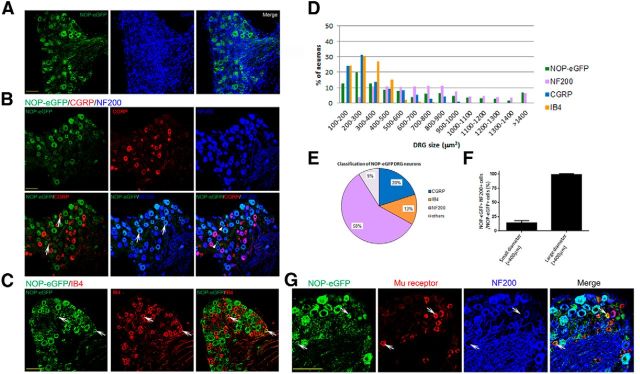
Various types of DRG neurons express NOP-eGFP receptors. To characterize the NOP-eGFP distribution in DRG neurons, sections were incubated with anti-GFP antibody together with anti-μ receptor antibody, DRG markers; anti-CGRP, -NF200 antibodies, or biotinylated IB4. A, NOP-eGFP expression in DRG neurons (green). Nuclei were stained with DAPI (blue). The NOP-eGFP containing DRG neurons were quantified by determining the percentage of eGFP-positive cells compared with the total number of sensory neurons (n = 1396). The total number of DRG sensory neurons was determined by counting the total number of DAPI-stained cells and excluding those from glia and connective tissues. Tissue sections were also costained with anti-CGRP and -NF200 antibodies, (B) in which the white arrowheads indicate NOP-eGFP+, CGRP-myelinated medium DRG neurons, or (C) biotinylated IB4. In each panel, white arrows indicate the cells where costaining occurs. D, Size profiling of DRG neurons that are expressing NOP-eGFP, CGRP, or NF200, or bind to IB4. E, Identity of NOP-eGFP+ DRG neurons. F, Percentage of medium NOP-eGFP+ DRG neurons that are myelinated and not coexpressing CGRP. Data are represented as mean ± SEM, G, colocalization of NOP-eGFP and μ receptors in DRG. Small unmyelinated neurons coexpress NOP-eGFP and μ receptors. White arrows depict the cells coexpressing NOP-eGFP and μ receptors. Scale bars, 100 μm.
Figure 7.
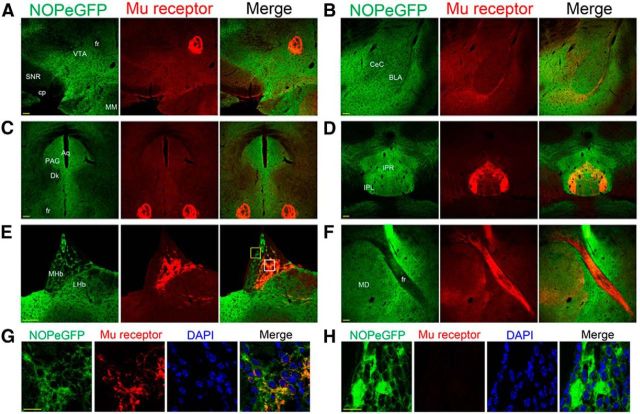
Colocalization of NOP-eGFP and μ receptors was examined in brain regions that are essential for pain and reward system. A, VTA and fr; B, amygdala; C, PAG; D, IPN; E, medial habenula; F, fr in a sagittal section, G, H, High-power images of the white and yellow squares respectively in image E. In the merged representative images, yellow indicates that NOP-eGFP colocalizes with μ receptor. Scale bars: A–F, 100 μm; G, H, 15 μm.
NOP receptors are predominantly distributed in the dorsal horn of the lumbar spinal cord
In addition to the NOP-eGFP expression in the brain, NOP-eGFP immunoreactivity was also observed in the spinal cord (Fig. 5). To determine the laminar location of spinal NOP receptors, immunostaining was performed with lamina markers. We found that the NOP-eGFP receptor immunoreactivity was present in superficial laminae I and II, where CGRP-positive and IB4-positive nociceptive primary afferents project (Fig. 5A). In addition, the intense immunoreactivity also extended into the ventral border of laminae II and III, where protein kinase C γ (PKCγ)-positive interneurons are located (Fig. 5B), suggesting that the NOP receptors might regulate injury-induced chronic mechanical allodynia (Malmberg et al., 1997). Consistent with this hypothesis, NOP receptors are distributed between laminae I through III in the dorsal horn, regions important for the regulation of pain, itch, and touch. In addition to the NOP distribution in the dorsal horn, strong immunoreactivity was also detected in lamina X (Fig. 5C) and a moderate fluorescent signal was observed throughout the intermediate zone and ventral horn, in general agreement with the location of the receptors by in vitro autoradiography in rats (Neal et al., 1999).
NOP receptors are expressed in myelinated, and peptidergic and nonpeptidergic unmyelinated DRG neurons
We next analyzed the distribution of NOP-eGFP receptors in DRG and found that numerous sensory neurons displayed NOP-eGFP immunoreactivity. This is in contrast to autoradiographic studies, which found no receptor binding in the DRG (Neal et al., 1999), but consistent with several electrophysiological and immunohistochemical studies that found NOP receptors and N/OFQ-mediated activities in DRG (Chen and Sommer, 2006; Murali et al., 2012). Approximately 43% of all DRG neurons express NOP-eGFP (Fig. 6A). Among the NOP-eGFP-positive neurons, 52–55% had small diameter cell bodies (<400 μm2), whereas 45–48% had large diameter cell bodies (>400 μm2; Fig. 6D). We next investigated the identity of NOP-eGFP-positive neurons using DRG markers defining functional classes of DRG neurons. We found that the majority of the NOP-eGFP+ cells (∼58%) coexpresses neurofilament 200 (NF200) a marker of neurons with myelinated axons (A-fibers; Fig. 6B,D,E). These results indicate that NOP-eGFP is mostly expressed by A fibers. Next, we addressed the NOP-eGFP positive cells that belong to C-nociceptors. Approximately 34% of NOP-eGFP+ small unmyelinated (NF200−) DRG neurons coexpress CGRP (Fig. 6B), indicating that NOP-eGFP receptors are present in peptidergic C nociceptors, which are essential to acute heat pain and injury-induced heat hyperalgesia (Cavanaugh et al., 2009). Peptidergic C-fibers project to laminae I and IIouter of the spinal cord (Basbaum et al., 2009) where a robust immunoreactivity of NOP-eGFP is observed (Fig. 5A). On the other hand, ∼20% of small unmyelinated (NF200−) NOP-eGFP+ DRG neurons bind IB4, indicating that NOP receptors are present in the nonpeptidergic DRG neurons, many of which respond to noxious mechanical stimuli and are essential for acute mechanical pain (Fig. 6C; Basbaum et al., 2009; Cavanaugh et al., 2009; Scherrer et al., 2009; Vrontou et al., 2013; Bardoni et al., 2014). Together, our immunohistochemical studies indicate the NOP receptors might regulate the function of two major classes of C nociceptors that are critical to heat and mechanical pain modalities. Interestingly, we noted that a small number (∼17%) of medium diameter myelinated (NF200+) DRG neurons express NOP-eGFP (Fig. 6B,F). These NOP-eGFP-positive neurons do not coexpress CGRP, suggesting that they are not typical A nociceptors but might rather be myelinated low-threshold mechanoreceptors (A LTMRs) that encode touch (Abraira and Ginty, 2013) and might contribute to injury-induced mechanical allodynia.
NOP-eGFP receptors colocalize with μ receptors in the brain and DRG neurons
As seen in Figure 7, strong immunoreactivity of NOP-eGFP is widely present throughout the brain regions including PAG and raphe nucleus that correspond to the descending pathway in pain management. These NOP-eGFP-positive brain regions are also known to express μ-opioid receptors; however, in the brain, activation of the NOP receptors attenuates opiate actions in analgesia and reward (Mogil et al., 1996b; Tian et al., 1997; Murphy et al., 1999; Ciccocioppo et al., 2000; Khroyan et al., 2007). To better understand the connection between the action of NOP and μ receptors in pain and reward systems, we investigated the colocalization of these receptors in the brain. Figure 7 shows that both NOP and μ receptors are expressed in several of the brain regions. The MHb has been demonstrated to have very high levels of μ receptors that might have a role in both pain and reward systems and our results correspond very well with both in vitro autoradiography and location of the μ-mCherry receptor (Kitchen et al., 1997; Gardon et al., 2014). We found a very intense staining of both NOP and μ receptors in the MHb (Fig. 7E), although the highest densities of NOP and μ receptors appear in different subregions within the MHb (Fig. 7G,H). μ Receptor staining was very intense in the basolateral part of the MHb, corresponding to results obtained with μ-mCherry knock-in mice (Gardon et al., 2014), whereas NOP-eGFP receptors were found in highest density in the apical part of the MHb. However, even in regions with the highest expression of μ receptors, all of the μ receptor-positive cells coexpress NOP receptors (Fig. 7G). NOP and μ receptors are also highly expressed in the IPN (Fig. 7D). NOP receptors appear all through the IPN, though levels are highest in the rostral (IPR) and lateral (IPL) parts of the IPN, regions that are also highest in μ receptors (Gardon et al., 2014). Interestingly, the immunoreactivity of NOP-eGFP was barely observed in the fasciculus retroflexus (fr); the fiber tract connecting the MHb and the IPN, whereas μ receptors are highly distributed in the fiber (Fig. 7A,C,F). Additionally, the expressions of NOP and μ receptors were also found in the PAG (Fig. 7C). The ratio of immunoreactivity between NOP and μ receptors is consistent with the previously reported electrophysiological studies (Pan et al., 2000), in that there are a greater number of NOP containing cells than μ containing cells. Note that the staining patterns with the μ receptor antibody in the brain regions shown in Figure 7 are also consistent with the recently reported μ receptor-positive brain regions in the μ-mCherry knock-in mice (Gardon et al., 2014; Erbs et al., 2015).
We also investigated the colocalization of NOP and μ receptors in the DRG neurons. Figure 6G shows that NOP-eGFP and μ-receptors colocalize in small unmyelinated (NF200−) cells. Since most μ-positive neurons are known to coexpress CGRP (Scherrer et al., 2009; Bardoni et al., 2014), NOP-eGFP and μ receptors are colocalized in peptidergic C-nociceptors. In addition, the percentage of NOP-eGFP+ cells expressing μ receptors is ∼30%, which is consistent with that of NOP-eGFP+ CGRP+ NF200− small cells described above.
Discussion
NOP receptor localization has been previously characterized using in vitro autoradiography, and NOP receptor mRNA-containing cells were determined with in situ hybridization (Neal et al., 1999; Mollereau and Mouledous, 2000; Sim-Selley et al., 2003). There have also been previous immunohistochemical studies using various antibodies (Anton et al., 1996). However, one paper was withdrawn because it was later determined that the antibodies provided an identical staining pattern in NOP receptor knock-out mice [Corrigendum (1999) J Comp Neurol 412:708], and there never have been NOP receptor antibodies appropriately validated in this way. As an alternative to in vitro autoradiography, and to provide greatly increased resolution, knock-in mice with eGFP attached to the C-terminal of the NOP receptor were developed.
In the knock-in NOP-eGFP mice, the NOP receptor mRNA level does not change in the three genotypes, however, receptor number does appear to increase, similar to what was found for the δ-eGFP receptor (Scherrer et al., 2006). These results suggest that the eGFP-containing receptor might be translated with greater efficiency or perhaps is more stable in the membrane. However, the location of NOP receptors in the NOP-eGFP mice corresponds well to the location determined by in situ hybridization and in vitro autoradiography, and the receptors function appropriately both in vitro and in vivo. Consistent with the increase in receptor number, the knock-in mice have increased N/OFQ stimulated [35S]GTPγS binding. Furthermore, application of N/OFQ reversibly inhibited field EPSP, indicating that it inhibited synaptic transmission. This result is consistent with a report that showed the reversible inhibitory effect of N/OFQ on field EPSP and LTP in hippocampal slices (Yu and Xie, 1998). Also consistent with the wild-type mouse, systemic administration of the NOP agonist SR16835 blocks morphine antinociception. Therefore, the presence of the eGFP fused to the NOP receptor carboxy terminal does not appear to affect receptor function in the knock-in mouse, and the fluorescently labeled receptor should be useful for determining the location and trafficking of the receptor.
Previous in vitro autoradiography and in situ hybridization provided a basic understanding of brain regions involved in NOP receptor activation. Experiments with NOP-eGFP mice mostly confirmed these initial characterizations, with some significant differences. For instance, in situ hybridization demonstrated a very large amount of NOP receptor mRNA in DRG, however in vitro autoradiography found no [3H]N/OFQ binding in these cells (Neal et al., 1999). Current studies demonstrated considerable NOP-eGFP fluorescence in DRG. This is consistent with previous immunohistochemical studies that had been performed with unvalidated antibodies (Chen and Sommer, 2006) and electrophysiological experiments showing activity of N/OFQ on DRG neurons (Murali et al., 2012).
The presence of this receptor in DRG neurons and location in the spinal cord is consistent with the profound effect of intrathecally administered NOP agonists on nociception. Our findings demonstrated that NOP-eGFP receptors are expressed in various subpopulations of DRG neurons; coexpressed with CGRP, μ receptors, NF-200, and in IB4+ cells. A recent electrophysiological study demonstrated that the majority (84%) of small IB4-DRG neurons (peptidergic C-nociceptors) are responsive to both N/OFQ and DAMGO, whereas a small percentage of small neurons were N/OFQ-responsive IB4+ cells (Murali et al., 2012). Our immunohistochemical studies are consistent with these and demonstrate that a small percentage of NOP-eGFP-positive cells are IB4+ (nonpeptidergic), with a larger number coexpressing CGRP (and therefore peptidergic). These data suggest that the NOP-N/OFQ system functions more by inhibition of peptidergic nociceptors, which are essential to acute heat pain and heat hyperalgesia. Furthermore, recent literature has shown that δ receptors are present on a subpopulation of IB4+ C-fibers, which modulate mechanical pain (Scherrer et al., 2009; Bardoni et al., 2014). These results might suggest that NOP receptor-containing IB4+ cells are nonpeptidergic C-nociceptors, which might cooperate with δ receptors to regulate acute mechanical pain. We also found a number of myelinated DRG neurons expressing NOP-eGFP. Our results demonstrated that medium myelinated primary afferents express NOP-eGFP receptors but not together with CGRP. These primary afferents are not typical A-δ nociceptors, suggesting the possible presence of NOP receptors on myelinated low-threshold mechanoreceptors (Luo et al., 2009; Abraira and Ginty, 2013; Bardoni et al., 2014). NOP-eGFP immunoreactivity was also observed in the ventral border of lamina IIinner, which receives low-threshold myelinated primary afferent inputs and is known to be involved in the development of injury-related allodynia (Malmberg et al., 1997; Neumann et al., 2008). These results support our previous finding on the anti-allodynic action of systemically administered small molecule NOP agonists (Khroyan et al., 2011). A-fibers can be identified by determining the neurotrophin receptors that they express (Bourane et al., 2009; Bardoni et al., 2014). Further molecular characterization using a variety of additional markers will be required to fully resolve the identity of these primary afferents.
As expected, NOP receptors are very highly expressed in a number of brain regions involved in both pain and reward. Pain related brain regions, in addition to laminae I and II of the spinal cord and DRG, include the vlPAG, thalamus, LC, and MHb, among others. Brain regions known to be involved in drug reward, including the VTA, NAc, MHb, IPN, Amy, and Hyp also all express high levels of NOP receptors (Fig. 4; Neal et al., 1999). The exact anatomy with relation to the μ receptor has not yet been fully characterized. This is important because, unlike in the spinal cord where NOP receptor activation resembles μ receptor activation with respect to physiological function (Xu et al., 1996; Jhamandas et al., 1998), in the brain N/OFQ and small molecule NOP agonists reverse the actions of morphine with respect to both pain and reward (Mogil et al., 1996a; Murphy et al., 1999; Sakoori and Murphy, 2004). In fact, the actions of N/OFQ are contrary to μ agonists in specific brain regions, despite colocalization. For instance, μ agonists morphine and DAMGO mediate an antinociceptive response when injected directly into the PAG (Lewis and Gebhart, 1977; Rossi et al., 1994). N/OFQ coinjected into the PAG blocks the morphine-mediated antinociception (Morgan et al., 1997). μ Receptors are present and modulate Ca2+ channels on ∼40% of the neurons in the vlPAG, whereas NOP receptors modulate Ca2+ currents on all cells in this region (Connor and Christie, 1998). Therefore, despite the fact that both receptors can be identified on the same cells, and transduce a similar signal, the additional NOP containing cells can attenuate the actions of the μ-containing cells. Studies are underway to better characterize the various cell types to understand the anti-opiate actions of N/OFQ in the PAG.
Morphine and other μ agonists have been postulated to induce reward in the VTA by inhibiting GABA interneurons thereby disinhibiting the dopaminergic cells projecting to the NAc (Johnson and North, 1992). In contrast, N/OFQ delivered into the VTA blocks the cocaine-induced increase in locomotor activity and extracellular dopamine in the NAc (Murphy and Maidment, 1999; Narayanan et al., 2004). Studies are underway to identify the NOP-containing cells in the VTA to determine the mechanism by which NOP and μ-receptor activation have such different outcomes. N/OFQ also blocks a cocaine-induced increase in extracellular dopamine in the NAc when reverse dialized into the NAc (Vazquez-DeRose et al., 2013), blocks release of hypocretin/orexin from hypocretin-containing cells in the lateral hypothalamus (Xie et al., 2008), and acts in opposition to CRF in the amygdala (Ciccocioppo et al., 2004). All of these actions can attenuate drug reward, and it might be the presence of NOP receptors in so many regions involved in drug abuse and stress-induced relapse that renders N/OFQ and small molecule NOP receptor agonists so effective in blocking CPP of so many abused drugs (Kotlinska et al., 2003; Kuzmin et al., 2003; Sakoori and Murphy, 2004).
One potential use of fluorescent-labeled receptors is to investigate agonist-induced trafficking. For the δ-eGFP receptor, a large fraction of the receptors appear on the plasma membrane, and with agonist stimulation internalization could be easily visualized (Scherrer et al., 2006; Pradhan et al., 2009). NOP-eGFP receptors seem to be more similar to the newly described μ-mCherry receptor, in which the receptors are found throughout the cell, with low fluorescence detectable at the cell surface (Erbs et al., 2015). Accordingly, although some cells and cell processes appear more punctate in primary culture after agonist administration, fluorescence changes upon receptor activation cannot be easily demonstrated or quantified under the conditions that we have used.
In conclusion, the eGFP tag on NOP receptors in knock-in mice provides a detailed description of the presence of NOP receptors in brain, spinal cord, and DRG sensory neurons that is consistent with the known pharmacology of NOP-mediated activity. A detailed examination of the cellular location of NOP receptors compared with μ receptors should provide explanations as to how NOP receptor activation attenuates μ-mediated actions with respect to both pain and reward when administered supraspinally. Overall, these mice should become a valuable tool to further examine the importance of NOP receptors to other sensory modalities in the spinal cord and peripheral nerves, as well as the involvement of supraspinal sites in modulating opioid analgesia and addiction.
Footnotes
This work was supported by NIH Grants DA023281 to L.T., DA031777 to G.S., DA027811 to N.T.Z., and the State of Florida Executive Office of the Governor's Department of Economic Opportunity. We thank Jennifer Schoch, Michelle Weger, and Kelly Gaiolini for excellent technical assistance, the Mouse Clinic Institute, Strasbourg France for making the knock-in mice, and the Imaging Facility at Vaccine & Gene Therapy Institute of Florida and the Optical Workshop and Light Microscopy Facility Center at the MAX PLANCK Florida institute for the use of the microscopes.
The authors declare no competing financial interests.
References
- Abraira VE, Ginty DD. The sensory neurons of touch. Neuron. 2013;79:618–639. doi: 10.1016/j.neuron.2013.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adapa ID, Toll L. Relationship between binding affinity and functional activity of nociceptin/orphanin FQ. Neuropeptides. 1997;31:403–408. doi: 10.1016/S0143-4179(97)90032-9. [DOI] [PubMed] [Google Scholar]
- Anton B, Fein J, To T, Li X, Silberstein L, Evans CJ. Immunohistochemical localization of ORL-1 in the central nervous system of the rat. J Comp Neurol. 1996;368:229–251. doi: 10.1002/(SICI)1096-9861(19960429)368:2<229::AID-CNE5>3.3.CO%3B2-G. [DOI] [PubMed] [Google Scholar]
- Bardoni R, Tawfik VL, Wang D, François A, Solorzano C, Shuster SA, Choudhury P, Betelli C, Cassidy C, Smith K, de Nooij JC, Mennicken F, O'Donnell D, Kieffer BL, Woodbury CJ, Basbaum AI, MacDermott AB, Scherrer G. Delta opioid receptors presynaptically regulate cutaneous mechanosensory neuron input to the spinal cord dorsal horn. Neuron. 2014;81:1312–1327. doi: 10.1016/j.neuron.2014.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basbaum AI, Bautista DM, Scherrer G, Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139:267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourane S, Garces A, Venteo S, Pattyn A, Hubert T, Fichard A, Puech S, Boukhaddaoui H, Baudet C, Takahashi S, Valmier J, Carroll P. Low-threshold mechanoreceptor subtypes selectively express MafA and are specified by Ret signaling. Neuron. 2009;64:857–870. doi: 10.1016/j.neuron.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Brewer GJ. Isolation and culture of adult rat hippocampal neurons. J Neurosci Methods. 1997;71:143–155. doi: 10.1016/S0165-0270(96)00136-7. [DOI] [PubMed] [Google Scholar]
- Cavanaugh DJ, Lee H, Lo L, Shields SD, Zylka MJ, Basbaum AI, Anderson DJ. Distinct subsets of unmyelinated primary sensory fibers mediate behavioral responses to noxious thermal and mechanical stimuli. Proc Natl Acad Sci U S A. 2009;106:9075–9080. doi: 10.1073/pnas.0901507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Sommer C. Nociceptin and its receptor in rat dorsal root ganglion neurons in neuropathic and inflammatory pain models: implications on pain processing. J Peripher Nerv Syst. 2006;11:232–240. doi: 10.1111/j.1529-8027.2006.0093.x. [DOI] [PubMed] [Google Scholar]
- Ciccocioppo R, Angeletti S, Sanna PP, Weiss F, Massi M. Effect of nociceptin/orphanin FQ on the rewarding properties of morphine. Eur J Pharmacol. 2000;404:153–159. doi: 10.1016/S0014-2999(00)00590-2. [DOI] [PubMed] [Google Scholar]
- Ciccocioppo R, Cippitelli A, Economidou D, Fedeli A, Massi M. Nociceptin/orphanin FQ acts as a functional antagonist of corticotropin-releasing factor to inhibit its anorectic effect. Physiol Behav. 2004;82:63–68. doi: 10.1016/j.physbeh.2004.04.035. [DOI] [PubMed] [Google Scholar]
- Connor M, Christie MJ. Modulation of Ca2+ channel currents of acutely dissociated rat periaqueductal grey neurons. J Physiol. 1998;509:47–58. doi: 10.1111/j.1469-7793.1998.047bo.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbani M, Gonindard C, Meunier JC. Ligand-regulated internalization of the opioid receptor-like 1: a confocal study. Endocrinology. 2004;145:2876–2885. doi: 10.1210/en.2004-0062. [DOI] [PubMed] [Google Scholar]
- Cox BM, Christie MJ, Devi L, Toll L, Traynor JR. Challenges for opioid receptor nomenclature: IUPHAR review 9. Br J Pharmacol. 2015;172:317–323. doi: 10.1111/bph.12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darland T, Heinricher MM, Grandy DK. Orphanin FQ/nociceptin: a role in pain and analgesia, but so much more. Trends Neurosci. 1998;21:215–221. doi: 10.1016/S0166-2236(97)01204-6. [DOI] [PubMed] [Google Scholar]
- Erbs E, Faget L, Scherrer G, Matifas A, Filliol D, Vonesch JL, Koch M, Kessler P, Hentsch D, Birling MC, Koutsourakis M, Vasseur L, Veinante P, Kieffer BL, Massotte D. A mu-delta opioid receptor brain atlas reveals neuronal co-occurrence in subcortical networks. Brain Struct Funct. 2015;220:677–702. doi: 10.1007/s00429-014-0717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flau K, Redmer A, Liedtke S, Kathmann M, Schlicker E. Inhibition of striatal and retinal dopamine release via nociceptin/orphanin FQ receptors. Br J Pharmacol. 2002;137:1355–1361. doi: 10.1038/sj.bjp.0704998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardon O, Faget L, Chu Sin Chung P, Matifas A, Massotte D, Kieffer BL. Expression of μ opioid receptor in dorsal diencephalic conduction system: new insights for the medial habenula. Neuroscience. 2014;277:595–609. doi: 10.1016/j.neuroscience.2014.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Watanabe M, Ichikawa T, Kobayashi T, Yano R, Kumanishi T. Distribution of prepro-nociceptin/orphanin FQ mRNA and its receptor mRNA in developing and adult mouse central nervous systems. J Comp Neurol. 1998;399:139–151. doi: 10.1002/(SICI)1096-9861(19980914)399:1<139::AID-CNE11>3.0.CO%3B2-C. [DOI] [PubMed] [Google Scholar]
- Jhamandas KH, Sutak M, Henderson G. Antinociceptive and morphine modulatory actions of spinal orphanin FQ. Can J Physiol Pharmacol. 1998;76:314–324. doi: 10.1139/y98-019. [DOI] [PubMed] [Google Scholar]
- Johnson SW, North RA. Opioids excite dopamine neurons by hyperpolarization of local interneurons. J Neurosci. 1992;12:483–488. doi: 10.1523/JNEUROSCI.12-02-00483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khroyan TV, Zaveri NT, Polgar WE, Orduna J, Olsen C, Jiang F, Toll L. SR 16435 [1-(1-(bicyclo[3.3.1]nonan-9-yl)piperidin-4-yl)indolin-2-one], a novel mixed nociceptin/orphanin FQ/mu-opioid receptor partial agonist: analgesic and rewarding properties in mice. J Pharmacol Exp Ther. 2007;320:934–943. doi: 10.1124/jpet.106.111997. [DOI] [PubMed] [Google Scholar]
- Khroyan TV, Polgar WE, Orduna J, Montenegro J, Jiang F, Zaveri NT, Toll L. Differential effects of nociceptin/orphanin FQ (NOP) receptor agonists in acute versus chronic pain: studies with bifunctional NOP/mu receptor agonists in the sciatic nerve ligation chronic pain model in mice. J Pharmacol Exp Ther. 2011;339:687–693. doi: 10.1124/jpet.111.184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen I, Slowe SJ, Matthes HW, Kieffer B. Quantitative autoradiographic mapping of mu-, delta- and kappa-opioid receptors in knock-out mice lacking the mu-opioid receptor gene. Brain Res. 1997;778:73–88. doi: 10.1016/S0006-8993(97)00988-8. [DOI] [PubMed] [Google Scholar]
- Kotlinska J, Rafalski P, Biala G, Dylag T, Rolka K, Silberring J. Nociceptin inhibits acquisition of amphetamine-induced place preference and sensitization to stereotypy in rats. Eur J Pharmacol. 2003;474:233–239. doi: 10.1016/S0014-2999(03)02081-8. [DOI] [PubMed] [Google Scholar]
- Kuzmin A, Sandin J, Terenius L, Ogren SO. Acquisition, expression, and reinstatement of ethanol-induced conditioned place preference in mice: effects of opioid receptor-like 1 receptor agonists and naloxone. J Pharmacol Exp Ther. 2003;304:310–318. doi: 10.1124/jpet.102.041350. [DOI] [PubMed] [Google Scholar]
- Lambert DG. The nociceptin/orphanin FQ receptor: a target with broad therapeutic potential. Nat Rev Drug Discov. 2008;7:694–710. doi: 10.1038/nrd2572. [DOI] [PubMed] [Google Scholar]
- Lewis VA, Gebhart GF. Evaluation of the periaqueductal central gray (PAG) as a morphine-specific locus of action and examination of morphine-induced and stimulation-produced analgesia at coincident PAG loci. Brain Res. 1977;124:283–303. doi: 10.1016/0006-8993(77)90886-1. [DOI] [PubMed] [Google Scholar]
- Luo W, Enomoto H, Rice FL, Milbrandt J, Ginty DD. Molecular identification of rapidly adapting mechanoreceptors and their developmental dependence on ret signaling. Neuron. 2009;64:841–856. doi: 10.1016/j.neuron.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg AB, Chen C, Tonegawa S, Basbaum AI. Preserved acute pain and reduced neuropathic pain in mice lacking PKCgamma. Science. 1997;278:279–283. doi: 10.1126/science.278.5336.279. [DOI] [PubMed] [Google Scholar]
- Meunier JC, Mollereau C, Toll L, Suaudeau C, Moisand C, Alvinerie P, Butour JL, Guillemot JC, Ferrara P, Monsarrat B. Isolation and structure of the endogenous agonist of opioid receptor-like ORL1 receptor. Nature. 1995;377:532–535. doi: 10.1038/377532a0. [DOI] [PubMed] [Google Scholar]
- Mogil JS, Pasternak GW. The molecular and behavioral pharmacology of the orphanin FQ/nociceptin peptide and receptor family. Pharmacol Rev. 2001;53:381–415. [PubMed] [Google Scholar]
- Mogil JS, Grisel JE, Zhangs G, Belknap JK, Grandy DK. Functional antagonism of mu-, delta- and kappa-opioid antinociception by orphanin FQ. Neurosci Lett. 1996a;214:131–134. doi: 10.1016/0304-3940(96)12917-7. [DOI] [PubMed] [Google Scholar]
- Mogil JS, Grisel JE, Reinscheid RK, Civelli O, Belknap JK, Grandy DK. Orphanin FQ is a functional anti-opioid peptide. Neuroscience. 1996b;75:333–337. doi: 10.1016/0306-4522(96)00338-7. [DOI] [PubMed] [Google Scholar]
- Mollereau C, Mouledous L. Tissue distribution of the opioid receptor-like (ORL1) receptor. Peptides. 2000;21:907–917. doi: 10.1016/S0196-9781(00)00227-8. [DOI] [PubMed] [Google Scholar]
- Morgan MM, Grisel JE, Robbins CS, Grandy DK. Antinociception mediated by the periaqueductal gray is attenuated by orphanin FQ. Neuroreport. 1997;8:3431–3434. doi: 10.1097/00001756-199711100-00003. [DOI] [PubMed] [Google Scholar]
- Murali SS, Napier IA, Rycroft BK, Christie MJ. Opioid-related (ORL1) receptors are enriched in a subpopulation of sensory neurons and prolonged activation produces no functional loss of surface N-type calcium channels. J Physiol. 2012;590:1655–1667. doi: 10.1113/jphysiol.2012.228429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy NP, Maidment NT. Orphanin FQ/nociceptin modulation of mesolimbic dopamine transmission determined by microdialysis. J Neurochem. 1999;73:179–186. doi: 10.1046/j.1471-4159.1999.0730179.x. [DOI] [PubMed] [Google Scholar]
- Murphy NP, Lee Y, Maidment NT. Orphanin FQ/nociceptin blocks acquisition of morphine place preference. Brain Res. 1999;832:168–170. doi: 10.1016/S0006-8993(99)01425-0. [DOI] [PubMed] [Google Scholar]
- Narayanan S, Lam H, Carroll FI, Lutfy K. Orphanin FQ/nociceptin suppresses motor activity through an action along the mesoaccumbens axis in rats. J Psychiatry Neurosci. 2004;29:116–123. [PMC free article] [PubMed] [Google Scholar]
- Neal CR, Jr, Mansour A, Reinscheid R, Nothacker HP, Civelli O, Akil H, Watson SJ., Jr Opioid receptor-like (ORL1) receptor distribution in the rat central nervous system: comparison of ORL1 receptor mRNA expression with (125)I-[(14)Tyr]-orphanin FQ binding. J Comp Neurol. 1999;412:563–605. doi: 10.1002/(SICI)1096-9861(19991004)412:4<563::AID-CNE2>3.3.CO%3B2-Q. [DOI] [PubMed] [Google Scholar]
- Neumann S, Braz JM, Skinner K, Llewellyn-Smith IJ, Basbaum AI. Innocuous, not noxious, input activates PKCgamma interneurons of the spinal dorsal horn via myelinated afferent fibers. J Neurosci. 2008;28:7936–7944. doi: 10.1523/JNEUROSCI.1259-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olianas MC, Dedoni S, Boi M, Onali P. Activation of nociceptin/orphanin FQ-NOP receptor system inhibits tyrosine hydroxylase phosphorylation, dopamine synthesis, and dopamine D(1) receptor signaling in rat nucleus accumbens and dorsal striatum. J Neurochem. 2008;107:544–556. doi: 10.1111/j.1471-4159.2008.05629.x. [DOI] [PubMed] [Google Scholar]
- Pan Z, Hirakawa N, Fields HL. A cellular mechanism for the bidirectional pain-modulating actions of orphanin FQ/nociceptin. Neuron. 2000;26:515–522. doi: 10.1016/S0896-6273(00)81183-6. [DOI] [PubMed] [Google Scholar]
- Pradhan AA, Becker JA, Scherrer G, Tryoen-Toth P, Filliol D, Matifas A, Massotte D, Gavériaux-Ruff C, Kieffer BL. In vivo delta opioid receptor internalization controls behavioral effects of agonists. PLoS One. 2009;4:e5425. doi: 10.1371/journal.pone.0005425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinscheid RK, Nothacker HP, Bourson A, Ardati A, Henningsen RA, Bunzow JR, Grandy DK, Langen H, Monsma FJ, Jr, Civelli O. Orphanin FQ: a neuropeptide that activates an opioidlike G-protein-coupled receptor. Science. 1995;270:792–794. doi: 10.1126/science.270.5237.792. [DOI] [PubMed] [Google Scholar]
- Rossi GC, Pasternak GW, Bodnar RJ. Mu and delta opioid synergy between the periaqueductal gray and the rostro-ventral medulla. Brain Res. 1994;665:85–93. doi: 10.1016/0006-8993(94)91155-X. [DOI] [PubMed] [Google Scholar]
- Sakoori K, Murphy NP. Central administration of nociceptin/orphanin FQ blocks the acquisition of conditioned place preference to morphine and cocaine, but not conditioned place aversion to naloxone in mice. Psychopharmacology. 2004;172:129–136. doi: 10.1007/s00213-003-1643-3. [DOI] [PubMed] [Google Scholar]
- Scherrer G, Tryoen-Tóth P, Filliol D, Matifas A, Laustriat D, Cao YQ, Basbaum AI, Dierich A, Vonesh JL, Gavériaux-Ruff C, Kieffer BL. Knockin mice expressing fluorescent delta-opioid receptors uncover G protein-coupled receptor dynamics in vivo. Proc Natl Acad Sci U S A. 2006;103:9691–9696. doi: 10.1073/pnas.0603359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherrer G, Imamachi N, Cao YQ, Contet C, Mennicken F, O'Donnell D, Kieffer BL, Basbaum AI. Dissociation of the opioid receptor mechanisms that control mechanical and heat pain. Cell. 2009;137:1148–1159. doi: 10.1016/j.cell.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim-Selley LJ, Vogt LJ, Childers SR, Vogt BA. Distribution of ORL-1 receptor binding and receptor-activated G-proteins in rat forebrain and their experimental localization in anterior cingulate cortex. Neuropharmacology. 2003;45:220–230. doi: 10.1016/S0028-3908(03)00155-2. [DOI] [PubMed] [Google Scholar]
- Spampinato S, Di Toro R, Alessandri M, Murari G. Agonist-induced internalization and desensitization of the human nociceptin receptor expressed in CHO cells. Cell Mol Life Sci. 2002;59:2172–2183. doi: 10.1007/s000180200016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian JH, Xu W, Fang Y, Mogil JS, Grisel JE, Grandy DK, Han JS. Bidirectional modulatory effect of orphanin FQ on morphine-induced analgesia: antagonism in brain and potentiation in spinal cord of the rat. Br J Pharmacol. 1997;120:676–680. doi: 10.1038/sj.bjp.0700942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynor JR, Nahorski SR. Modulation by mu-opioid agonists of guanosine-5′-O-(3-[35S]thio)triphosphate binding to membranes from human neuroblastoma SH-SY5Y cells. Mol Pharmacol. 1995;47:848–854. doi: 10.1016/S0026-895X(25)08634-1. [DOI] [PubMed] [Google Scholar]
- Ueda H, Inoue M, Takeshima H, Iwasawa Y. Enhanced spinal nociceptin receptor expression develops morphine tolerance and dependence. J Neurosci. 2000;20:7640–7647. doi: 10.1523/JNEUROSCI.20-20-07640.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-DeRose J, Stauber G, Khroyan TV, Xie XS, Zaveri NT, Toll L. Retrodialysis of N/OFQ into the nucleus accumbens shell blocks cocaine-induced increases in extracellular dopamine and locomotor activity. Eur J Pharmacol. 2013;699:200–206. doi: 10.1016/j.ejphar.2012.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrontou S, Wong AM, Rau KK, Koerber HR, Anderson DJ. Genetic identification of C fibres that detect massage-like stroking of hairy skin in vivo. Nature. 2013;493:669–673. doi: 10.1038/nature11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin JM, Statnick MA, Rorick-Kehn LM, Pintar JE, Ansonoff M, Chen Y, Tucker RC, Ciccocioppo R. The biology of nociceptin/orphanin FQ (N/OFQ) related to obesity, stress, anxiety, mood, and drug dependence. Pharmacol Ther. 2014;141:283–299. doi: 10.1016/j.pharmthera.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Lee MR, Choi S, Kim T, Choi DS. ENT1 regulates ethanol-sensitive EAAT2 expression and function in astrocytes. Alcohol Clin Exp Res. 2010;34:1110–1117. doi: 10.1111/j.1530-0277.2010.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Wisor JP, Hara J, Crowder TL, LeWinter R, Khroyan TV, Yamanaka A, Diano S, Horvath TL, Sakurai T, Toll L, Kilduff TS. Hypocretin/orexin and nociceptin/orphanin FQ coordinately regulate analgesia in a mouse model of stress-induced analgesia. J Clin Invest. 2008;118:2471–2481. doi: 10.1172/JCI35115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XJ, Hao JX, Wiesenfeld-Hallin Z. Nociceptin or antinociceptin: potent spinal antinociceptive effect of orphanin FQ/nociceptin in the rat. Neuroreport. 1996;7:2092–2094. [PubMed] [Google Scholar]
- Yu TP, Xie CW. Orphanin FQ/nociceptin inhibits synaptic transmission and long-term potentiation in rat dentate gyrus through postsynapticmechanisms. J Neurophysiol. 1998;80:1277–1284. doi: 10.1152/jn.1998.80.3.1277. [DOI] [PubMed] [Google Scholar]