Abstract
Cladosporium species are ubiquitous, saprobic, dematiaceous fungi, only infrequently associated with human and animal opportunistic infections. We have studied a large set of Cladosporium isolates recovered from clinical samples in the United States to ascertain the predominant species there in light of recent taxonomic changes in this genus and to determine whether some could possibly be rare potential pathogens. A total of 92 isolates were identified using phenotypic and molecular methods, which included sequence analysis of the internal transcribed spacer (ITS) region and a fragment of the large subunit (LSU) of the nuclear ribosomal DNA (rDNA), as well as fragments of the translation elongation factor 1 alpha (EF-1α) and actin (Act) genes. The most frequent species was Cladosporium halotolerans (14.8%), followed by C. tenuissimum (10.2%), C. subuliforme (5.7%), and C. pseudocladosporioides (4.5%). However, 39.8% of the isolates did not correspond to any known species and were deemed to comprise at least 17 new lineages for Cladosporium. The most frequent anatomic site of isolation was the respiratory tract (54.5%), followed by superficial (28.4%) and deep tissues and fluids (14.7%). Species of the two recently described cladosporiumlike genera Toxicocladosporium and Penidiella are reported for the first time from clinical samples. In vitro susceptibility testing of 92 isolates against nine antifungal drugs showed a variety of results but high activity overall for the azoles, echinocandins, and terbinafine.
INTRODUCTION
Cladosporium species are among the most common fungal inhabitants worldwide, being isolated from almost any environmental source and geographic location (1). The genus is characterized by the typical form of its conidiophores, which are erect, straight or geniculate, produce abundant branched acropetal chains of smooth to roughened dry conidia, and show a distinct darkened coronate hilum, i.e., conidial scar characterized by a thick rim surrounding a central convex dome (2, 3). The relatively small conidia are easily detached and disseminated by the wind, Cladosporium being one of the most frequently isolated airborne fungi (2, 4).
The most common Cladosporium species are primarily isolated from soil and plant material, where they are frequently encountered as saprobes or secondary invaders on follicular lesions, concomitant with other plant-pathogenic fungi (1, 5, 6). However, several species are important pathogens of plants and some are also able to affect animals, including humans (7–9). Cladosporium is usually associated with allergic rhinitis (10) or localized superficial or deep lesions (11–14) but, rarely, can cause disseminated infections (7, 15–17).
The genus Cladosporium has been shown to be both morphologically and phylogenetically heterogeneous (18). On the basis of molecular data, the true human-pathogenic species C. bantiana, C. carrionii, and C. devriesii, characterized by their thermotolerance and the absence of conidiophores with pigmented conidial scars, were transferred to Cladophialophora (1, 7, 18). More recently, Cladosporium underwent extensive revisions based on polyphasic approaches (1, 3, 19–21), which resulted in the delimitation of 169 species currently accepted in Cladosporium sensu stricto (Cladosporiaceae, Capnodiales). On the other hand, a great number of taxa were excluded from that genus, now being considered doubtful species or accommodated into several related new genera, such as Hyalodendriella, Ochrocladosporium, Rachicladosporium, Rhizocladosporium, Toxicocladosporium, and Verrucocladosporium (1, 3).
The diversity of Cladosporium species associated with human disease is currently reduced to four, i.e., C. cladosporioides, C. herbarum, C. oxysporum, and C. sphaerospermum (7). Most of these data, however, are based on a reduced number of clinical cases with the identification of the etiological agents not confirmed by reliable methods. Moreover, three of the clinically relevant species, C. cladosporioides, C. herbarum, and C. sphaerospermum, have been demonstrated to be species complexes (19–21) encompassing several morphologically sibling species that can only be distinguished by means of phylogenetic analyses (1, 7). The clinical significance of these phylogenetic species, however, has yet to be evaluated (22).
The objective of this work was to assess the diversity of Cladosporium species associated with human and animal disease by analyzing a large set of isolates from clinical specimens by means of phenotypic and DNA sequence data analyses. In addition, the in vitro susceptibility of these isolates was evaluated against nine clinically available antifungal drugs.
MATERIALS AND METHODS
Fungal isolates.
A total of 92 isolates tentatively identified as Cladosporium spp. were included in this study (Table 1). All of the isolates were obtained from human and animal clinical specimens, mostly from the United States, received in the Fungus Testing Laboratory at the University of Texas Health Science Center at San Antonio (UTHSC) from different parts of the country mainly for identification purposes.
TABLE 1.
Clinical isolates, type or reference strains, and sequences included in this study
| Species | Strain/isolate no.a | Origin (country)b | GenBank nucleotide accession no. for: |
|||
|---|---|---|---|---|---|---|
| ITS | LSU | EF-1α | Act | |||
| Cercospora beticola | CBS 116456T | Beta vulgaris (Italy) | NR121315 | GU214404 | AY840494 | AY840458 |
| Cercospora olivascens | CBS 253.67T | Unknown | NR111773 | |||
| Cladosporium allicinum | CBS 121624T | Hordeum vulgare (Belgium) | EF679350 | EF679425 | EF679502 | |
| UTHSC DI-13-173 | Human, lung (USA) | LN834353 | LN834449 | LN834537 | ||
| UTHSC DI-13-176 | Human, skin (USA) | LN834354 | LN834450 | LN834538 | ||
| UTHSC DI-13-266 | Canine, skin (USA) | LN834355 | LN834451 | LN834539 | ||
| Cladosporium angustisporum | CBS 125983T | Alloxylon wickhamii (Australia) | HM147995 | HM148236 | HM148482 | |
| UTHSC DI-13-240 | Human, toe nail (USA) | LN834356 | LN834452 | LN834540 | ||
| Cladosporium asperulatum | CBS 126339 | Eucalyptus leaf litter (India) | HM147997 | HM148238 | HM148484 | |
| CBS 126340T | Protea susannae (Portugal) | HM147998 | HM148239 | HM148485 | ||
| UTHSC DI-13-216 | Feline, nasal (USA) | LN834357 | LN834453 | LN834541 | ||
| Cladosporium cladosporioides | CBS 101367 | Soil (Brasil) | HM148002 | HM148243 | HM148489 | |
| CBS 112388T | Indoor air (Germany) | HM148003 | HM148244 | HM148490 | ||
| UTHSC DI-13-204 | Human, abdomen (USA) | LN834358 | LN834454 | LN834542 | ||
| UTHSC DI-13-209 | Human, pleural (USA) | LN834359 | LN834455 | LN834543 | ||
| UTHSC DI-13-215 | Human, sputum (USA) | LN834360 | LN834456 | LN834544 | ||
| Cladosporium colocasiae | CBS 386.64T | Colocasia antiquorum (Taiwan) | HM148067 | HM148310 | HM148555 | |
| CBS 119542 | Colocasia esculanta (Taiwan) | HM148066 | HM148309 | HM148554 | ||
| Cladosporium cucumerinum | CBS 171.52T | Fruit of Cucumis sativus (Netherlands) | HM148072 | HM148316 | HM148561 | |
| CBS 173.54 | Fruit of Cucumis sativus (Netherlands) | HM148074 | HM148318 | HM148563 | ||
| Cladosporium flabelliforme | CBS 126345T | Melaleuca cajuputi (Australia) | HM148092 | HM148336 | HM148581 | |
| UTHSC DI-13-267 | Human, sputum (USA) | LN834361 | LN834457 | LN834545 | ||
| Cladosporium funiculosum | CBS 122128 | Unknown | HM148093 | HM148337 | HM148582 | |
| CBS 122129T | Leaf of Vigna umbellata (Japan) | HM148094 | HM148338 | HM148583 | ||
| UTHSC DI-13-175 | Human, BAL fluid (USA) | LN834362 | LN834458 | LN834546 | ||
| UTHSC DI-13-223 | Human, BAL fluid (USA) | LN834363 | LN834459 | LN834547 | ||
| UTHSC DI-13-242 | Human, nasal wash (USA) | LN834364 | LN834460 | LN834548 | ||
| Cladosporium halotolerans | CBS 119416T | Hypersaline water (Namibia) | DQ780364 | JN906989 | EF101397 | |
| FMR 13493 | Human, unknown (Spain) | LN834365 | LN834461 | LN834549 | ||
| UTHSC DI-13-164 | Human, bone marrow (USA) | LN834366 | LN834462 | LN834550 | ||
| UTHSC DI-13-182 | Marine mammal, dermis (USA) | LN834367 | LN834463 | LN834551 | ||
| UTHSC DI-13-183 | Human, bronchus (USA) | LN834368 | LN834464 | LN834552 | ||
| UTHSC DI-13-206 | Human, BAL fluid (USA) | LN834369 | LN834465 | LN834553 | ||
| UTHSC DI-13-213 | Human, lymph node (USA) | LN834370 | LN834466 | LN834554 | ||
| UTHSC DI-13-221 | Human, bone marrow (USA) | LN834371 | LN834467 | LN834555 | ||
| UTHSC DI-13-231 | Catheter tip (USA) | LN834372 | LN834468 | LN834556 | ||
| UTHSC DI-13-249 | Human, nasal (USA) | LN834373 | LN834469 | LN834557 | ||
| UTHSC DI-13-250 | Human, scalp (USA) | LN834374 | LN834470 | LN834558 | ||
| UTHSC DI-13-252 | Human, toe nail (USA) | LN834375 | LN834471 | LN834559 | ||
| UTHSC DI-13-259 | Human, BAL fluid (USA) | LN834376 | LN834472 | LN834560 | ||
| UTHSC DI-13-263 | Human, BAL fluid (USA) | LN834377 | LN834473 | LN834561 | ||
| Cladosporium herbaroides | CBS 121626T | Hypersaline water (Israel) | EF679357 | EF679432 | EF679509 | |
| Cladosporium herbarum | CBS 121621T | Hordeum vulgare (Netherlands) | EF679363 | EF679440 | EF679516 | |
| UTHSC DI-13-220 | Human, BAL fluid (USA) | LN834378 | LN834474 | LN834562 | ||
| Cladosporium iranicum | CBS 126346T | Leaf of Citrus sinensis (Iran) | HM148110 | HM148354 | HM148599 | |
| Cladosporium iridis | CBS 138.40T | Leaf of Iris sp. (Netherlands) | EF679370 | EF679447 | EF679523 | |
| Cladosporium macrocarpum | CBS 121623T | Spinacia oleracea (USA) | EF679375 | EF679453 | EF679529 | |
| UTHSC DI-13-191 | Human, face (USA) | LN834379 | LN834475 | LN834563 | ||
| Cladosporium oxysporum | CBS 125991 | Soil (China) | HM148118 | HM148362 | HM148607 | |
| CBS 126351 | Indoor air (Venezuela) | HM148119 | HM148363 | HM148608 | ||
| Cladosporium perangustum | CBS 125996T | Cussonia sp. (South Africa) | HM148121 | HM148365 | HM148610 | |
| UTHSC DI-13-208 | Canine, BAL fluid (USA) | LN834380 | LN834476 | LN834564 | ||
| Cladosporium pseudocladosporioides | CBS 117153 | Leaf of Paeonia sp. (Germany) | HM148157 | HM148401 | HM148646 | |
| CBS 125993T | Outside air (Netherlands) | HM148158 | HM148402 | HM148647 | ||
| UTHSC DI-13-187 | Turtle, unknown (USA) | LN834381 | LN834477 | LN834565 | ||
| UTHSC DI-13-232 | Human, shoulder (USA) | LN834382 | LN834478 | LN834566 | ||
| UTHSC DI-13-233 | Human, BAL fluid (USA) | LN834383 | LN834479 | LN834567 | ||
| UTHSC DI-13-261 | Human, sputum (USA) | LN834384 | LN834480 | LN834568 | ||
| Cladosporium ramotenellum | CBS 121628T | Hypersaline water (Slovenia) | EF679384 | EF679462 | EF679538 | |
| UTHSC DI-13-166 | Human, nasal tissue (USA) | LN834385 | LN834481 | LN834569 | ||
| UTHSC DI-13-222 | Animal, Nasal (USA) | LN834386 | LN834482 | LN834570 | ||
| UTHSC DI-13-224 | Animal, Nasal (USA) | LN834387 | LN834483 | LN834571 | ||
| Cladosporium sinuosum | CBS 121629T | Fuchsia excorticata (New Zealand) | EF679386 | EF679464 | EF679540 | |
| Cladosporium sphaerospermum | CBS 193.54T | Human, nail (Netherlands) | DQ780343 | EU570261 | EU570269 | |
| UTHSC DI-13-184 | Frog, abscess (USA) | LN834388 | LN834484 | LN834572 | ||
| UTHSC DI-13-229 | Human, BAL fluid (USA) | LN834389 | LN834485 | LN834573 | ||
| UTHSC DI-13-237 | Human, BAL fluid (USA) | LN834390 | LN834486 | LN834574 | ||
| Cladosporium subinflatum | CBS 121630T | Hypersaline water (Slovenia) | EF679389 | EF679467 | EF679543 | |
| UTHSC DI-13-189 | Human, toe nail (USA) | LN834391 | LN834487 | LN834575 | ||
| Cladosporium subtilissimum | CBS 113754T | Grape berry (USA) | EF679397 | EF679475 | EF679551 | |
| Cladosporium subuliforme | CBS 126500T | Chamaedorea metallica (Thailand) | HM148196 | HM148441 | HM148686 | |
| UTHSC DI-13-171 | Human, CSF (USA) | LN834392 | LN834488 | LN834576 | ||
| UTHSC DI-13-180 | Human, BAL fluid (USA) | LN834393 | LN834489 | LN834577 | ||
| UTHSC DI-13-214 | Human, BAL fluid (USA) | LN834394 | LN834490 | LN834578 | ||
| UTHSC DI-13-254 | Human, BAL fluid (USA) | LN834395 | LN834491 | LN834579 | ||
| UTHSC DI-13-255 | Human, toe nail (USA) | LN834396 | LN834492 | LN834580 | ||
| Cladosporium tenuissimum | CBS 125995T | Fruits of Lagerstroemia sp. (USA) | HM148197 | HM148442 | HM148687 | |
| UTHSC DI-13-174 | Marine mammal, lung (USA) | LN834397 | LN834493 | LN834581 | ||
| UTHSC DI-13-177 | Human, skin (USA) | LN834398 | LN834494 | LN834582 | ||
| UTHSC DI-13-188 | Human, BAL fluid (USA) | LN834399 | LN834495 | LN834583 | ||
| UTHSC DI-13-205 | Human, BAL fluid (USA) | LN834400 | LN834496 | LN834584 | ||
| UTHSC DI-13-236 | Human, nasal (USA) | LN834401 | LN834497 | LN834585 | ||
| UTHSC DI-13-239 | Human, sputum (USA) | LN834402 | LN834498 | LN834586 | ||
| UTHSC DI-13-253 | Human, BAL fluid (USA) | LN834403 | LN834499 | LN834587 | ||
| UTHSC DI-13-258 | Human, thorancentesis fluid (USA) | LN834404 | LN834500 | LN834588 | ||
| UTHSC DI-13-274 | Human, toe (USA) | LN834405 | LN834501 | LN834589 | ||
| Cladosporium sp. | UTHSC DI-13-165 | Human, arm drainage (USA) | LN834406 | LN834502 | LN834590 | |
| UTHSC DI-13-168 | Human, BAL fluid (USA) | LN834407 | LN834503 | LN834591 | ||
| UTHSC DI-13-169 | Human, BAL fluid (USA) | LN834408 | LN834504 | LN834592 | ||
| UTHSC DI-13-170 | Human, toe nail (USA) | LN834409 | LN834505 | LN834593 | ||
| UTHSC DI-13-178 | Animal, abscess (USA) | LN834410 | LN834506 | LN834594 | ||
| UTHSC DI-13-179 | Human, hand (USA) | LN834411 | LN834507 | LN834595 | ||
| UTHSC DI-13-190 | Human, CSF (USA) | LN834412 | LN834508 | LN834596 | ||
| UTHSC DI-13-207 | Human, CSF (USA) | LN834413 | LN834509 | LN834597 | ||
| UTHSC DI-13-210 | Human, skin (USA) | LN834414 | LN834510 | LN834598 | ||
| UTHSC DI-13-211 | Human, BAL fluid (USA) | LN834415 | LN834511 | LN834599 | ||
| UTHSC DI-13-212 | Human, ethmoid sinus (USA) | LN834416 | LN834512 | LN834600 | ||
| UTHSC DI-13-217 | Human, nasal (USA) | LN834417 | LN834513 | LN834601 | ||
| UTHSC DI-13-218 | Human, BAL fluid (USA) | LN834418 | LN834514 | LN834602 | ||
| UTHSC DI-13-219 | Human, foot (USA) | LN834419 | LN834515 | LN834603 | ||
| UTHSC DI-13-225 | Animal, BAL fluid (USA) | LN834420 | LN834516 | LN834604 | ||
| UTHSC DI-13-226 | Human, BAL fluid (USA) | LN834421 | LN834517 | LN834605 | ||
| UTHSC DI-13-227 | Human, sputum (USA) | LN834422 | LN834518 | LN834606 | ||
| UTHSC DI-13-228 | Human, foot skin (USA) | LN834423 | LN834519 | LN834607 | ||
| UTHSC DI-13-234 | Human, sputum (USA) | LN834424 | LN834520 | LN834608 | ||
| UTHSC DI-13-235 | Human, BAL fluid (USA) | LN834425 | LN834521 | LN834609 | ||
| UTHSC DI-13-238 | Human, leg (USA) | LN834426 | LN834522 | LN834610 | ||
| UTHSC DI-13-241 | Human, foot (USA) | LN834427 | LN834523 | LN834611 | ||
| UTHSC DI-13-244 | Human, BAL fluid (USA) | LN834428 | LN834524 | LN834612 | ||
| UTHSC DI-13-245 | Human, toe (USA) | LN834429 | LN834525 | LN834613 | ||
| UTHSC DI-13-246 | Human, BAL fluid (USA) | LN834430 | LN834526 | LN834614 | ||
| UTHSC DI-13-247 | Human, BAL fluid (USA) | LN834431 | LN834527 | LN834615 | ||
| UTHSC DI-13-251 | Human, BAL fluid (USA) | LN834432 | LN834528 | LN834616 | ||
| UTHSC DI-13-257 | Human, sputum (USA) | LN834433 | LN834529 | LN834617 | ||
| UTHSC DI-13-262 | Dolphin, bronchus (USA) | LN834434 | LN834530 | LN834618 | ||
| UTHSC DI-13-265 | Human, BAL fluid (USA) | LN834435 | LN834531 | LN834619 | ||
| UTHSC DI-13-268 | Human, toe nail (USA) | LN834436 | LN834532 | LN834620 | ||
| UTHSC DI-13-269 | Human, BAL fluid (USA) | LN834437 | LN834533 | LN834621 | ||
| UTHSC DI-13-270 | Human, nail (USA) | LN834438 | LN834534 | LN834622 | ||
| UTHSC DI-13-271 | Human, BAL fluid (USA) | LN834439 | LN834535 | LN834623 | ||
| UTHSC DI-13-273 | Human, toe nails (USA) | LN834440 | LN834536 | LN834624 | ||
| Cladosporium variabile | CBS 121636T | Spinacia oleracea (USA) | EF679402 | EF679480 | EF679556 | |
| Penidiella sp. | UTHSC DI-13-256 | Human, nail (USA) | LN834441 | LN834445 | ||
| Toxicocladosporium banksiae | CBS 128215T | Leaf of Banksia emulata (Australia) | HQ599598 | HQ599599 | ||
| Toxicocladosporium chlamydosporum | CBS 124157T | Leaf of Eucalyptus camaldulensis (Madagascar) | FJ790283 | FJ790301 | ||
| Toxicocladosporium ficiniae | CBS 136406T | Leaf of Ficinia sp. (South Africa) | KF777190 | KF777241 | ||
| Toxicocladosporium irritans | CBS 185.58T | Moldy paint (Suriname) | EU040243 | EU040243 | ||
| UTHSC DI-13-181 | Human, blood (USA) | LN834442 | LN834446 | |||
| UTHSC DI-13-230 | Human, finger nail (USA) | LN834443 | LN834447 | |||
| Toxicocladosporium pini | CBS 138005T | Needles of Pinus sp. (China) | KJ869160 | KJ869217 | ||
| Toxicocladosporium posoqueriae | CBS 133583T | Leaf of Posoqueria latifolia (Australia) | NR121555 | KC005803 | ||
| Toxicocladosporium protearum | CBS 126499T | Leaf of Protea burchellii (South Africa) | HQ599586 | HQ599587 | ||
| Toxicocladosporium pseudoveloxum | CBS 128775T | Leaf of Phaenocoma prolifera (South Africa) | JF499847 | JF499867 | ||
| Toxicocladosporium rubrigenum | CBS 124158T | Leaf of Eucalyptus camaldulensis (Madagascar) | FJ790287 | FJ790305 | ||
| Toxicocladosporium sp. | UTHSC DI-13-172 | Human, BAL fluid (USA) | LN834444 | LN834448 | ||
| Toxicocladosporium strelitziae | CBS 132535T | Leaf of Strelitzia reginae (South Africa) | NR111765 | JX069858 | ||
| Toxicocladosporium veloxum | CBS 124159T | Leaf of Eucalyptus camaldunensis (Madagascar) | FJ790288 | FJ790306 | ||
CBS, CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands; FMR, Facultat de Medicina, Universitat Rovira i Virgili, Reus, Spain; UTHSC, Fungus Testing Laboratory at the University of Texas Health Science Center, San Antonio, TX, USA; T, ex-type strain.
BAL fluid, bronchoalveolar lavage fluid specimen; CSF, cerebrospinal fluid.
Phenotypic identification.
The isolates were morphologically characterized following the procedures outlined in Bensch et al. (1), Crous et al. (23), Schubert et al. (19), and Zalar et al. (20). Briefly, all of the isolates were grown on synthetic nutrient-poor agar (SNA) (1 g KH2PO4, 1 g KNO3, 0.5 g MgSO4 · 7H2O, 0.5 g KCl, 0.2 g glucose, 0.2 g sucrose, 1 liter water) and potato dextrose agar (PDA) (Pronadisa, Spain) for 7 days at 25°C. Microscopic observations were made from cultures on SNA mounted in Shear's solution (23). Colony characteristics were recorded from cultures on SNA and PDA. For the estimation of cardinal growth temperatures, the isolates were grown on PDA agar for 14 days at temperatures ranging from 15°C to 35°C at intervals of 5°C, as well as at 32°C and 37°C.
DNA extraction, amplification, and sequencing.
Total genomic DNA was extracted from mycelia obtained from colonies growing on PDA, using FastPrep (MP Biomedicals, Santa Ana, CA) according to the manufacturer's protocol. DNA was quantified using the NanoDrop 3000 (Thermo Scientific, Madrid, Spain).
The primers ITS5 and ITS4 (24) were used to amplify a region spanning internal transcribed spacer 1 (ITS1) and ITS2 and the 5.8S gene of the ribosomal DNA (rDNA); the primer pair LR0R/LR5 (25, 26) was used to amplify a fragment of the large subunit (LSU) gene of the rDNA; and the EF-728F/EF-986R and ACT-512F/ACT-783R primer pairs (27) were used for the translation elongation factor 1α gene (EF-1α) and the actin gene (Act), respectively.
Sequencing was performed in both directions using the same PCR primers at Macrogen Europe (Macrogen, Inc., Amsterdam, the Netherlands). Consensus sequences were obtained using SeqMan version 7.0.0 (DNAStar Lasergene, Madison, WI).
Molecular identification and phylogenetic analyses.
An initial presumptive generic identification of the isolates was performed based on BLAST searches of ITS and LSU sequences in the GenBank (http://www.ncbi.nlm.nih.gov/) and CBS (http://www.cbs.knaw.nl/) databases. Multiple sequence alignments of each locus were performed in MEGA version 6 (28) using the ClustalW application (29), refined with MUSCLE (30), and manually adjusted if necessary. Phylogenetic reconstructions were made using maximum-likelihood (ML) and Bayesian Inference (BI) under MEGA version 6 and MrBayes version 3.1.2 (31), respectively. The best nucleotide substitution model (generalized time-reversible model with gamma distribution and a portion of invariable sites [GTR+G+I] for the three independent data sets) was estimated using MrModelTest version 2.3 (32) following the Akaike criterion. Phylogenetic analyses using ML were at first made individually for each locus and compared in order to assess for any incongruent results between nodes with high statistical support. As no incongruences were observed, the four loci were combined as follows: ITS, EF-1α, and Act for members of Cladosporium sensu stricto and ITS combined with LSU for members of other cladosporiumlike genera. For the ML analysis, nearest-neighbor interchange (NNI) was used as the heuristic method for tree inference. Support for the internal branches was assessed by a search of 1,000 bootstrapped sets of data. A bootstrap support value of ≥70 was considered significant. For BI analysis, two simultaneous runs of 10,000,000 generations were performed and samples were stored every 1,000 generations. The 50% majority-rule consensus tree and posterior probability values (PP) were calculated after discarding the first 25% of the samples. A PP value of ≥0.95 was considered significant.
The first combined phylogenetic analysis with ITS, EF-1α, and Act sequences of clinical isolates belonging to Cladosporium sensu stricto and all the available type and reference strains was carried out following the alignments of Bensch et al. (1; data not shown). Only sequences of those species closely related (>95% similarity) to the clinical isolates tested here were included in the final analysis.
Antifungal susceptibility.
The antifungal susceptibility test was performed according to the CLSI M38-A2 standard (33) with slight modifications. The incubation temperature was set to 25°C, given the optimal growth requirements of Cladosporium and related taxa (1, 33). Nine antifungal agents were tested: amphotericin B (AMB), 5-fluorocytosine (5FC), itraconazole (ITC), posaconazole (PSC), voriconazole (VRC), terbinafine (TRB), anidulafungin (AFG), caspofungin (CFG), and micafungin (MFG). The minimal effective concentration (MEC), defined as the lowest drug concentration at which short, stubby, highly branched hyphae were observed, was determined at 24 h for the echinocandins, and the MIC was determined at 48 h for the remaining drugs. The MIC was defined as the lowest concentration exhibiting 100% inhibition of visible growth for AMB, ITC, PSC, and VRC or 50% and 80% reduction in growth for 5FC and TRB, respectively. Paecilomyces variotii ATCC MYA-3630 and Aspergillus fumigatus ATCC MYA-3626 were used as quality control strains. Statistical analyses of the MIC/MEC data were performed using the Mann-Whitney test in Prism version 6.0 (GraphPad Software, San Diego, CA).
Nucleotide sequence accession numbers.
DNA sequences determined in this study were deposited in GenBank under accession numbers LN834353 to LN834448 (rDNA), LN834449 to LN834536 (EF-1α), and LN834537 to LN834624 (Act) (Table 1).
RESULTS
Analysis of ITS and LSU sequences showed that 88 isolates (96%) belonged to Cladosporium sensu stricto, three isolates (3%) to the genus Toxicocladosporium, and one isolate (1%) to the genus Penidiella.
The phylogenetic analysis of Cladosporium sensu stricto included 121 taxa and 1,002 bp (447 bp for ITS, 337 bp for EF-1α, and 218 bp for Act), of which 485 bp were constant (347 bp for ITS, 60 bp for EF-1α, and 78 bp for Act), 496 were variable (97 bp for ITS, 260 bp for EF-1α, and 139 bp for Act) and 328 were parsimony informative (24 bp for ITS, 197 bp for EF-1α, and 107 bp for Act) (Fig. 1). The majority of isolates (57, 65%) nested into the C. cladosporioides complex: 28 belonged to nine species (i.e., C. angustisporum, C. asperulatum, C. cladosporioides, C. flabelliforme, C. funiculosum, C. perangustum, C. pseudocladosporioides, C. subuliforme, and C. tenuissimum), while 29 isolates clustered into 12 terminal subclades genetically distant from any currently known species of the genus. A total of 14 isolates were related to the C. herbarum complex (16%), mostly corresponding to five species (i.e., C. allicinum, C. herbarum, C. macrocarpum, C. ramotenellum, and C. subinflatum), while five isolates clustered into four new lineages in the genus. Seventeen isolates were nested within the C. sphaerospermum complex (19%) and mostly belonged to two species (i.e., C. halotolerans and C. sphaerospermum), while a single isolate represented a new lineage.
FIG 1.
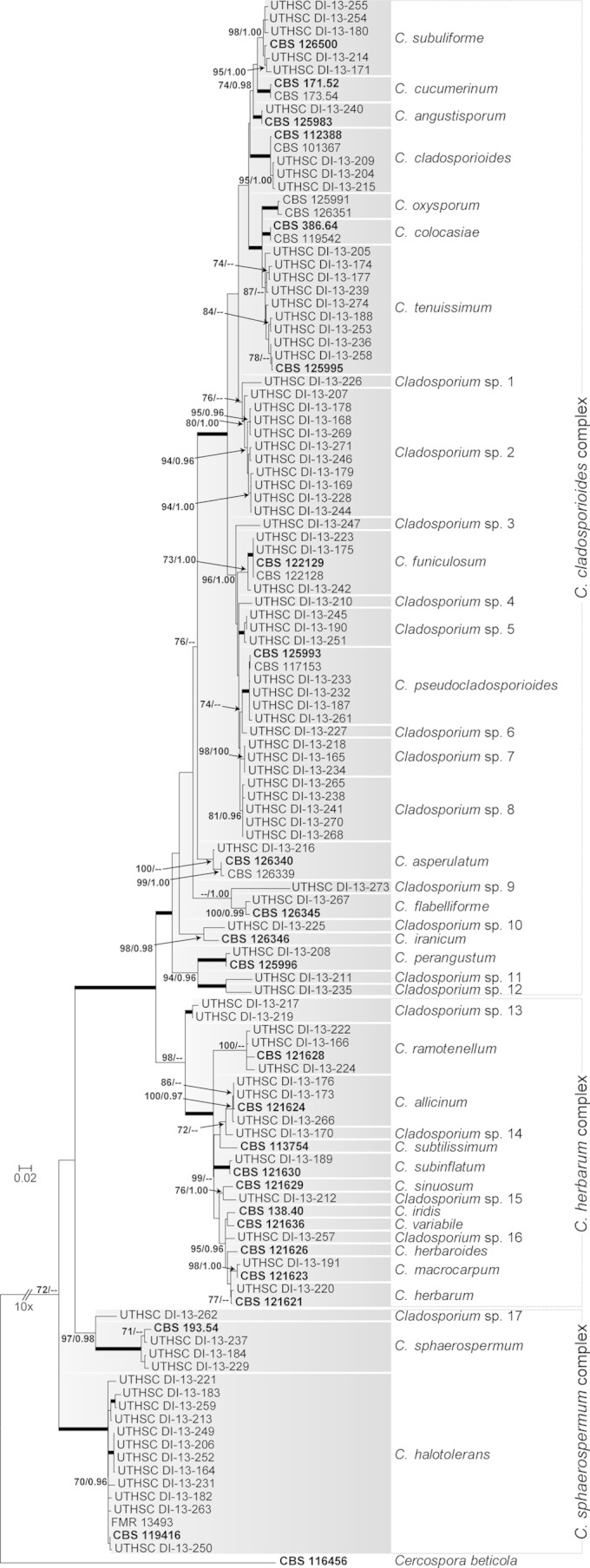
Maximum-likelihood (ML) tree inferred from combined ITS, EF-1α, and Act sequences of Cladosporium isolates. Branch lengths are proportional to phylogenetic distance. ML bootstrap support (BS) values of ≥70% and posterior probability (PP) values of ≥0.95 are shown above the branches. Thickened branches indicate BS of 100% and PP of 1.00. Cercospora beticola (CBS 116456) was used to root the tree. Type strains are indicated in bold font. CBS, CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands; FMR, Facultat de Medicina, Reus, Spain; UTHSC, Fungus Testing Laboratory at the University of Texas Health Science Center, San Antonio, TX.
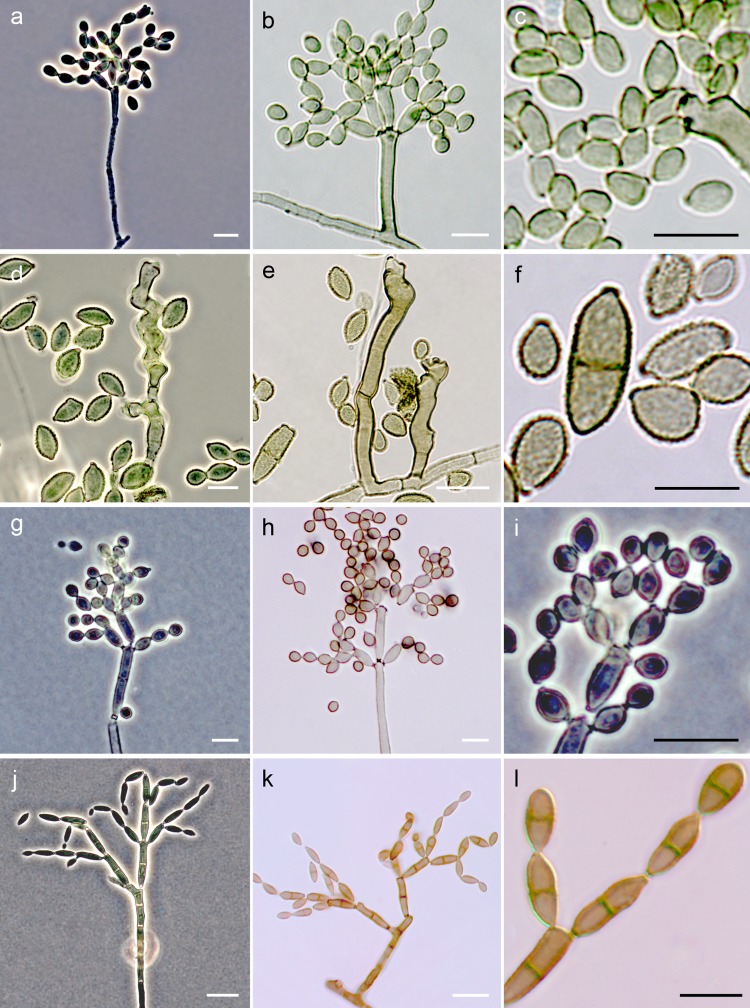
Distinct morphological features of isolates in the C. cladosporioides complex included the formation of mostly unbranched, cylindrical conidiophores, bearing ovoidal to ellipsoidal intercalary and terminal conidia, smooth or rarely showing fine ornamentation (Fig. 2a to c); the maximum temperatures for growth were 32°C for C. cladosporioides, C. flabelliforme, C. perangustum, and C. pseudocladosporioides and 35°C for C. angustisporum, C. funiculosum, C. subuliforme, and C. tenuissimum. Isolates of the C. herbarum complex exhibited mostly nodulose conidiophores, bearing distinctly ornamented globose to subglobose terminal conidia (Fig. 2d to f); none of the isolates of this complex were able to grow at temperatures above 32°C, and C. allicinum exhibited a maximum growth temperature of 30°C. Isolates of the C. sphaerospermum complex formed cylindrical and branched conidiophores, bearing globose to subglobose conidia, smooth or finely ornamented (Fig. 2g to i); the maximum temperatures for growth were 32°C for C. sphaerospermum and 35°C for C. halotolerans. None of the clinical isolates formed sexual morphs in culture.
FIG 2.
Conidiophores and conidia of fungi belonging to the C. cladosporioides complex (a to c), C. herbarum complex (d to f), C. sphaerospermum complex (g to i), and Toxicocladosporium spp. (j to l). White bars, 10 μm; black bars, 5 μm.
Overall, the most commonly identified species was C. halotolerans (14.8%), followed by C. tenuissimum (10.2%) and C. subuliforme (5.7%). However, 39.8% of isolates did not match with any known taxa and represent at least 17 putative new Cladosporium species (Fig. 1). The most common anatomical site of isolation was the respiratory tract (54.5%), mainly from bronchoalveolar lavage (BAL) fluid and nasal specimens, followed by superficial sites (28.4%); these percentages were similar for all of the species and species complexes identified.
Phylogenetic analysis of the Toxicocladosporium isolates included 15 taxa and 984 bp (530 bp for LSU and 454 bp for ITS), of which 826 bp were constant (464 bp for LSU and 362 bp for ITS), 155 were variable (66 bp for LSU and 89 bp for ITS), and 129 were parsimony informative (56 bp for LSU and 73 bp for ITS) (Fig. 3). Two clinical isolates belonged to Toxicocladosporium irritans, while the isolate UTHSC DI-13-172 formed an independent lineage, genetically related to Toxicocladosporium strelitziae but showing distinctive morphological features and probably corresponding to a new species. The main morphological characteristics of members of Toxicocladosporium were the presence of nonnodulose conidiophores with dark and thickened cell walls and septa, producing conidia without the typical coronate scars of Cladosporium (Fig. 2j to l), and a maximum temperature for growth of 35°C.
FIG 3.
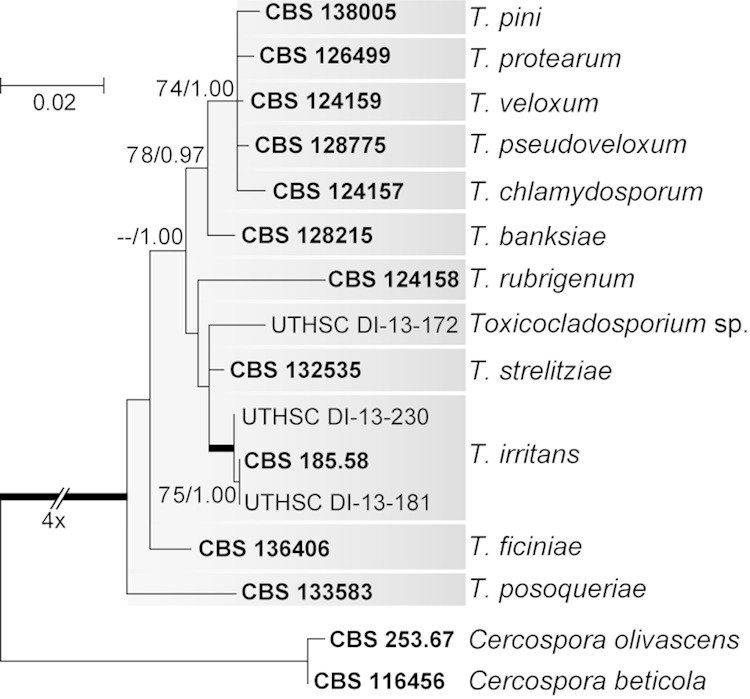
Maximum-likelihood (ML) tree inferred from combined ITS and LSU sequences of Toxicocladosporium isolates. Branch lengths are proportional to phylogenetic distance. ML bootstrap support (BS) values of ≥70% and posterior probability (PP) values of ≥0.95 are shown above the branches. Thickened branches indicate BS of 100% and PP of 1.00. Cercospora beticola (CBS 116456) and Cercospora olivascens (CBS 253.67) were used to root the tree. Type strains are indicated in bold font. CBS, CBS-KNAW Fungal Biodiversity Centre, the Netherlands; UTHSC, Fungus Testing Laboratory at the University of Texas Health Science Center, San Antonio, TX.
According to the LSU sequence analysis, a single isolate (UTHSC DI-13-256), originally identified as C. sphaerospermum, was related to but distant (<98.2% sequence similarity) from members of the genus Penidiella (i.e., Penidiella aggregata and Penidiella drakensbergensis; sequence accession numbers JF499862 and KC005792, respectively) (data not shown). However, its final identification was not possible given the scarcity of DNA sequences of the latter species for comparison. This isolate was characterized by restricted growth (3 to 4 mm at 25°C for 7 days) and the production of solitary penicillate conidiophores, composed of chains of ramoconidia with slightly pigmented and thickened conidiogenous scars.
The results of the antifungal susceptibility testing are summarized in Table 2. The overall results for Cladosporium species showed a geometric mean (GM) MIC and MIC90 for AMB of 0.64 μg/ml and 2 μg/ml, respectively. Among the azoles, ITC and PSC were the most active, with both drugs having a GM MIC of 0.43 μg/ml and respective MIC90s of 0.5 μg/ml and 1 μg/ml, while VRC showed a GM MIC and MIC90 of 1.68 μg/ml and 4 μg/ml, respectively. Flucytosine showed variable activity and had a GM MIC and MIC90 of 1.37 μg/ml and 4 μg/ml, respectively. TRB exhibited the most potent activity, with a GM MIC and MIC90 of 0.09 μg/ml and 1 μg/ml, respectively. With the exception of CFG, the echinocandins exhibited strong in vitro activity, with GM MIC values of 0.19 μg/ml and 0.12 μg/ml for AFG and MFG. All of the Cladosporium species tested showed similar susceptibility patterns except for C. sphaerospermum, where the three isolates tested exhibited higher MIC and MEC values, especially for the azoles, AMB, AFG, and MFG (P < 0.001). Comparison of antifungal susceptibility by species complex (Table 2) showed that AMB exhibited more potent activity against members of the C. herbarum complex, with GM MIC and MIC90 values of 0.18 μg/ml and 1 μg/ml (P < 0.002), while members of the C. sphaerospermum complex exhibited higher GM MIC and MIC90 values for AMB, PSC, ITC, and CSP (P < 0.003). Toxicocladosporium and Penidiella isolates exhibited similar susceptibility patterns, with mostly low GM MIC and MIC90 values against all antifungals tested but without statistically significant differences.
TABLE 2.
Results of in vitro antifungal susceptibility testing of the 92 clinical isolates included in the study
| Genus | Species (no. of isolates tested) | MIC/MEC parametera | Result (μg/ml) forb: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMB | 5FC | VRC | PSC | ITC | TBF | CFGc | AFGc | MFGc | |||
| Cladosporium | C. cladosporioides complex (57) | Range | 0.06–2 | 0.06–>32 | 0.25–16 | <0.03–1 | <0.03–2 | <0.03–4 | 0.125–8 | 0.03–0.5 | 0.03–0.5 |
| GM | 0.73 | 1.20 | 1.65 | 0.40 | 0.34 | 0.12 | 2.78 | 0.19 | 0.11 | ||
| MIC90 | 1 | 4 | 4 | 0.5 | 0.5 | 1 | 8 | 0.5 | 0.25 | ||
| C. tenuissimum (9) | Range | 0.5–1 | 1–>16 | 1–4 | 0.25–0.5 | 0.25–0.5 | 0.06–1 | 4–8 | 0.125–0.5 | 0.125–0.25 | |
| GM | 0.93 | 2.72 | 1.85 | 0.37 | 0.29 | 0.18 | 4.67 | 0.37 | 0.15 | ||
| MIC90 | 1.00 | 4.00 | 2.00 | 0.50 | 0.50 | 0.25 | 8.00 | 0.50 | 0.25 | ||
| C. subuliforme (5) | Range | 1–2 | 0.25–2 | 0.25–2 | 0.25–0.5 | 0.25 | 0.06–1 | 4–8 | 0.06–0.5 | 0.06–0.25 | |
| GM | 1.15 | 1.00 | 0.66 | 0.29 | 0.25 | 0.28 | 5.28 | 0.16 | 0.12 | ||
| MIC90 | 1.00 | 2.00 | 1.00 | 0.25 | 0.25 | 1.00 | 8.00 | 0.25 | 0.13 | ||
| C. pseudocladosporioides (4) | Range | 0.5–1 | 0.5–1 | 2–4 | 0.25–0.5 | 0.5–1 | 0.03–2 | 0.25–8 | 0.03–0.25 | 0.03–0.125 | |
| GM | 0.59 | 0.71 | 2.38 | 0.42 | 0.59 | 0.21 | 2.38 | 0.15 | 0.07 | ||
| MIC90 | |||||||||||
| C. cladosporioides (3) | Range | 0.5–1 | 1–2 | 0.5–16 | 0.25–1 | 0.25–0.5 | 0.5–2 | 1–4 | 0.125–0.25 | 0.125 | |
| GM | 0.79 | 1.26 | 1.59 | 0.40 | 0.31 | 1.00 | 2.00 | 0.16 | 0.13 | ||
| MIC90 | |||||||||||
| C. funiculosum (3) | Range | 0.06–1 | 0.5–1 | 0.5–2 | 0.25–0.5 | 0.125–0.25 | 0.03–0.06 | 4 | 0.06–0.25 | 0.06–0.125 | |
| GM | 0.31 | 0.63 | 1.00 | 0.31 | 0.20 | 0.04 | 4.00 | 0.12 | 0.08 | ||
| MIC90 | |||||||||||
| C. angustisporum (1) | Range | 1.00 | 1.00 | 4.00 | 0.50 | 0.50 | 2.00 | 4.00 | 0.13 | 0.06 | |
| GM | |||||||||||
| MIC90 | |||||||||||
| C. asperulatum (1) | Range | 1 | 0.25 | 2 | 0.25 | 0.5 | 0.03 | 8 | 0.25 | 0.125 | |
| GM | |||||||||||
| MIC90 | |||||||||||
| C. flabelliforme (1) | Range | 2 | 2 | 2 | 0.5 | 0.25 | 0.03 | 4 | 0.25 | 0.125 | |
| GM | |||||||||||
| MIC90 | |||||||||||
| C. perangustum (1) | Range | 0.5 | 4 | 4 | 0.5 | 0.5 | 0.03 | 4 | 0.125 | 0.06 | |
| GM | |||||||||||
| MIC90 | |||||||||||
| Cladosporium sp. (29) | Range | 0.125–2 | 0.06–>16 | 0.25–8 | <0.03–1 | <0.03–2 | <0.03–4 | 0.125–8 | 0.03–0.5 | 0.03–0.5 | |
| GM | 0.67 | 1.10 | 1.73 | 0.43 | 0.36 | 0.09 | 2.00 | 0.18 | 0.10 | ||
| MIC90 | 1.00 | 4.00 | 4.00 | 0.50 | 0.50 | 1.00 | 8.00 | 0.50 | 0.25 | ||
| C. herbarum complex (14) | Range | <0.03–2 | 0.5–>16 | 0.5–8 | 0.06–0.5 | 0.06–1 | <0.03–0.125 | 0.125–8 | 0.06–1 | 0.06–0.5 | |
| GM | 0.18 | 2.97 | 1.81 | 0.37 | 0.35 | 0.05 | 0.67 | 0.23 | 0.15 | ||
| MIC90 | 1 | 8 | 4 | 0.5 | 0.5 | 0.125 | 2 | 0.5 | 0.5 | ||
| C. allicinum (3) | Range | <0.03–0.125 | 2–4 | 2–4 | 0.5 | 0.25–0.5 | 0.03–0.06 | 1.00 | 0.25–0.5 | 0.125–0.25 | |
| GM | 0.05 | 2.52 | 3.17 | 0.50 | 0.40 | 0.05 | 1.00 | 0.31 | 0.20 | ||
| MIC90 | |||||||||||
| C. ramotenellum (3) | Range | 1–2 | 4–>16 | 1–2 | 0.5 | 0.5–1 | <0.03–0.125 | 0.5–8 | 0.25–1 | 0.125–0.5 | |
| GM | 1.26 | 4.00 | 1.59 | 0.50 | 0.63 | 0.05 | 2.00 | 0.40 | 0.20 | ||
| MIC90 | |||||||||||
| C. herbarum (1) | Range | 0.06 | 8 | 2 | 0.5 | 0.5 | 0.03 | 0.5 | 0.125 | 0.125 | |
| GM | |||||||||||
| MIC90 | |||||||||||
| C. macrocarpum (1) | Range | 0.5 | 2 | 1 | 0.25 | 0.5 | 0.125 | 1 | 0.125 | 0.5 | |
| GM | |||||||||||
| MIC90 | |||||||||||
| C. subinflatum (1) | Range | 0.5 | 0.5 | 4 | 0.5 | 0.5 | 0.03 | 0.5 | 0.5 | 0.25 | |
| GM | |||||||||||
| MIC90 | |||||||||||
| Cladosporium sp. (5) | Range | 0.06–0.5 | 2–4 | 0.5–8 | 0.06–0.5 | 0.06–0.5 | <0.03–0.125 | 0.125–0.5 | 0.06–0.5 | 0.06–0.125 | |
| GM | 0.11 | 2.30 | 1.32 | 0.25 | 0.19 | 0.05 | 0.29 | 0.14 | 0.08 | ||
| MIC90 | 1.00 | 4.00 | 4.00 | 0.50 | 0.50 | 1.00 | 8.00 | 0.50 | 0.25 | ||
| C. sphaerospermum complex (17) | Range | 0.125–2 | 0.06–4 | 0.5–16 | 0.06–4 | 0.25–>16 | <0.03–1 | 0.06–4 | <0.03–1 | 0.06–1 | |
| GM | 1.13 | 1.13 | 1.70 | 0.64 | 1.13 | 0.06 | 1.27 | 0.15 | 0.13 | ||
| MIC90 | 2 | 2 | 8 | 2 | 32 | 0.5 | 4 | 0.25 | 0.125 | ||
| C. halotolerans (13) | Range | 0.125–2 | 0.06–4 | 0.5–2 | 0.06–1 | 0.25–2 | <0.03–1 | 0.06–4 | <0.03–0.25 | 0.06–0.125 | |
| GM | 1.00 | 0.90 | 1.11 | 0.47 | 0.56 | 0.08 | 1.00 | 0.11 | 0.10 | ||
| MIC90 | 2.00 | 2.00 | 2.00 | 1.00 | 2.00 | 1.00 | 4.00 | 0.25 | 0.13 | ||
| C. sphaerospermum (3) | Range | 1–2 | 2–4 | 8–16 | 2–4 | >16 | <0.03–0.125 | 2–4 | 0.25–1 | 0.125–1 | |
| GM | 1.59 | 2.52 | 10.08 | 2.52 | >16 | 0.03 | 3.17 | 0.50 | 0.40 | ||
| MIC90 | |||||||||||
| Cladosporium sp. (1) | Range | 2 | 2 | 2 | 0.5 | 0.5 | <0.03 | 2 | 0.25 | 0.125 | |
| GM | |||||||||||
| MIC90 | |||||||||||
| Overall (88) | Range | <0.03–2 | 0.06–>16 | 0.25–16 | <0.03–4 | <0.03–>16 | <0.03–4 | 0.06–8 | <0.03–1 | 0.03–1 | |
| GM | 0.64 | 1.37 | 1.68 | 0.43 | 0.43 | 0.09 | 1.91 | 0.19 | 0.12 | ||
| MIC90 | 2.00 | 4.00 | 4.00 | 0.50 | 1.00 | 1.00 | 8.00 | 0.50 | 0.25 | ||
| Toxicocladosporium | T. irritans (2) | Range | 0.5–1 | 0.25–2 | 0.25 | 0.25–1 | 0.5 | <0.03–0.06 | 0.125–2 | 0.06–0.25 | 0.06–0.5 |
| GM | 0.71 | 0.71 | 0.25 | 0.50 | 0.50 | 0.03 | 0.50 | 0.12 | 0.17 | ||
| MIC90 | |||||||||||
| Toxicocladosporium sp. (1) | Range | 0.5 | 0.125 | 0.25 | 0.125 | 0.125 | <0.03 | 1 | 0.125 | 0.06 | |
| GM | |||||||||||
| MIC90 | |||||||||||
| Overall (3) | Range | 0.5–1 | 0.125–2 | 0.25 | 0.125–1 | 0.125–0.5 | <0.03–0.06 | 0.125–2 | 0.06–0.25 | 0.06–0.5 | |
| GM | 0.63 | 0.40 | 0.25 | 0.31 | 0.31 | 0.02 | 0.63 | 0.12 | 0.12 | ||
| MIC90 | |||||||||||
| Penidiella | Penidiella sp. (1) | Range | 2 | 0.06 | 0.125 | 0.06 | >0.03 | >0.03 | 0.25 | 0.25 | 0.25 |
| GM | |||||||||||
| MIC90 | |||||||||||
GMs and MIC90s are shown only for species with ≥5 isolates. GM, geometric mean.
AMB, amphotericin B; 5FC, flucytosine; VRC, voriconazole; PSC, posaconazole; ITC, itraconazole; CFG, caspofungin; AFG, anidulafungin; MFG, micafungin; TRB, terbinafine.
These columns include MEC data.
DISCUSSION
Members of Cladosporium are relatively easy to identify to genus and species complex based on their typical conidiogenous structures. However, morphological identification of Cladosporium species is difficult given the high morphological similarity between closely related species. In light of our results, it is strongly recommended that phenotypic identifications be confirmed with DNA sequencing. Several authors have demonstrated the usefulness of the EF-1α and Act loci to allow a good species delimitation in Cladosporium (1, 19, 21). This is especially important for members of the C. cladosporioides complex, which demonstrated the greatest species diversity, the highest number of species associated with clinical samples, and also, the greatest number of undescribed species. Moreover, we found that C. cladosporioides, the species most frequently cited as being clinically relevant, was poorly represented in our set of isolates, while C. asperulatum, C. funiculosum, C. flabelliforme, C. pseudocladosporioides, C. subuliforme, and C. tenuissimum are described for the first time from clinical samples. Similarly, in the C. sphaerospermum complex, most of the isolates morphologically identified as C. sphaerospermum were genetically reidentified as belonging to the phenotypically similar species C. halotolerans, which according to our data, emerged as the most common species from clinical origins. The latter species has never been associated with human infection; however, some isolates had been reported from human or animal clinical samples (1). In the case of the C. herbarum complex, 13 of the 14 isolates morphologically identified as C. herbarum, also considered a clinically relevant species, were found to belong to other species of this complex (i.e., C. allicinum, C. macrocarpum, and C. ramotenellum). While C. macrocarpum has been identified as the causative agent of human infections (17), C. allicinum and C. ramotenellum have never been reported before in the clinical setting, although some isolates have been recorded as obtained from human samples (1). However, due to the lack of clinical histories and histopathological findings, it was impossible for us to confirm a pathogenic role of the species reported here for the first time from clinical specimens.
It is remarkable that our phylogenetic analysis was unable to provide species-level identification of a high number of Cladosporium isolates (39.8%) that were originally considered to belong to several common morphospecies. Instead, those unidentified isolates were grouped into 5 terminal clades and 12 monotypic lineages, representing a large variety of phylogenetic species. It is probable that many of these clades and monotypic lineages represent new species; however, further studies combining phenotypic and molecular data would be necessary to confirm these findings. We report also for the first time the isolation of Toxicocladosporium and Penidiella species from clinical specimens. Isolates of these recently proposed genera were only known from leaves of several plants and from environmental sources (3, 34). According to our data, the vast majority of isolates were obtained from respiratory specimens, including BAL fluid, nasal, and sputum samples. This is not rare, because Cladosporium is preponderant in the airborne mycobiota (35), being considered one of the most important respiratory allergenic fungi, after Alternaria (10, 34, 36).
Reports of invasive infections by Cladosporium are extremely rare. Bentz and Sautter (37) reported a mixed disseminated infection by Aspergillus fumigatus and C. cladosporioides in an immunocompromised patient. Cladosporium cladosporioides and C. macrocarpum have been reported from two clinical cases involving the central nervous system (15, 17), while C. sphaerospermum was isolated from an intrabronchial infection (38). However, in none of these cases was the etiology of the infection supported by histopathological studies. The isolation of Cladosporium species from deep tissues seems improbable considering the inability of these organisms to grow at temperatures exceeding 35°C, and thermotolerance being one of the most important virulence factors for invasive or disseminated infections (39). In our study, less than half of the isolates exhibited very limited growth at 35°C, while none was able to grow at 37°C. However, surprisingly, several of our isolates were obtained from deep tissue samples, including bone marrow, cerebrospinal fluid (CSF), and lung and lymphatic tissue samples, among others. Isolation of these fungi from invasive infections may have been due to environmental contamination of the samples; however, occasionally isolates that fail to grow in culture at 37°C have been reported to cause invasive disease in immunocompromised individuals (40).
There is a paucity of information regarding antifungal susceptibility patterns for Cladosporium species. Most data are from a few reported clinical cases (7, 13, 15, 41). Our study provides the first in vitro data for a large set of clinical isolates, including several species obtained from diverse anatomical sites and not previously reported from clinical samples. Case reports have shown a favorable outcome using azole-based therapies. ITC has shown efficacy in the treatment of superficial infections caused by C. cladosporioides, C. sphaerospermum, and C. oxysporum (8, 14, 37, 41–44), while VRC was effective against C. macrocarpum in a brain abscess (17). This agrees with our in vitro data, which demonstrated that the azoles, particularly ITC and PSC, have good activity against Cladosporium species, although VRC displayed variable activity. AMB has been shown to be ineffective against C. cladosporioides (41) and C. sphaerospermum (38) in cases of skin and intrabronchial infections, respectively. Our results, however, suggest that this drug might be effective, especially against members of the C. herbarum complex. Kantarcioğlu and Yücel (45) reported potent in vitro activity of TRB against a set of unidentified Cladosporium species. Our data confirmed the results of that study, with TRB showing significant activity against all of the species tested. Echinocandin activity against Cladosporium species has not been previously evaluated; however, we observed that both AFG and MFG exhibited notable in vitro activity against all of our isolates, indicating that they could represent an important alternative for the treatment of infections by these fungi pending further confirmatory studies.
In conclusion, our study has significantly expanded the diversity of Cladosporium species seen in clinical specimens as a result of the molecular characterization of these isolates. We were unable, however, to document these organisms as etiologic agents in human or animal disease due to the lack of clinical information and/or histopathological findings. It is also important to note that most reported cases of Cladosporium infections lack molecular confirmation, and in those cases where they have been so characterized, the strains are not available. Given that many journals require the public availability of DNA sequence data, we recommend that clinical strains be deposited in international culture collections, thereby making them available for reidentification and further study.
ACKNOWLEDGMENTS
This study was supported by the Spanish Ministerio de Economía y Competitividad, grants CGL2011-27185 and CGL2013-43789-P.
REFERENCES
- 1.Bensch K, Braun U, Groenewald JZ, Crous PW. 2012. The genus Cladosporium. Stud Mycol 72:1–401. doi: 10.3114/sim0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David JC. 1997. A contribution to the systematics of Cladosporium. Revision of the fungi previously referred to Heterosporium. Mycological papers no. 172. CAB International, Wallingford, United Kingdom. [Google Scholar]
- 3.Crous PW, Braun U, Schubert K, Groenewald JZ. 2007. Delimiting Cladosporium from morphologically similar genera. Stud Mycol 58:33–56. doi: 10.3114/sim.2007.58.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellis D, Davis S, Alexiou H, Handke R, Bartley R (ed). 2007. Descriptions of medical fungi, 2nd ed Nexus Print Solutions, Underdale, Australia. [Google Scholar]
- 5.Ellis MB. 1971. Dematiaceous hyphomycetes. Commonwealth Mycological Institute, Kew, England. [Google Scholar]
- 6.Ellis MB. 1976. More dematiaceous hyphomycetes. Commonwealth Mycological Institute, Kew, England. [Google Scholar]
- 7.de Hoog GS, Guarro J, Gené J, Figueras MJ. 2011. Atlas of clinical fungi, electronic version 3.1. CBS-KNAW Fungal Biodiversity Centre, Utrecht, Netherlands. [Google Scholar]
- 8.Ma X, Gu Y, Liu X, Li D, Ling S, Hou J, Wang C, Cao S, Huang X, Wen X, Ruan J, Dong C, Li C, Tong Y. 2013. Phaeohyphomycotic dermatitis in a giant panda (Ailuropoda melanoleuca) caused by Cladosporium cladosporioides. Med Mycol Case Rep 2:119–121. doi: 10.1016/j.mmcr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercier E, Peters IR, Billen F, Battaille G, Clercx C, Day MJ, Peeters D. 2013. Potential role of Alternaria and Cladosporium species in canine lymphoplasmacytic rhinitis. J Small Anim Pract 54:179–183. doi: 10.1111/jsap.12049. [DOI] [PubMed] [Google Scholar]
- 10.Sellart-Altisent M, Torres-Rodríguez JM, Gómez de Ana S, Alvarado-Ramírez E. 2007. Nasal fungal microbiota in allergic and healthy subjects. Rev Iberoam Micol 24:125–130. (In Spanish) doi: 10.1016/S1130-1406(07)70027-X. [DOI] [PubMed] [Google Scholar]
- 11.Drabick JJ, Gomatos PJ, Solis JB. 1990. Cutaneous cladosporiosis as a complication of skin testing in a man positive for human immunodeficiency virus. J Am Acad Dermatol 22:135–136. doi: 10.1016/S0190-9622(08)80019-9. [DOI] [PubMed] [Google Scholar]
- 12.Gugnani HC, Sood N, Singh B, Makkar R. 2000. Case Report. Subcutaneous phaeohyphomycosis due to Cladosporium cladosporioides. Mycoses 43:85–87. [DOI] [PubMed] [Google Scholar]
- 13.Gugnani HC, Ramesh V, Sood N, Guarro J, Moin-Ul-Haq Paliwal-Joshi A, Singh B, Makkar R. 2006. Cutaneous phaeohyphomycosis caused by Cladosporium oxysporum and its treatment with potassium iodide. Med Mycol 44:285–288. doi: 10.1080/13693780500294824. [DOI] [PubMed] [Google Scholar]
- 14.Sang H, Zheng XE, Zhou WQ, He W, Lv GX, Shen YN, Kong QT, Liu WD. 2012. A case of subcutaneous phaeohyphomycosis caused by Cladosporium cladosporioides and its treatment. Mycoses 55:195–197. doi: 10.1111/j.1439-0507.2011.02057.x. [DOI] [PubMed] [Google Scholar]
- 15.Kantarcioglu AS, Yücel A, de Hoog GS. 2002. Case report. Isolation of Cladosporium cladosporioides from cerebrospinal fluid. Mycoses 45:500–503. [DOI] [PubMed] [Google Scholar]
- 16.Kantarcioglu AS, de Hoog GS. 2004. Infections of the central nervous system by melanized fungi: a review of cases presented between 1999 and 2004. Mycoses 47:4–13. doi: 10.1046/j.1439-0507.2003.00956.x. [DOI] [PubMed] [Google Scholar]
- 17.Lalueza A, López-Medrano F, del Palacio A, Alhambra A, Alvarez E, Ramos A, Pérez A, Lizasoain M, Meije Y, García-Reyne A, Aguado JM. 2011. Cladosporium macrocarpum brain abscess after endoscopic ultrasound-guided celiac plexus block. Endoscopy 43:E9–E10. doi: 10.1055/s-0030-1255804. [DOI] [PubMed] [Google Scholar]
- 18.de Hoog GS, Guého E, Masclaux F, Gerrits van den Ende AH, Kwon-Chung KJ, McGinnis MR. 1995. Nutritional physiology and taxonomy of human-pathogenic Cladosporium-Xylohypha species. J Med Vet Mycol 33:339–347. doi: 10.1080/02681219580000661. [DOI] [PubMed] [Google Scholar]
- 19.Schubert K, Groenewald JZ, Braun U, Dijksterhuis J, Starink M, Hill CF, Zalar P, de Hoog GS, Crous PW. 2007. Biodiversity in the Cladosporium herbarum complex (Davidiellaceae, Capnodiales), with standardisation of methods for Cladosporium taxonomy and diagnostics. Stud Mycol 58:105–156. doi: 10.3114/sim.2007.58.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zalar P, de Hoog GS, Schroers HJ, Crous PW, Groenewald JZ, Gunde-Cimerman N. 2007. Phylogeny and ecology of the ubiquitous saprobe Cladosporium sphaerospermum, with descriptions of seven new species from hypersaline environments. Stud Mycol 58:157–183. doi: 10.3114/sim.2007.58.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bensch K, Groenewald JZ, Dijksterhuis J, Starink-Willemse M, Andersen B, Summerell BA, Shin HD, Dugan FM, Schroers HJ, Braun U, Crous PW. 2010. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Stud Mycol 67:1–94. doi: 10.3114/sim.2010.67.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Hoog GS, Chaturvedi V, Denning DW, Dyer PS, Frisvad JC, Geiser D, Gräser Y, Guarro J, Haase G, Kwon-Chung KJ, Meis JF, Meyer W, Pitt JI, Samson RA, Taylor JW, Tintelnot K, Vitale RG, Walsh TJ, Lackner M, ISHAM working group on Nomenclature of Medical Fungi. 2015. Name changes in medically important fungi and their implications for clinical practice. J Clin Microbiol 53:1056–1062. doi: 10.1128/JCM.02016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crous PW, Verkley GJM, Groenewald JZ, Samson RA (ed). 2009. Fungal biodiversity. CBS Laboratory Manual Series 1. Centraalbureau voor Schimmelcultures, Utrecht, Netherlands. [Google Scholar]
- 24.White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY. [Google Scholar]
- 25.Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rehner SA, Samuels GJ. 1994. Taxonomy and phylogeny of Gliocladium analyzed from nuclear large subunit ribosomal DNA sequences. Mycol Res 98:625–634. doi: 10.1016/S0953-7562(09)80409-7. [DOI] [Google Scholar]
- 27.Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556. doi: 10.2307/3761358. [DOI] [Google Scholar]
- 28.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 32.Nylander JA. 2004. MrModeltest v2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden. [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed CLSI document M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 34.Crous PW, Groenewald JZ. 2011. Why everlastings don't last. Persoonia 26:70–84. doi: 10.3767/003158511X574532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Ana SG, Torres-Rodríguez JM, Ramírez EA, García SM, Belmonte-Soler J. 2006. Seasonal distribution of Alternaria, Aspergillus, Cladosporium and Penicillium species isolated in homes of fungal allergic patients. J Investig Allergol Clin Immunol 16:357–363. [PubMed] [Google Scholar]
- 36.Torras MA, Artigas JG, Fernandez GS. 1981. Air-borne fungi in the air of Barcelona (Spain). IV. The genus Cladosporium. Mycopathologia 74:19–24. [DOI] [PubMed] [Google Scholar]
- 37.Bentz MS, Sautter RL. 1993. Disseminated infection with Aspergillus fumigatus and Cladosporium cladosporioides in an immunocompromised host, abstr F-37, p 533 Abstr 93rd Gen Meet Am Soc Microbiol. American Society for Microbiology, Washington, DC. [Google Scholar]
- 38.Yano S, Koyabashi K, Kato K. 2003. Intrabronchial lesion due to Cladosporium sphaerospermum in a healthy, non-asthmatic woman. Mycoses 46:348–350. [DOI] [PubMed] [Google Scholar]
- 39.Seyedmousavi S, Guillot J, de Hoog GS. 2013. Phaeohyphomycoses, emerging opportunistic diseases in animals. Clin Microbiol Rev 26:19–35. doi: 10.1128/CMR.00065-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tucker DL, Beresford CH, Sigler L, Rogers K. 2004. Disseminated Beauveria bassiana infection in a patient with acute lymphoblastic leukemia. J Clin Microbiol 42:5412–5414. doi: 10.1128/JCM.42.11.5412-5414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vieira MR, Milheiroy A, Pacheco FA. 2001. Phaeohyphomycosis due to Cladosporium cladosporioides. Med Mycol 39:135–137. doi: 10.1080/mmy.39.1.135.137. [DOI] [PubMed] [Google Scholar]
- 42.Romano C, Bilenchi R, Alessandrini C, Miracco C. 1999. Case report. Cutaneous phaeohyphomycosis caused by Cladosporium oxysporum. Mycoses 42:111–115. [DOI] [PubMed] [Google Scholar]
- 43.Duquia RP, de Almeida HL Jr, Vettorato G, Rocha NM, de Castro LA. 2010. Ecthyma-like phaeohyphomycosis caused by Cladosporium cladosporioides. Mycoses 53:541–543. doi: 10.1111/j.1439-0507.2009.01745.x. [DOI] [PubMed] [Google Scholar]
- 44.Sosa EE, Cohen PR, Tschen JA. 2012. Cladosporium scalp infection. Skinmed 10:393–394. [PubMed] [Google Scholar]
- 45.Kantarcioğlu AS, Yücel A. 2002. In-vitro activities of terbinafine, itraconazole and amphotericin B against Aspergillus and Cladosporium species. J Chemother 14:562–567. doi: 10.1179/joc.2002.14.6.562. [DOI] [PubMed] [Google Scholar]