Abstract
A quantitative comparison between discriminatory indexes and concordance among multilocus variable-number tandem-repeat analysis (MLVA), pulsed-field gel electrophoresis (PFGE), automated ribotyping, and phage typing has been performed, testing 238 Salmonella enterica serotype Enteritidis isolates not epidemiologically correlated. The results show that MLVA is the best choice, but each typing method provides a piece of information for establishing clonal relationships between the isolates.
TEXT
Salmonella enterica serotype Enteritidis is the most frequently reported Salmonella serovar in humans in the European Union (1). This study evaluated the discriminatory power and congruence among typing results collected by applying multilocus variable-number tandem-repeat analysis (MLVA), pulsed-field gel electrophoresis (PFGE), and automated ribotyping, with a new combination of restriction enzymes, to a collection of 238 S. Enteritidis isolates not correlated from an epidemiological point of view. They were isolated in 19 regions in Italy from humans, animals, foods, and environments between 2008 and 2012 (Table 1). For all isolates, except those isolated from humans, the phage type (PT) was kindly provided by the Italian Reference Laboratory for Salmonella and included in the analysis of typing results (Table 1).
TABLE 1.
Source, isolation year, and phage type of the S. Enteritidis isolates tested
| Source (no. of isolates) | Isolation year, phage type (no. of isolates if >1) [region(s)]a |
|---|---|
| Environmental isolates | |
| River water (1) | 2011, PT4 [U] |
| Dust of poultry farm (17) | 2008, PT4 [FR], PT21 (2) [TA, V] |
| 2009, PT4 (2) [SA,T], PT4A [LO], PT4B [V], PT7 [P] | |
| 2010, PT6 [SI], PT8 [FR], PT21 (2) [MA, MA] | |
| 2011, PT3A [SI], PT7 [V], PT8 [LO], PT14B [LO], PT21 [U] | |
| Swab of poultry farm (7) | 2008, PT14B [PU] |
| 2009, PT6 [V], PT8 [FR] | |
| 2010, PT1 [A], PT4 [TA], PT4B [CL] | |
| 2011, PT4 [V] | |
| Poultry farm material (1) | 2011, PT1 [V] |
| Environmental poultry farm material (6) | 2008, PT4 [P] |
| 2010, PT4 [LO], PT14B [LO] | |
| 2011, PT6A [MA], PT35 [MA], PT59 [MA] | |
| Turkey farm material (1) | 2009, PT4 [ER] |
| Animal isolates | |
| Boar (2)b | 2011, PT1B [LO], PT4 [LO] |
| Bovine (4)b | 2008, PT4 [V] |
| 2010, PT8 [V] | |
| 2011, PT1 [LO], PT4 [MA] | |
| Canary (1)b | 2011, PT4 [V] |
| Guinea hen (1)b | 2009, PT4 [ER] |
| Hedgehog (1)b | 2011, PT11 [ER] |
| Ovine (2)b | 2008, PT4 [P], PT8 [T] |
| Poultry (28)b | 2008, PT1 (3) [PU, V, V], PT2 (2) [V, LO], PT4 (3) [CA, FR, V], PT21 (3) [MA, V, T] |
| 2009, PT3 [V], PT4 [U], PT 8 [CA], PT14B [V] | |
| 2010, PT1 (2) [LA], PT4 (4) [ER, LO, V, V], PT14B [V] | |
| 2011, PT4 (3) [V, LO, LO], PT7 [V], PT13 [U], PT14B [V] | |
| Quail (1)b | 2011, PT4 [V] |
| Swine (3)b | 2008, PT4 [LO], PT4B [U], PT21 [LO] |
| Unknown (4)b | 2010, PT1 [ER], PT1B [LO], PT8 [LA] |
| 2011, PT8 [LO] | |
| Poultry (4)c | 2008, PT1 [U], PT4 (2) [SA, P], PT6 [P] |
| Bovine (2)d | 2009, PT4 [V] |
| 2011, PT11 [T] | |
| Crow (1)d | 2011, PT13 [LA] |
| Dog (1)d | 2011, PT4 [U] |
| Duck (2)d | 2008, PT4 [V] |
| 2009, PT1 [V] | |
| Poultry (54)d | 2008, PT4 [V], PT8 (2) [TA, A], PT14B [U], PT21 (2) [V, SA], PT37 [PU], PT51 [P] |
| 2009, PT1 (3) [V, LO, FR], PT3 [ER], PT4 (4) [ER, P, U, V], PT4B [MA], PT6 [V], PT6A [MA], PT7 [A], PT7A [U], PT13A [V], PT21 (2) [U, V] | |
| 2010, PT1 (2) [ER, LO], PT3 [V], PT3A [SI], PT4 (2) [CA, LA], PT6A (3) [SI, MA, MA], PT8 (3) [ER, LA, LO], PT12 [U], PT14B (2) [LO, LO] | |
| 2011, PT1B [V], PT4 (3) [U, V, FR], PT4A [V], PT6C [V], PT7 [LO], PT8 (2) [MA, LA], PT13 [U], PT14B (2) [LO, CA], PT21 (2) [LA, MA], PT59 [V] | |
| Ovine (1)e | 2010, PT13A [PU] |
| Food/feed isolates | |
| Poultry feed (3) | 2008, PT8 [ER] |
| 2010, PT4 [MA] | |
| 2011, PT8 [P] | |
| Fresh beef meat (2) | 2008, PT4 [ER] |
| 2009, PT1 [LO] | |
| Fresh poultry meat (14) | 2008, PT1 (3) [T, V, V], PT4 [ER], PT8 [U], PT21 [V] |
| 2009, PT13A [V], PT25 [CA] | |
| 2010, PT3 [ER], PT6 (5) [ER, FR, V, V, V] | |
| Fresh turkey meat (1) | RDNCf [U] |
| Processed beef meat (4) | 2008, PT4 [CA], PT8 [P] |
| 2009, PT6 [V], PT51 [P] | |
| Processed poultry meat (2) | 2008, PT1 [V] |
| 2010, PT6 [V] | |
| Processed swine meat (2) | 2009, PT6A [P] |
| 2010, PT21 [T] | |
| Processed turkey meat (2) | 2008, PT4 [ER], PT13A [V] |
| Processed meat of unknown origin (2) | 2010, PT4 [LI] |
| 2011, PT14B [V] | |
| Cheese (2) | 2010, PT1 [V] |
| 2011, PT14B [TA] | |
| Milk (2) | 2008, PT4 [CA] |
| 2010, PT1 [V] | |
| Eggs and egg products (15) | 2008, PT1 [CA], PT4 (4) [FR, U, LO, LO], PT14B [PU], PT21 [LO] |
| 2009, PT4 (2) [A, LO], PT14B [LO], PT59 [B] | |
| 2010, PT7 [A], PT14B [LO], PT21 [A] | |
| 2011, PT14B [V] | |
| Fish products/shellfish (6) | 2008, PT1 [V] |
| 2010, PT6C (2) [ER, V], PT8 [CA] | |
| 2011, PT1 [ER], PT59 [CA] | |
| Fresh pastry (1), cream biscuit (1), Tiramisu (1) | 2010, PT14B [T] |
| 2011, PT14B [FR] | |
| 2011, PT8 [V] | |
| Ham (1) | 2009, PT4 [LA] |
| Unknown food product (2) | 2008, PT21 [PU] |
| 2009, PT14B [CA] | |
| Human isolates | |
| Feces (30) | 2012, Phage type not available [CA (2), LA (14), MA (2), MO (3), TA (9)] |
Region: A, Abruzzo; B, Basilicata; CA, Campania; CL, Calabria; ER, Emilia-Romagna; FR, Friuli Venezia Giulia; LA, Lazio; LI, Liguria; LO, Lombardia; MA, Marche; MO, Molise; P, Piemonte; PU, Puglia; SA, Sardina; SI, Sicily; T, Tuscany; TA, Trentino Alto Adige; U, Umbria; V, Veneto.
Organ/tissue.
Unknown
Feces.
Carcass.
RDNC, reaction does not conform.
MLVA was performed according to the protocol published by Hopkins et al. (2) based on the loci SE3, SENTR4, SENTR5, SENTR6, and SENTR7. The five loci were amplified in one multiplex PCR (25-μl volume) containing 5 pmol of each primer, using a multiplex PCR kit (Qiagen). The amplification products were diluted 1:50 in sterile distilled water, and 1 μl of each dilution was mixed with 10 μl of Hi-Di formamide (Applied Biosystems) and 0.4 μl GeneScan LIZ 600 size standard (Applied Biosystems) before being subjected to capillary electrophoresis using POP-7 polymer on a 3130 genetic analyzer (Applied Biosystems). An MLVA type, labeled as a number, was assigned to each isolate based on the difference in the variable-number tandem repeat (VNTR) profile in at least one locus. Minimum spanning trees (MST) based on the MLVA profiles were built in BioNumerics 7.5 (Applied Maths) using the categorical coefficient. Distances between MLVA profiles were calculated based on the numbers of different loci between profiles, irrespective of their within-locus differences in the number of repeats.
PFGE was performed according to the PulseNet protocol (3), digesting the plug with 50 U of XbaI (Fermentas). The Salmonella Braenderup strain H9812 PulseNet standard was used as a molecular weight marker. The fingerprinting profiles were analyzed using BioNumerics 7.1 and compared by cluster analysis using the Dice coefficient and the unweighted pair group method with arithmetic means (UPGMA), with a position tolerance limit and optimization of 1%. Isolates showing a PFGE similarity level of ≥95% were assigned to the same pulsotype. The pulsotypes were labeled as numbers.
Automated ribotyping was performed with the RiboPrinter according to the manufacturer's instructions (4), using a mixture of 1,250 U of PvuII (Qualicon) and 1.250 U of PstI (New England BioLabs). The restriction digestion was performed at 37°C for 20 min. The characterization consisted of combining profiles within a similarity range (as calculated using the RiboPrinter's proprietary algorithm) >0.93 to form a dynamic ribogroup (RIBO) labeled with an alphanumeric code (4).
The Comparing Partitions website (http://darwin.phyloviz.net/ComparingPartitions/index.php?link=Toll) was used to calculate the discriminatory index (DI) of each typing method through Simpson's diversity index (5, 6), the unidirectional concordance between methods by applying the Wallace (W) and adjusted Wallace (AW) coefficients (7), and the bidirectional concordance using the adjusted Rand (AR) coefficient (8).
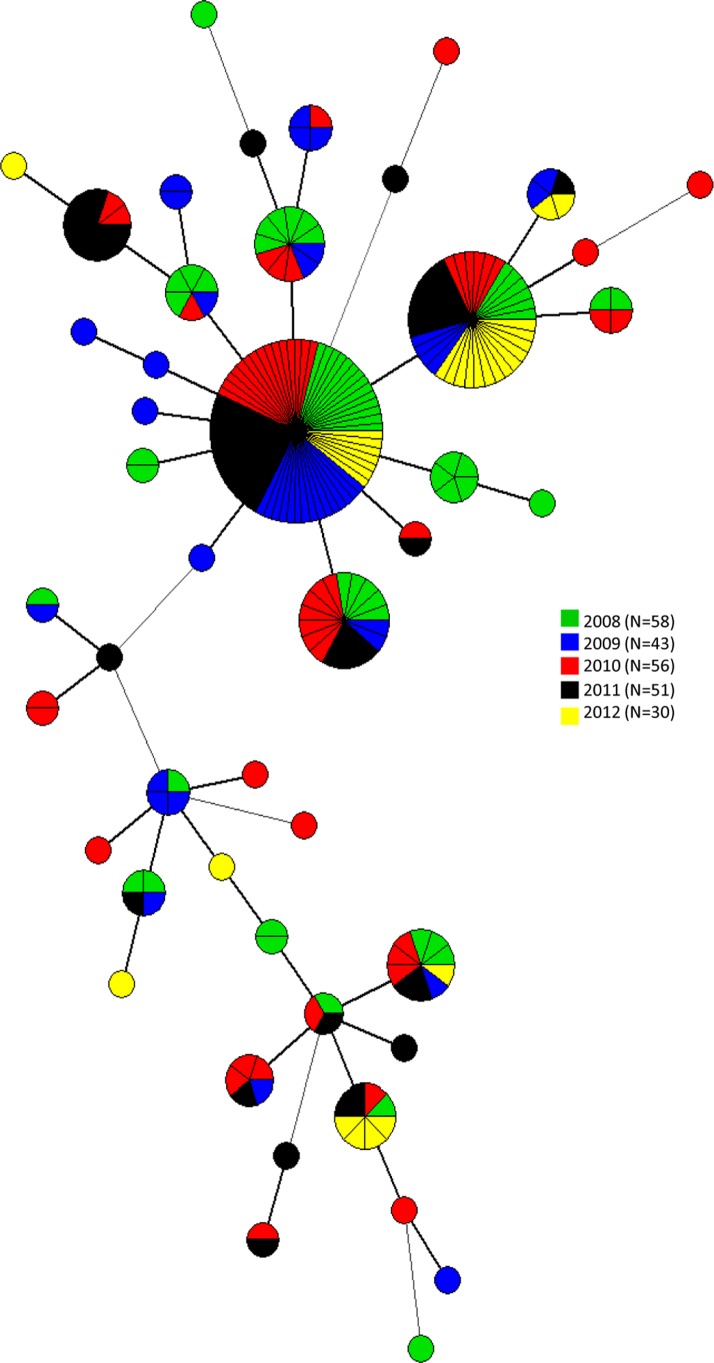
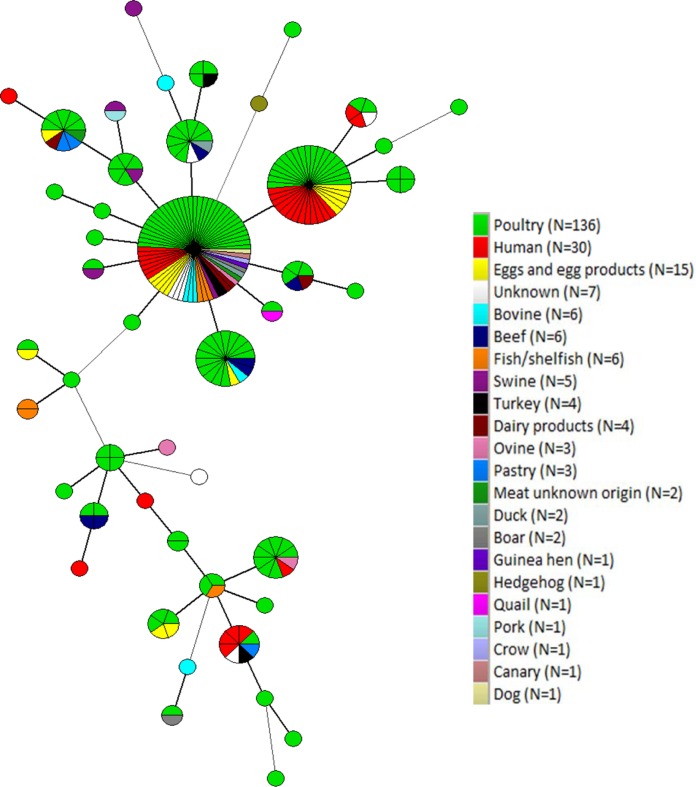
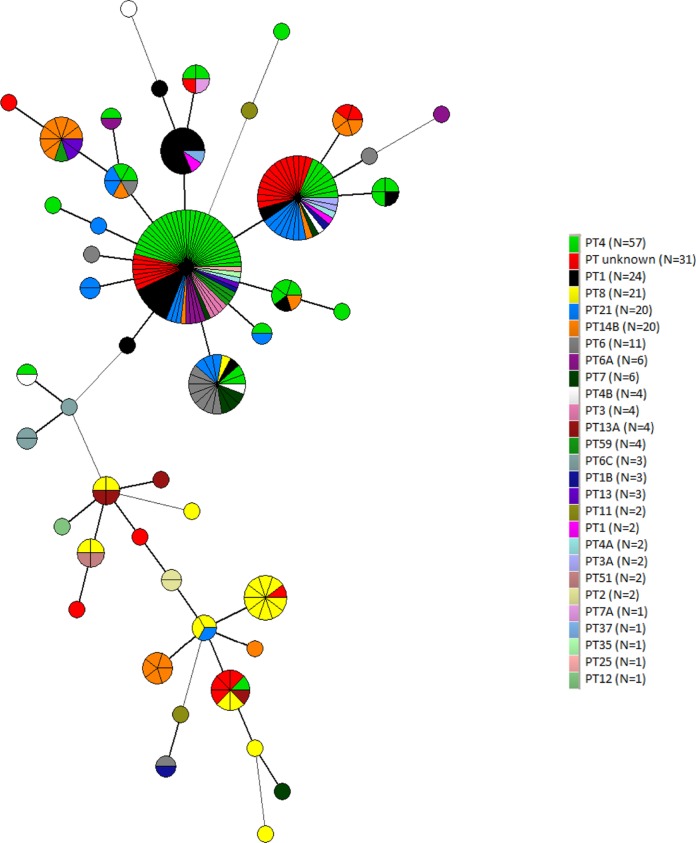
Overall, 21 MLVA types were each associated to a single isolate, whereas 23 comprised between 2 and 67 isolates. The two largest MLVA types grouped isolates collected from all sampling years (Fig. 1). However, small clusters of isolates collected only in specific years were also identified (Fig. 1). One of the two main MLVA types clustered together human, poultry, and egg isolates. Furthermore, the same cluster included isolates from other sources, confirming that S. Enteritidis can be transmitted between humans, wildlife, livestock, and pets (Fig. 2). In relation to the PT, an MST was built, including the MLVA types of human isolates and the reaction does not conform (RDNC) isolate (in red). Human isolates clustered in the two main groups with isolates belonging to the PT identified in S. Enteritidis isolates collected in Italy from confirmed cases of human salmonellosis (Fig. 3).
FIG 1.
Minimum spanning tree calculated for MLVA profiles of 238 S. Enteritidis isolates collected between 2008 and 2012 in 19 regions in Italy.
FIG 2.
Minimum spanning tree calculated for MLVA profiles of 238 S. Enteritidis isolates collected from environmental (n = 33), animal (n = 112), food/feed (n = 63), and human (n = 30) sources.
FIG 3.
Phage type distribution among the minimum spanning tree calculated for MLVA profiles of S. Enteritidis isolates.
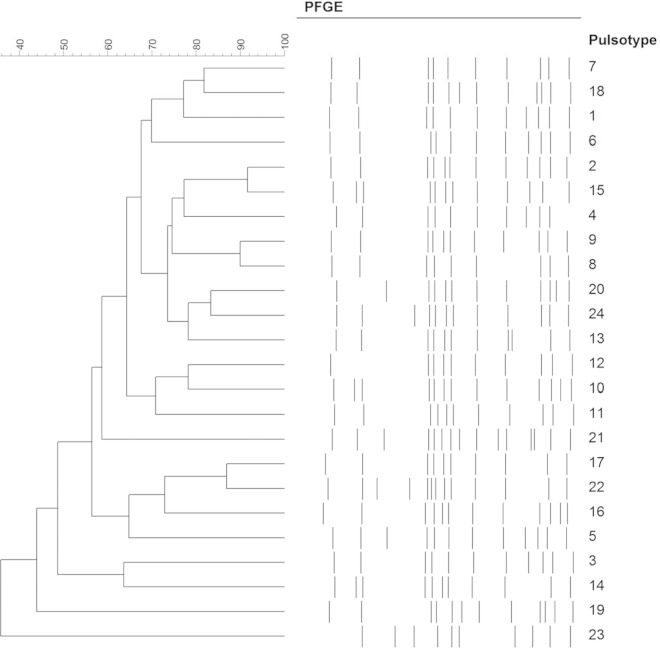
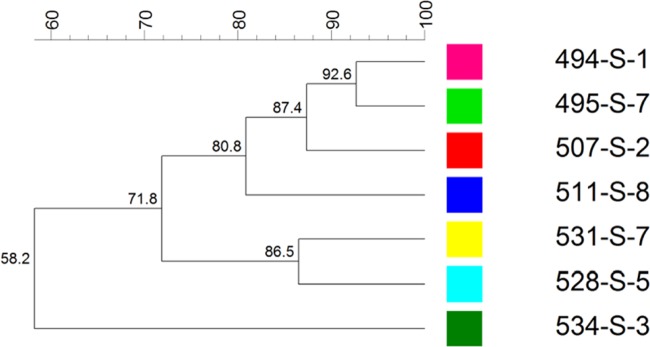
The S. Enteritidis isolates were characterized by 23 pulsotypes, with bands between 20 and 900 kbp, showing a similarity level ranging between 36 and 92% (Fig. 4), and seven ribotyping profiles, characterized by 7 to 10 bands, with molecular weight ranging between 2 and 15 kbp (Fig. 5). The ribotyping profiles 153-494-S-1 and 153-507-S-2 were identified in 57.1 and 36.5% of the isolates, respectively. PFGE is the current gold standard to assess relatedness among Salmonella isolates from different sources (9, 10) and for outbreak investigations (11, 12). However, according to the literature, PFGE exhibits limited discriminatory power for S. Enteritidis (13). This aspect was confirmed in a multicountry outbreak of S. Enteritidis recently reported in Europe (14, 15). Therefore, the European Centre for Disease Prevention and Control (ECDC) decided to promote the use of MLVA to subtype S. Enteritidis isolates (14). Since the European Food Safety Authority (EFSA) stated that there is currently no comprehensive collection of comparable background on MLVA typing data for S. Enteritidis available at the European Union level, this paper should help to start filling this gap (14).
FIG 4.
Pulsotypes identified among the 238 S. Enteritidis isolates.
FIG 5.
Ribotyping profiles associated with the 238 S. Enteritidis isolates.
For each typing method applied alone, the DI ranged between 0.54 for automated ribotyping and 0.88 for MLVA (Table 2), whereas combining two or three methods resulted in a DI ranging between 0.88 and 0.97 (Table 2). Since S. Enteritidis is one of the most genetically homogeneous serotypes of Salmonella (16) it is not surprising that in this study no typing method applied alone reached a DI of >0.90. The combination MLVA-PT has been identified as the best option by Cho et al. (17) and Dewaele et al. (18), even though they applied a different MLVA scheme.
TABLE 2.
Discriminatory power of each typing method applied alone or in combination with other methods on the S. Enteritidis isolates tested
| Method/combination (no. of isolates) | No. types | No. unique isolates | No. clustered isolates | Cluster size | DI (95% CI)a |
|---|---|---|---|---|---|
| MLVA (238) | 44 | 21 | 217 | 2–67 | 0.88 (0.85–0.91) |
| PFGE (238) | 24 | 10 | 228 | 2–109 | 0.74 (0.69–0.79) |
| RIBO (238) | 7 | 3 | 235 | 3–136 | 0.54 (0.50–0.57) |
| PT (207) | 25 | 5 | 202 | 2–57 | 0.87 (0.84–0.90) |
| MLVA-PT (207) | 85 | 51 | 156 | 2–32 | 0.96 (0.95–0.97) |
| PFGE-PT (207) | 64 | 41 | 166 | 2–40 | 0.93 (0.91–0.95) |
| RIBO-PT (207) | 50 | 22 | 185 | 2–33 | 0.94 (0.92–0.95) |
| MLVA-PFGE (238) | 81 | 55 | 183 | 2–39 | 0.95 (0.94–0.97) |
| MLVA-RIBO (238) | 64 | 30 | 208 | 2–41 | 0.94 (0.93–0.96) |
| PFGE-RIBO (238) | 38 | 19 | 219 | 2–59 | 0.88 (0.86–0.91) |
| MLVA-PFGE-RIBO (238) | 104 | 66 | 172 | 2–25 | 0.97 (0.97–0.98) |
DI, Discriminatory index; CI, confidence interval.
All methods tested, including PT, showed a weak directional concordance (i.e., <0.5) (Table 3). However, MLVA showed an AW of >0.16 with PFGE and PT. Moreover, PT exhibited an AW of >0.18 with MLVA and PFGE. The bidirectional concordance between methods was also very low and was ≥0.19 for MLVA-PT and PFGE-PT only (Table 4). The same combinations of typing methods reached the highest AW values. The low congruence between the applied typing methods demonstrated that they all provide a piece of information for establishing possible clonal relationships among the isolates tested. Such relationships concerned 42.4% of the isolates forming 32 clusters containing 2 to 16 isolates sharing the same type strain, as defined according to the MLVA, PFGE, RIBO, and PT profiles (Table 5). Overall, 65.6% of these clusters grouped isolates of common origin (e.g., poultry), whereas 34.4% isolates were classically not correlated (e.g., poultry and cheese) (Table 5).
TABLE 3.
Unidirectional concordance of the typing methods applied
| No. of isolates | Typing method A | Typing method B | WA→B (95% CI) | AWA→B (95% CI) |
|---|---|---|---|---|
| 238 | MLVA | PFGE | 0.377 (0.297–0.457) | 0.164 (0.057–0.272) |
| 238 | MLVA | RIBO | 0.465 (0.396–0.533) | 0.010 (0.000–0.0137) |
| 207 | MLVA | PT | 0.300 (0.212–0.388) | 0.201 (0.101–0.302) |
| 238 | PFGE | MLVA | 0.173 (0.127–0.219) | 0.064 (0.011–0.116) |
| 238 | PFGE | RIBO | 0.446 (0.397–0.494) | 0.000 (0.000–0.090) |
| 207 | PFGE | PT | 0.240 (0.183–0.297) | 0.133 (0.068–0.198) |
| 238 | RIBO | MLVA | 0.118 (0.086–0.150) | 0.002 (0.000–0.038) |
| 238 | RIBO | PFGE | 0.247 (0.199–0.295) | 0.000 (0.000–0.064) |
| 207 | RIBO | PT | 0.130 (0.101–0.159) | 0.007 (0.000–0.040) |
| 207 | PT | MLVA | 0.276 (0.193–0.358) | 0.183 (0.090–0.276) |
| 207 | PT | PFGE | 0.506 (0.408–0.604) | 0.332 (0.199–0.465) |
| 207 | PT | RIBO | 0.480 (0.422–0.538) | 0.043 (0.000–0.150) |
TABLE 4.
Bidirectional concordance of the typing methods applied
| No. of isolates | Typing method A | Typing method B | AR (95% CI) |
|---|---|---|---|
| 238 | MLVA | PFGE | 0.092 (0.043–0.141) |
| 238 | MLVA | RIBO | 0.003 (0.000–0.030) |
| 207 | MLVA | PT | 0.192 (0.112–0.274) |
| 238 | PFGE | RIBO | 0.000 (0.000–0.014) |
| 207 | PFGE | PT | 0.190 (0.125–0.257) |
| 207 | RIBO | PT | 0.012 (0.000–0.033) |
TABLE 5.
Type strains identified
| Cluster label | Type strain MLVA-PFGE(RIBO)PT (no. of isolates) | Source (no. of isolates if >1) | Isolation year (no. of isolates if >1) [region(s)]a |
|---|---|---|---|
| 1 | 8-1 (494-S-1) PT8 (8) | Poultry feces (5) | 2008 [TA], 2010 [ER, LO], 2011 [LA, MA] |
| Fresh poultry meat | 2008 [U] | ||
| Swab poultry farm | 2009 [FR] | ||
| Dust poultry farm | 2010 [FR] | ||
| 2 | 9-1(507-S-2)PT13A (2) | Poultry feces | 2009 [V] |
| Fresh poultry meat | 2009 [V] | ||
| 3 | 13-18(494-S-1)PT6C (2) | Fish products (2) | 2010 [ER, V] |
| 4 | 18-1(494-S-1)PT8 (2) | Fish product | 2010 [CA] |
| Dust poultry farm | 2011 [LO] | ||
| 5 | 21-1(494-S-1)PT14B (4) | Poultry feces | 2010 [LO] |
| Dust poultry farm | 2011 [LO] | ||
| Eggs/egg products (2) | 2009 [LO], 2010[LO] | ||
| 6 | 21-1(507-S-2)PT14B (2) | Poultry feces (2) | 2010 [LO], 2011 [LO] |
| 7 | 28-10(494-S-1)PT21 (2) | Fresh poultry meat | 2008 [V] |
| Poultry feces | 2011 [LA] | ||
| 8 | 28-9(494-S-1)PT6 (3) | Fresh poultry meat (2) | 2010 [ER, V] |
| Processed beef meat | 2009 [V] | ||
| 9 | 28-9(507-S-2)PT6 (3) | Fresh poultry meat (2) | 2010 [V, V] |
| Poultry feces | 2009 [V] | ||
| 10 | 28-10(507-S-2)PT7 (2) | Poultry feces | 2011 [LO] |
| Dust poultry farm | 2011 [V] | ||
| 11 | 30-9(507-S-2)PT4 (2) | Processed beef meat | 2008 [CA] |
| Milk | 2008 [CA] | ||
| 12 | 31-9(494-S-1)PT1 (3) | Feces of poultry | 2009 [V] |
| Cheese | 2010 [V] | ||
| Milk | 2010 [V] | ||
| 13 | 31-10(494-S-1)PT1 (2) | Swab poultry farm | 2010 [A] |
| Fish product | 2011 [ER] | ||
| 14 | 31-10(494-S-1)PT3 (2) | Organ/tissue poultry | 2009 [V] |
| Fresh poultry meat | 2010 [ER] | ||
| 15 | 31-9(494-S-1)PT4 (16) | Organ/tissue bovine (2) | 2009 [V], 2011 [MA] |
| Organ/tissue poultry | 2008 [V] | ||
| Organ/tissue swine | 2008 [LO] | ||
| Duck feces | 2008 [V] | ||
| Unknown poultry farm | 2008 [P] | ||
| Eggs/egg products (2) | 2008 [LO, LO] | ||
| Processed turkey meat | 2008 [ER] | ||
| Bovine feces | 2009 [V] | ||
| Poultry feces | 2009 [P] | ||
| Dust poultry farm | 2009 [T] | ||
| Turkey farm material | 2008 [ER] | ||
| Organ/tissue canary | 2011 [V] | ||
| Organ/tissue boar | 2011 [LO] | ||
| River water | 2011 [U] | ||
| 16 | 31-13(494-S-1)PT4 (2) | Organ/tissue poultry | 2008 [FR] |
| Organ/tissue Guinea hen | 2009 [ER] | ||
| 17 | 31-17(494-S-1)PT4 (3) | Poultry feces | 2010 [CA] |
| Organ/tissue poultry (2) | 2010 [V], 2011 [LO] | ||
| 18 | 31-9(507-S-2)PT4 (5) | Organ/tissue ovine | 2008 [P] |
| Eggs/egg product | 2008 [FR] | ||
| Processed meat | 2010 [LI] | ||
| Dog feces | 2011 [U] | ||
| Poultry feces | 2011 [U] | ||
| 19 | 31-10(494-S-1)PT59 (2) | Poultry farm | 2011 [MA] |
| Fish product | 2011 [CA] | ||
| 20 | 31-17(507-S-2)PT4 (3) | Organ/tissue poultry (2) | 2010 [V], 2011 [V] |
| Eggs/egg products | 2009 [A] | ||
| 21 | 31-10(507-S-2)PT6A (2) | Poultry feces (2) | 2010 [MA, MA] |
| 22 | 33-9(494-S-1)PT1 (3) | Processed poultry meat | 2008 [V] |
| Fresh poultry meat | 2008 [V] | ||
| Fresh beef meat | 2009 [LO] | ||
| 23 | 33-9(507-S-2)PT1 (2) | Fresh poultry meat | 2008 [V] |
| Poultry feces | 2010 [ER] | ||
| 24 | 35-9(494-S-1)PT14B (3) | Food unknown origin | 2009 [CA] |
| Organ/tissue poultry | 2009 [V] | ||
| Poultry feces | 2011 [CA] | ||
| 25 | 37-10(494-S-1)PT21 (4) | Poultry feces (2) | 2008 [V], 2009 [U] |
| Organ/tissue poultry (2) | 2008 [MA, V] | ||
| 26 | 37-14(494-S-1)PT21 (2) | Eggs/egg products | 2010 [A] |
| Dust poultry farm | 2011 [U] | ||
| 27 | 37-9(494-S1)PT4 (2) | Unknown poultry farm | 2010 [LO] |
| Swab poultry farm | 2011 [V] | ||
| 28 | 37-9(507-S-2)PT4 (3) | Poultry feces | 2009 [V] |
| Eggs/egg products | 2009 [LO] | ||
| Organ/tissue poultry | 2011 [V] | ||
| 29 | 37-17(507-S-2)PT4 (2) | Organ/tissue poultry | 2010 [LO] |
| Poultry feces | 2011 [V] | ||
| 30 | 39-9(494-S-1)PT14B (4) | Organ/tissue poultry | 2010 [V] |
| Eggs/egg products | 2011 [V] | ||
| Pastry products (2) | 2010 [T], 2011 [FR] | ||
| 31 | 39-9(507-S-2)PT14B (2) | Organ/tissue poultry | 2011 [V] |
| Cheese | 2011 [TA] | ||
| 32 | 40-9(494-S-1)PT21 (2) | Dust poultry farm | 2008 [V] |
| Processed pig meat | 2010 [T] |
Region: A, Abruzzo; CA, Campania; ER, Emilia-Romagna; FR, Friuli Venezia Giulia; LA, Lazio; LI, Liguria; LO, Lombardia; MA, Marche; P, Piemonte; T, Tuscany; TA, Trentino Alto Adige; U, Umbria; V, Veneto.
In conclusion, the results of this study allow a quantitative comparison between different typing methods for S. Enteritidis in terms of discriminatory power and concordance. These results, along with execution time, cost effectiveness, and the level of complexity of data interpretation and sharing, should help in making a critical choice on the most appropriate method to apply for typing S. Enteritidis during both outbreak investigations and longtime surveillance. Even if the most promising routine epidemiological typing tool for Salmonella enterica seems to be whole-genome sequencing (WGS) (19), when an alternative method must be selected, MLVA is confirmed to be the best choice. In fact, it is highly discriminatory, reproducible, fast, and easy to perform. Moreover, it produces results easy to interpret and analyze, which can be shared using international databases.
REFERENCES
- 1.European Food Safety Authority and European Centre for Disease Prevention and Control. 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J 13(1):3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hopkins KL, Peters TM, de Pinna E, Wain J. 2011. Standardisation of multilocus variable-number tandem repeat analysis (MLVA) for subtyping of Salmonella enterica serovar Enteritidis. Euro Surveill 16(32):pii=19942 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19942. [PubMed] [Google Scholar]
- 3.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 4.Bruce J. 1996. Automated system rapidly identifies and characterizes micro-organisms in foods. Food Technol 50:77–78. [Google Scholar]
- 5.Hunter PR. 1990. Reproducibility and indices of discriminatory of discriminatory power of microbial typing methods. J Clin Microbiol 28:1903–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Severiano A, Pinto FR, Ramirez M, Carriço JA. 2011. Adjusted Wallace coefficient as a measure of congruence between typing methods. J Clin Microbiol 49:3997–4000. doi: 10.1128/JCM.00624-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carriço JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, Almeida JS, Ramirez M. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J Clin Microbiol 44:2524–2532. doi: 10.1128/JCM.02536-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynne AM, Kaldhone P, David D, White DG, Foley SL. 2009. Characterization of antimicrobial resistance in Salmonella enterica serotype Heidelberg isolated from food animals. Foodborne Pathog Dis 6:207–215. doi: 10.1089/fpd.2008.0172. [DOI] [PubMed] [Google Scholar]
- 10.Ponce E, Khan AA, Cheng CM, Summage-West C, Cerniglia CE. 2008. Prevalence and characterization of Salmonella enterica serovar Weltevreden from imported seafood. Food Microbiol 25:29–35. doi: 10.1016/j.fm.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2010. Investigation update: multistate outbreak of human Salmonella enteritidis infections associated with shell eggs. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/salmonella/Enteritidis. [Google Scholar]
- 12.Jacobs W, Kuiling S, van der Zwaluw K. 2014. Molecular typing of Salmonella strains isolated from food, feed and animals: state of play and standard operating procedures for pulsed field gel electrophoresis (PFGE) and multiple-locus variable number tandem repeat Analysis (MLVA) typing, profiles interpretation and curation. EFSA supporting publication 2014:EN-703. European Food Safety Authority, Parma, Italy. [Google Scholar]
- 13.Boxrud D, Pederson-Gulrud K, Wotton J, Medus C, Lyszkowicz E, Besser J, Bartkus JM. 2007. Comparison of multiple-locus variable-number tandem repeat analysis, pulsed-field gel electrophoresis, and phage typing for subtype analysis of Salmonella enterica serotype Enteritidis. J Clin Microbiol 45:536–543. doi: 10.1128/JCM.01595-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.European Food Safety Authority and European Centre for Disease Prevention and Control. 2014. Multi-country outbreak of Salmonella Enteritidis infections associated with consumption of eggs from Germany. EFSA supporting publication 2014:EN-646. http://www.efsa.europa.eu/it/supporting/doc/646e.pdf [Google Scholar]
- 15.Peters TM, Berghold C, Brown D, Coia J, Dionisi AM, Echeita A, Fisher IS, Gatto AJ, Gill N, Green J, Gerner-Smidt P, Heck M, Lederer I, Lukinmaa S, Luzzi I, Maguire C, Prager R, Usera M, Siitonen A, Threlfall EJ, Torpdahl M, Tschäpe H, Wannet W, Zwaluw WK. 2007. Relationship of pulsed-field profiles with key phage types of Salmonella enterica serotype Enteritidis in Europe: results of an international multicenter study. Epidemiol Infect 135:1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saeed AM, Walk ST, Arshad M, Whittam TS. 2006. Clonal structure and variation in virulence of Salmonella Enteritidis isolated from mice, chickens, and humans. J AOAC Int 9:504–511. [PubMed] [Google Scholar]
- 17.Cho S, Whittam TS, Boxrud DJ, Bartkus JM, Rankin SC, Wilkins MJ, Somsel P, Downes FP, Musser KA, Root TP, Warnick LD, Wiedmann M, Saeed AM. 2010. Use of multiple-locus variable number tandem repeat analysis and phage typing for subtyping of Salmonella Enteritidis from sporadic human cases in the United States. J Appl Microbiol 108:859–867. doi: 10.1111/j.1365-2672.2009.04492.x. [DOI] [PubMed] [Google Scholar]
- 18.Dewaele I, Rasschaert G, Bertrand S, Wildemauwe C, Wattiau P, Imberechts H, Herman L, Ducatelle R, De Reu K, Heyndrickx M. 2012. Molecular characterization of Salmonella Enteritidis: comparison of an optimized multi-locus variable-number of tandem repeat analysis (MLVA) and pulsed-field gel electrophoresis. Foodborne Pathog Dis 9:885–895. doi: 10.1089/fpd.2012.1199. [DOI] [PubMed] [Google Scholar]
- 19.Leekitcharoenphon P, Nielsen EM, Kaas RS, Lund O, Aarestrup FM. 2014. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS One 9:e87991. doi: 10.1371/journal.pone.0087991. [DOI] [PMC free article] [PubMed] [Google Scholar]