Abstract
Outbreaks of bloody diarrhea in swine herds in the late 2000s signaled the reemergence of an economically significant disease, swine dysentery, in the United States. Investigations confirmed the emergence of a novel spirochete in swine, provisionally designated “Brachyspira hampsonii,” with two genetically distinct clades. Although it has since been detected in swine and migratory birds in Europe and North America, little is known about its genetic diversity or its relationships with other Brachyspira species. This study characterizes B. hampsonii using a newly developed multilocus sequence typing (MLST) approach and elucidates the diversity, distribution, population structure, and genetic relationships of this pathogen from diverse epidemiological sources globally. Genetic characterization of 81 B. hampsonii isolates, originating from six countries, with our newly established MLST scheme identified a total of 20 sequence types (STs) belonging to three clonal complexes (CCs). B. hampsonii showed a heterogeneous population structure with evidence of microevolution locally in swine production systems, while its clustering patterns showed associations with its epidemiological origins (country, swine production system, and host species). The close genetic relatedness of B. hampsonii isolates from different countries and host species highlights the importance of strict biosecurity control measures. A comparative analysis of 430 isolates representing seven Brachyspira species (pathogens and commensals) from 19 countries and 10 host species depicted clustering by microbial species. It revealed the close genetic relatedness of B. hampsonii with commensal Brachyspira species and also provided support for the two clades of B. hampsonii to be considered a single species.
INTRODUCTION
Outbreaks of bloody mucoid diarrhea were reported in North American swine herds in the late 2000s. This diarrheal condition was accompanied by inappetance, weight loss/reduced weight gain, and reduced feed conversion efficiency, all suggesting the reemergence of swine dysentery (1). Swine dysentery is a mucohemorrhagic diarrheal disease that until recently was known to be caused by an anaerobic spirochete Brachyspira hyodysenteriae, which predominantly affects the cecum and colon of grower-finisher pigs. The disease has caused major economic losses to swine-producing countries globally due to production losses (associated with morbidity and mortality) and the cost of treatment and control of the disease (2). In 2007, a mucohemorrhagic diarrheal disease similar to swine dysentery and caused by a novel pathogen “Brachyspira suanatina” was reported in swine and mallards in Sweden and Denmark (3). Since then, B. suanatina has not been identified in other countries (4), and consequently B. hyodysenteriae has continued to be the main pathogen of interest to swine producers and diagnostic laboratories across the world for finisher pigs with clinical signs of mucohemorrhagic diarrhea.
Although widely prevalent until the late 1980s, swine dysentery has been rarely reported in North America since the early 1990s. It is likely that a change from continuous to an all-in-all-out management system and other animal husbandry practices have played a role in controlling the disease (5). This changed in the late 2000s when outbreaks of bloody mucoid diarrhea were reported in North American swine herds (1). Surprisingly, the spirochetes isolated from diseased swine often tested negative for B. hyodysenteriae by diagnostic tests such as PCR. Analysis of the characteristic NADH oxidase (nox) gene and 16S rRNA gene indicated the emergence of a novel species (1) that was most closely related to a single atypical Brachyspira isolated from a pig in the United Kingdom in the early 1990s (6). Since its identification, clinical trials have experimentally reproduced swine dysentery-like disease (7–9), thus fulfilling Koch's postulates of disease causation. This novel pathogenic species, comprising two genetically diverse clades (I and II) in North America, has been designated “Brachyspira hampsonii” (1). Since its discovery, it has been identified in migratory waterfowl in Europe (10) and North America (11). Interestingly, it has also been detected in commercial pigs transported between European countries, including the Czech Republic, Belgium, and Germany (12, 13).
Despite increasing international interest in B. hampsonii, little is known about the genotypic diversity of this pathogen within the United States and globally. Strain characterization can advance understanding of the genetic relationships between B. hampsonii isolates from commercial pigs and migratory wild birds as well as potentially correlate phenotypic features such as virulence and antimicrobial susceptibility with genotypes. It can also allow monitoring of the strains circulating in a population and tracking of strains in outbreak situations (5). Gel-based molecular typing methods used previously for other Brachyspira species are known to be time-consuming with limited discriminatory power (14–18). Multilocus sequence typing (MLST) is a high-resolution method that uses nucleotide sequence differences between conserved housekeeping genes to characterize and differentiate genotypes of a pathogen (19). This technique has been used for local and global molecular epidemiological studies of various Brachyspira species (20), including B. hyodysenteriae (5, 21, 22), Brachyspira pilosicoli (23), Brachyspira intermedia (24), Brachyspira murdochii, and Brachyspira innocens (25), thus warranting its application for the characterization of B. hampsonii.
The objectives of the current study are (i) to develop an MLST scheme for B. hampsonii, (ii) to characterize and compare genotypes of B. hampsonii in the United States and globally, (iii) to elucidate the diversity, distribution, microevolution, and population structure of B. hampsonii, (iv) to identify the genetic relatedness of B. hampsonii isolates infecting pigs and migratory wild birds, and (v) to compare the diversity and relatedness of B. hampsonii with other pathogenic and commensal Brachyspira species commonly isolated from swine.
MATERIALS AND METHODS
B. hampsonii isolates.
A total of 81 B. hampsonii isolates originating from different host species from North America and Europe were evaluated in this study (Table 1). Of these, 66 B. hampsonii isolates (45 clade I and 21 clade II) were obtained from the University of Minnesota Veterinary Diagnostic Laboratory (UMN-VDL) to represent strains currently circulating in the United States. These isolates were confirmed to the species level by a strongly hemolytic pattern on blood agar and sequencing of the characteristic nox gene. The isolates originated from commercial pigs in 45 sites, 17 systems, and 6 states (Iowa, Illinois, Minnesota, Missouri, North Carolina, and Arkansas) across the United States from 2009 to 2014. Although most sites were represented by a single isolate, 12 of these sites were represented by multiple isolates at a single time point. A swine site refers to the swine farm from which the isolates originated, and a swine system refers to the production company that owns multiple such swine sites. An additional eight B. hampsonii swine-origin isolates were obtained from Canada (n = 2) (8, 9), Belgium (n = 1) (12), Germany (n = 4) (13), and the United Kingdom (n = 1) (6). Finally, another seven isolates originating from European migratory waterfowl wintering in Spain (10) were included in this study. All isolates from countries outside the United States were generously provided by colleagues for inclusion in this study. For the Brachyspira genus-wide comparison, allele number and sequences for genes est, glpK, pgm, and thi for 430 Brachyspira isolates were downloaded from the PubMLST database (http://pubmlst.org/brachyspira/), and these included seven species of Brachyspira (B. hampsonii, B. hyodysenteriae, B. pilosicoli, B. intermedia, B. murdochii, B. innocens, and B. suanatina). These isolates originated from a total of 19 countries (Australia, Belgium, Canada, Denmark, Finland, France, Germany, Hungary, Italy, Japan, Netherlands, New Zealand, Papua New Guinea, Portugal, Serbia, Spain, Sweden, the United Kingdom, and the United States), 10 host species (chicken, dog, duck, goose, horse, human, mallard, mouse, rhea, and swine), and 5 decades (1970s to 2010s).
TABLE 1.
Epidemiological and genotypic information of 81 B. hampsonii isolates evaluated in this studya
| Isolate name | Alleles of six MLST loci (NT/AA)b |
Genetic characterization |
Epidemiological information |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| adh | gdh | thi | pgm | glpK | est | ST/AAT | CC | GGc | Sitee | Systemf | State | Country | Host | |
| G12 | 3/3 | 1/1 | 4/4 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | I | M2 | M | MO | USA | pig |
| G13 | 3/3 | 1/1 | 4/4 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | I | M4 | M | MO | USA | pig |
| G16 | 3/3 | 1/1 | 4/4 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | I | L1 | L | NC | USA | pig |
| G19 | 3/3 | 1/1 | 4/4 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | I | V2 | V | IL | USA | pig |
| G20 | 3/3 | 1/1 | 4/4 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | I | V3 | V | IL | USA | pig |
| G21 | 3/3 | 1/1 | 4/4 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | I | V4 | V | IL | USA | pig |
| G22C | 3/3 | 1/1 | 4/4 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | I | V1 | V | IL | USA | pig |
| G28A | 3/3 | 1/1 | 4/4 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | I | W1 | W | IL | USA | pig |
| G28B | 3/3 | 1/1 | 4/4 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | I | W2 | W | IL | USA | pig |
| G28C | 3/3 | 1/1 | 4/4 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | I | W3 | W | IL | USA | pig |
| G46B | 3/3 | 1/1 | 4/4 | 1/1 | 1/1 | 1/1 | 1/1 | 1/1 | I | W4 | W | IL | USA | pig |
| G38 | 2/1 | 2/2 | 1/1 | 5/3 | 3/3 | 3/3 | 10/10 | 2/2 | II | H1 | H | MO | USA | pig |
| G39 | 2/1 | 2/2 | 1/1 | 5/3 | 3/3 | 3/3 | 10/10 | 2/2 | II | H3 | H | MO | USA | pig |
| G44C | 2/1 | 2/2 | 1/1 | 5/3 | 3/3 | 3/3 | 10/10 | 2/2 | II | H2 | H | MO | USA | pig |
| G10B | 4/2 | 2/2 | 1/1 | 6/4 | 7/3 | 3/3 | 11/9 | 3/3 | II | R2 | R | IA | USA | pig |
| 30599 | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 6/6 | 12/12 | 1/1 | I | 6 | 6 | AB | Canada | pig |
| 5364 | 5/3 | 4/1 | 6/6 | 7/5 | 6/5 | 4/4 | 13/13 | */* | I | 2 | 2 | Un.d | Germany | pig |
| 5366 | 5/3 | 4/1 | 6/6 | 7/5 | 6/5 | 4/4 | 13/13 | */* | I | 2 | 2 | Un. | Germany | pig |
| 5369 | 5/3 | 4/1 | 6/6 | 7/5 | 6/5 | 4/4 | 13/13 | */* | I | 2 | 2 | Un. | Germany | pig |
| D52 | 5/3 | 4/1 | 6/6 | 7/5 | 6/5 | 4/4 | 13/13 | */* | I | 1 | 1 | Un. | Belgium | pig |
| 3824 | 6/4 | 5/3 | 7/5 | 8/6 | 8/6 | 5/5 | 14/14 | */* | III | 3 | 3 | SN | Germany | pig |
| P280 | 7/5 | 6/3 | 8/8 | 9/7 | 9/7 | 7/7 | 15/11 | */* | III | 4 | 4 | Un. | UK | pig |
| AIS50 | 7/5 | 7/3 | 9/8 | 11/11 | 10/10 | 8/8 | 16/16 | */* | III | 5 | 5 | ZA | Spain | goose |
| AIS72 | 8/6 | 10/4 | 10/10 | 10/10 | 12/11 | 9/9 | 17/17 | */* | IV | 5 | 5 | ZA | Spain | mallard |
| AIS166 | 9/3 | 9/1 | 12/7 | 13/9 | 13/9 | 10/10 | 18/18 | */* | I | 5 | 5 | ZA | Spain | goose |
| AIS198 | 10/7 | 8/1 | 13/7 | 14/9 | 14/8 | 10/10 | 19/19 | */* | I | 5 | 5 | ZA | Spain | goose |
| AIS8 | 10/7 | 8/1 | 13/7 | 14/9 | 14/8 | 10/10 | 19/19 | */* | I | 5 | 5 | ZA | Spain | mallard |
| G26 | 3/3 | 1/1 | 5/5 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | I | B2 | B | MO | USA | pig |
| G27A | 3/3 | 1/1 | 5/5 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | I | B3 | B | MO | USA | pig |
| G27B | 3/3 | 1/1 | 5/5 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | I | B3 | B | MO | USA | pig |
| G27C | 3/3 | 1/1 | 5/5 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | I | B3 | B | MO | USA | pig |
| G37 | 3/3 | 1/1 | 5/5 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | I | F1 | F | NC | USA | pig |
| G43A | 3/3 | 1/1 | 5/5 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | I | B5 | B | MO | USA | pig |
| G5A | 3/3 | 1/1 | 5/5 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | I | N1 | N | NC | USA | pig |
| G5B | 3/3 | 1/1 | 5/5 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | I | N1 | N | NC | USA | pig |
| G6B | 3/3 | 1/1 | 5/5 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | I | N2 | N | NC | USA | pig |
| G7A | 3/3 | 1/1 | 5/5 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | I | N3 | N | NC | USA | pig |
| NSH4 | 3/3 | 1/1 | 5/5 | 1/1 | 1/1 | 1/1 | 2/2 | 1/1 | I | N1 | N | NC | USA | pig |
| AIS159 | 11/6 | 11/4 | 11/9 | 12/8 | 11/11 | 11/9 | 20/15 | */* | IV | 5 | 5 | ZA | Spain | goose |
| AIS85 | 11/6 | 11/4 | 11/9 | 12/8 | 11/11 | 11/9 | 20/15 | */* | IV | 5 | 5 | ZA | Spain | goose |
| G1 | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | B1 | B | MO | USA | pig |
| G14A | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | M1 | M | MO | USA | pig |
| G14B | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | M1 | M | MO | USA | pig |
| G14C | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | M1 | M | MO | USA | pig |
| G14D | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | M1 | M | MO | USA | pig |
| G15 | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | M5 | M | MO | USA | pig |
| G23 | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | B4 | B | MO | USA | pig |
| G2B | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | U1 | U | IA | USA | pig |
| G2C | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | U2 | U | IA | USA | pig |
| G2D | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | U3 | U | IA | USA | pig |
| G35 | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | A2 | A | MO | USA | pig |
| G42 | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | S1 | S | MO | USA | pig |
| G49A | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | S2 | S | MO | USA | pig |
| G57 | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | U4 | U | IA | USA | pig |
| G58 | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | A3 | A | AR | USA | pig |
| NSH-16 | 3/3 | 1/1 | 4/4 | 1/1 | 2/2 | 1/1 | 3/3 | 1/1 | I | M3 | M | MO | USA | pig |
| G30A | 3/3 | 1/1 | 3/3 | 2/1 | 5/1 | 1/1 | 4/4 | */1 | I | X1 | X | MO | USA | pig |
| G30B | 3/3 | 1/1 | 3/3 | 2/1 | 5/1 | 1/1 | 4/4 | */1 | I | X1 | X | MO | USA | pig |
| G31A | 3/3 | 1/1 | 3/3 | 2/1 | 5/1 | 1/1 | 4/4 | */1 | I | X2 | X | MO | USA | pig |
| G31B | 3/3 | 1/1 | 3/3 | 2/1 | 5/1 | 1/1 | 4/4 | */1 | I | X2 | X | MO | USA | pig |
| G31C | 3/3 | 1/1 | 3/3 | 2/1 | 5/1 | 1/1 | 4/4 | */1 | I | X2 | X | MO | USA | pig |
| G36 | 3/3 | 1/1 | 3/3 | 2/1 | 5/1 | 1/1 | 4/4 | */1 | I | T1 | T | MO | USA | pig |
| G48A | 3/3 | 1/1 | 3/3 | 2/1 | 5/1 | 1/1 | 4/4 | */1 | I | X3 | X | MO | USA | pig |
| G25A | 1/1 | 3/2 | 2/2 | 4/2 | 4/4 | 2/2 | 5/5 | */* | II | A1 | A | MO | USA | pig |
| G25B | 1/1 | 3/2 | 2/2 | 4/2 | 4/4 | 2/2 | 5/5 | */* | II | A1 | A | MO | USA | pig |
| G8 | 2/1 | 2/2 | 1/1 | 3/2 | 3/3 | 3/3 | 6/6 | 2/2 | II | J5 | J | MN | USA | pig |
| G9 | 2/1 | 2/2 | 1/1 | 3/2 | 3/3 | 3/3 | 6/6 | 2/2 | II | J7 | J | MN | USA | pig |
| NSH-24 | 2/1 | 2/2 | 1/1 | 3/2 | 3/3 | 3/3 | 6/6 | 2/2 | II | J6 | J | MN | USA | pig |
| 30446 | 2/1 | 2/2 | 1/1 | 3/2 | 3/3 | 2/2 | 7/7 | 2/2 | II | 7 | 7 | SK | Canada | pig |
| G32A | 2/1 | 2/2 | 1/1 | 3/2 | 3/3 | 2/2 | 7/7 | 2/2 | II | J1 | J | MN | USA | pig |
| G33 | 2/1 | 2/2 | 1/1 | 3/2 | 3/3 | 2/2 | 7/7 | 2/2 | II | J4 | J | MN | USA | pig |
| G34A | 2/1 | 2/2 | 1/1 | 3/2 | 3/3 | 2/2 | 7/7 | 2/2 | II | J3 | J | MN | USA | pig |
| G34B | 2/1 | 2/2 | 1/1 | 3/2 | 3/3 | 2/2 | 7/7 | 2/2 | II | J3 | J | MN | USA | pig |
| G34C | 2/1 | 2/2 | 1/1 | 3/2 | 3/3 | 2/2 | 7/7 | 2/2 | II | J3 | J | MN | USA | pig |
| G41 | 2/1 | 2/2 | 1/1 | 3/2 | 3/3 | 2/2 | 7/7 | 2/2 | II | J4 | J | MN | USA | pig |
| G51A | 2/1 | 2/2 | 1/1 | 3/2 | 3/3 | 2/2 | 7/7 | 2/2 | II | J2 | J | MN | USA | pig |
| G17 | 4/2 | 2/2 | 1/1 | 6/4 | 3/3 | 2/2 | 8/8 | 3/3 | II | Z1 | Z | MN | USA | pig |
| G10A | 4/2 | 2/2 | 1/1 | 6/4 | 7/3 | 2/2 | 9/8 | 3/3 | II | R1 | R | IA | USA | pig |
| G40 | 4/2 | 2/2 | 1/1 | 6/4 | 7/3 | 2/2 | 9/8 | 3/3 | II | Y2 | Y | MN | USA | pig |
| G50A | 4/2 | 2/2 | 1/1 | 6/4 | 7/3 | 2/2 | 9/8 | 3/3 | II | Y1 | Y | MN | USA | pig |
| G50B | 4/2 | 2/2 | 1/1 | 6/4 | 7/3 | 2/2 | 9/8 | 3/3 | II | Y1 | Y | MN | USA | pig |
All alleles, molecular strains (ST/AAT), and clonal complex (CC) information is provided in the nucleotide/amino acid format. *, singletons that do not group into a clonal complex.
NT, nucleotide; AA, amino acid.
GG, genetic group.
Un., unknown.
Site names are represented by an alphanumeric code for confidentiality purposes.
System names are represented by an alphabetic code for client confidentiality purposes.
DNA extraction.
Frozen B. hampsonii isolates obtained from the UMN-VDL were passaged twice on tryptic soy agar containing commercial 5% defibrinated sheep blood (I-Tek Medical Technologies, MN, USA) before further use. Chromosomal DNA was extracted from culture plates using PrepMan Ultra sample preparation reagent (Applied Biosystems) per the manufacturer's instruction.
Development of a B. hampsonii multilocus sequence typing scheme.
The six housekeeping genes targeted for use as loci for MLST were the same as those used previously for other Brachyspira species (20). The PCR primers for four of these genes, viz. esterase (est), glycerol kinase (glpK), phosphoglucomutase (pgm), and acetyl coenzyme A (acetyl-CoA) acetyltransferase (thi) genes, were the same as those used previously for B. hyodysenteriae (5, 20, 21) and are listed in Table 2. New primers for the alcohol dehydrogenase (adh) and glutamate dehydrogenase (gdh) genes are described in this study and are listed in Table 2. Based on our investigation of the draft genome of representative B. hampsonii clade I and clade II isolates, we were unable to detect the alkaline phosphatase (alp) gene in B. hampsonii. Given the absence of the alp gene in B. hampsonii and that previous MLST studies for Brachyspira have suggested that even five loci can provide sufficient resolution (21, 24), this MLST scheme does not include alp and thus utilizes a total of six loci. The HotStarTaq master mix kit (Qiagen, Valencia, CA, USA) was used to perform PCRs of the six loci in a 40-μl reaction volume, which included 20 μl HotStarTaq master mix, 1.6 μl of each primer (forward and reverse) at a concentration of 10 μM, 14.8 μl RNase-Free water, and 2 μl (∼500 ng) of DNA template. The PCR conditions were optimized using ATCC reference isolates Brachyspira hampsonii NSH-16 (ATCC BAA-2463) and Brachyspira hampsonii NSH-24 (ATCC BAA-2464) of clade I and clade II, respectively (1). The optimized PCR conditions were as follows: initialization at 95°C for 15 min, 38 cycles of denaturation at 94°C for 30 s, annealing at 48°C for 1 min, and extension at 72°C for 1 min, a final extension step at 72°C for 10 min, and a cold hold step at 4°C. PCR products were visualized on a 1.2% agarose gel to confirm amplification of loci.
TABLE 2.
Details of PCR primers for B. hampsonii MLST
| Gene | Primer name | Primer |
|---|---|---|
| est | EST-F229 | 5′-GATGCTTCAGGCGGAGTTATG-3′ |
| EST-R847 | 5′-CCACACTCATAGCATAAATACTG-3′ | |
| glpK | GLP-F123 | 5′-AGGCTGGGTAGAACATAATGC-3′ |
| GLP-R1158 | 5′-TCTTTACTTTGATAAGCAATAGC-3′ | |
| pgm | PGM-F172 | 5′-GTTGGTACTAACAGAATGAATA-3′ |
| PGM-R1220 | 5′-CCGTCTTTATCGCGTACATT-3′ | |
| thi | THI-F163 | 5′-TGTGTTATACAATCAGCACTTC-3′ |
| THI-R1079 | 5′-GTAGTAAGTATTCTAGCTCCAG-3′ | |
| adh | ADH-2F | 5′-GATATACACTCAGCAAGAAGCG-3′ |
| ADH-2R | 5′-ATGTTATCGGTAAAGAGGCGG-3′ | |
| gdh | GDH-2F | 5′-GGAGTTGGCGGAAGAGAAATA-3′ |
| GDH-2R | 5′-ATCCCATTTATCCATAGAAGCA-3′ |
Multilocus sequence typing.
PCR amplification of all six loci of each B. hampsonii isolate, positive controls (strains NSH-16 for clade I and NSH-24 for clade II), and a negative control (ultrapure water) were performed as mentioned above. The purification and Sanger sequencing of PCR amplicons, alignment of nucleotide sequences, and translation of nucleotides into predicted amino acid sequences were performed as mentioned previously (5).
Analyses.
All B. hampsonii isolates with unique nucleotide and amino acid sequences were assigned unique nucleotide and amino acid allele numbers, respectively. Isolates with a unique combination of nucleotide and amino acid alleles at the six loci were given unique sequence type (ST) and amino acid type (AAT) numbers, respectively (Table 1). The isolates were evaluated on multiple levels, including similarities/differences of genotypes within a site, between sites within a system, between countries, and between host species. Simpson's index of diversity (DI) (26) was used to determine the diversity of genotypes within the United States and globally, on a clade and species level. Similarly, the START2 program was used to detect recombination between the loci of isolates within the United States and globally by the index of association (27), using isolates and STs as the unit of analysis. The START2 program was also used to detect the ratio of the nonsynonymous to synonymous mutations in U.S. and global data sets (28). The eBURST v3 program (29) was used to determine the population snapshot analysis of STs, wherein isolates were grouped into clonal complexes (CCs) of single locus variants (SLVs) to predict putative founder types. BioNumerics v7.1 software was used to generate a minimum spanning tree (MST) to depict the relatedness and predicted microevolution of STs. The MST was color-coded to represent the country, swine system, host species, and year of origin. All six loci were concatenated for each isolate using R software (30). The concatenated sequences were used to generate a maximum parsimony tree using MEGA6 (31) software to depict the relatedness of STs, with comparisons being made between clades, host species, and countries of origin.
Comparison of seven Brachyspira species.
For the genus-wide comparison, arbitrary numbers were assigned to each unique allelic profile as observed by using only four loci (genes est, glpK, thi, and pgm) commonly identified in all seven Brachyspira species evaluated, resulting in a total of 326 diverse Brachyspira types. The sequences of all available alleles for each locus were aligned using the ClustalW function of the MEGA6 software (31), and the ends were trimmed to ensure the same length and region of all isolates were being used for comparison in downstream analyses. The four loci were concatenated for each isolate using R software (30), and the concatenated sequences of all 326 unique Brachyspira types were aligned once again using the ClustalW function of MEGA6 software. Finally, a consensus maximum likelihood tree was generated to depict the relatedness and clustering of the 326 Brachyspira types and color-coded to represent unique Brachyspira species.
Nucleotide sequence accession numbers.
The coding regions for genes adh and gdh needed for amino acid translation have been made available on NCBI GenBank under accession numbers KR559025 and KR559026.
RESULTS
The 81 B. hampsonii isolates evaluated represented a total of 20 STs and 19 AATs (Table 1). Of these, isolates of United States origin represented 11 STs and 10 AATs, of which ST1 and ST3 were most frequently isolated. An additional nine STs and nine AATs represented isolates from countries outside the United States, although one ST was present in the United States and Canada. Data concerning all B. hampsonii isolates evaluated, including nucleotide allele numbers, sequence type numbers, and nucleotide sequences of unique alleles have been made publicly available on the PubMLST database (http://pubmlst.org/brachyspira/) and are listed in Table 1. The mean ratio of the number of nonsynonymous to synonymous mutations (dN/dS), DI, and index of association (IA) values for global and U.S. isolates by clade and species are listed in Table 3.
TABLE 3.
Measures of genetic diversity and recombination for global and U.S. B. hampsonii isolates
| Level of discrimination | Level of analysis | Mean dN/dSa | Simpson's DI | IA (P value) |
|---|---|---|---|---|
| Global isolates | Species | 0.045 | 0.921 | 3.444 (<0.0001) |
| Clade I | 0.040 | 0.820 | 2.852 (<0.0001) | |
| Clade II | 0.026 | 0.843 | 1.469 (<0.0001) | |
| U.S. isolates | Species | 0.031 | 0.879 | 3.228 (<0.0001) |
| Clade I | 0.031 | 0.746 | 0.841 (<0.0001) | |
| Clade II | 0.026 | 0.860 | 1.485 (<0.0001) |
The eBURST analysis grouped 20 STs into three CCs based on similarities between SLVs. Clade I consisted of one CC (CC1) and four singletons, clade II consisted of two CCs (CC2 and CC3) with one singleton, and an additional five singletons were identified that were not clearly grouped into either of the two previously described clades (1). No founder types were predicted with high bootstrap values for B. hampsonii within North America or globally. Since the number of STs and AATs differed by only one, the resulting analyses (eBURST analysis, maximum parsimony tree, and minimum spanning tree analysis) using AATs showed no significant difference in results compared to those obtained by using STs (data not shown).
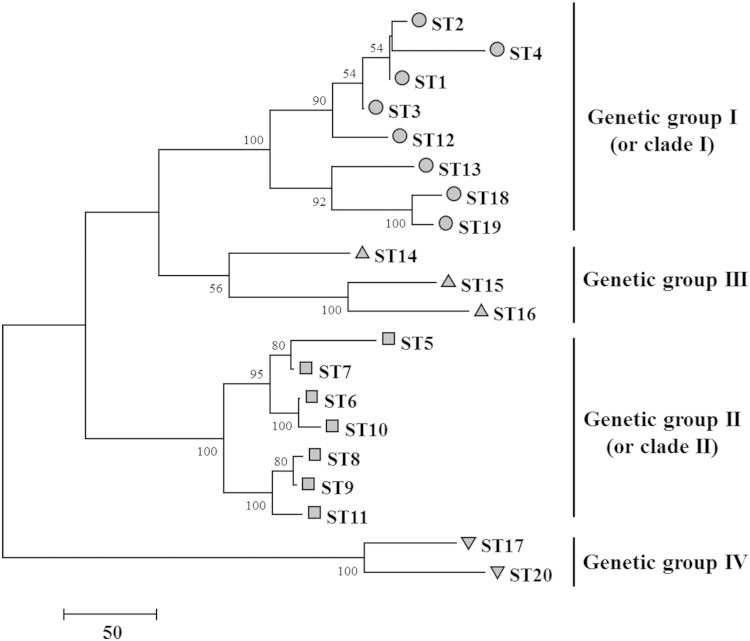
The genetic relatedness of evaluated global B. hampsonii isolates determined using nucleotide differences is depicted in Fig. 1. Based on nucleotide sequences, four major genetic groups of STs (I, II, III, and IV) were identified, of which genetic groups I and II represented clades I and II, respectively. The genotypes in genetic groups I and II were grouped into one and two CCs, respectively, as identified by eBURST analysis. The genotypes of two Canadian isolates were clustered along with genetic group I (ST12) and genetic group II (ST7) from the United States. The porcine-origin genetic group I isolates from Germany and Belgium were characterized as the same genotype (ST13), and although they broadly clustered with other genetic group I genotypes, they formed a distinct subgroup with some migratory wild bird-origin genetic group I genotypes (ST18 and ST19) from Spain. Genetic group III consisting of porcine-origin genotypes from Germany and the United Kingdom (ST14 and ST15), and a migratory wild bird-origin genotype (ST16) from Spain did not consistently group with either of the two previously described clades (1). Migratory wild bird-origin isolates characterized as ST17 and ST20 formed a genetic group IV, which was clearly distinct from the other three genetic groups.
FIG 1.
Maximum parsimony bootstrap consensus tree (1,000 replicates) depicting relatedness of global B. hampsonii sequence types. The subtree-pruning-regrafting algorithm was used to compute the genetic similarity in a total of 4,113 nucleotide positions of 20 sequence types. The scale unit represents 50 nucleotide substitutions, and bootstrap values greater than 50% are shown at the nodes. Four main genetic groups (I, II, III, and IV) were identified and were represented by gray shapes as follows: genetic group I (circle), genetic group II (square), genetic group III (triangle), and genetic group IV (inverted triangle). Genetic group I and genetic group II included clade I and clade II sequence types, respectively, which represented isolates from North America and Europe. The genetic groups III and IV included sequence types representing European isolates that did not fall under either clade I or clade II.
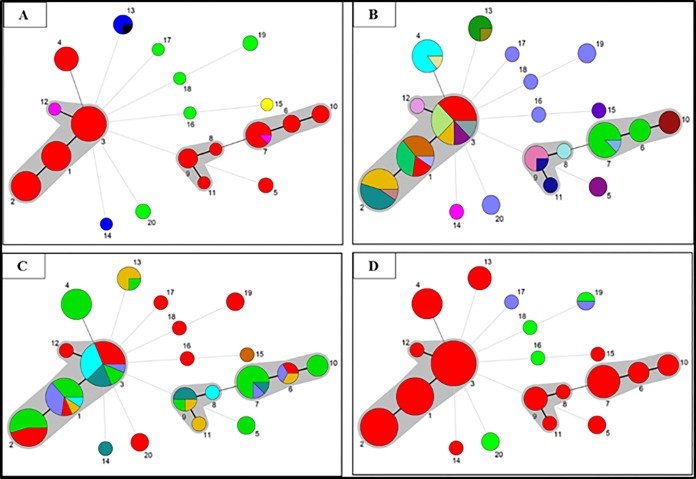
The basic clustering of 81 B. hampsonii isolates using nucleotide allelic similarities depicted by eBURST and maximum parsimony analysis was also depicted by the MST analysis (Fig. 2), wherein most clade I and II isolates were clustered into CCs, and most other isolates were distributed as singletons. Figure 2A depicts the geographical origins of 81 isolates characterized as 20 STs, wherein the observed CCs included North American isolates only. Although most genotypes were specific to the country from which they were isolated, ST7 was found in the United States and Canada, and ST13 was found in Belgium and Germany. Figure 2B depicts the production systems from which these 81 B. hampsonii isolates originated. Within the United States, clade I genotypes were found in multiple systems, whereas those of clade II were usually restricted to a single system. Figure 2C illustrates the year of isolation of these genotypes, wherein the U.S. genotypes, particularly those of clade I, are maintained within the population for a few years. Finally, Fig. 2D depicts the host species from which these isolates originated, wherein the genotypes were uniquely either of porcine or avian host origin.
FIG 2.
Minimum spanning tree (MST) analyses depicting the epidemiological origins of 20 sequence types (STs) representing 81 B. hampsonii isolates. In the MST analyses, each circle represents a unique ST, its size represents the relative number of isolates, and its color represents a specific epidemiological characteristic. The width of the lines reflects the genetic distance between STs with darker/thicker lines connecting more similar STs and lighter/thinner lines connecting less similar STs. STs that were single locus variants of at least one other ST were grouped together in a clonal complex (shaded in gray). (A) MST analysis depicting the country of origin of these 81 B. hampsonii isolates. Colors represent countries as follows: United States (red), Spain (light green), Germany (navy blue), Canada (pink), United Kingdom (yellow), and Belgium (black). (B) MST analysis depicting the swine system of origin of these 81 B. hampsonii isolates. Each of the 23 unique colors represents a different swine system, while one color (medium blue) represents the avian origin isolates that did not fall within a swine system. (C) MST analysis depicting the year in which these 81 B. hampsonii isolates were recovered. Colors represent the year of isolation as follows: 2014 (turquoise), 2013 (light green), 2012 (mustard), 2011 (red), 2010 (dark cyan), 2009 (lavender), and 1994 (light brown). (D) MST analysis depicting the host species from which these 81 B. hampsonii isolates originated. Colors represent the host species of origin as follows: pig (red), goose (light green), and mallard (lavender).
In general, a single genotype was found circulating within a swine site at a single time point. Within a swine system, either one single or two clonally related genotypes were detected, and in general, a specific clade was detected. Additionally, genotypes were either specific to a country or were detected in two countries within a common geographical region/continent, such as in North America (the United States and Canada) or Europe (Belgium and Germany).
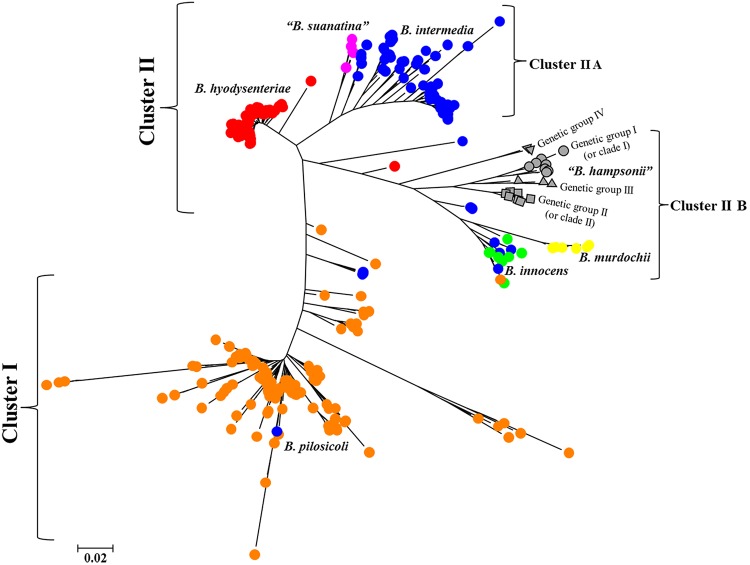
The genetic relatedness and clustering of the 326 Brachyspira types of 430 Brachyspira isolates determined using nucleotide differences among four loci is depicted as a radial tree in Fig. 3. As seen in Fig. 3, isolates cluster by the Brachyspira species they represent, wherein one main cluster (I) consisted of B. pilosicoli isolates, and the other main cluster (II) consisted of the other six evaluated Brachyspira species. Within cluster II, two main groups were identified: subcluster IIA mostly consisted of B. hyodysenteriae, B. suanatina, and B. intermedia, while subcluster IIB mostly consisted of B. hampsonii, B. murdochii, and B. innocens. A few occasional Brachyspira isolates, particularly those of B. intermedia, unexpectedly clustered with groups of other Brachyspira species. Based on this analysis, B. hampsonii was observed to have higher diversity than that of B. hyodysenteriae, a much lower diversity than that of B. pilosicoli, and a diversity that was most similar to that of the main B. intermedia cluster.
FIG 3.
Radial tree of maximum likelihood analysis portraying the clustering of 326 Brachyspira types (430 isolates) representing seven Brachyspira species. Each unique type was depicted by a colored shape to represent its taxonomic identity. The genetic groups of B. hampsonii were represented by the following filled gray shapes: genetic group I (gray circle), genetic group II (gray square), genetic group III (gray triangle), and genetic group IV (gray inverted triangle). The other six Brachyspira species were represented by filled colored circles as follows: B. hyodysenteriae (red circle), B. suanatina (pink circle), B. intermedia (blue circle), B. murdochii (yellow circle), B. innocens (light green circle), and B. pilosicoli (orange circle). A total of 2,807 positions of 326 Brachyspira types were used to compute the genetic similarity, and the scale unit represents 2 substitutions per 100 nucleotide positions. Two major clusters were identified: cluster I consisting of B. pilosicoli and cluster II consisting of another six Brachyspira species. Cluster II showed two smaller subclusters as cluster IIA (B. hyodysenteriae, B. suanatina, and B. intermedia) and cluster IIB (B. hampsonii, B. murdochii, and B. innocens).
DISCUSSION
This study was undertaken with the objective of developing a MLST scheme for B. hampsonii to characterize and compare its genotypes and to elucidate its genetic diversity, distribution, and population structure. The inclusion of B. hampsonii isolates from different commercial production systems, countries, and host species allowed us to investigate its epidemiological distribution within the United States and globally.
Heterogeneity in B. hampsonii was detected in the population by high diversity values (Table 3) globally and within the United States, although the United States showed lower diversity values than the global population. Within the United States, B. hampsonii clade I showed a lower diversity value than that of clade II or the species as a whole, despite clade I being more frequently isolated in the United States. However, when the diversity of genotypes by clades is analyzed on a global level, clade I appears to be as diverse as clade II (Table 3). This is partly because the only isolates that can clearly be characterized as clade II originated from North America; thus, clade II does not contribute any further diversity in the global data set analysis. Specifically, clade I and II genotypes included in this study originated from swine in North America, some clade I genotypes originated from swine and migratory wild birds in Europe, and some genotypes that were not clearly classified as belonging to either clade originated from swine and migratory wild birds in Europe. Further, in a previous study (11), two isolates originating from migratory wild birds in North America were shown to cluster most closely with B. hampsonii clade II. It therefore appears that B. hampsonii clade I is widespread globally (in swine and migratory wild birds in North America and Europe), which based on current knowledge is in contrast to clade II, which as of now has been detected only in North America. Several genotypes representing isolates of swine and migratory wild bird origins in Europe cannot clearly be classified as either clade I or clade II, and likely represent additional genetic groups of B. hampsonii (Fig. 1). It is possible that further detection of B. hampsonii from across the world may result in the characterization of more genotypes and potentially more subgroups/outliers. A previous study (1) suggested that B. hampsonii clade II might have evolved from ancestral groups that contained isolate P280/1, and clade I might have evolved independently of clade II. In the current study, we have identified additional swine and migratory wild bird isolates that cluster with isolate P280/1; however, this cluster does not consistently fall within either clade I or clade II (Fig. 1), and it is unclear whether either of these clades have evolved from isolate P280/1 (6).
Some factors that might explain why the B. hampsonii U.S. data set shows clade I having a lower diversity than that of clade II in this study, in contrast to a previous study (1), include the time at which the previous study was carried out (i.e., soon after the disease emergence) and the sample size (limited by a smaller sample size originating from few epidemiological sources). Given the clinical emergence of the two clades at the same time in North America, it is unknown why clade I is homogeneous relative to clade II within the United States. It can be hypothesized that since the disease has only recently emerged, clade II might include many of the diversifying genotypes that have emerged from an ancestor of clade I. A previous study (1) discussed whether all B. hampsonii isolates represent a single monophyletic taxon having a single common ancestor or if they represent a polyphyletic taxon with more than one ancestor (i.e., one for each clade). To our knowledge, no common ancestral genotypes have yet been identified for all B. hampsonii; thus, B. hampsonii may potentially represent a polyphyletic taxon that consists of two or more monophyletic clades.
The mean dN/dS ratios for B. hampsonii (Table 3) indicated the fact that most mutations were synonymous in nature, which is expected for housekeeping genes that are under selective pressure to prevent deleterious changes that may affect their functions. Despite the heterogeneity observed in the population, evidence of clonality was provided by the detection of significant linkage disequilibrium (P < 0.0001) in the global and U.S. B. hampsonii data sets, irrespective of the level of analysis (species or clade) (Table 3). Within swine systems, genotypes have the opportunity to undergo mutations to form new but closely related genotypes by a process known as microevolution (32), thus forming clonal groups within systems. Therefore, it appears that B. hampsonii is heterogeneous on a population level, and within swine systems, the genotypes diversify clonally.
The similar number of STs and AATs of B. hampsonii identified within the United States and globally may indicate an actively diversifying population, and it is possible that over long periods of time, some of these genotypes might be eliminated from the population. Alternatively, the similar number of STs and AATs of B. hampsonii may indicate that it is truly a relatively diverse species, which may also be supported by the detection of additional clusters that do not clearly fall within the two clades. Future studies over longer periods of time may provide further support for either of the potential scenarios explaining the genetic diversity of B. hampsonii. It is interesting to note that although B. intermedia and particularly B. pilosicoli have a similar number of STs as they have AATs (23, 24), B. hyodysenteriae shows a lower number of AATs compared to STs (5). It is thought that the high plasticity, recombination, and genetic diversity of B. pilosicoli may contribute to its ability to infect and cause disease in multiple host species, including swine, chicken, and human beings (23). This is in contrast to B. hyodysenteriae that shows a strong clonal distribution in a diverse population and is known to cause disease most often in swine (5). Although B. intermedia is clonal compared to the highly recombinant B. pilosicoli, its pattern of high diversity in the population with smaller groups of clusters/clones particularly within swine systems resembles that of B. hampsonii observed in this study. B. intermedia isolates are known to cause disease in chickens, and although they are frequently isolated from swine, they are not usually associated with disease in this host species (24). Similarly, although B. hampsonii has been isolated from birds (10, 11), to our knowledge it is only known to cause disease in swine (1).
B. hampsonii genotypes clearly identified as clade I or II by nox gene sequencing prior to this study were also identified as belonging to these clades based on the maximum parsimony analysis using six housekeeping genes (Fig. 1). The four genetic groups in the dendrogram were associated with their genetic identity, geographical origin, and host species from which these B. hampsonii isolates originated (Fig. 1). For instance, ST7 represented clade II isolates from the United States and Canada, and ST12 represented a clade I isolate from Canada that most closely grouped with clade I isolates from the United States. Similarly, two closely related migratory wild bird-origin genotypes (ST18 and ST19) from Europe clustered close to a swine origin genotype (ST13) from Europe (Fig. 1). This suggests that, in this study, although B. hampsonii isolates originating from migratory wild birds were found to represent distinct genotypes from those of swine, they do show a close relationship with the swine origin genotypes within their geographical region, i.e., Europe rather than North America. Interestingly, the swine-origin clade I genotype ST13, referred to above, represented two previously reported (12, 13) B. hampsonii isolates from swine in Europe (Fig. 1). One was identified in Belgium from gilts recently imported from the Czech Republic that showed soft, watery, nonhemorrhagic colonic content and mild diarrhea (12). The other was identified in Germany from swine recently purchased from Belgium with a history of mild to moderate diarrhea (13). This is in contrast to the mucohemorrhagic diarrhea typically reported with other clade I genotypes in North America (1); however, no experimental trials have been conducted in order to understand whether or not these European B. hampsonii isolates are pathogenic to swine.
The B. hampsonii isolates originating from migratory wild waterfowl exhibited diverse genotypes and clusters (Fig. 1). For instance, based on the use of the six housekeeping gene loci, genotypes ST18 and ST19 clustered in clade I (or genetic group I), while others were not clearly grouped into either of the two previously described clades of B. hampsonii (1), and instead clustered within additional genetic groups. B. hampsonii genotype ST16 closely clustered along with isolate P280/1 (originally from a pig in the United Kingdom in the 1990s) and isolate 3824-15x/14 (originally from a pig in Germany in 2014) (Fig. 1). Although previous studies have suggested that this cluster falls within clade II (or genetic group II), since we did not obtain any high bootstrap values or consistency in its grouping within any clade, we have considered this to be a separate group (genetic group III). Finally, the most diverse genotypes ST17 and ST20 formed their own distinct cluster (genetic group IV) in the dendrogram (Fig. 1). The clustering pattern of these migratory wild bird-origin B. hampsonii isolates generally resembled that seen by the use of the nox gene segment, as reported in a previous study (10). However, those B. hampsonii isolates that grouped with isolate P280/1 are not classified as either genetic group I or II in this study, as their relative positions were inconsistent on multiple generations of dendrograms generated using the six housekeeping genes (Fig. 1). Interestingly, genotype ST19 represented isolates from wild geese and mallards (Fig. 2D). Wild waterfowl are known to serve as carriers for known Brachyspira pathogens of swine and poultry (33). The migratory patterns of wild birds, their close contact with livestock, and contamination of drinking water sources by their feces may allow the spread of and exposure of livestock to infectious agents, thus suggesting their potential role as reservoirs (34). Additionally, a previous study (11) reported colonization without clinical signs in an experimental inoculation of a B. hampsonii clade II (or genetic group II) isolate that was isolated from a migratory bird (snow goose). Therefore, the clustering of migratory wild bird-origin B. hampsonii isolates with those of swine origin in this study provides further support for the hypothesis of B. hampsonii transmission between these host species.
The three CCs of B. hampsonii included only North American genotypes since they mostly represented multiple swine systems in a single country (United States) (Fig. 1 and 2). Most singletons were those from Europe, since they mostly represented individual isolates that were obtained from different host species, countries, or periods of time (Fig. 1 and 2). The presence of the same or clonally related genotypes in the United States and Canada suggests the transmission of B. hampsonii between the two countries (Fig. 2A). This may be facilitated by transport of livestock, contact with contaminated fomites, or vectors/carriers of Brachyspira such as migratory wildbirds. The presence of the same genotype from Belgium and Germany (Fig. 2A) also suggests the role of transport of infected animals, especially because in each of the cases the swine had recently been imported from neighboring countries (12, 13). Additionally, although swine and migratory wild birds had different genotypes, those from the same geographical region were closely related, suggesting some potential for exposure between the different host species (Fig. 1). The distribution of B. hampsonii genotypes within swine systems in the United States suggested that clade I genotypes are widespread and present in multiple systems, while clade II genotypes are present in fewer systems (Fig. 2B). The observation of a single B. hampsonii genotype being detected within a swine site at a single point of time suggests a single source of infection rather than multiple different sources. Further, a system that tests positive for a particular genotype, and especially a particular clade of B. hampsonii, if infected for long durations, usually will continue to isolate the same genotypic cluster or clade over time. For instance, one system had reported a history of severe mucohemorrhagic diarrhea in different sites (separate sites for each stage of production) within their system since the emergence of disease in 2009. The different stages of production of swine included a single flow, i.e., the gilts would be transported into sow farms, and the piglets from the sow farm would be transported to finisher farms. Despite an effort to treat the infection, the genotypic characterization detected the same two related genotypes and the same clade of B. hampsonii within the different sites of the system from 2009 to 2014, suggesting that one of the sites in the system is likely serving as a source of contamination for the other sites. The likely transmission of B. hampsonii between farms or between countries highlights the importance of strict biosecurity measures to prevent introduction of the pathogen to naive herds.
Comparison of four housekeeping gene segments of isolates of seven Brachyspira species from diverse host species, geographical origins, and years of isolation provides a unique opportunity to use the multilocus sequence typing technique to further understand the genetic relationships of the different species. This approach, known as multilocus sequence analysis (MLSA) (35), has been suggested to be particularly useful for identifying and differentiating less understood bacterial species within a genus. The use of a single, highly conserved gene such as 16S rRNA or nox may not be able to capture the true extent of genetic diversification of all isolates within a genus. Single mutations in a single gene can exaggerate the overall genetic difference between two isolates or clusters of isolates. In the Brachyspira genus MLSA analysis, inclusion of the alkaline phosphatase gene (which is missing only in B. hampsonii) might have exaggerated the overall genetic distance of B. hampsonii from the other six Brachyspira species. Therefore, the use of multiple conserved genes consistently present in all evaluated microbes provides a balanced and more reliable approach to understanding the clustering of genotypes into potential species. The use of a higher number of genes increases the resolution of the MLSA analysis; thus, analysis of more genes may provide a higher resolution.
The diversity of and relationships between different Brachyspira species commonly isolated from swine were best illustrated by the differential clustering of species in this study (Fig. 3). Although most species clearly clustered only with isolates of their own species, occasional isolates, particularly those of B. intermedia, did not cluster into these species-based groups. Several isolates that were identified as B. intermedia in a previous study (24) instead clustered directly within or as outliers to groups of other weakly beta-hemolytic species such as B. innocens, B. pilosicoli, and B. murdochii in our study. This previous study (24) concluded that given the high diversity and numerous outliers identified, it is likely that all of these isolates are too diverse to be classified as B. intermedia and likely represent other Brachyspira species. Similarly, one B. hyodysenteriae isolate clustered as an outlier to a large cluster containing B. innocens, B. murdochii, and B. hampsonii isolates, and may warrant further investigation. It may be important to confirm whether the positions of these isolates in the current dendrogram stay consistent when an increased number of housekeeping genes are evaluated.
Interestingly, the B. hampsonii isolates formed a tightly clustered species group, irrespective of their geographical region or host species of origin with only two isolates clustering as outliers within the core group (Fig. 3). Despite the identification of two or more diverse genetic groups, this interspecies comparison depicts a close clustering of all genetic groups of B. hampsonii isolates compared with that of other Brachyspira species. Additionally, phenotypic (hemolysis pattern, biochemical assays, pathogenicity) and ecological (host specificity, tissue specificity) differences between these clusters should also be considered while determining whether a cluster should be designated a unique species. Since the two clades of B. hampsonii exhibit strong hemolysis on culture, are not always distinguishable by biochemical tests, cause a mucohemorrhagic diarrheal disease affecting the colon of swine, and form a single cluster compared to other swine Brachyspira species, it would be a judicious approach to consider B. hampsonii a single species, with several genetically diverse groups.
The high diversity and the genetic difference of B. pilosicoli from other Brachyspira species in the dendrogram (Fig. 3) can be explained by its highly recombinant nature, which is thought to aid in its ability to infect and cause disease in a variety of host species, including poultry, swine, and humans. B. suanatina, reported to cause mucohemorrhagic diarrhea in swine and mallards (3), was most closely related to B. intermedia, a commensal spirochete in swine that causes disease in poultry. These two pathogens clustered close to B. hyodysenteriae, the primary causative agent of swine dysentery globally. Interestingly, B. hampsonii was found to be most closely related to the commensal species B. murdochii and B. innocens. The close relationship of B. hampsonii to these commensal species has been reported in earlier studies (1, 10) that have used other genes for comparison, including the nox and 16S rRNA genes. The swine colon may provide an ideal environment and opportunity for genomic exchange and recombination between commensal and pathogenic Brachyspira through a prophage that facilitates horizontal gene transfer events (36, 37). Since a variety of different Brachyspira species have been isolated from migratory wild birds/waterfowl, their intestinal tracts likely serve as another ideal environment for exchange of genomic material between pathogenic and commensal species. Such genetic events may lead to the emergence of new and potentially pathogenic species, such as B. hampsonii. Further whole-genome sequencing studies comparing the genomes of various Brachyspira species will provide a better understanding of the evolution of B. hampsonii in relation to other species.
This study identified 20 genotypes of B. hampsonii from diverse epidemiological sources using a newly developed MLST scheme. B. hampsonii showed a heterogeneous population structure, with microevolution identified locally within swine production systems. These findings underscore the importance of strict biosecurity measures in preventing the spread of pathogens, such as B. hampsonii, between swine herds and also potentially between migratory birds and swine. It also highlights the utility of MLST as a tool for routine surveillance and monitoring of B. hampsonii within commercial swine systems and possibly in migratory bird populations. Further, this study identified four genetic groups of B. hampsonii, two of which represented previously identified clades (1). The investigation of multiple Brachyspira species confirmed the close genetic relatedness of B. hampsonii with the commensal species B. murdochii and B. innocens and provided support for the two clades of B. hampsonii to be considered a single species. Based on the findings that B. hampsonii consists of genetically diverse groups that form a single cluster distinct from clusters of other Brachyspira species, we suggest that the term “genetic group” supplant the term “clade” for B. hampsonii. Future studies will help expand our understanding of the genetic diversity and host range of B. hampsonii, the origin of its diverse genetic groups, how it emerged in North American swine herds to cause severe mucohemorrhagic diarrhea, and its evolutionary relationships with other Brachyspira species.
ACKNOWLEDGMENTS
This work was supported by the Minnesota Agricultural Experiment Station Signature Program project MIN-63-111 and Emerging and Zoonotic Disease Signature Program Graduate Fellowship (to N.S.M.) and the University of Minnesota Discovery Research and Innovation Economy (MnDRIVE) Global Food Venture Fellowship (to N.S.M.).
We thank the University of Minnesota Veterinary Diagnostic Laboratory for providing us with B. hampsonii field isolates originating from the United States. We are grateful to Judith Rohde (University of Veterinary Medicine, Hannover, Germany), Maxime Mahu (Ghent University, Belgium), Pedro Rubio (University of Leon, Spain), David Hampson (Murdoch University, Australia), and Janet Hill (University of Saskatchewan, Canada) for providing us with DNA extracts of international B. hampsonii isolates included in this study. We thank Jesus Osorio for providing us with sequence files of Spanish isolates for inclusion in the Brachyspira genus comparison. We thank Tom La for uploading the isolates and maintaining the “B. hampsonii” PubMLST database and all our colleagues for their contributions to the database that made the genus-wide Brachyspira comparison possible.
REFERENCES
- 1.Chander Y, Primus A, Oliveira S, Gebhart CJ. 2012. Phenotypic and molecular characterization of a novel strongly hemolytic Brachyspira species, provisionally designated “Brachyspira hampsonii.” J Vet Diagn Invest 24:903–910. [DOI] [PubMed] [Google Scholar]
- 2.Hampson DJ. 2012. Brachyspiral colitis. Diseases of swine, p 680–696. Wiley-Blackwell, Chichster, United Kingdom. [Google Scholar]
- 3.Rasback T, Jansson DS, Johansson KE, Fellstrom C. 2007. A novel enteropathogenic, strongly haemolytic spirochaete isolated from pig and mallard, provisionally designated ‘Brachyspira suanatina’ sp. nov. Environ Microbiol 9:983–991. doi: 10.1111/j.1462-2920.2006.01220.x. [DOI] [PubMed] [Google Scholar]
- 4.Rohde J, Habighorst-Blome K. 2012. An up-date on the differentiation of Brachyspira species from pigs with nox-PCR-based restriction fragment length polymorphism. Vet Microbiol 158:211–215. doi: 10.1016/j.vetmic.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Mirajkar NS, Gebhart CJ. 2014. Understanding the molecular epidemiology and global relationships of Brachyspira hyodysenteriae from swine herds in the United States: a multi-locus sequence typing approach. PLoS One 9:e107176. doi: 10.1371/journal.pone.0107176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neef NA, Lysons RJ, Trott DJ, Hampson DJ, Jones PW, Morgan JH. 1994. Pathogenicity of porcine intestinal spirochetes in gnotobiotic pigs. Infect Immun 62:2395–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrough ER, Strait EL, Kinyon JM, Bower LP, Madson DM, Wilberts BL, Schwartz KJ, Frana TS, Songer JG. 2012. Comparative virulence of clinical Brachyspira spp. isolates in inoculated pigs. J Vet Diagn Invest 24:1025–1034. doi: 10.1177/1040638712457927. [DOI] [PubMed] [Google Scholar]
- 8.Costa MO, Hill JE, Fernando C, Lemieux HD, Detmer SE, Rubin JE, Harding JC. 2014. Confirmation that “Brachyspira hampsonii” clade I (Canadian strain 30599) causes mucohemorrhagic diarrhea and colitis in experimentally infected pigs. BMC Vet Res 10:129. doi: 10.1186/1746-6148-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubin JE, Costa MO, Hill JE, Kittrell HE, Fernando C, Huang Y, O'Connor B, Harding JC. 2013. Reproduction of mucohaemorrhagic diarrhea and colitis indistinguishable from swine dysentery following experimental inoculation with “Brachyspira hampsonii” strain 30446. PLoS One 8:e57146. doi: 10.1371/journal.pone.0057146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Lobo FJ, Hidalgo A, Garcia M, Arguello H, Naharro G, Carvajal A, Rubio P. 2013. First identification of “Brachyspira hampsonii” in wild European waterfowl. PLoS One 8:e82626. doi: 10.1371/journal.pone.0082626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin JE, Harms NJ, Fernando C, Soos C, Detmer SE, Harding JC, Hill JE. 2013. Isolation and characterization of Brachyspira spp. including “Brachyspira hampsonii” from lesser snow geese (Chen caerulescens caerulescens) in the Canadian Arctic. Microb Ecol 66:813–822. doi: 10.1007/s00248-013-0273-5. [DOI] [PubMed] [Google Scholar]
- 12.Mahu M, de Jong E, De Pauw N, Vande Maele L, Vandenbroucke V, Vandersmissen T, Miry C, Pasmans F, Haesebrouck F, Martel A, Boyen F. 2014. First isolation of “Brachyspira hampsonii” from pigs in Europe. Vet Rec 174:47. doi: 10.1136/vr.101868. [DOI] [PubMed] [Google Scholar]
- 13.Rohde J, Habighorst-Blome K, Seehusen F. 2014. “Brachyspira hampsonii” clade I isolated from Belgian pigs imported to Germany. Vet Microbiol 168:432–435. doi: 10.1016/j.vetmic.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Atyeo RF, Oxberry SL, Hampson DJ. 1999. Analysis of Serpulina hyodysenteriae strain variation and its molecular epidemiology using pulsed-field gel electrophoresis. Epidemiol Infect 123:133–138. doi: 10.1017/S0950268899002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Combs BG, Hampson DJ, Harders SJ. 1992. Typing of Australian isolates of Treponema hyodysenteriae by serology and by DNA restriction endonuclease analysis. Vet Microbiol 31:273–285. doi: 10.1016/0378-1135(92)90085-8. [DOI] [PubMed] [Google Scholar]
- 16.Dugourd D, Jacques M, Bigras-Poulin M, Harel J. 1996. Characterization of Serpulina hyodysenteriae isolates of serotypes 8 and 9 by random amplification of polymorphic DNA analysis. Vet Microbiol 48:305–314. doi: 10.1016/0378-1135(95)00149-2. [DOI] [PubMed] [Google Scholar]
- 17.Fisher LN, Mathiesen MR, Duhamel GE. 1997. Restriction fragment length polymorphism of the periplasmic flagellar flaA1 gene of Serpulina species. Clin Diagn Lab Immunol 4:681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JI, Hampson DJ, Combs BG, Lymbery AJ. 1993. Genetic relationships between isolates of Serpulina (Treponema) hyodysenteriae, and comparison of methods for their subspecific differentiation. Vet Microbiol 34:35–46. doi: 10.1016/0378-1135(93)90005-R. [DOI] [PubMed] [Google Scholar]
- 19.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasback T, Johansson KE, Jansson DS, Fellstrom C, Alikhani MY, La T, Dunn DS, Hampson DJ. 2007. Development of a multilocus sequence typing scheme for intestinal spirochaetes within the genus Brachyspira. Microbiology 153:4074–4087. doi: 10.1099/mic.0.2007/008540-0. [DOI] [PubMed] [Google Scholar]
- 21.La T, Phillips ND, Harland BL, Wanchanthuek P, Bellgard MI, Hampson DJ. 2009. Multilocus sequence typing as a tool for studying the molecular epidemiology and population structure of Brachyspira hyodysenteriae. Vet Microbiol 138:330–338. doi: 10.1016/j.vetmic.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 22.Osorio J, Carvajal A, Naharro G, La T, Phillips ND, Rubio P, Hampson DJ. 2012. Dissemination of clonal groups of Brachyspira hyodysenteriae amongst pig farms in Spain, and their relationships to isolates from other countries. PLoS One 7:e39082. doi: 10.1371/journal.pone.0039082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neo E, La T, Phillips ND, Alikani MY, Hampson DJ. 2013. The pathogenic intestinal spirochaete Brachyspira pilosicoli forms a diverse recombinant species demonstrating some local clustering of related strains and potential for zoonotic spread. Gut Pathog 5:24. doi: 10.1186/1757-4749-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips ND, La T, Amin MM, Hampson DJ. 2010. Brachyspira intermedia strain diversity and relationships to the other indole-positive Brachyspira species. Vet Microbiol 143:246–254. doi: 10.1016/j.vetmic.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 25.Osorio J, Carvajal A, Naharro G, Rubio P, La T, Phillips ND, Hampson DJ. 2013. Identification of weakly haemolytic Brachyspira isolates recovered from pigs with diarrhoea in Spain and Portugal and comparison with results from other countries. Res Vet Sci 95:861–869. doi: 10.1016/j.rvsc.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith JM, Smith NH, O'Rourke M, Spratt BG. 1993. How clonal are bacteria? Proc Natl Acad Sci U S A 90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gojobori T, Ishii K, Nei M. 1982. Estimation of average number of nucleotide substitutions when the rate of substitution varies with nucleotide. J Mol Evol 18:414–423. doi: 10.1007/BF01840889. [DOI] [PubMed] [Google Scholar]
- 29.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.RCore Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- 31.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morschhauser J, Kohler G, Ziebuhr W, Blum-Oehler G, Dobrindt U, Hacker J. 2000. Evolution of microbial pathogens. Philos Trans R Soc Lond B Biol Sci 355:695–704. doi: 10.1098/rstb.2000.0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jansson DS, Persson M, Zimmerman U, Johansson KE. 2011. Phenotypic and genetic diversity among intestinal spirochaetes (genus Brachyspira) in free-living wild mallards (Anas platyrhynchos) sampled in southern Sweden. Syst Appl Microbiol 34:566–575. doi: 10.1016/j.syapm.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Lillehaug A, Monceyron Jonassen C, Bergsjo B, Hofshagen M, Tharaldsen J, Nesse LL, Handeland K. 2005. Screening of feral pigeon (Colomba livia), mallard (Anas platyrhynchos) and graylag goose (Anser anser) populations for Campylobacter spp., Salmonella spp, avian influenza virus and avian paramyxovirus. Acta Vet Scand 46:193–202. doi: 10.1186/1751-0147-46-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanage WP, Fraser C, Spratt BG. 2006. Sequences, sequence clusters and bacterial species. Philos Trans R Soc Lond B Biol Sci 361:1917–1927. doi: 10.1098/rstb.2006.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanton TB, Thompson MG, Humphrey SB, Zuerner RL. 2003. Detection of bacteriophage VSH-1 svp38 gene in Brachyspira spirochetes. FEMS Microbiol Lett 224:225–229. doi: 10.1016/S0378-1097(03)00438-5. [DOI] [PubMed] [Google Scholar]
- 37.Motro Y, La T, Bellgard MI, Dunn DS, Phillips ND, Hampson DJ. 2009. Identification of genes associated with prophage-like gene transfer agents in the pathogenic intestinal spirochaetes Brachyspira hyodysenteriae, Brachyspira pilosicoli and Brachyspira intermedia. Vet Microbiol 134:340–345. doi: 10.1016/j.vetmic.2008.09.051. [DOI] [PubMed] [Google Scholar]