Abstract
The impact of the HCV genotype 1b core amino acid (aa) 70 mutant on the cumulative rate of hepatocellular carcinoma following eradication of HCV RNA by antiviral therapy was investigated with the Q-Invader assay. Multivariate analysis based on 649 patients indicated that a core aa70 Q-Invader mutant level ≥20% is a predictor of hepatocellular carcinoma.
TEXT
Previous reports indicated that amino acid (aa) substitutions at position 70 in the hepatitis C virus (HCV) core region in patients infected with HCV genotype 1b (HCV-1b) are pretreatment predictors of poor virological response to antiviral therapy (1, 2) and also affect hepatocarcinogenesis, regardless of treatment response (3–7). In particular, hepatocellular carcinoma (HCC) still occurs even after eradication of HCV RNA by antiviral therapy (8–12), and substitutions of aa70 at the start of antiviral therapy also affect hepatocarcinogenesis following eradication of HCV RNA (13).
Recently, Tadokoro et al. (14) reported the results of a comparative quantitative analysis of aa70 substitution in the HCV-1b core region by the real-time PCR monitoring Invader reaction (Q-Invader assay; BML, Inc., Saitama, Japan). This highly sensitive assay (based on the Q-Invader technology using mutant-specific primers) was developed to determine the relative ratio of the aa70 mutant (glutamine/histidine) to the aa70 wild-type (arginine) virus. The assay in that study (14) detected a minor type plasmid that constituted only 1% of a mixture of plasmids containing wild-type and mutant sequences. In this study, we evaluated the associations between core aa70 Q-Invader mutant levels at the start of antiviral therapy and HCC following eradication of HCV RNA.
Among 5,023 consecutive patients infected with HCV in whom antiviral therapy (interferon monotherapy, interferon plus ribavirin combination therapy, or triple therapy of NS3/4A protease inhibitor plus peginterferon plus ribavirin) was initiated between February 1987 and June 2013 at Toranomon Hospital, 2,121 patients infected with a single genotype of HCV-1b, -2a, or -2b were selected for this retrospective study. All patients achieved sustained virological response, defined as negative HCV RNA at 24 weeks after cessation of antiviral therapy, based on HCV RNA qualitative analysis (Amplicor; Roche Diagnostics, Manheim, Germany) or by the Cobas TaqMan HCV test (Roche Diagnostics, Tokyo, Japan), and they were free of HCC before and during interferon therapy. The cumulative rate of HCC was calculated using the Kaplan-Meier technique; differences between the HCC rate curves were tested using the log-rank test. The period between the end of antiviral therapy and the diagnosis of HCC was used for group-based statistical analysis of HCC. Stepwise Cox regression analysis (multivariate analysis) was used to determine independent factors associated with HCC. This study protocol was in compliance with the Good Clinical Practice guidelines and the 1975 Declaration of Helsinki, and it was approved by the institutional review board at Toranomon Hospital.
During the follow-up of 2,121 patients, 43 patients (2.0%) developed HCC. The median interval between the end of antiviral therapy and detection of HCC was 2.4 years (range, 0.0 to 15.9 years). The cumulative HCC rates were 3.1%, 4.9%, 7.3%, and 11.1% at the end of 5, 10, 15, and 20 years, respectively, from the end of antiviral therapy.
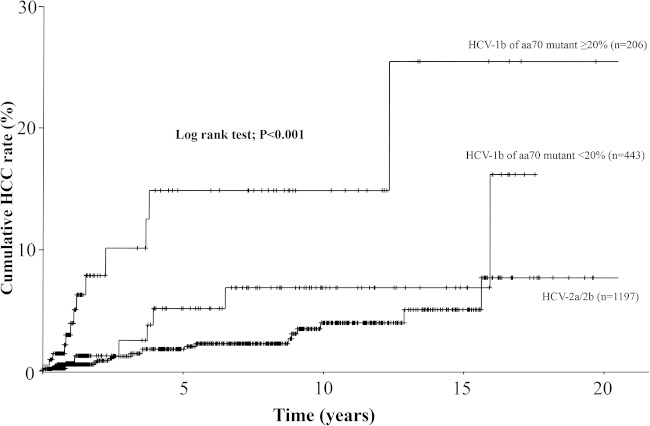
The levels of core aa70 mutant could be quantitated in 649 patients with HCV-1b. Figure 1 shows the cumulative HCC rates for three HCV subgroups, including two HCV genotypes, and the level of HCV-1b core aa70 Q-Invader mutant. The HCC rates were significantly different among the three HCV subgroups (P < 0.001). In particular, the HCC rate for HCV-1b with levels of aa70 mutant ≥20% was significantly higher than for HCV-1b with levels of aa70 mutant <20% (P = 0.005) and HCV-2a/2b (P < 0.001), but the rate for HCV-1b with levels of aa70 mutant <20% was not significantly higher than that for HCV-2a/2b (P = 0.178).
FIG 1.
Cumulative HCC rate for patients with two different levels of HCV-1b core aa70 Q-Invader mutant (≥20% and <20%) and those infected with HCV-2a/2b genotype. The HCC rates were significantly different among the three HCV subgroups (log-rank test; P < 0.001).
The data of 649 patients with HCV-1b in whom the level of core aa70 mutant was quantitated by Q-Invader assay were analyzed to determine those factors that could predict HCC. Univariate analysis showed a significant correlation between eight parameters and HCC at the start of antiviral therapy; these parameters were age (≥51 years; P < 0.001), gender (male; P = 0.002), aspartate aminotransferase (AST) (≥50 IU/liter; P = 0.014), platelet count (<15.0 × 104/mm3; P = 0.019), AST-to-platelet ratio index (APRI) (≥1.5; P = 0.003), high-density lipoprotein cholesterol (<50 mg/dl; P = 0.012), triglycerides (≥151 mg/dl; P = 0.021), and level of core aa70 Q-Invader mutant (≥20%; P = 0.005). All eight factors were entered into multivariate analysis, which identified three parameters that significantly and independently influenced the HCC rate; age (≥51 years; P = 0.007), AST (≥50 IU/liter; P = 0.022), and level of core aa70 Q-Invader mutant (≥20%; P = 0.026) (Table 1).
TABLE 1.
Factors associated with hepatocarcinogenesis in 649 patients infected with HCV genotype 1b of sustained virological response, identified by multivariate analysis
| Factor and category | Hazard ratio | 95% confidence interval | P |
|---|---|---|---|
| Age (yr) | |||
| <51 | 1 | ||
| ≥51 | 19.5 | 2.28–167.3 | 0.007 |
| Serum aspartate aminotransferase (IU/liter) | |||
| <50 | 1 | ||
| ≥50 | 13.5 | 1.45–125.5 | 0.022 |
| Level of core aa70 Q-Invader mutant (%) | |||
| <20.0 | 1 | ||
| ≥20.0 | 3.62 | 1.16–11.2 | 0.026 |
This study is the first report to indicate the associations between the level of core aa70 mutant (≥20%) at the start of antiviral therapy and the HCC rate following eradication of HCV RNA, suggesting the oncogenic potential of the mutant irrespective of persistence of HCV RNA. Kobayashi et al. (15) indicated that the core aa70 mutation was associated with severe liver disease accompanied by high AST and γ-glutamyl transpeptidase levels as well as by the development of HCC. Furthermore, Miura et al. (16) recently showed that the core aa70 mixture ratio, determined by deep sequencing, reflected the status of liver disease and demonstrated a significant association between core aa70 and disease progression in patients infected with HCV-1b. These findings suggest that high levels of the core aa70 mutant could be a useful marker for the high oncogenic potential of HCV-1b in patients with chronic liver disease. The present study showed that platelet count and APRI (i.e., markers of fibrosis stage) did not have significant associations with the core aa70 mutant level, though the fibrosis stage could not be evaluated by liver biopsy samples (data not shown).
The number of HCV-1 patients who achieve sustained virological response by interferon-free regimens, such as direct-acting antiviral agents, is expected to increase in the future (17–21). However, the reported HCC annual rate after eradication of HCV RNA by antiviral therapy is ≤1% (9, 12, 13). Based on the results of the present study, we recommend measurement of the core aa70 mutant level of HCV-1b before the start of antiviral therapy for eradication of HCV RNA. In particular, even after achievement of sustained virological response by patients with high levels of core aa70 mutant (≥20%), blood tests and imaging studies should be conducted at regular intervals for early detection of HCC. Further studies are needed to investigate the effect of any interaction between viral factor (core aa70 mutant) and various host factors (e.g., status of underlying liver disease) on the oncogenic potential of such a mutant on the development of HCC following eradication of HCV RNA.
ACKNOWLEDGMENTS
We thank Kenichi Tadokoro, BML, Inc., for comparative quantitative analysis with the Q-Invader assay.
This study was supported in part by a grant-in-aid from the Ministry of Health, Labor, and Welfare, Japan. Hiromitsu Kumada received honoraria from MSD K.K., Bristol-Myers Squibb, Janssen Pharmaceutical K.K., and GlaxoSmithKline K.K. and holds rights for royalty from SRL, Inc. Fumitaka Suzuki received an honorarium from Bristol-Myers Squibb. Y.S. received an honorarium from Bristol-Myers Squibb. Yasuji Arase received an honorarium from MSD K.K. Kenji Ikeda received honoraria from Dainippon Sumitomo Pharma, and Eisai Co., Ltd. All other authors declare no conflict of interest.
Norio Akuta, Fumitaka Suzuki, Masahiro Kobayashi, Hitomi Sezaki, Yusuke Kawamura, Tetsuya Hosaka, Mariko Kobayashi, Satoshi Saitoh, Yoshiyuki Suzuki, Yasuji Arase, Kenji Ikeda, and Hiromitsu Kumada contributed to this work. Norio Akuta and Masahiro Kabayashi analyzed the data. Norio Akuta wrote the paper. Norio Akuta, Fumitaka Suzuki, Masahiro Kobayashi, Hitomi Sezaki, Yusuke Kawamura, Tetsuya Hosaka, Mariko Kobayashi, Satoshi Saitoh, Yoshiyuki Suzuki, Yasuji Arase, Kenji Ikeda, and Hiromitsu Kumada provided the samples.
REFERENCES
- 1.Akuta N, Suzuki F, Sezaki H, Suzuki Y, Hosaka T, Someya T, Kobayashi M, Saitoh S, Watahiki S, Sato J, Matsuda M, Kobayashi M, Arase Y, Ikeda K, Kumada H. 2005. Association of amino acid substitution pattern in core protein of hepatitis C virus genotype 1b high viral load and nonvirological response to interferon-ribavirin combination therapy. Intervirology 48:372–380. doi: 10.1159/000086064. [DOI] [PubMed] [Google Scholar]
- 2.Donlin MJ, Cannon NA, Yao E, Li J, Wahed A, Taylor MW, Belle SH, Di Bisceglie AM, Aurora R, Tavis JE. 2007. Pretreatment sequence diversity differences in the full-length hepatitis C virus open reading frame correlate with early response to therapy. J Virol 81:8211–8224. doi: 10.1128/JVI.00487-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Arase Y, Ikeda K, Kumada H. 2007. Amino acid substitutions in the hepatitis C virus core region are the important predictor of hepatocarcinogenesis. Hepatology 46:1357–1364. doi: 10.1002/hep.21836. [DOI] [PubMed] [Google Scholar]
- 4.Fishman SL, Factor SH, Balestrieri C, Fan X, Dibisceglie AM, Desai SM, Benson G, Branch AD. 2009. Mutations in the hepatitis C virus core gene are associated with advanced liver disease and hepatocellular carcinoma. Clin Cancer Res 15:3205–3213. doi: 10.1158/1078-0432.CCR-08-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Z, Muroyama R, Kowatari N, Chang J, Omata M, Kato N. 2009. Characteristic mutations in hepatitis C virus core gene related to the occurrence of hepatocellular carcinoma. Cancer Sci 100:2465–2468. doi: 10.1111/j.1349-7006.2009.01338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamoto S, Imazeki F, Fukai K, Fujiwara K, Arai M, Kanda T, Yonemitsu Y, Yokosuka O. 2010. Association between mutations in the core region of hepatitis C virus genotype 1 and hepatocellular carcinoma development. J Hepatol 52:72–78. doi: 10.1016/j.jhep.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Akuta N, Suzuki F, Seko Y, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Hara T, Kobayashi M, Saitoh S, Arase Y, Ikeda K, Kumada H. 2012. Complicated relationships of amino acid substitution in hepatitis C virus core region and IL28B genotype influencing hepatocarcinogenesis. Hepatology 56:2134–2141. doi: 10.1002/hep.25949. [DOI] [PubMed] [Google Scholar]
- 8.Ikeda K, Kobayashi M, Saitoh S, Someya T, Hosaka T, Akuta N, Suzuki F, Tsubota A, Suzuki Y, Arase Y, Kumada H. 2003. Recurrence rate and prognosis of patients with hepatocellular carcinoma that developed after elimination of hepatitis C virus RNA by interferon therapy. A closed cohort study including matched control patients. Oncology 65:204–210. doi: 10.1159/000074472. [DOI] [PubMed] [Google Scholar]
- 9.Tokita H, Fukui H, Tanaka A, Kamitsukasa H, Yagura M, Harada H, Okamoto H. 2005. Risk factors for the development of hepatocellular carcinoma among patients with chronic hepatitis C who achieved a sustained virological response to interferon therapy. J Gastroenterol Hepatol 20:752–758. doi: 10.1111/j.1440-1746.2005.03800.x. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda M, Fujiyama S, Tanaka M, Sata M, Ide T, Yatsuhashi H, Watanabe H. 2005. Risk factors for development of hepatocellular carcinoma in patients with chronic hepatitis C after sustained response to interferon. J Gastroenterol 40:148–156. doi: 10.1007/s00535-004-1519-2. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi S, Takeda T, Enomoto M, Tamori A, Kawada N, Habu D, Sakaguchi H, Kuroda T, Kioka K, Kim SR, Kanno T, Ueda T, Hirano M, Fujimoto S, Jomura H, Nishiguchi S, Seki S. 2007. Development of hepatocellular carcinoma in patients with chronic hepatitis C who had a sustained virological response to interferon therapy: a multicenter, retrospective cohort study of 1,124 patients. Liver Int 27:186–191. doi: 10.1111/j.1478-3231.2006.01406.x. [DOI] [PubMed] [Google Scholar]
- 12.Hirakawa M, Ikeda K, Arase Y, Kawamura Y, Yatsuji H, Hosaka T, Sezaki H, Akuta N, Kobayashi M, Saitoh S, Suzuki F, Suzuki Y, Kumada H. 2008. Hepatocarcinogenesis following HCV RNA eradication by interferon in chronic hepatitis patients. Intern Med 47:1637–1643. doi: 10.2169/internalmedicine.47.1087. [DOI] [PubMed] [Google Scholar]
- 13.Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Saitoh S, Arase Y, Ikeda K, Kumada H. 2011. Amino acid substitutions in hepatitis C virus core region predict hepatocarcinogenesis following eradication of HCV RNA by antiviral therapy. J Med Virol 83:1016–1022. doi: 10.1002/jmv.22094. [DOI] [PubMed] [Google Scholar]
- 14.Tadokoro K, Kobayashi M, Suzuki F, Tanaka C, Yamaguchi T, Nagano M, Egashira T, Kumada H. 2013. Comparative quantitative analysis of hepatitis C mutations at amino acids 70 and 91 in the core region by the Q-Invader assay. J Virol Methods 189:221–227. doi: 10.1016/j.jviromet.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi M, Akuta N, Suzuki F, Hosaka T, Sezaki H, Kobayashi M, Suzuki Y, Arase Y, Ikeda K, Watahiki S, Mineta R, Iwasaki S, Miyakawa Y, Kumada H. 2010. Influence of amino-acid polymorphism in the core protein on progression of liver disease in patients infected with hepatitis C virus genotype 1b. J Med Virol 82:41–48. doi: 10.1002/jmv.21629. [DOI] [PubMed] [Google Scholar]
- 16.Miura M, Maekawa S, Takano S, Komatsu N, Tatsumi A, Asakawa Y, Shindo K, Amemiya F, Nakayama Y, Inoue T, Sakamoto M, Yamashita A, Moriishi K, Enomoto N. 2013. Deep-sequencing analysis of the association between the quasispecies nature of the hepatitis C virus core region and disease progression. J Virol 87:12541–12551. doi: 10.1128/JVI.00826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chayama K, Takahashi S, Toyota J, Karino Y, Ikeda K, Ishikawa H, Watanabe H, McPhee F, Hughes E, Kumada H. 2012. Dual therapy with the nonstructural protein 5A inhibitor, daclatasvir, and the nonstructural protein 3 protease inhibitor, asunaprevir, in hepatitis C virus genotype 1b-infected null responders. Hepatology 55:742–748. doi: 10.1002/hep.24724. [DOI] [PubMed] [Google Scholar]
- 18.Kumada H, Suzuki Y, Ikeda K, Toyota J, Karino Y, Chayama K, Kawakami Y, Ido A, Yamamoto K, Takaguchi K, Izumi N, Koike K, Takehara T, Kawada N, Sata M, Miyagoshi H, Eley T, McPhee F, Damokosh A, Ishikawa H, Hughes E. 2014. Daclatasvir plus asunaprevir for chronic HCV genotype 1b infection. Hepatology 59:2083–2091. doi: 10.1002/hep.27113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afdhal N, Zeuzem S, Kwo P, Chojkier M, Gitlin N, Puoti M, Romero-Gomez M, Zarski JP, Agarwal K, Buggisch P, Foster GR, Bräu N, Buti M, Jacobson IM, Subramanian GM, Ding X, Mo H, Yang JC, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Mangia A, Marcellin P; ION-1 Investigators. 2014. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N Engl J Med 370:1889–1898. doi: 10.1056/NEJMoa1402454. [DOI] [PubMed] [Google Scholar]
- 20.Afdhal N, Reddy KR, Nelson DR, Lawitz E, Gordon SC, Schiff E, Nahass R, Ghalib R, Gitlin N, Herring R, Lalezari J, Younes ZH, Pockros PJ, Di Bisceglie AM, Arora S, Subramanian GM, Zhu Y, Dvory-Sobol H, Yang JC, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Sulkowski M, Kwo P, ION-2 Investigators . 2014. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. N Engl J Med 370:1483–1493. doi: 10.1056/NEJMoa1316366. [DOI] [PubMed] [Google Scholar]
- 21.Kowdley KV, Gordon SC, Reddy KR, Rossaro L, Bernstein DE, Lawitz E, Shiffman ML, Schiff E, Ghalib R, Ryan M, Rustgi V, Chojkier M, Herring R, Di Bisceglie AM, Pockros PJ, Subramanian GM, An D, Svarovskaia E, Hyland RH, Pang PS, Symonds WT, McHutchison JG, Muir AJ, Pound D, Fried MW, ION-3 Investigators . 2014. Ledipasvir and sofosbuvir for 8 or 12 weeks for chronic HCV without cirrhosis. N Engl J Med 370:1879–1888. doi: 10.1056/NEJMoa1402355. [DOI] [PubMed] [Google Scholar]