Abstract
Mycoplasma pneumoniae is a leading cause of community-acquired pneumonia (CAP) across patient populations of all ages. We have developed a loop-mediated isothermal amplification (LAMP) assay that enables rapid, low-cost detection of M. pneumoniae from nucleic acid extracts and directly from various respiratory specimen types. The assay implements calcein to facilitate simple visual readout of positive results in approximately 1 h, making it ideal for use in primary care facilities and resource-poor settings. The analytical sensitivity of the assay was determined to be 100 fg by testing serial dilutions of target DNA ranging from 1 ng to 1 fg per reaction, and no cross-reactivity was observed against 17 other Mycoplasma species, 27 common respiratory agents, or human DNA. We demonstrated the utility of this assay by testing nucleic acid extracts (n = 252) and unextracted respiratory specimens (n = 72) collected during M. pneumoniae outbreaks and sporadic cases occurring in the United States from February 2010 to January 2014. The sensitivity of the LAMP assay was 88.5% tested on extracted nucleic acid and 82.1% evaluated on unextracted clinical specimens compared to a validated real-time PCR test. Further optimization and improvements to this method may lead to the availability of a rapid, cost-efficient laboratory test for M. pneumoniae detection that is more widely available to primary care facilities, ultimately facilitating prompt detection and appropriate responses to potential M. pneumoniae outbreaks and clusters within the community.
INTRODUCTION
Mycoplasma pneumoniae is a human pathogen that accounts for an estimated 15% to 20% of cases of community-acquired pneumonia (CAP) (1, 2). An atypical bacterial pneumonia, M. pneumoniae infection can result in both pulmonary and extrapulmonary manifestations, including pharyngitis, encephalitis, Stevens-Johnson syndrome, septic arthritis, and pericarditis, among others (3). Ongoing endemic M. pneumoniae infection is overlaid with cyclical epidemics of disease, typically occurring every 3 to 5 years (1). Although symptoms of M. pneumoniae infection are generally mild or subclinical, the organism accounts for up to 33% of hospitalizations among children and adults with bacterial pneumonia (4). In the absence of a diagnostic result, empirical therapy can be misdirected, resulting in inappropriate use of antibiotics and continued spread of M. pneumoniae in the community. This may promote the evolution of antibiotic resistance and leads to negative economic impact due to health care costs and missed school and work days.
Traditional diagnostics for M. pneumoniae infections exist but are underused due to their cost, time-intensive nature, or requirement for specialized expertise and equipment. M. pneumoniae is fastidious and slow-growing, with isolation from liquid culture requiring up to 8 weeks and often resulting in low yield (5, 6). Appropriate interpretation of serological data requires paired acute-phase and convalescent-phase serum samples, constraining their use to retrospective investigations. Thus, these methods are better suited for epidemiological studies than for treatment decision-making. While rapid diagnostic serology kits based on IgM response offer results within 2 days, sensitivity and specificity are poor, intertest variability is high, and IgM responses can vary significantly by patient age and sample collection time (7–9).
More-advanced diagnostics include nucleic acid amplification techniques, such as real-time PCR, that specifically amplify M. pneumoniae DNA from specimen nucleic acid extracts, enabling fast, sensitive detection of pathogen DNA (10–12). With the exception of the FDA-cleared FilmArray respiratory panel (Biofire Diagnostics, Salt Lake City, UT), existing M. pneumoniae real-time PCR assays are not standardized and may differ in levels of sensitivity (13). These methods also require an expensive thermal cycler and fluorescent oligonucleotide probe and are therefore not routinely performed, especially in point-of-care settings. For improved clinical decision-making, there is a need for a low-cost, simple, and sensitive M. pneumoniae assay that can be easily implemented in this type of setting.
Loop-mediated isothermal amplification (LAMP) relies on four to six primers and a strand-displacing DNA polymerase to drive autocycling amplification of target DNA at a constant temperature, eliminating the need for instrumentation beyond a water bath or heat block (14). Rapid amplification initiates from stem-loops introduced upstream and downstream of a target sequence, producing massive amounts of concatenated amplicons in under an hour. The pyrophosphate ion byproduct of this reaction combines with Mg2+ ions in the amplification buffer, yielding a magnesium pyrophosphate precipitate that can be observed visually in the reaction tube. Turbidity and metal ion indicators that exploit this change in soluble ion concentration over the course of the reaction can be used as a simple visual readout, further decreasing the time to results (15–17). LAMP has been widely implemented as a rapid diagnostic method for various pathogens and is amenable to use in point-of-care settings (18–20).
Although LAMP assays for M. pneumoniae detection have been developed, including the FDA-cleared illumigene Mycoplasma kit (Meridian Bioscience, Inc., Cincinnati, OH), they are not intended for point-of-care use, require an expensive real-time turbidimeter for readout, and display variable levels of performance with respect to specificity and sensitivity (21, 22). Further, these assays all require an extraction step using a prescribed kit or automated system. In contrast, we have developed, optimized, and evaluated a novel LAMP assay that detects M. pneumoniae directly from clinical specimens and their nucleic acid extracts and have assessed the sensitivity and specificity of this assay in comparison to a validated real-time PCR assay.
MATERIALS AND METHODS
Bacterial strains and clinical specimens.
To assess the sensitivity and specificity of the LAMP assay, genomic DNA was prepared from 44 reference strains representing 17 members of the family Mycoplasmataceae and 27 common respiratory agents (Table 1). A variety of respiratory clinical specimens (nasopharyngeal [NP]/oropharyngeal [OP] swabs, bronchoalveolar lavage fluid samples, and nasal washes) collected during domestic outbreaks and sporadic case investigations from February 2010 to January 2014 were transported to the CDC in universal transport medium (UTM; Copan Diagnostics, Murrieta, CA) and stored at a temperature of −70°C or lower.
TABLE 1.
Reference strains tested to evaluate sensitivity and specificity of M. pneumoniae LAMP
| Strain and category |
| Mycoplasmataceae family members |
| Mycoplasma amphoriforme |
| Mycoplasma arginine |
| Mycoplasma buccale |
| Mycoplasma faucium |
| Mycoplasma fermentans |
| Mycoplasma genitalium |
| Mycoplasma hominis |
| Mycoplasma hyorhinis |
| Mycoplasma lipophilum |
| Mycoplasma orale |
| Mycoplasma penetrans |
| Mycoplasma pirum |
| Mycoplasma pneumoniae |
| Mycoplasma salivarium |
| Mycoplasma volis |
| Ureaplasma parvum |
| Ureaplasma urealyticum |
| Non-Mycoplasmataceae testing panel |
| Bordetella pertussis |
| Candida albicans |
| Chlamydia trachomatis |
| Chlamydophila pneumoniae |
| Chlamydophila psittaci |
| Corynebacterium diphtheriae |
| Escherichia coli |
| Haemophilus influenzae type b |
| Homo sapiens |
| Human parainfluenza virus type 1 |
| Human parainfluenza virus type 2 |
| Human parainfluenza virus type 3 |
| Human parainfluenza virus type 4 |
| Human rhinovirus |
| Klebsiella pneumoniae |
| Lactobacillus plantarum |
| Legionella longbeachae |
| Legionella pneumophila |
| Neisseria elongata |
| Neisseria meningitidis |
| Pseudomonas aeruginosa |
| Respiratory syncytial virus |
| Staphylococcus aureus |
| Staphylococcus epidermidis |
| Streptococcus agalactiae |
| Streptococcus pneumoniae |
| Streptococcus pyogenes |
| Streptococcus salivarius |
Specimen processing and nucleic acid isolation.
Total nucleic acid was extracted from a 400-μl specimen or reference strain culture with a MagNA Pure Compact instrument and Nucleic Acid Isolation kit I (Roche Applied Science, Indianapolis, IN) and eluted in 100 μl. For reference strains, extracted nucleic acid was quantitated on a Nanodrop instrument (Thermo Scientific, Wilmington, DE) and normalized to 2 ng/μl. A subset of specimens was selected for direct testing with LAMP (i.e., each specimen was added directly to the reaction without nucleic acid extraction). Immediately prior to LAMP, cells in these specimens were pelleted by centrifugation at 12,000 rpm for 10 min at 4°C on a microcentrifuge and resuspended in an equivalent volume of 1× isothermal amplification buffer [20 mM Tris-HCl, 10 mM (NH4)2SO4, 50 mM KCl, 2 mM MgSO4, 0.1% Tween 20, pH 8.8 at 25°C; New England BioLabs, Ipswich, MA]. Resuspended specimens were then heated at 95°C for 10 min prior to addition to the LAMP reaction mixture.
Primer design.
LAMP primers (Table 2) specific to the community-acquired respiratory distress syndrome (CARDS) toxin gene (GenBank accession no. DQ447750.1) of M. pneumoniae M129 were designed using the EIKEN Primer Explorer web tool (http://primerexplorer.jp). The forward loop primer (LF) was manually designed (23). Primers were synthesized by Integrated DNA Technologies (Coralville, IA), and the forward inner primer (FIP) and backward inner primer (BIP) were subjected to additional purification by high-performance liquid chromatography (HPLC).
TABLE 2.
Primers used for LAMP of the M. pneumoniae CARDS toxin gene
| Primer | Name | Sequence (5′ to 3′) |
|---|---|---|
| F3 | Forward outer primer | CCACCTAGTGATTTGGAAGA |
| B3 | Backward outer primer | GGACAAAGAAGATTTTCGAAGTT |
| FIP | Forward inner primer | GCTGAACATCAACAAAGAAGGTGCATTGTTGATGAATGTACTACCCA |
| BIP | Backward inner primer | ATACCCCACAATTAAGTGGTTGATTCATAGAATATCTGTCCATCTGG |
| LF | Forward loop primer | CTGCACGCATAGTAACAAACTG |
LAMP reaction.
LAMP reactions were performed in a 25-μl volume consisting of 5 μl template, 8 U Bst 2.0 DNA polymerase (New England BioLabs, Ipswich, MA), 20 mM Tris-HCl, 10 mM (NH4)2SO4, 50 mM KCl, 5.5 mM MgSO4, 0.1% Tween 20, 0.8 M betaine, 1.4 mM (each) deoxynucleoside triphosphates (dNTPs), and 25 μM calcein. Primers were used at the following concentrations: 0.2 μM (each) F3 and B3, 1.6 μM (each) FIP and BIP, and 0.8 μM LF. For LAMP reactions performed with an unextracted respiratory specimen as the template, all reaction components were added as described above, except using 10.0 μl of specimen to occupy the remaining available volume in the 25-μl reaction mixture, as would be the case in point-of-care use. Unless otherwise noted, for reaction optimization experiments, MnCl2 was added to reach a final concentration of 1.375 mM. Amplification was monitored in real time on the 6-carboxyfluorescein (FAM) channel of an ABI 7500 Fast thermal cycler, with 60 cycles of 62°C for 15 s and 63°C for 45 s to simulate isothermal conditions. Reactions were terminated by a subsequent enzyme inactivation at 80°C for 2 min. The excitation and emission wavelengths of calcein (495 nm and 515 nm, respectively) are very close to those of the FAM fluorophore detected by most real-time instruments (494 nm and 518 nm). Thus, the reaction readout was determined by the presence of an exponential amplification curve above the baseline in the FAM channel or by exposing the reaction tube to UV light of wavelength 365 nm and visually inspecting for green fluorescence.
Real-time PCR.
An established multiplex real-time PCR also targeting the CARDS toxin gene of M. pneumoniae was used as a benchmark comparison for LAMP (24). The assay was described in prior work that established the CARDS toxin gene as a sensitive and specific target for M. pneumoniae detection (10). The master mix contained 12.5 μl of PerfeCTa multiplex quantitative PCR (qPCR) SuperMix (Quanta Biosciences, Gaithersburg, MD) and previously published concentrations of primers targeting M. pneumoniae, Chlamydophila pneumoniae, Legionella pneumophila, and human RNase P, assembled with 5 μl template DNA in a final volume of 25 μl. Amplification was monitored in real time on an ABI 7500 Fast thermal cycler, with an initial denaturation at 95°C for 5 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min.
Analysis of LAMP products.
The LAMP amplicon was purified from positive reactions using a GeneClean Turbo DNA purification kit (MP Biomedicals, Santa Ana, CA) following the manufacturer's instructions. A 1-μg volume of purified DNA was digested with 10 U MboI (New England BioLabs, Ipswich, MA) in a 50-μl reaction mixture for 1 h at 37°C according to the protocol. A 1-μl volume of purified amplicon and a 1-μl purified digest volume were run on a Bioanalyzer (Agilent Technologies, Santa Clara, CA) to visualize the amplified products pre- and postrestriction digestion and to assess reaction specificity.
Evaluation of LAMP.
To evaluate sensitivity, serial 10-fold dilutions of M. pneumoniae M129 genomic DNA extract from 0.2 ng/μl to 0.2 fg/μl were prepared, assayed in eight replicates with LAMP, and compared to results obtained with real-time PCR. Analytical sensitivity, or the limit of detection (LOD), was defined as the lowest concentration at which ≥50% of replicates were positive by the assay. To test for potential cross-reactivity of the M. pneumoniae LAMP primers, total nucleic acid (TNA) extracted from a panel of other bacterial and viral respiratory pathogens (Table 1) was tested with LAMP in triplicate at a concentration of 2 ng/μl. Human genomic DNA (catalog no. G3041; Promega, Madison, WI) was also included in this evaluation at a concentration of 20 ng/μl.
RESULTS
Sensitivity and specificity of LAMP.
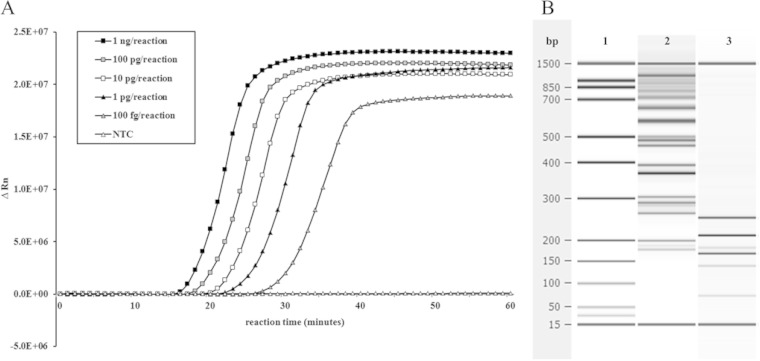
The analytical sensitivity of the LAMP assay targeting the CARDS toxin gene was determined by testing 10-fold serial dilutions of M. pneumoniae M129 genomic DNA. The LOD was determined to be 100 fg/reaction, or ∼110 genome copies/reaction (Fig. 1A), compared to the 50-fg limit of the real-time PCR assay. In some cases, LAMP was capable of detecting M. pneumoniae at concentrations as low as 10 fg/reaction, or ∼11 genome copies/reaction.
FIG 1.
Target amplification with the CARDS toxin LAMP assay. (A) Representative amplification of serial dilutions of M. pneumoniae nucleic acid using the LAMP assay targeting the CARDS toxin gene. LAMP reactions with calcein were monitored in real-time on the FAM channel of an ABI 7500 instrument. As the less-abundant Mn2+ precipitates from the reaction with a pyrophosphate ion, Mg2+ binds to calcein, and it fluoresces upon excitation with 495-nm-wavelength light. A no-template control reaction (NTC) served as the baseline. ΔRn is defined as the magnitude of the fluorescence signal generated during the PCR at each time point. (B) Restriction digest with MboI, which recognizes a site within the M. pneumoniae CARDS toxin amplicon, cut all products into the expected identical component parts, thereby confirming specificity. 1, Bioanalyzer ladder; 2, LAMP amplicon species, 3, LAMP amplicons digested with MboI.
No cross-reactivity was observed with M. pneumoniae LAMP primers tested on any of the other species of Mycoplasmataceae, respiratory near neighbors (Table 1), or human nucleic acid. Additionally, extensive (n = 300) no-template control (NTC) testing consisting of 5 μl water added to LAMP reactions in place of a template revealed no false-positive amplification in a 1-h reaction time.
The restriction endonuclease MboI recognizes a site inside the CARDS toxin amplicon sequence internal to the stem-loop introduced by the backward inner primer (BIP) during LAMP amplification. A LAMP amplicon digested with this enzyme was completely resolved into the expected identical component parts of its long, incremented concatemers (Fig. 1B), indicating that the amplified product was derived from the intended M. pneumoniae sequence and thereby confirming the specificity of the reaction.
Optimization of colorimetric readout with calcein.
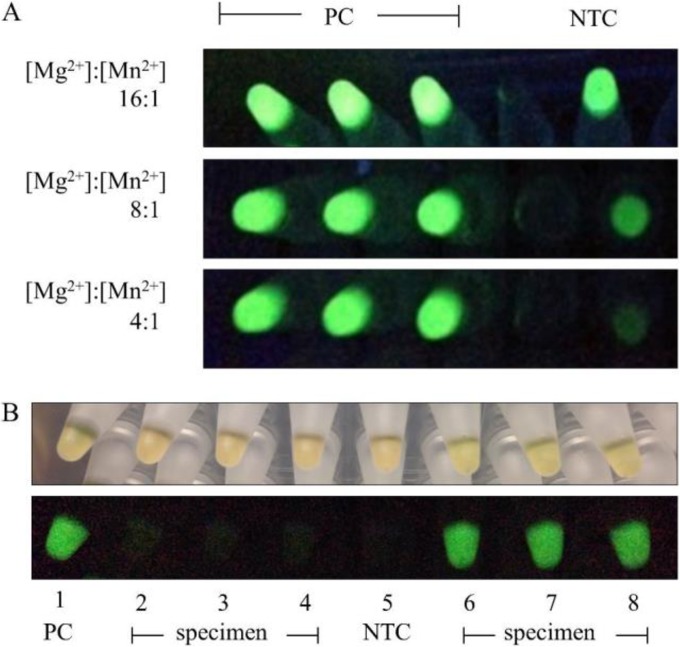
To determine the optimal ratio of Mg2+ to Mn2+ for calcein quenching in negative LAMP reactions and fluorescence in positive LAMP reactions, Mn2+ was added to the reaction mix in various concentrations relative to an Mg2+ concentration of 5.5 mM. The choice of this Mg2+ concentration was the result of independent optimization experiments to achieve an early amplification time in positive controls without false-positive amplification in negative controls (data not shown). Mixes were tested in triplicate with 1 ng of M. pneumoniae genomic DNA as the template. Negative reactions with 16:1 and 8:1 Mg2+/Mn2+ ion ratios had high background fluorescence under UV light compared to true positives, while a 4:1 ratio of Mg2+ to Mn2+ possessed sufficient quenching to eliminate background signal and produced a superior signal-to-noise ratio (Fig. 2).
FIG 2.
Endpoint fluorescent detection of M. pneumoniae LAMP amplification with calcein. (A) Using calcein with a 16:1 Mg2+/Mn2+ ratio as previously described (16) resulted in high background fluorescence in negative controls when reactions were excited with UV light (365-nm wavelength). Increased quenching with added Mn2+ decreased this background to provide a clear visual distinction between positive and negative reactions, optimal at a 4:1 ratio of Mg2+/Mn2+. PC, positive control; NTC, no-template control. (B) Examples of visual results for unextracted specimens tested with the M. pneumoniae LAMP assay in 96-well plate format (top row, ambient light; bottom row, UV light). Results of positive-control (PC) and negative-control (NTC) reactions performed in duplicate are shown in columns 1 and 5, respectively. Four specimens were tested in triplicate: two per row in wells 2, 3, and 4 and wells 6, 7, and 8. Two of the four extracts were positive for M. pneumoniae.
M. pneumoniae detection in clinical specimen extracts with LAMP versus real-time PCR.
A total of 252 respiratory specimen extracts were tested in triplicate with the M. pneumoniae LAMP assay and by real-time PCR (Table 3). Of these, 201 (79.7%) were positive in the LAMP assay and 226 (89.7%) were positive by real-time PCR; one of the specimens was positive in LAMP but not by real-time PCR, resulting in an analytical sensitivity of 88.5% (200/226) and a specificity of 96% (25/26) compared to the “gold standard,” real-time PCR. These results yielded positive and negative predictive values for the LAMP assay of 99.5% and 49.0%, respectively. Of 26 extracts positive by real-time PCR but negative in LAMP, 25 (96%) had a real-time cycle threshold (CT) value above 30. Thus, for extracts with a CT value of ≤30 (n = 153), the sensitivity of the LAMP assay is nearly identical to that of real-time PCR at 99.3% (152/153).
TABLE 3.
Correlation between LAMP and real-time PCR for M. pneumoniae detection from specimen extracts
| LAMP result | No. (%) of specimens |
|||
|---|---|---|---|---|
| qPCR CT < 30 (n = 153) | qPCR CT ≥ 30 (n = 73) | qPCR negative (n = 26) | Total | |
| Positive | 152 (99.3) | 48 (65.8) | 1 (0.04) | 201 (79.8) |
| Negative | 1 (0.7) | 25 (34.2) | 25 (96.2) | 51 (20.2) |
| Total | 226 (89.7) | 26 (10.3) | 252 (100) | |
Direct testing of primary specimens with LAMP.
A subset of clinical specimens comprised of nasopharyngeal (NP) swabs, oropharyngeal (OP) swabs, and combined NP/OP swabs, all in UTM (n = 72), was subjected to direct specimen testing in triplicate with the LAMP assay (Table 4). In order to simulate point-of-care testing conditions (i.e., no need for specialized transport media), specimens were pelleted by centrifugation, resuspended in 1× LAMP buffer, and briefly heated to facilitate rapid detection. Of these specimens, 39 were known to be M. pneumoniae positive based on prior testing by real-time PCR; direct specimen testing with LAMP identified 32 of these specimens as M. pneumoniae positive. With minimal processing, input of specimen directly into the LAMP reaction gives an analytical sensitivity of 82.1% (32/39) and a specificity of 100% (33/33) compared to the real-time PCR. These results yield positive and negative predictive values for direct specimen testing in LAMP of 100% and 82.5%, respectively.
TABLE 4.
M. pneumoniae detection directly from clinical specimens with LAMP
| LAMP result | No. (%) of specimens |
||
|---|---|---|---|
| qPCR-positive extract (n = 39) | qPCR-negative extract (n = 33) | Total | |
| Positive | 32 (82.1) | 0 (0) | 32 (44.4) |
| Negative | 7 (17.9) | 33 (100) | 40 (55.6) |
| Total | 39 (54.2) | 33 (45.8) | 72 (100) |
DISCUSSION
We designed and evaluated a LAMP assay that targets the CARDS toxin gene of M. pneumoniae and developed a modified version of a published reaction mixture (16) to achieve reduced background calcein fluorescence in a simple visual readout. This assay is specific for M. pneumoniae and is 88.5% as sensitive as a validated real-time PCR assay targeting the same gene tested on nucleic acid extracted from clinical specimens. In extracts from specimens with a relatively high M. pneumoniae load, defined at a real-time PCR CT value of ≤30, the sensitivity of the LAMP assay is 99.3% compared to this established assay. We also evaluated our current LAMP assay in a simulated point-of-care setting by directly testing specimens without automated nucleic acid extraction. In this format, the assay is 82.1% as sensitive as real-time PCR and provides results within 70 min from specimen collection as opposed to the days that would be required for shipment to and processing at a remote, specialized facility.
Although two LAMP assays targeting M. pneumoniae have been previously described (21, 22), including a commercially available FDA-cleared assay (illumigene), we were prompted to design a new assay for several reasons. First, although the illumigene assay is validated (25), it is intended for use only in hospital or reference laboratories rather than point-of-care settings and requires a prescribed nucleic acid extraction procedure. Additionally, while one of the published assays has been used in subsequent clinical studies (26, 27), the other had been tested on only 6 M. pneumoniae-positive specimens (21). To best understand the performance characteristics of an isothermal LAMP assay, we sought to evaluate its ability to amplify the CARDS toxin target from patient specimens in direct comparison to our validated real-time PCR test.
Finally, the LAMP reaction is widely accepted to have greater stringency for specificity than real-time PCR assays due to the four required LAMP primers (F3, B3, FIP, and BIP) having to recognize six sites on the target sequence, as opposed to the three required by 5′ hydrolysis chemistry. In practice, we have found that many potential LAMP primer sets are prone to false-positive amplification and must be extensively tested to confirm specificity. Despite the initial six-site specificity requirement, once LAMP autocycling begins, DNA synthesis takes place independently of the organism-derived template, regardless of whether it is due to specific or false priming. Driven by self-priming of generated stem-loops, spurious LAMP-like amplification can result from nonspecific binding events and produce a convincing false-positive signal (28). This risk is compounded by high concentrations of up to six oligonucleotides in the reaction, two of which are typically >40 bp in length, and high Mg2+ concentrations in standard reaction mixture formulations (16). Because of these risks, LAMP assays require significant optimization and validation, and these steps are not consistently performed or reported. In our evaluation of previously published primer sets, we observed an ∼15% false-positive rate in reaction mixtures containing only water as the template. We extensively tested the current primer set targeting the CARDS toxin of M. pneumoniae to confirm no false-positive amplification within the 1-h reaction time.
Although our M. pneumoniae LAMP assay enables rapid (<1-h) detection, it is 2-fold less sensitive in tests of the analytical limit of detection and 88.5% as sensitive for clinical specimen extracts as a validated real-time PCR targeting the same gene. Reports of LAMP sensitivity in comparison to real-time PCR assays vary: in some cases, the limit of detection with LAMP is shown to be between 10- and 100-fold better than that with PCR (19, 29), while in others, the reverse is true (21). Due to the aforementioned false-positive amplification rate observed in some published assays, it is difficult to determine a true sensitivity value for this method. There are several potential explanations for the decreased relative sensitivity of the current M. pneumoniae LAMP assay. For instance, in real-time PCR, the CARDS toxin has been previously shown to be a superior target for detection of M. pneumoniae from clinical specimens (10); the LAMP primers amplify a different region of the gene, perhaps introducing sequence-specific variations in sensitivity. Additionally, differences in the formulation of the LAMP reaction mix may affect sensitivity. Multiple groups have reported that the presence of calcein and Mn2+ in the reaction mix results in reduced sensitivity compared to other detection methods (17, 30). Given the high concentration of Mn2+ that we found necessary to reduce background calcein fluorescence, it is unsurprising that the LAMP assay does not achieve the same sensitivity as the benchmark real-time PCR method. Further optimization to reduce the Mn2+ concentration or implement an alternative detection method such as the use of hydroxynaphthol blue or pH-sensitive dyes could improve results to achieve comparable sensitivity (17, 31).
Additional procedural modifications may also prove advantageous. For example, an alternative method that includes denaturation of the template DNA at 95°C for 5 min prior to the reaction has been reported to increase sensitivity by as much as 200-fold, although others report no change (32, 33). A thermostable strand-displacing DNA polymerase that would enable denaturation after enzyme addition for ease of reaction assembly has recently been developed and may increase the efficiency and fidelity of LAMP assays in general (34).
Reduced sensitivity may also be attributed to low bacterial load in real-time PCR-positive but LAMP-negative patient specimens, a phenomenon that has been described previously (35). This idea is supported by results showing that 96% of LAMP false negatives have a real-time CT value of ≥30. Additionally, specimen extracts were tested immediately after extraction with real-time PCR but retrospectively with LAMP, sometimes more than a year later, thereby negatively impacting the performance of the LAMP assay. We plan to assess the M. pneumoniae LAMP assay and real-time PCR in a side-by-side manner in future outbreaks, resulting in a more accurate comparison. Furthermore, the swab specimens used in this study were typically received in a volume of 2 to 3 ml of transport media, which dilutes the specimen and leads to decreased sensitivity. If M. pneumoniae LAMP were to be used as a primary-care diagnostic method, the specimen could be concentrated by collection in a significantly smaller (∼200-μl) volume of water or buffer, likely resulting in decreased time to detection and improved sensitivity. Finally, the percentage of false negatives and, thus, the low (49.0%) negative predictive value of this assay for specimen extracts (Table 3) might be improved by evaluating a broader range of respiratory disease specimens shipped to the CDC (i.e., not only those already suspected to be M. pneumoniae positive) to reduce selection bias among the specimens tested.
The Bst DNA polymerase used in LAMP is resistant to inhibitors typically found in clinical specimens (36, 37). As a consequence, previous work has demonstrated that LAMP can amplify target DNA directly from unextracted specimen, rendering nucleic acid extraction unnecessary (38, 39). The lack of a cell wall in M. pneumoniae makes it an especially promising candidate for this approach, as it renders the bacterium more susceptible to lysis during specimen collection or transport, when heated, or in the presence of detergent contained in the reaction buffer. Amplification of M. pneumoniae DNA directly from unextracted respiratory specimens with LAMP demonstrates the utility of this approach. Further optimization will likely improve the initial results of direct specimen testing. If LAMP reagents were lyophilized, the specimen could be added in a volume equal to that of the final reaction mixture, simultaneously resuspending the reagents and contributing a roughly 2.5-fold increase of the input volume of template used in this work (40). These approaches will be the focus of future studies and will greatly benefit by our undertaking this effort jointly with an industry partner in order to streamline operation of the assay and commercialize it for widely accessible point-of-care use.
With these improvements, the LAMP assay will enable rapid, low-cost detection of M. pneumoniae cases and earlier recognition of outbreaks by medical providers. The inclusion of an unambiguous visual readout further simplifies an assay already amenable to point-of-care use, and proof of principle with direct specimen testing shows that time to diagnosis can be reduced to meet the time constraints of a patient visit. Implementation of this assay will significantly improve response times to M. pneumoniae infections and enable members of the medical community to make informed decisions when prescribing antibiotics for treatment of CAP.
ACKNOWLEDGMENT
The findings and conclusions in this report are ours and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.Foy HM. 1993. Infections caused by Mycoplasma pneumoniae and possible carrier state in different populations of patients. Clin Infect Dis 17(Suppl 1):S37–S46. [DOI] [PubMed] [Google Scholar]
- 2.Winchell JM. 2013. Mycoplasma pneumoniae—a national public health perspective. Curr Pediatr Rev 9:324–333. doi: 10.2174/15733963113099990009. [DOI] [Google Scholar]
- 3.Waites KB, Talkington DF. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev 17:697–728. doi: 10.1128/CMR.17.4.697-728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marston BJ, Plouffe JF, File TM Jr, Hackman BA, Salstrom SJ, Lipman HB, Kolczak MS, Breiman RF. 1997. Incidence of community-acquired pneumonia requiring hospitalization. Results of a population-based active surveillance study in Ohio. The Community-Based Pneumonia Incidence Study Group. Arch Intern Med 157:1709–1718. [PubMed] [Google Scholar]
- 5.Tully JG, Rose DL, Whitcomb RF, Wenzel RP. 1979. Enhanced isolation of Mycoplasma pneumoniae from throat washings with a newly-modified culture medium. J Infect Dis 139:478–482. doi: 10.1093/infdis/139.4.478. [DOI] [PubMed] [Google Scholar]
- 6.She RC, Thurber A, Hymas WC, Stevenson J, Langer J, Litwin CM, Petti CA. 2010. Limited utility of culture for Mycoplasma pneumoniae and Chlamydophila pneumoniae for diagnosis of respiratory tract infections. J Clin Microbiol 48:3380–3382. doi: 10.1128/JCM.00321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorigo-Zetsma JW, Zaat SA, Wertheim-van Dillen PM, Spanjaard L, Rijntjes J, van Waveren G, Jensen JS, Angulo AF, Dankert J. 1999. Comparison of PCR, culture, and serological tests for diagnosis of Mycoplasma pneumoniae respiratory tract infection in children. J Clin Microbiol 37:14–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ieven MM, Loens K. 2013. Should serology be abolished in favor of PCR for the diagnosis of Mycoplasma pneumoniae infections? Curr Pediatr Rev 9:304–313. doi: 10.2174/157339630904131223110501. [DOI] [Google Scholar]
- 9.Beersma MF, Dirven K, van Dam AP, Templeton KE, Claas EC, Goossens H. 2005. Evaluation of 12 commercial tests and the complement fixation test for Mycoplasma pneumoniae-specific immunoglobulin G (IgG) and IgM antibodies, with PCR used as the “gold standard”. J Clin Microbiol 43:2277–2285. doi: 10.1128/JCM.43.5.2277-2285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winchell JM, Thurman KA, Mitchell SL, Thacker WL, Fields BS. 2008. Evaluation of three real-time PCR assays for detection of Mycoplasma pneumoniae in an outbreak investigation. J Clin Microbiol 46:3116–3118. doi: 10.1128/JCM.00440-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michelow IC, Olsen K, Lozano J, Duffy LB, McCracken GH, Hardy RD. 2004. Diagnostic utility and clinical significance of naso- and oropharyngeal samples used in a PCR assay to diagnose Mycoplasma pneumoniae infection in children with community-acquired pneumonia. J Clin Microbiol 42:3339–3341. doi: 10.1128/JCM.42.7.3339-3341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardegger D, Nadal D, Bossart W, Altwegg M, Dutly F. 2000. Rapid detection of Mycoplasma pneumoniae in clinical samples by real-time PCR. J Microbiol Methods 41:45–51. doi: 10.1016/S0167-7012(00)00135-4. [DOI] [PubMed] [Google Scholar]
- 13.Poritz MA, Blaschke AJ, Byington CL, Meyers L, Nilsson K, Jones DE, Thatcher SA, Robbins T, Lingenfelter B, Amiott E, Herbener A, Daly J, Dobrowolski SF, Teng DH, Ririe KM. 2011. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One 6:e26047. doi: 10.1371/journal.pone.0026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori Y, Kitao M, Tomita N, Notomi T. 2004. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J Biochem Biophys Methods 59:145–157. doi: 10.1016/j.jbbm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Tomita N, Mori Y, Kanda H, Notomi T. 2008. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 17.Goto M, Honda E, Ogura A, Nomoto A, Hanaki K. 2009. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 46:167–172. doi: 10.2144/000113072. [DOI] [PubMed] [Google Scholar]
- 18.Lucchi NW, Demas A, Narayanan J, Sumari D, Kabanywanyi A, Kachur SP, Barnwell JW, Udhayakumar V. 2010. Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS One 5:e13733. doi: 10.1371/journal.pone.0013733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parida M, Posadas G, Inoue S, Hasebe F, Morita K. 2004. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J Clin Microbiol 42:257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boehme CC, Nabeta P, Henostroza G, Raqib R, Rahim Z, Gerhardt M, Sanga E, Hoelscher M, Notomi T, Hase T, Perkins MD. 2007. Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J Clin Microbiol 45:1936–1940. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito R, Misawa Y, Moriya K, Koike K, Ubukata K, Okamura N. 2005. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of Mycoplasma pneumoniae. J Med Microbiol 54:1037–1041. doi: 10.1099/jmm.0.46071-0. [DOI] [PubMed] [Google Scholar]
- 22.Yoshino M, Annaka T, Kojima T, Ikedo M. 2008. Sensitive and rapid detection of Mycoplasma pneumoniae by loop-mediated isothermal amplification. Kansenshogaku Zasshi 82:168–176. doi: 10.11150/kansenshogakuzasshi1970.82.168. [DOI] [PubMed] [Google Scholar]
- 23.Nagamine K, Hase T, Notomi T. 2002. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol Cell Probes 16:223–229. doi: 10.1006/mcpr.2002.0415. [DOI] [PubMed] [Google Scholar]
- 24.Thurman KA, Warner AK, Cowart KC, Benitez AJ, Winchell JM. 2011. Detection of Mycoplasma pneumoniae, Chlamydia pneumoniae, and Legionella spp. in clinical specimens using a single-tube multiplex real-time PCR assay. Diagn Microbiol Infect Dis 70:1–9. doi: 10.1016/j.diagmicrobio.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratliff AE, Duffy LB, Waites KB. 2014. Comparison of the illumigene Mycoplasma DNA amplification assay and culture for detection of Mycoplasma pneumoniae. J Clin Microbiol 52:1060–1063. doi: 10.1128/JCM.02913-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kakuya F, Kinebuchi T, Fujiyasu H, Tanaka R, Kano H. 2014. Genetic point-of-care diagnosis of Mycoplasma pneumoniae infection by LAMP assay. Pediatr Int 56:547–552. doi: 10.1111/ped.12327. [DOI] [PubMed] [Google Scholar]
- 27.Aizawa Y, Oishi T, Tsukano S, Taguchi T, Saitoh A. 2014. Clinical utility of loop-mediated isothermal amplification for rapid diagnosis of Mycoplasma pneumoniae in children. J Med Microbiol 63:248–251. [DOI] [PubMed] [Google Scholar]
- 28.Kimura Y, de Hoon MJ, Aoki S, Ishizu Y, Kawai Y, Kogo Y, Daub CO, Lezhava A, Arner E, Hayashizaki Y. 2011. Optimization of turn-back primers in isothermal amplification. Nucleic Acids Res 39:e59. doi: 10.1093/nar/gkr041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Seo DJ, Lee MH, Choi C. 2014. Comparison of conventional PCR, multiplex PCR, and loop-mediated isothermal amplification assays for rapid detection of Arcobacter species. J Clin Microbiol 52:557–563. doi: 10.1128/JCM.02883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wastling SL, Picozzi K, Kakembo AS, Welburn SC. 2010. LAMP for human African trypanosomiasis: a comparative study of detection formats. PLoS Negl Trop Dis 4:e865. doi: 10.1371/journal.pntd.0000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanner NA, Zhang Y, Evans TC Jr. 2015. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques 58:59–68. [DOI] [PubMed] [Google Scholar]
- 32.Nagamine K, Watanabe K, Ohtsuka K, Hase T, Notomi T. 2001. Loop-mediated isothermal amplification reaction using a nondenatured template. Clin Chem 47:1742–1743. [PubMed] [Google Scholar]
- 33.Aryan E, Makvandi M, Farajzadeh A, Huygen K, Bifani P, Mousavi SL, Fateh A, Jelodar A, Gouya MM, Romano M. 2010. A novel and more sensitive loop-mediated isothermal amplification assay targeting IS6110 for detection of Mycobacterium tuberculosis complex. Microbiol Res 165:211–220. doi: 10.1016/j.micres.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Ignatov KB, Barsova EV, Fradkov AF, Blagodatskikh KA, Kramarova TV, Kramarov VM. 2014. A strong strand displacement activity of thermostable DNA polymerase markedly improves the results of DNA amplification. Biotechniques 57:81–87. [DOI] [PubMed] [Google Scholar]
- 35.Dittrich S, Castonguay-Vanier J, Moore CE, Thongyoo N, Newton PN, Paris DH. 2014. Loop-mediated isothermal amplification for Rickettsia typhi (the causal agent of murine typhus): problems with diagnosis at the limit of detection. J Clin Microbiol 52:832–838. doi: 10.1128/JCM.02786-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaneko H, Kawana T, Fukushima E, Suzutani T. 2007. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J Biochem Biophys Methods 70:499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 37.Francois P, Tangomo M, Hibbs J, Bonetti EJ, Boehme CC, Notomi T, Perkins MD, Schrenzel J. 2011. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol Med Microbiol 62:41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- 38.Hill J, Beriwal S, Chandra I, Paul VK, Kapil A, Singh T, Wadowsky RM, Singh V, Goyal A, Jahnukainen T, Johnson JR, Tarr PI, Vats A. 2008. Loop-mediated isothermal amplification assay for rapid detection of common strains of Escherichia coli. J Clin Microbiol 46:2800–2804. doi: 10.1128/JCM.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poon LL, Wong BW, Ma EH, Chan KH, Chow LM, Abeyewickreme W, Tangpukdee N, Yuen KY, Guan Y, Looareesuwan S, Peiris JS. 2006. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem 52:303–306. [DOI] [PubMed] [Google Scholar]
- 40.Hayashida K, Kajino K, Hachaambwa L, Namangala B, Sugimoto C. 2015. Direct blood dry LAMP: a rapid, stable, and easy diagnostic tool for human African trypanosomiasis. PLoS Negl Trop Dis 9:e0003578. doi: 10.1371/journal.pntd.0003578. [DOI] [PMC free article] [PubMed] [Google Scholar]