LETTER
In feedlot cattle, the occurrence of liver abscesses is a complex interplay between an aggressive grain feeding program and a number of dietary and management factors. The incidence of liver abscesses ranges from 12 to 32% (1). Fusobacterium necrophorum, an anaerobic bacterium that originates from the rumen, is the primary etiologic agent (2, 3). The second most common bacterial species isolated is Trueperella (formerly Arcanobacterium) pyogenes (4). We analyzed liver abscess samples collected at slaughter from 10 feedlot cattle in the summer of 2013 to determine the dominant bacterial species involved. The 10 samples were collected randomly from a lot of Holstein steers that originated from the same feedlot in the Midwest. In addition to F. necrophorum (100%; 10/10) and T. pyogenes (40%; 4/10), for the first time, we isolated Salmonella enterica from all 10 samples that were plated on a blood agar plate and incubated for 24 h in an anaerobic glove box (Thermo Fisher Scientific, Waltham, MA). The species identification was confirmed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Bruker Daltonics Inc., Billerica, MA). Interestingly, on a blood agar plate incubated anaerobically, Salmonella appeared as whitish, large, round, and somewhat rough colonies (Fig. 1A). However, when the colony was picked and restreaked on a blood agar plate and incubated aerobically, colonies appeared small, round, smooth, and gray (Fig. 1B). All 10 isolates agglutinated with Salmonella polyvalent O antiserum but did not agglutinate with B, C1, C2, D1, D2, and E serogroup antisera. All isolates were PCR positive for the invA gene (5). The strains were submitted to the National Veterinary Services Laboratories (Ames, IA) for serotyping and were identified as 6,7:g,m,s:e,n,z15, a novel serotype. The pulsed-field gel electrophoresis (PFGE) analysis, done per the CDC's PulseNet protocol (6), indicated that all 10 Salmonella isolates had 100% Dice similarity.
FIG 1.
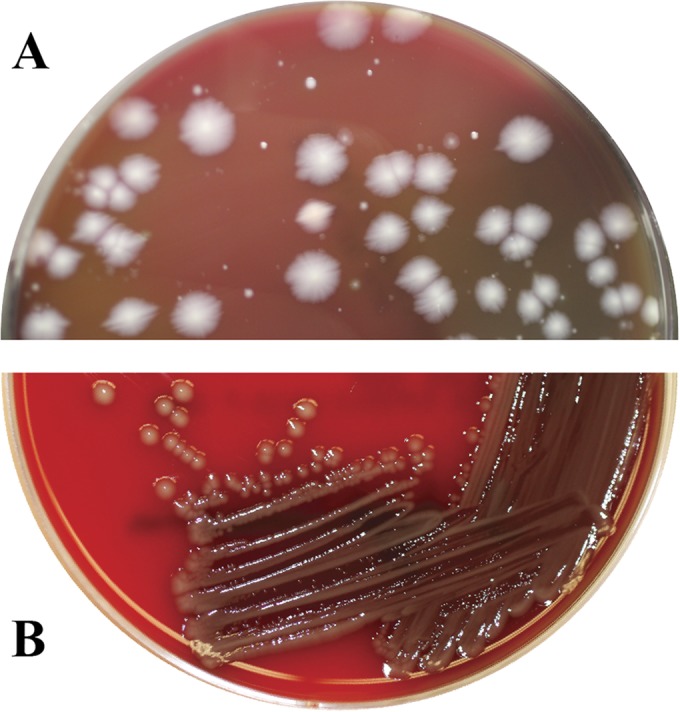
Colony morphology of Salmonella on an anaerobic blood agar plate from initial inoculation of liver abscess sample (A) and on an aerobic-growth blood agar plate (B).
This is the first report of Salmonella isolation from liver abscesses of cattle. There are reports of Salmonella isolation from liver abscesses in humans (7, 8). It is not known whether S. enterica is one of the etiologic agents or whether it entered, via blood or lymph, into an abscess initiated by F. necrophorum in the liver and survived. It is possible that Salmonella present in the gut could cross the gut epithelial barrier, enter the portal circulation, and get trapped in the portal capillary system of the liver to initiate infection. The entry through the gut epithelium may be facilitated by inflammation of the ruminal epithelium, and possibly the mucous membrane of the hindgut, associated with ruminal and hindgut acidosis (1). Salmonella bacteria have been isolated from lymph nodes of healthy cattle at the time of slaughter (9). Recently, Bugarel et al. (10) have published draft genome sequences of two novel strains of Salmonella, isolated from lymph nodes of slaughtered cattle, which match the antigenic designation of our strains from liver abscesses, and have named the serotype Lubbock. Interestingly, the PFGE banding patterns of the two Lubbock strains from lymph nodes were 100% identical to the banding patterns of the 10 liver abscess strains (data not shown). However, the identical banding pattern may not necessarily mean strain relatedness. Although PFGE is routinely used to assess strain relatedness in outbreak investigations, in the case of Salmonella, XbaI restriction, at least with some serotypes, has proven to be serotype specific (11).
Salmonella bacteria are facultative intracellular pathogens that have the ability to adapt quickly to diverse environments, including fluctuations in oxygen concentrations. It is well known that Salmonella bacteria are capable of robust growth under anaerobic conditions (12). In fact, Salmonella bacteria grown under anaerobic conditions are more invasive and virulent and adhere better to mammalian cells than do aerobically grown cells (13, 14). Fink et al. (15) have demonstrated participation of the oxygen-sensing, global regulator Fnr in regulating anaerobic metabolism, flagellar biosynthesis, motility, chemotaxis, and virulence in Salmonella enterica serotype Typhimurium. Therefore, it is possible that Salmonella could be contributing to the formation of abscesses. However, further studies are needed to evaluate the prevalence and importance of Salmonella in liver abscesses of feedlot cattle.
Footnotes
This article is contribution no. 15-409-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Nagaraja TG, Lechtenberg KF. 2007. Liver abscesses in feedlot cattle. Vet Clin North Am Food Anim Pract 23:351–369. doi: 10.1016/j.cvfa.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Scanlan CM, Hathcock TL. 1983. Bovine rumenitis-liver abscess complex: a bacteriological review. Cornell Vet 73:288–297. [PubMed] [Google Scholar]
- 3.Nagaraja TG, Chengappa MM. 1998. Liver abscesses in feedlot cattle: a review. J Anim Sci 76:287–298. [DOI] [PubMed] [Google Scholar]
- 4.Lechtenberg KF, Nagaraja TG, Leipold HW, Chengappa MM. 1988. Bacteriologic and histologic studies of hepatic abscesses in cattle. Am J Vet Res 49:58–62. [PubMed] [Google Scholar]
- 5.Alam MJ, Renter DG, Ives SE, Thomson DU, Sanderson MW, Hollis LC, Nagaraja TG. 2009. Potential associations between fecal shedding of Salmonella in feedlot cattle treated for apparent respiratory disease and subsequent adverse health outcomes. Vet Res 40:2. doi: 10.1051/vetres:2008040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhry R, Mahajan RK, Diwan A, Khan S, Singhal R, Chandel DS, Hans C. 2003. Unusual representation of enteric fever: three cases of splenic and liver abscesses due to Salmonella typhi and Salmonella paratyphi A. Trop Gastroenterol 24:198–199. [PubMed] [Google Scholar]
- 8.Qu F, Fan Z, Cui E, Zhang W, Bao C, Chen S, Mao Y, Zhou D. 2013. First report of liver abscess caused by Salmonella enterica serovar Dublin. J Clin Microbiol 51:3140–3142. doi: 10.1128/JCM.01034-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arthur TM, Brichta-Harhay DM, Bosilevac JM, Guerini MN, Kalchayanand N, Wells JE, Shackelford SD, Wheeler TL, Koohmaraie M. 2008. Prevalence and characterization of Salmonella in bovine lymph nodes potentially destined for use in ground beef. J Food Prot 71:1685–1688. [DOI] [PubMed] [Google Scholar]
- 10.Bugarel M, den Bakker HC, Nightingale KK, Brichta-Harhay DM, Edrington TS, Loneragan GH. 2015. Two draft genome sequences of a new serovar of Salmonella enterica, serovar Lubbock. Genome Announc 3:e00215-15. doi: 10.1128/genomeA.00215-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou W, Lin WJ, Foley SL, Chen CH, Nayak R, Chen JJ. 2010. Evaluation of pulsed-field gel electrophoresis profiles for identification of Salmonella serotypes. J Clin Microbiol 48:3122–3126. doi: 10.1128/JCM.00645-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto NY, Droffner ML. 1985. Mechanisms determining aerobic or anaerobic growth in the facultative anaerobe Salmonella typhimurium. Proc Natl Acad Sci U S A 82:2077–2081. doi: 10.1073/pnas.82.7.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee CA, Falkow S. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci U S A 87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schiemann DA, Shope SR. 1991. Anaerobic growth of Salmonella typhimurium results in increased uptake by Henle 407 epithelial and mouse peritoneal cells in vitro and repression of a major outer membrane protein. Infect Immun 59:437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fink RC, Evans MR, Porwollik S, Vasquez-Torres A, Jones-Carson J, Troxell B, Libby SJ, McClelland M, Hassan HM. 2007. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028). J Bacteriol 189:2262–2273. doi: 10.1128/JB.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


