Abstract
We compared the Remel Spectra CRE agar plate to CDC standard methodology for the isolation of carbapenem-resistant Enterobacteriaceae (CRE) from 300 rectal swab specimens obtained from patients residing in a long-term-care facility (LTCF). Multiplex PCR experiments were performed on isolates to identify specific Klebsiella pneumoniae carbapenemases (KPC) and additional β-lactamases. Of the 300 patients, 72 (24%) harbored CRE and were PCR positive for KPC enzymes. The Remel Spectra CRE plates detected KPC-type CRE in isolates from 70 of 72 patients (97.2%), while the CDC method detected CRE in 56 of 72 (77.8%). CRE identification results were available in 18 h compared to 36 h for the CDC method. Remel Spectra CRE agar plates can provide useful means for a fast and reliable method for detecting KPC-type CRE and for accelerated institution of appropriate infection control precautions.
INTRODUCTION
Klebsiella pneumoniae carbapenemases (KPC) are predominantly found in Klebsiella pneumoniae worldwide and have become endemic in major cities of the United States (1–4). They have also been characterized in Klebsiella oxytoca, Escherichia coli, Enterobacter spp., Citrobacter spp., Serratia spp., Acinetobacter baumannii, Salmonella spp., and Pseudomonas spp (1–3). Although KPC-type carbapenemases are the leading cause of carbapenem resistance among Enterobacteriaceae (CRE) in the United States, carbapenem resistance can be mediated by many other enzymes alone or in combination with other resistance mechanisms, including efflux and porin mutations (3, 5). Infections with these organisms are associated with higher mortality rates than those infections with more susceptible strains (6, 7). Patients who are asymptomatic carriers can serve as a source for nosocomial spread of these carbapenem-resistant organisms, necessitating the prompt detection of carriers and institution of appropriate infection control measures (8–11). Surveillance for CRE is especially useful among immunocompromised patients, including those in intensive care and transplant units (11, 12). Screening residents of long-term-care facilities (LTCF) prior to admission and/or transfer to a hospital may be of increasing importance, as they may harbor antibiotic-resistant isolates (13–15). Although they are rapid, PCR-based technologies can be expensive and limited, as they only interrogate specific genes that are being screened and a positive result does not always predict the expression of resistance. While traditional culture methods are slow and may lack sensitivity, the use of chromogenic media to culture CRE and other multidrug-resistant microorganisms (MDRO) may facilitate the detection of these organisms in a timelier manner (16–18). The purpose of this investigation was to compare Remel Spectra CRE agar plates to the standard CDC method for the isolation of CRE from rectal swab specimens obtained from patients residing in an LTCF. Remel Spectra CRE agar is a selective and differential agar intended for use in the qualitative and presumptive identification of carbapenem-resistant Enterobacteriaceae directly from clinical specimens.
MATERIALS AND METHODS
Culture screening methods.
Rectal swab specimens using double swabs were obtained from patients residing at an affiliated LTCF consisting of 320 beds with an 80-bed ventilator unit as part of a facility-wide infection control initiative. Specimens were obtained from patients without signs or symptoms of diarrhea or active or recent Clostridium difficile infection from September 2013 through January 2014 (19). Specimens were collected using double swabs, placed into Stuart's media, and transported to the clinical microbiology laboratory for processing. The swab specimens were used to inoculate Remel Spectra CRE plates (Remel, Thermo Fisher, Lenexa, KS) directly and a 5-ml Trypticase soy broth (TSB) (Remel) containing a 10-μg ertapenem disk (BD Diagnostics, Sparks, MD) (CDC method) (20). Broth tubes and plated media were incubated at 37°C in ambient air. The TSB cultures were incubated for 18 h and subcultured onto a MacConkey agar plate (Remel), which was incubated at 37°C in ambient air for an additional 18 h. Remel Spectra CRE plates were examined for growth after 18, 24, and 48 h of incubation. Isolates were obtained after 18 h of incubation for analysis. Colonies of Klebsiella pneumoniae appeared blue (Fig. 1), while Escherichia coli appeared pink on CRE plates (Fig. 2). Colonies of Enterobacter spp., Serratia spp., and Citrobacter spp. will appear blue on this medium, while Acinetobacter spp. and Stenotrophomonas maltophilia will appear white. Pseudomonas aeruginosa will appear either as white or turquoise-green colonies. All colonies growing on MacConkey agar or Spectra CRE plates were identified, and susceptibility testing performed using Vitek 2 (bioMérieux, Inc., Durham, NC). Isolates were identified directly from the Spectra CRE plates. This was validated prior to the initiation of the study. Isolates testing nonsusceptible to ertapenem (MIC, ≥1 μg/ml) were tested for carbapenem inactivation using the modified Hodge test (21). The Remel Spectra CRE plates are pending FDA approval and are currently available as a research-use-only product.
FIG 1.
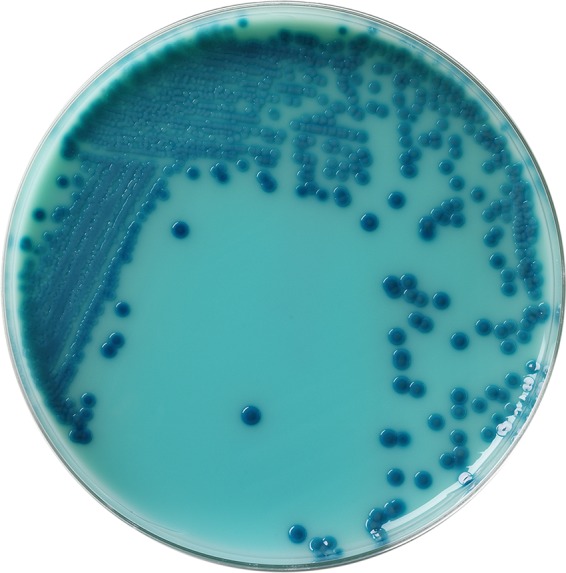
Klebsiella pneumoniae on a Spectra CRE agar plate.
FIG 2.
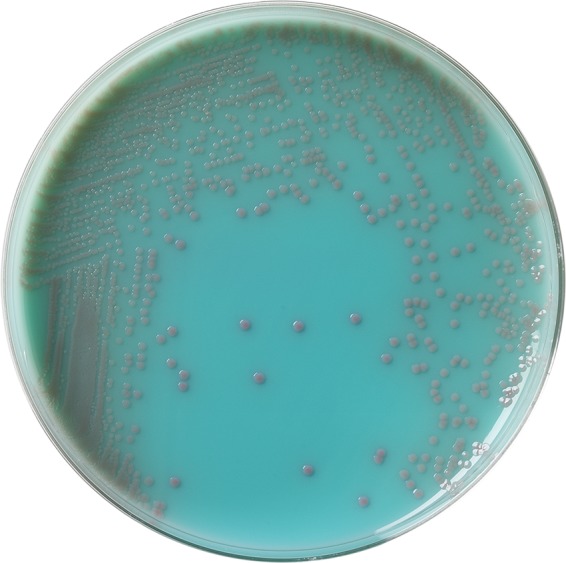
E. coli on a Spectra CRE agar plate.
PCR methods.
Molecular testing for CRE and extended-spectrum β-lactamase (ESBL) genes, including blaKPC, blaNDM-1, blaVIM, blaIMP, blaCTX-M, blaSHV, blaTEM, and blaOXA-48, was performed by a molecular beacon real-time PCR (MB-PCR) assay (22, 23).
RESULTS
A total of 300 rectal swab specimens from 300 patients were received and included in this study. The identification of presumptive carbapenemase-harboring organisms was available in 18 h with the Remel Spectra CRE plate assay compared to 36 h with the standard CDC method. Seventy-eight CRE isolates were obtained from 72 patients, and 24% harbored KPC-type CRE. Remel Spectra CRE plates detected isolates from 70 of 72 (97.2%) patients with CRE and in 76 of 78 (97.4%) isolates. In contrast, the CDC method detected CRE in 56 of 72 (77.8%) patients. Specific organisms and KPC genotypes are shown in Table 1. Non-Enterobacteriaceae carbapenem-resistant organisms detected by Remel Spectra CRE plates are shown in Table 2. Carbapenem resistance mechanisms of these isolates were not characterized.
TABLE 1.
KPC and other β-lactamases associated with carbapenem-resistant Enterobacteriaceae recovered by each method
| Organism | No. of isolates | KPC enzyme no./genotype | No./name of associated β-lactamases | No. of isolates by Spectra CRE plate | No. of isolates by CDC method |
|---|---|---|---|---|---|
| Klebsiella pneumoniae | 51 | 33/KPC-3 | 49/SHV | 50 | 47 |
| 41/TEM | |||||
| 18/KPC-2 | 12/CTX-M-1 group | ||||
| Escherichia coli | 22 | 13/KPC-3 | 3/SHV | 21 | 13 |
| 14/TEM | |||||
| 9/KPC-2 | 3/CTX-M-1 group | ||||
| Enterobacter cloacae | 4 | 4/KPC-2 | 4/ACT/MIR | 4 | 0 |
| Enterobacter aerogenes | 1 | 1/KPC-2 | 1/TEM | 1 | 0 |
TABLE 2.
Non-Enterobacteriaceae multidrug-resistant organisms isolated by each method
| Organism | Total no. of isolates | No. of isolates by CRE plate | No. of isolates by CDC method |
|---|---|---|---|
| Acinetobacter baumannii | 14 | 14 | 7 |
| Pseudomonas aeruginosa | 16 | 15 | 14 |
| Stenotrophomonas maltophilia | 2 | 2 | 2 |
Of the 78 isolates that were suspected of harboring a carbapenemase, all but 4 Klebsiella pneumoniae isolates were positive using the modified Hodge test. All 78 strains tested positive for blaKPC and negative for blaNDM-1, blaVIM, blaIMP, and blaOXA-48. Of 51 K. pneumoniae isolates, 19 harbored the KPC-2 gene, while 33 contained KPC-3. Of the 22 E. coli isolates, 9 contained the KPC-2 gene and 13 carried KPC-3, and all 5 Enterobacter spp. harbored the KPC-2 gene (Table 1).
There were an additional 39 isolates found on the Remel Spectra CRE plates that were susceptible to the carbapenems but appeared to harbor ESBL-type β-lactamases based on their antibiotic susceptibility profiles. There were no differences in the representative pigmentation between CRE and non-CRE isolates. These were all negative for blaKPC, blaNDM-1, blaVIM, blaIMP, and blaOXA-48. Among these, 19 K. pneumoniae and 12 E. coli isolates were positive for CTX-M group 1 genes. One Citrobacter freundii isolate contained SHV-1 and CMY2, LAT, and CEF β-lactamases. An additional 7 E. coli isolates, all with elevated MICs to cefoxitin and cefepime, were negative for ESBL- and AmpC-type β-lactamases tested. Five of these isolates contained TEM-1 β-lactamases, while 2 isolates were negative for all other β-lactamase genes tested.
DISCUSSION
This study evaluated the utility of Remel Spectra CRE agar for detecting CRE from rectal swabs obtained during active surveillance from patients residing in an affiliated LTCF. Many commercial chromogenic media, including Supercarba, Remel Spectra CRE, CHROMagar KPC, chromID Carba, and Brilliance CRE, have been evaluated for the detection of CRE and isolates with reduced susceptibility to carbapenems (16–18, 24, 25). Two studies compared chromID Carba media with the standard CDC method during fecal screening surveillance programs, and sensitivities were 96.5% and 96.9%, respectively (17, 18). In another analysis for the detection of blaKPC, Remel Spectra CRE had an overall sensitivity of 97.8%; CHROMagar KPC, 76.6%; and a direct ertapenem disk method, 83.0% (16). In our study, Remel Spectra CRE plates detected CRE in isolates from 70 of 72 (97.2%) patients, while the standard CDC method detected rectal carriage of CRE in 56 of 72 (77.8%). This is in agreement with previous studies which showed poorer isolation of CRE using the CDC method (16, 17). The CDC method was especially poor in detecting KPC-producing E. coli and Enterobacter spp. Although not an indication, the Spectra CRE plates detected twice as many A. baumannii isolates than did the conventional CDC method.
Although not a focus of this investigation, an additional 39 specimens yielded characteristically pigmented colonies that were determined not to be CRE. Thirty-one of these isolates were Klebsiella pneumoniae and E. coli, all harboring CTX-M-type ESBLs. There was 1 Citrobacter freundii isolate which contained several β-lactamases, and 7 additional E. coli isolates with elevated MICs to the extended-spectrum cephalosporins did not contain any of the β-lactamase genes tested. This emphasizes the need to confirm the susceptibility profiles of those isolates growing on selective chromogenic media prior to reporting the results from the chromogenic screening medium.
This medium has been shown by the manufacturer to be capable of isolating organisms containing the NDM-1 metallo-β-lactamase. Although this medium should be capable of growing other metallo-β-lactamases and oxacillinases, none were recovered in this study, so no claims can be made as to the ability to recover organisms containing these enzymes on this medium.
Long-term-care facilities have become a major focus of investigations because the patients harbor resistant organisms (14, 15). Obtaining information about resistant bacteria from these facilities has been problematic for various reasons (14–15, 26). There is no consistent support for active surveillance in nonoutbreak situations, and LTCFs are grossly underfunded and understaffed for these types of undertakings. Molecular techniques are not routinely performed in most clinical microbiology laboratories, and bacteria harboring resistant genes have been revealed after the fact, with silent dissemination (8). In conclusion, Remel Spectra CRE media detected more CRE and MDRO than did the standard CDC method, and results were available faster with Remel Spectra CRE than with the CDC method. Analysis of the isolates by molecular identification confirms and supports the use of chromogenic media for the identification of CRE. Such media can provide useful means for the fast and reliable detection of CRE. In addition, the ease of introduction into clinical microbiology laboratories should accelerate the institution of appropriate infection control precautions.
ACKNOWLEDGMENTS
This project was funded by Thermo Fisher Scientific. This study was supported by a grant (R01AI090155 to B.N.K.) from the National Institutes of Health.
B.N.K. holds two patents that focus on using DNA sequencing to identify bacterial pathogens.
REFERENCES
- 1.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. 2013. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev 25:682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J, Gniadkowski M, Hayden MK, Kumarawamy K, Livermore DM, Maya JJ, Nordmann P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N, Quinn JP. 2013. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13:785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel G, Bonomo RA. 2013. Stormy waters ahead: global emergence of carbapenemases. Front Microbiol 4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaiser RM, Castanheira M, Jones RN, Tenover F, Lynfield R. 2013. Trends in Klebsiella pneumoniae carbapenemase-positive K. pneumoniae in U.S. hospitals: report from the 2007 to 2009 SENTRY Antimicrobial Surveillance Program. Diagn Microbiol Infect Dis 76:356–360. doi: 10.1016/j.diagmicrobio.2013.03.032. [DOI] [PubMed] [Google Scholar]
- 5.Urban C, Rahal JJ. 2004. Mechanisms and detection of carbapenem resistance in Pseudomonas aeruginosa, Klebsiella pneumoniae, and in Acinetobacter baumannii. Rev Med Microbiol 15:63–72. doi: 10.1097/01.revmedmi.0000131426.36224.82. [DOI] [Google Scholar]
- 6.Falagas ME, Tansarli GS, Karageorgopoulos DE, Vardakas KZ. 2014. Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg Infect Dis 20:1170–1175. doi: 10.3201/eid2007.121004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwaber MJ, Klarfeld-Lidji S, Navon-Venezia S, Schwartz D, Leavitt A, Carmeli Y. 2008. Predictors of carbapenem-resistant Klebsiella pneumoniae acquisition among hospitalized adults and effect of acquisition on mortality. Antimicrob Agents Chemother 52:1028–1033. doi: 10.1128/AAC.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viau RA, Hujer AM, Marshall SH, Perez F, Hujer KM, Briceno DF, Dul M, Jacobs MR, Grossberg R, Toltzis P, Bonomo RA. 2012. Silent dissemination of Klebsiella pneumoniae isolates bearing K. pneumoniae carbapenemase in a long-term care facility for children and young adults in northeast Ohio. Clin Infect Dis 54:1314–1321. doi: 10.1093/cid/cis036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schechner V, Kotlovsky T, Kazma M, Mishali H, Schwartz D, Navon-Venezia S, Schwaber MJ, Carmeli Y. 2013. Asymptomatic rectal carriage of blaKPC producing carbapenem-resistant Enterobacteriaceae: who is prone to become clinically infected? Clin Microbiol Infect 19:451–456. doi: 10.1111/j.1469-0691.2012.03888.x. [DOI] [PubMed] [Google Scholar]
- 10.O'Fallon E, Gautam S, D'Agata MC. 2009. Colonization with multidrug resistant gram-negative bacteria: prolonged duration and frequent cocolonization. Clin Infect Dis 49:1375–1381. doi: 10.1086/598194. [DOI] [PubMed] [Google Scholar]
- 11.Banach DB, Francois J, Blash S, Patel G, Jenkins SG, LaBombardi V, Kreiswirth BN, Srinivasan A, Calfee DP. 2014. Active surveillance for carbapenem-resistant Enterobacteriaceae using stool specimens submitted for testing for Clostridium difficile. Infect Control Hosp Epidemiol 35:82–84. doi: 10.1086/674391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel G, Rana MM, Huprikar S. 2013. Multidrug-resistant bacteria in organ transplantation: an emerging threat with limited therapeutic options. Curr Infect Dis Rep 15:504–513. doi: 10.1007/s11908-013-0371-z. [DOI] [PubMed] [Google Scholar]
- 13.Urban C, Bradford PA, Tuckman M, Segal-Maurer S, Wehbeh W, Grenner L, Colon-Urban R, Mariano N, Rahal JJ. 2008. Carbapenem-resistant Escherichia coli harboring Klebsiella pneumoniae carbapenemase beta-lactamases associated with long-term care facilities. Clin Infect Dis 46:e127–e130. doi: 10.1086/588048. [DOI] [PubMed] [Google Scholar]
- 14.Moro ML, Gagliotti C. 2013. Antimicrobial resistance and stewardship in long-term care facilities. Future Microbiol 8:1011–1025. doi: 10.2217/fmb.13.75. [DOI] [PubMed] [Google Scholar]
- 15.Urban C, Wehbeh W, Rahal JJ. 2008. Antibacterial resistance associated with long-term care facilities. Rev Med Microbiol 19:47–55. doi: 10.1097/MRM.0b013e32831a4125. [DOI] [Google Scholar]
- 16.Vasoo S, Lolans K, Li H, Prabaker K, Hayden MK. 2014. Comparison of the CHROMagar KPC, Remel Spectra CRE, and a direct ertapenem disk method for the detection of KPC-producing Enterobacteriaceae from perirectal swabs. Diagn Microbiol Infect Dis 78:356–359. doi: 10.1016/j.diagmicrobio.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Papadimitriou-Olivgeris M, Bartzavali C, Christofidou M, Bereksi N, Hey J, Zambardi G, Spiliopoulou I. 2014. Performance of chromID Carba medium for carbapenemases-producing Enterobacteriaceae detection during rectal screening. Eur J Clin Microbiol Infect Dis 33:35–40. doi: 10.1007/s10096-013-1925-6. [DOI] [PubMed] [Google Scholar]
- 18.Vrioni G, Daniil I, Voulgari E, Ranellou K, Koumaki V, Ghirardi S, Kimouli M, Zambardi G, Tsakris A. 2012. Comparative evaluation of a prototype chromogenic medium (ChromID Carba) for detecting carbapenemase-producing Enterobacteriaceae in surveillance rectal swabs. J Clin Microbiol 50:1841–1846. doi: 10.1128/JCM.06848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labaze G, Kopacz J, Prasad N, Chwa S, Platis D, Motiwala F, Thakore K, Pan C, Russo D, Labombardi V, Osorio G, Pollack S, Urban C, Segal-Maurer S. 2014. Rectal colonization with carbapenem-resistant Enterobacteriaceae and Clostridium difficile among residents of a long term-care facility, poster C 77. American Geriatrics Society, Orlando, FL, 14 to 17 May 2014. [Google Scholar]
- 20.Centers for Disease Control and Prevention (CDC). 2013. Laboratory protocol for detection of carbapenem-resistant or carbapenemase-producing Klebsiella spp. and E. coli from rectal swabs. CDC, Atlanta, GA. http://www.cdc.gov/hai/pdfs/labsettings/klebsiella_or_ecoli.pdf. [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial susceptibility testing; 22nd informational supplement. CLSI M100-S22. Clinical and Laboratory Standards Institute. Wayne, PA. [Google Scholar]
- 22.Chen L, Mediavilla JR, Endimiani A, Rosenthal ME, Zhao Y, Bonomo RA, Kreiswirth BN. 2011. Multiplex real-time PCR assay for detection and classification of Klebsiella pneumoniae carbapenemase gene (blaKPC) variants. J Clin Microbiol 49:579–585. doi: 10.1128/JCM.01588-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hujer A, Evans S, Jiang H, Jacobs M, Kreiswirth BN, Fowler V, Chambers HF, Bonomo RA. 2014. Can rapid molecular diagnostics assist in the choice of beta-lactam antibiotics? An analysis of data from primers-I, P1150a, 24th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID 2014). Barcelona, Spain. [Google Scholar]
- 24.Pence MA, Hink T, Burnham CAD. 2015. Comparison of Chromogenic media for recovery of carbapenemase-producing Enterobacteriaceae (CPE) and evaluation of CPE prevalence at a tertiary care academic medical center. J Clin Microbiol 53:663–666. doi: 10.1128/JCM.03208-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girlich D, Poirel L, Nordmann P. 2013. Comparison of the Supercarba, CHROMagar KPC, and Brilliance CRE screening media for detection of Enterobacteriaceae with reduced susceptibility to carbapenems. Diagn Microbiol Infect Dis 75:214–217. doi: 10.1016/j.diagmicrobio.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Kindschuh W, Russo D, Kariolis I, Wehbeh W, Urban C. 2012. Comparison of a hospital-wide antibiogram with that of an associated long-term care facility. J Am Geriatr Soc 60:798–800. doi: 10.1111/j.1532-5415.2011.03881.x. [DOI] [PubMed] [Google Scholar]


