Abstract
Serotype IV group B Streptococcus (GBS) is emerging in Canada and the United States with rates as high as 5% of the total burden of adult invasive GBS disease. To understand this emergence, we studied the population structure and assessed the antimicrobial susceptibility of serotype IV isolates causing adult invasive infection in Manitoba and Saskatchewan, Canada, between 2010 and 2014. Whole-genome sequencing was used to determine multilocus sequence typing information and identify genes encoding antimicrobial resistance in 85 invasive serotype IV GBS strains. Antimicrobial susceptibility testing was performed by standard methods. Strain divergence was assessed using genome-wide single-nucleotide polymorphism analysis. Serotype IV strains were responsible for 16.9% of adult invasive GBS infections in Manitoba and Saskatchewan during the period. The majority of serotype IV isolates (89%) were clonally related, tetracycline-, erythromycin-, and clindamycin-resistant sequence type 459 (ST459) strains that possessed genes tetM and ermTR. Genome comparisons between ST459 and serotype V ST1 GBS identified several areas of recombination in an overall similar genomic background. Serotype IV ST459 GBS strains are expanding and causing a substantial percentage of adult invasive GBS disease. This emergence may be linked to the acquisition of resistance to tetracycline, macrolides, and lincosamides.
INTRODUCTION
Group B Streptococcus (GBS, also known as Streptococcus agalactiae) is a leading cause of sepsis and meningitis in neonates (1) and also causes invasive diseases in adult populations, particularly in older adults and immunocompromised individuals (2, 3). GBS adult diseases include bacteremia, skin and soft tissue infections, and more rarely, meningitis and endocarditis and can have mortality rates as high as 25% (3–6). GBS strains are classified in 10 serotypes (Ia, Ib, and II to IX) based on a serological reaction against the capsular polysaccharide (7–9). Strains of serotype V have been associated with the marked increase of adult invasive infections observed in North America since the 1990s (3, 10–12). Strains of this serotype, particularly those identified by multilocus sequence typing (MLST) as sequence type 1 (ST1), reportedly cause more invasive disease and have reduced susceptibility to macrolide and lincosamide antibiotics (13–15).
Historically, serotype IV GBS has been rarely isolated in North America, but its prevalence is increasing (11, 16). In the United States, longitudinal surveillance suggests that the proportion of serotype IV GBS isolates from nonpregnant adults increased from 0.2% in 1998 to 1999 to 5.7% in 2005 to 2006 (2). In 2010, serotype IV strains were shown to be responsible for 16% of early onset neonatal infections in the state of Minnesota (17, 18). Consistent with this emergence, when we characterized a collection of 600 invasive strains from the province of Ontario, Canada, we found that serotype IV isolates were responsible for 6.2% of cases of GBS invasive disease (19). Further characterization of invasive GBS strains from Toronto uncovered two main circulating STs among serotype IV isolates: ST452, included in clonal complex 23 (CC23), and ST459, included in CC1 (20).
Here, we sought to test the hypothesis that serotype IV strains are also emerging among GBS adult invasive disease in the province of Manitoba, Canada, which shares borders with Ontario and Minnesota, and in the province of Saskatchewan, directly west of Manitoba. In addition, we investigated whether one or multiple serotype IV clones were associated with cases of adult human disease in these areas. We report the highest yet recorded prevalence of serotype IV among adult invasive GBS infections (19% in Saskatchewan and 16% in Manitoba). We also show that highly clonal strains of ST459 serotype IV GBS, which are resistant to erythromycin, clindamycin, and tetracycline, are the main drivers of this emergence.
MATERIALS AND METHODS
Isolate collection and serotyping.
A total of 549 GBS strains isolated from normally sterile sites in adult patients were collected by the Cadham Provincial Laboratory in Winnipeg, Manitoba, and by the Saskatchewan Disease Control Laboratory in Regina, Saskatchewan (355 and 194 isolates, respectively), from January 2010 to May 2014. In Manitoba, the strains represented almost all invasive GBS isolates recovered province-wide from adult patients. In Saskatchewan, organisms were collected from adult patients in the two major urban centers of Saskatoon and Regina. Available strain metadata were limited to patient age, geographic descriptors, and anatomical source of the isolate. Species identification was carried out by standard methods (21, 22). The provincial laboratories then submitted the isolates to the National Microbiology Laboratory (Winnipeg, MB) for serotyping by latex agglutination (SSI Diagnostica, Statens Serum Institute, Copenhagen, Denmark) (23). Of the 93 serotype IV GBS strains, 85 were available for further characterization, including 47 from blood cultures, 18 from tissue, 5 from synovial fluid, and 15 from other normally sterile sites (e.g., peritoneal or pleural fluid). These 85 isolates were received by Public Health Ontario for whole-genome sequencing (WGS) (see Table S1 in the supplemental material). Of them, 79 were isolated from presumptive unique patients. In three cases, two strains from a single patient (isolated from different sterile sites) were used (see Table S1 in the supplemental material). There was no clear evidence of localized outbreaks caused by serotype IV GBS during the collection period.
WGS and bioinformatics analysis.
Bacteria were grown overnight in Todd-Hewitt broth supplemented with 0.2% yeast extract. DNA was prepared from overnight liquid cultures using the QIAamp DNA minikit (Qiagen, Toronto, ON, Canada) following the manufacturers' protocol for Gram-positive organisms. Paired-end libraries were prepared using the Illumina Nextera XT kit (Illumina, San Diego, CA) and sequenced on Illumina HiSeq (101 + 101 bp) or MiSeq (150 + 150 bp) instruments. Parsing of the multiplexed sequencing reads and removal of barcode information was performed using Illumina onboard software. MLST was determined directly from short reads using SRST2 (24). Ambiguous or novel alleles were confirmed by PCR and Sanger sequencing using previously described primers and conditions described in the GBS MLST scheme (25). Novel sequence types have been submitted to PubMLST. Neighbor-joining phylogenetic trees (1,000 bootstrap replications) were generated with SplitsTree4 (26) using the concatenated sequences of the seven loci used in the GBS MLST scheme. Illumina short-reads were then aligned to the genomes of reference strains NGBS061 (serotype IV, ST459; GenBank accession number CP007631.2) and SGBS001 (serotype V, ST1; GenBank accession number CP010867) using Mosaik (https://code.google.com/p/mosaik-aligner/). These two reference strains are the only ST459 and ST1 closed genomes isolated from humans, and they were each assembled using a combination of long reads obtained by PacBio instrument and paired-end Illumina short-reads (20, 27). NGBS061 was isolated in 2010 in the greater Toronto area from a case of adult disease. Strain SGBS001 was isolated in 1993 in Houston, TX, USA. The average coverage of all 85 strains was 206× (see Table S1 in the supplemental material). Polymorphisms were identified using VAAL (28) as previously described (29, 30). A matrix file containing the genotype of all strains at each polymorphic locus relative to either reference genome was created from the VAAL polymorphism output data using a custom script. Then, for each individual strain, single-nucleotide polymorphisms (SNPs) were concatenated in order of occurrence relative to the genome of the corresponding reference strain and converted to multiFASTA format. Neighbor-joining phylogenetic trees (1,000 bootstrap replications) were generated from the multiFASTA files using SplitsTree4. For comparative purposes, the genome sequences of 76 previously characterized ST1 strains from Toronto (27) and 3 previously characterized ST459 strains from France and the Czech Republic (31) were downloaded from NCBI's Sequence Read Archive. SRA accession numbers for these isolates are provided in Table S2 in the supplemental material. Areas of recombination were defined using BRATNextGen software (32) and run with 20 iterations and 100 replicates using a P value of 0.05 as the significance cutoff. Genome visualizations were created using BRIG (33) and edited using Adobe Illustrator.
Antimicrobial drug susceptibility.
We used the SRST2 database listing 1,913 variants of genes encoding antimicrobial resistance (24) to test for the presence or absence of genetic determinants of antibiotic resistance in the genomes of the GBS strains and confirmed the genes with manual inspection of the sequences. All serotype IV GBS strains were tested by agar dilution for susceptibility to tetracycline, ampicillin, clindamycin, erythromycin, cefotaxime, penicillin, and vancomycin following Clinical and Laboratory Standards Institute guidelines (34). Technicians performing the phenotypic tests were blind to the genetic results.
Short-read whole-genome sequencing data accession number.
Data have been deposited at NCBI's Sequence Read Archive under accession number PRJNA286872.
RESULTS
High prevalence of serotype IV strains among adult invasive GBS disease in Manitoba and Saskatchewan.
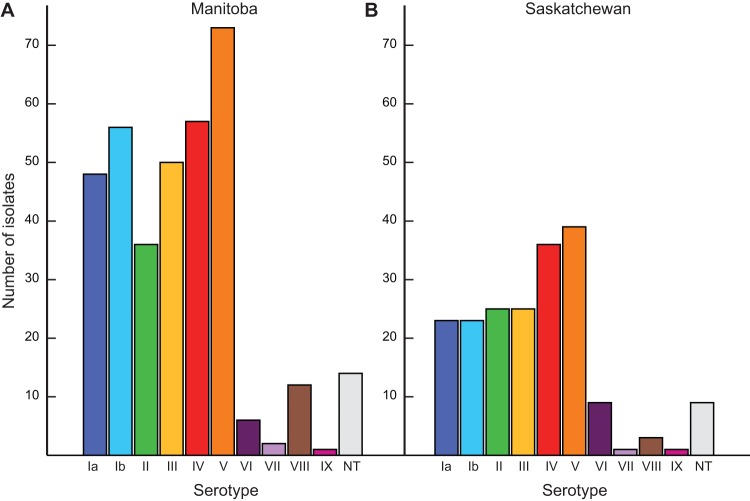
We first serotyped all 549 strains recovered from adult GBS invasive infections in Manitoba and Saskatchewan between 2010 and 2014 (see Table S3 in the supplemental material). In Manitoba, the three most common serotypes were V (73/355, 21%), IV (57/355, 16%), and Ib (56/355, 16%) (Fig. 1A). In Saskatchewan, serotype V was the most prevalent (39/194, 20%) and was followed by serotype IV (36/194, 19%) and serotypes II and III (each 25/194, 13%) (Fig. 1B). Thus, the overall prevalence of serotype IV strains among cases of GBS in adult invasive disease was 16.9% (93/549) during the period under consideration. Although the frequency of isolation of serotype IV GBS strains was relatively constant during each of the 5 years studied, the overall serotype IV GBS prevalence in Manitoba and Saskatchewan was higher than that previously reported elsewhere in North America (2, 4, 11, 18) and greatly exceeded the 6% prevalence recently reported in Toronto, Ontario, over a time period (2010 to 2012) that partially overlaps with the current data set (19). The median age of patients with invasive GBS infection was 60 years in Saskatchewan and Manitoba. The percentage of invasive serotype IV GBS disease in adults aged 18 to 59 was 19.1% (34/178) in Manitoba and 19.1% (18/94) in Saskatchewan, while in older adults (aged ≥60) serotype IV strains represented 13.0% (23/177) of the Manitoba cases and 18% (18/100) of the Saskatchewan cases.
FIG 1.
Distribution of group B Streptococcus (GBS) serotypes among cases of adult invasive disease in the provinces of Manitoba (A) and Saskatchewan (B) during the years 2010 to 2014. Serotype V GBS strains were the most frequently isolated in the two provinces among cases of adult invasive disease (patients ≥18 years old). Strains of serotype IV GBS were the second most frequently isolated in the two provinces (16% of cases in Manitoba and 19% in Saskatchewan).
The vast majority of serotype IV GBS isolates causing adult invasive disease in Manitoba and Saskatchewan are ST459.
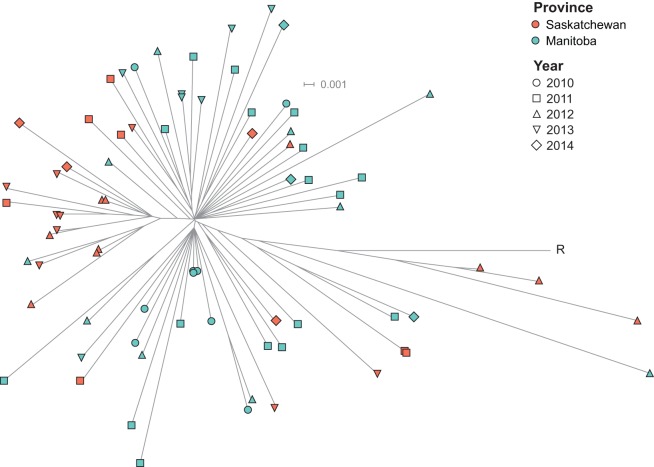
We used Illumina short-reads and the read-mapping-based tool SRST2 (24) to derive MLST information for each isolate directly from WGS data (STs are presented in Table S1 in the supplemental material). The vast majority of isolates belonged to CC1 ST459 (87% or 47/54 serotype IV isolates in Manitoba and 94% or 29/31 in Saskatchewan) (Fig. 2). The remaining isolates included five CC1 strains: two ST3, one ST711 (a novel single locus variant of ST3), one ST196, and one ST710 (a novel single locus variant of ST459). We also identified four CC23 ST452 isolates. Table S4 in the supplemental material lists the number of locus variants between the different STs identified. The proportion of ST459 isolates among invasive serotype IV GBS in the two provinces (89%) was significantly higher than that recently described in Toronto, Ontario, where approximately half of serotype IV isolates were ST459 (19/37 or 51%, P < 0.001) (20).
FIG 2.
Inferred genetic relationships between strains of serotype IV group B Streptococcus (GBS) causing invasive disease in adults in Manitoba and Saskatchewan. A neighbor-joining phylogenetic tree was constructed using the concatenated sequences of the 7 genes (adhP, pheS, tkt, sdhA, glcK, glnA, atr) used in the multilocus sequence typing scheme. Colored circles represent the six different sequence types (STs) identified among serotype IV strains isolated from adult patients in Manitoba and Saskatchewan. Circle size is proportional to the number of isolates (ST3, n = 2; ST196, n = 1; ST452, n = 4; ST459, n = 76; ST710, n = 1; ST711, n = 1).
Antibiotic susceptibility in serotype IV GBS strains.
All serotype IV GBS strains were susceptible to all tested β-lactams and vancomycin. Eighty-four percent (71/85) serotype IV strains were resistant to tetracycline (Table 1). Resistance to tetracycline was observed among strains of all STs except ST452 and ST711. Most ST459 isolates (68/76) contained the tetM gene, but 8 isolates did not and were susceptible to tetracycline (see Table S1 in the supplemental material). One additional tetracycline-susceptible ST459 isolate contained the tetM gene with mutations that are predicted to result in loss of function (see Table S1 in the supplemental material). Erythromycin and clindamycin resistance was observed in 77/85 strains (91%). All but one of the ST459 strains was resistant to clindamycin and erythromycin (Table 1). Using SRST2 (24), we identified that the ermTR gene was present in the genomes of all strains, including the susceptible isolate (see Table S1 in the supplemental material). We later discovered a transposon insertion in the ermTR gene that presumably results in loss of function (data not shown). The ST710 isolate possessed a functional ermTR and was resistant to the two agents. The ST711 isolate was also resistant to clindamycin and erythromycin, and we identified genes ermTR and ermT. On the other hand, ST452, ST3, and ST196 isolates were susceptible to erythromycin and clindamycin.
TABLE 1.
Resistance to selected antimicrobials among serotype IV group B Streptococcus strains of different sequence types
| Clonal complex | Sequence type | Susceptibility of isolates (no.) to:a |
|||||
|---|---|---|---|---|---|---|---|
| Tetracycline |
Erythromycin |
Clindamycin |
|||||
| S | R | S | R | S | R | ||
| 23 | ST452 | 4 | 0 | 4 | 0 | 4 | 0 |
| 1 | ST3 | 0 | 2 | 2 | 0 | 2 | 0 |
| 1 | ST196 | 0 | 1 | 1 | 0 | 1 | 0 |
| 1 | ST459 | 9 | 67 | 1 | 75 | 1 | 75 |
| 1 | ST710 | 0 | 1 | 0 | 1 | 0 | 1 |
| 1 | ST711 | 1 | 0 | 0 | 1 | 0 | 1 |
| Total | 14 | 71 | 8 | 77 | 8 | 77 | |
S, sensitive; R, resistant.
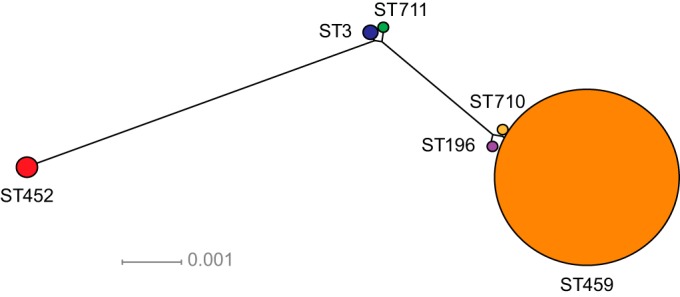
The population structure of ST459 isolates suggests clonal expansion.
Recently, we described that clonal ST459 GBS strains were responsible for more than 50% of serotype IV GBS invasive disease in Toronto, Ontario (20). Inasmuch as our data seem to indicate that ST459 isolates are also emerging in Manitoba and Saskatchewan, we hypothesized that the circulating strains are part of the same clone that is expanding country-wide. To test this hypothesis, we used whole-genome SNP analysis to study the genetic relationships between ST459 isolates causing invasive disease in the three geographical areas. Among the 76 ST459 strains included in the study, we identified a total of 1,140 phylogenetically informative SNP loci relative to the core genome (i.e., the 1.90 Mbp region of the genome that excludes mobile genetic elements) of reference strain NGBS061, isolated in 2010 from an adult in the greater Toronto area. Consistent with the hypothesis, neighbor-joining phylogenetic analysis showed that ST459 isolates are genetically very closely related (Fig. 3), with an average number of 39 SNPs separating any two ST459 strains. No unambiguous temporal clustering was identified, and the pattern of radial divergence is suggestive of clonal expansion. However, despite this limited diversity, we identified at least one clear geographic pattern of diversification, i.e., a conspicuous clade formed mostly by strains isolated in Saskatchewan (Fig. 3, left). SNP analysis also showed that the ST459 strains circulating in Manitoba and Saskatchewan are closely related to the strains described in Toronto (see Fig. S1 in the supplemental material). Furthermore, genome data for three ST459 strains identified in Europe showed a high degree of relatedness to the ST459 clones circulating in Canada, suggesting a recent common ancestor (see Fig. S1 in the supplemental material).
FIG 3.
Inferred phylogenetic relationships between ST459 group B Streptococcus (GBS) strains causing adult disease in Manitoba and Saskatchewan. A neighbor-joining phylogenetic tree was constructed using the concatenated sequences of 1,140 unique (nonredundant) single-nucleotide polymorphic loci relative to ST459 reference strain NGBS061 identified among 76 serotype IV ST459 strains from Manitoba and Saskatchewan. The average number of polymorphisms between each strain and the reference was 59 or 0.003% of the interrogated bases. The radial structure of the tree suggests clonal expansion. No clear temporal grouping was observed. R, reference strain NGBS061.
Large scale recombination differentiates ST459 strains from other CC1 GBS clones historically associated with disease in adults.
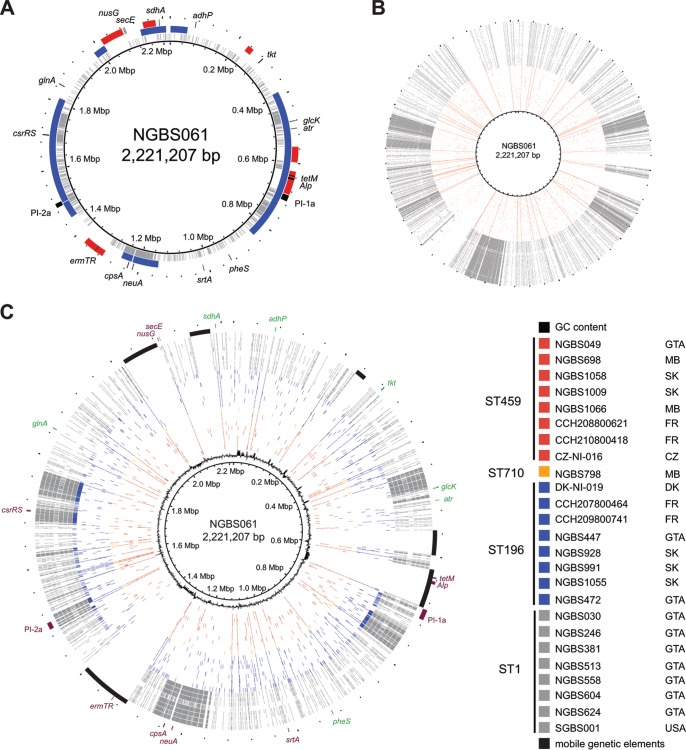
Recently, Flores et al. analyzed a large collection of geographically and temporally disparate serotype V isolates causing bacteremia in nonpregnant adults and discovered that the overwhelming majority of isolates belonged to a single ST1 clone (27). Despite the different capsule, ST1 and ST459 belong to CC1 and share some traits: They are most frequently isolated from nonpregnant adults, are predominantly resistant to tetracycline and, in most cases, to erythromycin and clindamycin, and, overall, the genetic diversity within each population is limited (20, 27, 35). We thus performed genomic comparisons to test the hypothesis that ST459 strains are derived from ST1 strains that have acquired a serotype IV capsule by means of a single recombination event. Consistent with the hypothesis, genome comparisons revealed that serotype IV ST459 and serotype V ST1 strains share a genome backbone, which comprises approximately 50% of their genome. We also discovered that reference strains of serotype V ST1 (strain SGBS001) and serotype IV ST459 (strain NGBS061) differ at a region of approximately 123 kb, which contains the cps genes involved in capsule biosynthesis (Fig. 4A). However, contrary to our hypothesis of a single recombination event but in line with a recent report by Da Cunha et al. (31), we also identified several other discrete areas of genomic divergence between strains of these two STs (Fig. 4A) that likely were also acquired by recombination. Together, these areas of divergence extend over ∼1 Mbp and include genes that have been involved or suggested to be involved in GBS virulence such as csrRS and pili. When we extended our analysis beyond reference strains and compared the genomes of all 76 ST459 strains to those of 76 ST1 strains isolated in Toronto, we observed similar results (Fig. 4B).
FIG 4.
Genome comparisons between ST459 and other CC1 strains causing disease in adults identifies recombination and differences in antimicrobial resistance gene content. (A) Polymorphisms identified in serotype V ST1 strain SGBS001 relative to serotype IV ST459 strain NGBS061 are plotted in gray against the genome of strain NGBS061. Overabundance of polymorphisms at discrete areas of the genome correlated with recombination identified between the two strains using Bayesian analysis of recombination (BRATNextGen), indicated in blue. Genome landmarks, including MLST genes (adhP, atr, tkt, glcK, sdhA, glnA, pheS) are provided for reference. Mobile genetic elements are shown in red. (B) A similar polymorphism analysis was performed using 76 serotype V ST1 strains (gray) and all 76 serotype ST459 (red). Similar genome areas with an overabundance of polymorphisms suggestive of recombination were identified between all ST1 relative the reference strain but not in ST459 strains. (C) Polymorphisms identified relative to strain NGBS061 in 8 selected ST459 are depicted in red. Despite their different geographic origin (three Canadian provinces and two European countries), polymorphisms were evenly distributed across the genome of the reference strain. Strain NGBS0798 (orange), belonging to a novel ST710 (a single locus variant of ST459), shows a similar pattern of polymorphisms. Strains of sequence type ST196 (shown in blue) similarly have relatively few polymorphisms throughout the genome, with the exception of strain NGBS472, which showed a pattern of polymorphisms much more similar to ST1 strains (depicted in gray). In NGBS472 and the ST1 strains, polymorphisms were concentrated in discrete areas of the genome. GTA, greater Toronto area; MB, Manitoba; SK, Saskatchewan; FR, France; CZ, Czech Republic; DK, Denmark.
Acquisition of macrolide and lincosamide resistance by ST459 serotype IV and ST1 serotype V strains contributes to clonal expansion.
As reported (27, 31), resistance to macrolides and lincosamides in ST1 strains is encoded by gene ermB in most strains, while in ST459 isolates the ermTR gene drives resistance (20). In the case of ST459, we identified that ermTR is carried in a mobile genetic element, which in strain NGBS061 is integrated between 1,323,845 and 1,367,722 bp (Fig. 4C; see also Fig. S2 in the supplemental material). BLAST comparisons identified high homology (97% at the nucleotide level) between this mobile genetic element and group A Streptococcus integrative and conjugative element Sp1108 (ICESp1108) (36). In contrast, in ST1 strains, gene ermB is carried by Tn917 integrated into an ICE also carrying gene tetM (31), although some ST1 isolates do contain the ermTR gene. In a small number of these ST1 isolates, ermTR is carried in an ICESp1108-like mobile genetic element (data not shown). Thus, overall, observed differences suggest independent acquisition of resistance to macrolides and lincosamides in ST459 and most ST1 GBS. Interestingly, while these two clonal populations are expanding, other CC1 clones are not. For example, strains of ST196 (a single locus variant of ST459 isolates belonging to CC1) causing invasive disease were found in low numbers in this and previous work (18, 20). None of the ST196 strains carried ermB or ermTR, and all were sensitive to macrolides and lincosamides. Thus, taken together, our results support the hypothesis of clonal expansion being driven by acquisition of resistance against these two classes of antimicrobials (31).
DISCUSSION
Population-based studies have shown that rates of invasive GBS disease have been increasing over the past 25 years in nonpregnant adults, with a significant part of the rise due to serotype V GBS strains (2, 4, 12, 19, 27, 37). In contrast, the emergence of adult serotype IV GBS invasive disease has only recently begun to be recognized. In a longitudinal study in the United States, the proportion of serotype IV GBS isolated from nonpregnant adult patients increased from 0.2% in 1998 to 1999 to 5.7% in 2005 to 2006 (2). In Canada, a country-wide survey conducted in 1996 found no cases of serotype IV in 79 nonpregnant adults (11), whereas more recent data from Toronto, Ontario identified that 6% of the total GBS invasive burden was due to serotype IV strains (19). Here, we report that for the period of 2010 to 2014 serotype IV strains were the second most common cause of adult invasive GBS in the provinces of Manitoba (16%) and Saskatchewan (19%). To our knowledge, this proportion is the highest ever reported for serotype IV GBS adult invasive disease. While we did not characterize the GBS serotype distribution among colonized pregnant women (these studies are under way in our laboratories), others have observed increasing numbers of serotype IV strains among colonizing isolates (17, 18, 38–40). Studies aimed at developing a capsular polysaccharide antigen-based GBS vaccine are ongoing worldwide, with several formulations that do not include serotype IV already under clinical trials (41). Given the emergence of serotype IV observed by us and others (2, 17–20), it seems apparent that conjugate vaccines that are being developed should include polysaccharide and/or associated GBS virulence proteins of serotype IV, or they risk not being completely protective.
WGS has enabled the investigation of large and small scale genetic changes in comprehensive collections of GBS strains, thus permitting enhanced understanding of the diversity of the organism and its ability to acquire novel adaptive traits, including genetic elements encoding antibiotic resistance (20, 27, 31, 42, 43). Recent work that analyzed ST17, ST1, and ST23 strains using WGS found that a few specific GBS clones, rather than a diverse array of strains, account for the vast majority of human GBS disease (31). In line with these findings, one of our key discoveries was that clonal ST459 strains dominate the serotype IV population of strains causing disease in adults. Phylogenetic analysis using the wealth of WGS data discovered expansion of the clone with minimal temporal diversification. Our phylogenetic analysis also indicates that ST459 strains causing invasive disease in Ontario, France, and the Czech Republic (31) also belong to this clone (see Fig. S1 in the supplemental material), suggesting global emergence. It seems apparent that investigation of the factors associated with the dominance of specific GBS clones such as ST459 should be considered in prospective international studies.
Adaptation of unique GBS clonal complexes to a particular niche(s) is not without precedent. In fact, it is widely recognized that the CC17 lineage of GBS (mostly composed of clonal serotype III strains) has adapted to the neonatal niche, particularly late-onset neonatal disease (44). Data from several independent studies suggest that the CC1 genomic background is associated with adult invasive disease (27, 31, 45). Recently, it was shown that the vast majority of serotype V strains causing disease in adults in Houston, TX, and Toronto, Canada, belonged to an ST1 clone with limited genetic diversity (27). Here, we show that serotype IV ST459 clonal strains are also expanding rapidly among the adult invasive infection niche. ST459 strains share a genomic backbone with ST1 strains but present genomic differences attributable to independent events of homologous recombination involving the capsular locus and other areas of the genome encoding known and putative virulence factors.
Almost all serotype IV CC1 strains analyzed here were resistant to tetracycline. It has recently been suggested that GBS clones infecting humans have been selected and fixed by extensive use of tetracycline (31). In addition and although recent data showed that many (up to 30% in some cases) ST1 serotype V strains do not carry genetic determinants conferring resistance to macrolides (27), it has been hypothesized that acquisition of erm-mediated macrolide and lincosamide resistance contributed to the emergence of serotype V ST1 GBS infections in adults (31). Consistent with this hypothesis, our data show that all strains of emerging ST459 GBS carry a copy of gene ermTR, conferring resistance to these antibiotic classes. Although we cannot disregard that clonal expansion may also be driven by other factors, for example, herd immunity or enhanced virulence of the clones that had undergone recombination, it is worth noting that, also consistent with the hypothesis of macrolide resistance acquisition leading to expansion of GBS clones causing disease in adults, strains of other serotype IV CC1 STs, such as ST196, are not expanding among the adult population. The ST196 genomes are either ST1-like or ST459-like (Fig. 4C), and thus, likely contain the same virulence factors as either ST1 or ST459 strains. However, supporting the hypothesis that acquisition of resistance to these antibiotic classes is the main driver of the emergence of selected CC1 GBS clones among adult disease, ST196 strains were sensitive to macrolides and lincosamides. Although the use of erythromycin is no longer recommended in the treatment of GBS infections (4, 11, 18), the use of macrolides for respiratory tract and/or other infections may be contributing to increased selective pressure for resistance in bacterial organisms (46).
In summary, we describe here the emergence of serotype IV among cases of invasive GBS disease in adults. We demonstrate that, for the most part, this emergence is driven by the expansion of a single ST459 clone that caused dozens of infections over a 4-year period in Manitoba and Saskatchewan. The clone is resistant to tetracycline, macrolides, and lincosamides, and strains show limited genetic diversity. Increased surveillance for serotype IV ST459 GBS infections is warranted as the continued expansion of this clone may seriously impact planned GBS vaccination efforts.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dax Torti (Donelly Centre, University of Toronto) and the staff at Public Health Ontario (PHO) Genome Core for Illumina sequencing of our strains. We are grateful to Samir Patel (PHO) and Charles Keown-Stoneman (University of Guelph) for antimicrobial testing of selected GBS isolates and help with statistical analysis, respectively. We are grateful to Jonathan Gubbay (PHO) for critical reading of an earlier version of the manuscript.
This publication made use of the Streptococcus agalactiae MLST website (http://pubmlst.org/sagalactiae/) developed by Keith Jolley and sited at the University of Oxford (47). The development of this site has been funded by the Wellcome Trust.
This work was supported by Public Health Ontario.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01128-15.
REFERENCES
- 1.Baker CJ. 1997. Group B streptococcal infections. Clin Perinatol 24:59–70. [PubMed] [Google Scholar]
- 2.Skoff TH, Farley MM, Petit S, Craig AS, Schaffner W, Gershman K, Harrison LH, Lynfield R, Mohle-Boetani J, Zansky S, Albanese BA, Stefonek K, Zell ER, Jackson D, Thompson T, Schrag SJ. 2009. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990-2007. Clin Infect Dis 49:85–92. doi: 10.1086/599369. [DOI] [PubMed] [Google Scholar]
- 3.Farley MM. 2001. Group B streptococcal disease in nonpregnant adults. Clin Infect Dis 33:556–561. doi: 10.1086/322696. [DOI] [PubMed] [Google Scholar]
- 4.Phares CR, Lynfield R, Farley MM, Mohle-Boetani J, Harrison LH, Petit S, Craig AS, Schaffner W, Zansky SM, Gershman K, Stefonek KR, Albanese BA, Zell ER, Schuchat A, Schrag SJ, Active Bacterial Core surveillance/ Emerging Infections Program Network. 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999-2005. JAMA 299:2056–2065. doi: 10.1001/jama.299.17.2056. [DOI] [PubMed] [Google Scholar]
- 5.Huang PY, Lee MH, Yang CC, Leu HS. 2006. Group B streptococcal bacteremia in non-pregnant adults. J Microbiol Immunol Infect 39:237–241. [PubMed] [Google Scholar]
- 6.Lambertsen L, Ekelund K, Skovsted IC, Liboriussen A, Slotved HC. 2010. Characterisation of invasive group B Streptococci from adults in Denmark 1999 to 2004. Eur J Clin Microbiol Infect Dis 29:1071–1077. doi: 10.1007/s10096-010-0941-z. [DOI] [PubMed] [Google Scholar]
- 7.Ferrieri P, Flores AE. 1997. Surface protein expression in group B streptococcal invasive isolates. Adv Exp Med Biol 418:635–637. doi: 10.1007/978-1-4899-1825-3_148. [DOI] [PubMed] [Google Scholar]
- 8.Cieslewicz MJ, Chaffin D, Glusman G, Kasper D, Madan A, Rodrigues S, Fahey J, Wessels MR, Rubens CE. 2005. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect Immun 73:3096–3103. doi: 10.1128/IAI.73.5.3096-3103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slotved HC, Kong F, Lambertsen L, Sauer S, Gilbert GL. 2007. Serotype IX, a proposed new Streptococcus agalactiae serotype. J Clin Microbiol 45:2929–2936. doi: 10.1128/JCM.00117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison LH, Dwyer DM, Johnson JA. 1995. Emergence of serotype V group B streptococcal infection among infants and adults. J Infect Dis 171:513. doi: 10.1093/infdis/171.2.513. [DOI] [PubMed] [Google Scholar]
- 11.Tyrrell GJ, Senzilet LD, Spika JS, Kertesz DA, Alagaratnam M, Lovgren M, Talbot JA. 2000. Invasive disease due to group B streptococcal infection in adults: results from a Canadian, population-based, active laboratory surveillance study–1996. Sentinel Health Unit Surveillance System site coordinators. J Infect Dis 182:168–173. [DOI] [PubMed] [Google Scholar]
- 12.Lamagni TL, Keshishian C, Efstratiou A, Guy R, Henderson KL, Broughton K, Sheridan E. 2013. Emerging trends in the epidemiology of invasive group B streptococcal disease in England and Wales, 1991–2010. Clin Infect Dis 57:682–688. doi: 10.1093/cid/cit337. [DOI] [PubMed] [Google Scholar]
- 13.Domelier AS, van der Mee-Marquet N, Arnault L, Mereghetti L, Lanotte P, Rosenau A, Lartigue MF, Quentin R. 2008. Molecular characterization of erythromycin-resistant Streptococcus agalactiae strains. J Antimicrob Chemother 62:1227–1233. doi: 10.1093/jac/dkn388. [DOI] [PubMed] [Google Scholar]
- 14.Bergseng H, Afset JE, Radtke A, Loeseth K, Lyng RV, Rygg M, Bergh K. 2009. Molecular and phenotypic characterization of invasive group B Streptococcus strains from infants in Norway 2006-2007. Clin Microbiol Infect 15:1182–1185. doi: 10.1111/j.1469-0691.2009.02789.x. [DOI] [PubMed] [Google Scholar]
- 15.Salloum M, van der Mee-Marquet N, Valentin-Domelier AS, Quentin R. 2011. Diversity of prophage DNA regions of Streptococcus agalactiae clonal lineages from adults and neonates with invasive infectious disease. PLoS One 6:e20256. doi: 10.1371/journal.pone.0020256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaleznik DF, Rench MA, Hillier S, Krohn MA, Platt R, Lee ML, Flores AE, Ferrieri P, Baker CJ. 2000. Invasive disease due to group B Streptococcus in pregnant women and neonates from diverse population groups. Clin Infect Dis 30:276–281. doi: 10.1086/313665. [DOI] [PubMed] [Google Scholar]
- 17.Diedrick MJ, Flores AE, Hillier SL, Creti R, Ferrieri P. 2010. Clonal analysis of colonizing group B Streptococcus, serotype IV, an emerging pathogen in the United States. J Clin Microbiol 48:3100–3104. doi: 10.1128/JCM.00277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrieri P, Lynfield R, Creti R, Flores AE. 2013. Serotype IV and invasive group B Streptococcus disease in neonates, Minnesota, USA, 2000-2010. Emerg Infect Dis 19:551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teatero S, McGeer A, Low DE, Li A, Demczuk W, Martin I, Fittipaldi N. 2014. Characterization of invasive group B Streptococcus from the greater Toronto area, Canada. J Clin Microbiol 52:1441–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teatero S, McGeer A, Li A, Gomes J, Seah C, Demczuk W, Martin I, Wasserscheid J, Dewar K, Melano RG, Fittipaldi N. 2015. Population structure and antimicrobial resistance of invasive serotype IV group B Streptococcus, Toronto, Ontario, Canada. Emerg Infect Dis 21:585–591. doi: 10.3201/eid2014.140759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker CJ, Clark DJ, Barrett FF. 1973. Selective broth medium for isolation of group B Streptococci. Appl Microbiol 26:884–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heelan JS, Struminsky J, Lauro P, Sung CJ. 2005. Evaluation of a new selective enrichment broth for detection of group B Streptococci in pregnant women. J Clin Microbiol 43:896–897. doi: 10.1128/JCM.43.2.896-897.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slotved HC, Elliott J, Thompson T, Konradsen HB. 2003. Latex assay for serotyping of group B Streptococcus isolates. J Clin Microbiol 41:4445–4447. doi: 10.1128/JCM.41.9.4445-4447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome medicine 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones N, Bohnsack JF, Takahashi S, Oliver KA, Chan MS, Kunst F, Glaser P, Rusniok C, Crook DW, Harding RM, Bisharat N, Spratt BG. 2003. Multilocus sequence typing system for group B Streptococcus. J Clin Microbiol 41:2530–2536. doi: 10.1128/JCM.41.6.2530-2536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. [DOI] [PubMed] [Google Scholar]
- 27.Flores AR, Galloway-Pena J, Sahasrabhojane P, Saldana M, Yao H, Su X, Ajami NJ, Holder ME, Petrosino JF, Thompson E, Margarit YRI, Rosini R, Grandi G, Horstmann N, Teatero S, McGeer A, Fittipaldi N, Rappuoli R, Baker CJ, Shelburne SA. 2015. Sequence type 1 group B Streptococcus, an emerging cause of invasive disease in adults, evolves by small genetic changes. Proc Natl Acad Sci U S A 112:6431–6436. doi: 10.1073/pnas.1504725112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nusbaum C, Ohsumi TK, Gomez J, Aquadro J, Victor TC, Warren RM, Hung DT, Birren BW, Lander ES, Jaffe DB. 2009. Sensitive, specific polymorphism discovery in bacteria using massively parallel sequencing. Nat Methods 6:67–69. doi: 10.1038/nmeth.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long SW, Beres SB, Olsen RJ, Musser JM. 2014. Absence of patient-to-patient intrahospital transmission of Staphylococcus aureus as determined by whole-genome sequencing. MBio 5:e01692-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shea PR, Beres SB, Flores AR, Ewbank AL, Gonzalez-Lugo JH, Martagon-Rosado AJ, Martinez-Gutierrez JC, Rehman HA, Serrano-Gonzalez M, Fittipaldi N, Ayers SD, Webb P, Willey BM, Low DE, Musser JM. 2011. Distinct signatures of diversifying selection revealed by genome analysis of respiratory tract and invasive bacterial populations. Proc Natl Acad Sci U S A 108:5039–5044. doi: 10.1073/pnas.1016282108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Da Cunha V, Davies MR, Douarre PE, Rosinski-Chupin I, Margarit I, Spinali S, Perkins T, Lechat P, Dmytruk N, Sauvage E, Ma L, Romi B, Tichit M, Lopez-Sanchez MJ, Descorps-Declere S, Souche E, Buchrieser C, Trieu-Cuot P, Moszer I, Clermont D, Maione D, Bouchier C, McMillan DJ, Parkhill J, Telford JL, Dougan G, Walker MJ, DEVANI Consortium, Holden MT, Poyart C, Glaser P. 2014. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun 5:4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marttinen P, Hanage WP, Croucher NJ, Connor TR, Harris SR, Bentley SD, Corander J. 2012. Detection of recombination events in bacterial genomes from large population samples. Nucleic Acids Res 40:e6. doi: 10.1093/nar/gkr928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI M100–S23 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 35.Sadowy E, Matynia B, Hryniewicz W. 2010. Population structure, virulence factors and resistance determinants of invasive, non-invasive and colonizing Streptococcus agalactiae in Poland. J Antimicrob Chemother 65:1907–1914. doi: 10.1093/jac/dkq230. [DOI] [PubMed] [Google Scholar]
- 36.Brenciani A, Tiberi E, Bacciaglia A, Petrelli D, Varaldo PE, Giovanetti E. 2011. Two distinct genetic elements are responsible for erm(TR)-mediated erythromycin resistance in tetracycline-susceptible and tetracycline-resistant strains of Streptococcus pyogenes. Antimicrob Agents Chemother 55:2106–2112. doi: 10.1128/AAC.01378-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tazi A, Morand PC, Reglier-Poupet H, Dmytruk N, Billoet A, Antona D, Trieu-Cuot P, Poyart C. 2011. Invasive group B streptococcal infections in adults, France (2007-2010). Clin Microbiol Infect 17:1587–1589. doi: 10.1111/j.1469-0691.2011.03628.x. [DOI] [PubMed] [Google Scholar]
- 38.Jannati E, Roshani M, Arzanlou M, Habibzadeh S, Rahimi G, Shapuri R. 2012. Capsular serotype and antibiotic resistance of group B Streptococci isolated from pregnant women in Ardabil, Iran. Iran J Microbiol 4:130–135. [PMC free article] [PubMed] [Google Scholar]
- 39.Kiely RA, Cotter L, Mollaghan AM, Cryan B, Coffey A, Lucey B. 2011. Emergence of group B Streptococcus serotype IV in women of child-bearing age in Ireland. Epidemiol Infect 139:236–238. doi: 10.1017/S0950268810001275. [DOI] [PubMed] [Google Scholar]
- 40.Palmeiro JK, Dalla-Costa LM, Fracalanzza SE, Botelho AC, Da Silva Nogueira K, Scheffer MC, de Almeida Torres RS, de Carvalho NS, Cogo LL, Madeira HM. 2010. Phenotypic and genotypic characterization of group B streptococcal isolates in southern Brazil. J Clin Microbiol 48:4397–4403. doi: 10.1128/JCM.00419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schrag SJ, Verani JR. 2013. Intrapartum antibiotic prophylaxis for the prevention of perinatal group B streptococcal disease: experience in the United States and implications for a potential group B streptococcal vaccine. Vaccine 31(Suppl):D20–D26. doi: 10.1016/j.vaccine.2012.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci U S A 102:13950–13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brochet M, Rusniok C, Couve E, Dramsi S, Poyart C, Trieu-Cuot P, Kunst F, Glaser P. 2008. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc Natl Acad Sci U S A 105:15961–15966. doi: 10.1073/pnas.0803654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Musser JM, Mattingly SJ, Quentin R, Goudeau A, Selander RK. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B Streptococcus) causing invasive neonatal disease. Proc Natl Acad Sci U S A 86:4731–4735. doi: 10.1073/pnas.86.12.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luan SL, Granlund M, Sellin M, Lagergard T, Spratt BG, Norgren M. 2005. Multilocus sequence typing of Swedish invasive group B Streptococcus isolates indicates a neonatally associated genetic lineage and capsule switching. J Clin Microbiol 43:3727–3733. doi: 10.1128/JCM.43.8.3727-3733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glass-Kaastra SK, Finley R, Hutchinson J, Patrick DM, Weiss K, Conly J. 2014. Provincial and temporal variation in macrolide and lincosamide antimicrobial use by outpatients in Canada, 1995 to 2010. Can J Infect Dis Med Microbiol 25:103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.