Abstract
The respiratory mycobiome is an important but understudied component of the human microbiota. Like bacteria, fungi can cause severe lung diseases, but their infection rates are much lower. This study compared the bacterial and fungal communities of sputum samples from a large cohort of 56 adult patients with cystic fibrosis (CF) during nonexacerbation periods and under continuous antibiotic treatment. Molecular fingerprinting based on single-strand conformation polymorphism (SSCP) analysis revealed fundamental differences between bacterial and fungal communities. Both groups of microorganisms were taxonomically classified by identification of gene sequences (16S rRNA and internal transcript spacer), and prevalences of single taxa were determined for the entire cohort. Major bacterial pathogens were frequently observed, whereas fungi of known pathogenicity in CF were detected only in low numbers. Fungal species richness increased without reaching a constant level (saturation), whereas bacterial richness showed saturation after 50 patients were analyzed. In contrast to bacteria, a large number of fungal species were observed together with high fluctuations over time and among patients. These findings demonstrated that the mycobiome was dominated by transient species, which strongly suggested that the main driving force was their presence in inhaled air rather than colonization. Considering the high exposure of human airways to fungal spores, we concluded that fungi have low colonization abilities in CF, and colonization by pathogenic fungal species may be considered a rare event. A comprehensive understanding of the conditions promoting fungal colonization may offer the opportunity to prevent colonization and substantially reduce or even eliminate fungus-related disease progression in CF.
INTRODUCTION
Humans are constantly exposed to environmental fungi, and many body surfaces, e.g., the human upper respiratory tract, are colonized by fungal commensals (1). Already in the 1970s, culturable fungi were observed in >80% of sputum and lung samples (2). A healthy oral mycobiome was recently identified, and the first comparisons with lung transplant recipients were made (3, 4). Apparently, the mycobiome has a significant impact on human health, but our understanding of the underlying mechanisms ruling the mycobiome structure in different body sites and under different pathological conditions lags behind understanding of the bacterial microbiome (5). The same is true for the overall role played by fungal species as part of the human airway microbiome in health and disease (6).
Lung diseases due to respiratory infections are the major cause of death in patients with cystic fibrosis (CF). CF is a systemic inherited metabolic disorder that predisposes the human body to microbial colonization of the airways. This in turn results in tissue damage and then lethal respiratory failure (7). Bacteria, such as Pseudomonas aeruginosa and Staphylococcus aureus, are considered to be responsible for the majority of respiratory tract infections in adult CF patients (8, 9). In addition, fungi exhibit the potential to cause lung diseases, but the mycobiome was only recently addressed in CF microbiome studies (5, 10). Fungi considered pathogens in CF are mainly filamentous species from the classes Sordariomycetes and Eurotiomycetes, namely, Aspergillus fumigatus, Scedosporium apiospermum (S. apiospermum complex, including Pseudallescheria boydii), Rasamsonia argillacea, and Exophiala dermatitidis (8, 11, 12). However, despite high prevalence rates, the numbers of reported fungal lung infections are remarkably low. Therefore, the impact of fungal pathogens and the cause of fungus-related lung disease progression are still matters of debate (13). Fungi are part of the biosphere, and being orders of magnitude smaller than the bacterial microbiome in terms of total cell numbers of the human microbiome, the structure and diversity of the mycobiome is poorly understood (5). Comparative analyses of the community structures between fungi and bacteria in human airways are rare, and comprehensive data sets from large cohort studies are missing. It is our understanding that culture-independent molecular fingerprinting targeting only abundant bacteria and fungi in the individual respiratory specimen will reveal the nature and structure of both communities in a comparable manner. In single-strand conformation polymorphism (SSCP) electrophoresis, identification and detection accuracy of sequence variation consists of two elements: (i) measurable differences in mobility through the gel and (ii) sequence analyses of the individual bands.
Characterization of fungal and bacterial communities in sputum from CF patients has been minimal (10, 14). This study provides the first taxonomic survey of the structure and composition of bacterial and fungal communities in a large cohort of adult CF patients. Prevalence of the most abundant species and dynamics of communities are presented, thereby offering the opportunity to directly compare both parts of the airway microbiome. Conclusions on colonization and infection abilities of fungi in lower respiratory tracts were drawn based on correlations with fungal prevalences in ambient air.
MATERIALS AND METHODS
Patient cohort and sample collection.
Ethical clearance and sputum sample collection from CF patients enrolled in a longitudinal study of microbial diagnostics were described in detail in previous publications (15, 16). In brief, 72 sputum samples were collected in sterile containers from 56 CF patients recruited in the CF ambulatory center of the Hannover Medical School (MHH; Hannover, Germany). This includes a subcohort of 13 patients who provided sputum two (n = 10) or three (n = 3) times within the 2-year sampling period. Patients were between 18 and 51 years old, and a median age of 31 years was calculated for the cohort. The genders were equally represented (48.2% female, 51.8% male). No pulmonary exacerbation was observed by the time of sputum collection. Sputum samples were collected in duplicate and stored at −20°C. All patients were in clinically stable condition during the time of our study. Administration of antibiotics to the CF patients followed the recommendations of the International Standard of Care for adult CF patients.
Sample preparation and DNA extraction.
Microbial DNA from sputum was extracted according to the protocol described by Kramer et al. (16). Briefly, sputum sample preparation consisted of cysteine buffer (2% NaOH, 1.45% sodium citrate, and 0.5% N-acetylcysteine) and enzymatic lysis, including lyticase (Sigma-Aldrich, Germany). DNA extractions were performed with the GeneMatrix tissue and bacteria DNA purification kit (EurX Roboklon; Berlin, Germany).
PCR amplification and preparation of single-strand DNA (ssDNA).
For bacteria, the primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 521R (5′-ACCGTGGCTGCTGGCAC-3′) were used to amplify parts of the 16S rRNA gene (17), including the variable regions V1 to V3 of the small subunit of the ribosome (SSU), with an amplicon size between 459 bp for Nocardia sp. and 505 bp for Veillonella sp. PCR was carried out using 50 ng DNA of the sputum extractions in a final volume of 50 μl, starting with an initial denaturation for 15 min at 95°C. A total of 30 cycles (1 min at 95°C, 40 s at 56°C, and 1 min at 72°C) was followed by a final elongation for 10 min at 72°C. For fungi, a seminested PCR strategy was chosen, using the forward primer internal transcript spacer 1 (ITS1) (5′-TCCGTAGGTGAACCTTGCGG-3′) and the reverse primers ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) (18). Length heterogeneity for the ITS region is known, and amplicon sizes in the current study varied from 134 bp for Yarrowia lipolytica to 475 bp for Candida glabrata (19). The first PCR was carried out using 50 ng DNA from sputum extract in a final volume of 50 μl. By adding 3 μl of MgCl2 (25 mM), PCR efficiency was substantially improved. Starting with an initial denaturation for 15 min at 95°C, 20 cycles (1 min at 95°C, 1 min at 58°C, and 1 min at 72°C) were followed by a final elongation for 10 min at 72°C. Amplification of the final target region, the ITS between the 18S and 5.8S rRNA genes, was achieved by using 1 μl of the previous PCR in a final volume of 50 μl. Initial denaturation for 15 min at 95°C was followed by a total of 30 cycles (30 s at 95°C, 20 s at 62°C, and 30 s at 72°C) and a final elongation for 10 min at 72°C. A total of 1.5 U HotStarTaq DNA polymerase was used for all amplifications (Qiagen Hilden, Germany). For single-strand DNA (ssDNA) preparation, the reverse primers 521R and ITS2 were 5′-biotin labeled, and magnetic streptavidin-coated beads (Promega, Madison, WI) were applied to obtain ssDNA from the PCR amplicons according to Eichler et al. (20).
SSCP fingerprints and sequencing of individual ssDNA bands.
Dried pellets of ssDNA were resuspended in 7 μl of gel loading buffer (95% formamide, 10 mM NaOH, 0.25% bromophenol blue, 0.25% xylene cyanol). After incubation for 10 min at 95°C, the ssDNA samples were stored on ice and loaded onto a nondenaturing polyacrylamide-like gel (0.6× MDE gel solution; Cambrex BioScience, Rockland, ME) for SSCP electrophoresis. SSCP fingerprints of bacterial and fungal DNA were obtained at 20°C and 400 V for 21 h on 20-cm glass plates and at 20°C and 700 V for 20 h DNA on 55-cm glass plates, respectively. The gel was silver stained according to the method described by Bassam et al. (21). All bands with an estimated intensity of ≥0.5% of the total lane were considered for further analysis and excised from the SSCP gel, boiled in Tris buffer (10 mM Tris-HCl, 5 mM MgCl2, 5 mM KCl, 0.1% Triton X-100, pH 9.0), and reamplified according to the conditions described above. Data of only the most abundant species of the bacterial and fungal microbiome (≥0.5% of at least one sample) were considered in this comparative study. Purified amplicons were sequenced by cycle sequencing (ABI Prism BigDye Terminator cycle sequencing ready reaction kit; Applied Biosystems, Foster City, CA). Products were purified again with the BigDye Terminator purification kit (Qiagen Hilden, Germany) and analyzed using capillary electrophoresis with fluorescence detection (ABI Prism 3100 genetic analyzer). More details on the SSCP fingerprints and gene sequencing were published by Eichler et al. (20).
Data processing and phylogenetic analysis.
Phylogenetic analysis of sequences was performed with the NCBI tool BLAST/blastn and the Ribosomal Database Project (RDP) Seqmatch tool (only for bacteria) (22). The closest taxonomic groups were determined by sequence similarity and the height of ssDNA bands in the molecular fingerprints. Operational taxonomic units (OTUs) with ambiguous BLAST results were defined according to the closest shared taxonomic level. Taxonomic classification of fungi followed the outline of fungal phylogeny published by Hibbett et al. (23). For identification at the species level, sequence similarity was defined as a minimum of 97%. Additionally, ssDNA bands with equal running distances were affiliated to the same species. Phylogenetic distances of sequences were calculated with the Jukes-Cantor algorithm and displayed in neighbor-joining trees using MEGA version 5 (24). Species accumulation curves were calculated using AccuCurve 1.0 (25). Incidence-based data sets were applied and permutated 250 times, and the resultant curves were averaged.
Accession numbers.
The complete ITS sequence data set for the identification of fungal OTUs is available in the European Nucleotide Archive (ENA) under accession numbers LN835855to LN836015. Bacterial 16S rRNA sequence data are available in the supplemental material of Kramer et al. (16).
RESULTS
Comparison of fungal and bacterial species richness in the CF cohort.
The prevalence of bacterial and fungal species was assessed in 72 individual sputum samples collected over a period of 2 years from 56 CF patients (sputum from 13 patients was collected repeatedly, at different time points). Molecular fingerprints of the bacterial and fungal communities were generated using SSCP gel electrophoresis, followed by sequencing of all major bands (see Fig. S1 in the supplemental material). In total, 60 fungi and 38 bacteria were considered OTUs, and species affiliation was identified by comparative sequence analysis (see Tables S1 and S2 in the supplemental material). Figure 1 illustrates the individual species counts per sample, showing a median of 4 bacterial and 2 fungal OTUs per sputum. In most of the 72 sputum samples, 3 to 6 bacterial species and 1 to 3 fungal species were observed. For bacteria and fungi, a maximum number of 11 species in one sample was detected. At least 1 bacterial OTU was detected in each sample, whereas 3 sputum samples did not contain any fungal species.
FIG 1.
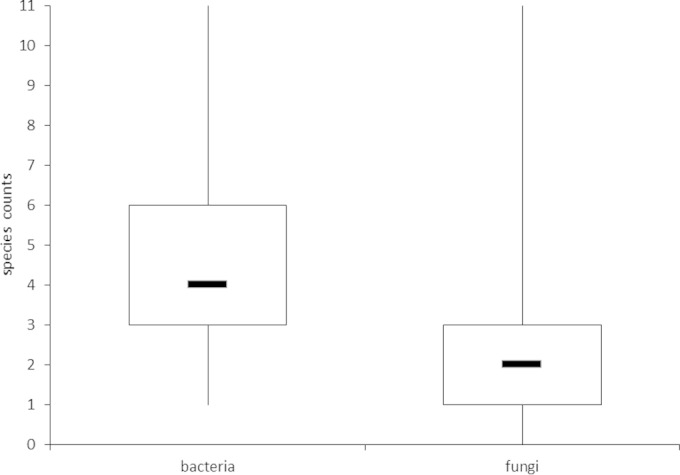
Boxplots of species counts per sample for bacteria and fungi. Boxes are defined by upper and lower quartiles, whereas bars within boxes indicate the median number of OTUs. The ends of the whiskers represent the minimum and maximum species counts found in individual samples.
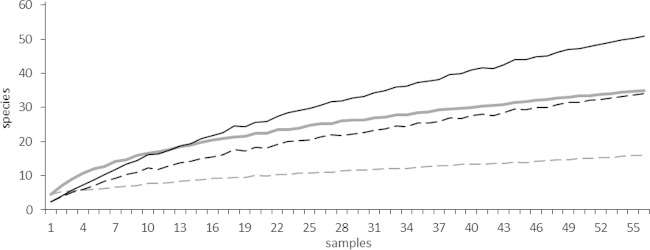
Species accumulation curves were calculated using observed species counts (richness) and counts for species only occurring in one sample (unique species) to determine the total number of bacterial and fungal species in the cohort (Fig. 2). Overall, opposing trends were observed for fungi and bacteria. For bacteria, the curve strongly rose at the beginning, indicating a high number of bacterial species in a relatively small number of samples. The curve flattened with increasing number of samples analyzed, indicating, in the end, only a small incremental increase in species number when more samples were analyzed. For fungi, no flattening of the curve was observed. A small number of samples showed a low incidence of species, but a continuous rise of the curve indicated a steady increase in fungal OTUs. Considering that the number of unique species should be as low as possible in a comprehensive sampling, the trends for the curves of unique species are more informative for the estimation of species richness. For bacteria, a flat curve similarly shows only a small increase in unique species when taking more samples into account, indicating, in the end, no substantial increase of unique species with further sampling. In 56 sputum samples (only the first collected sample from each patient), 16 bacterial species were occurring only in one sample (unique species). Again, a rather different picture was observed for fungi: the curve for unique species showed a continuous rise with increasing sample numbers. After analyzing 56 sputum samples, we found that 34 out of 51 fungal OTUs occurred only once. Overall, the number of bacterial species counts seemed to stabilize beyond 56 samples, whereas new fungal species had to be continuously considered with increasing sample numbers. Species accumulation curves for all collected sputum samples (n = 72) were likewise calculated and confirmed these results (see Fig. S2 in the supplemental material). In conclusion, a greater richness in fungal species in contrast to a relatively smaller number of bacteria was determined in this CF cohort study.
FIG 2.
Species accumulation curves are shown for bacterial and fungal OTUs detected for 56 CF patients. Only the first sample of each patient was considered. Averaged curves show the number of detected species versus the number of samples. Gray curves, bacteria; black curves, fungi; solid lines, observed species counts (richness); dashed lines, unique species counts.
Comparing individual species richness at two time points.
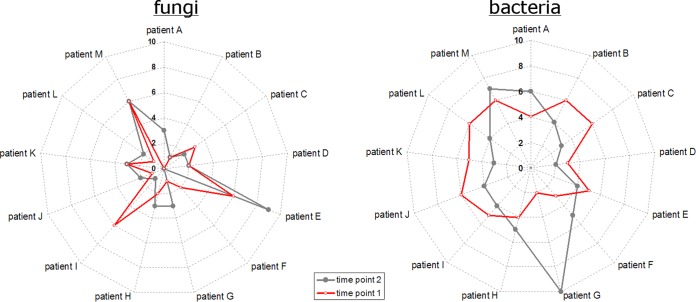
From a subcohort of 13 CF patients, sputum samples were repeatedly collected for 2 years. Comparing the species richness in the individual samples of this subgroup, again a different picture emerged for fungi and bacteria, as illustrated in Fig. 3. Differences were observed among the individual patients and between different time points for the same patients. For fungi, the starlike appearance of the curves indicated major differences in the number of observed OTUs. Individual numbers were rather diverse among the 13 patients and between the different time points of the same patient. In contrast, the circlelike appearance of the radar plots for bacteria indicated a relatively stable number of OTUs among the patients and between the different time points of the same patient. One exception was patient G, in whom samples showed only 2 bacterial OTUs at the first collection, whereas 10 OTUs were detected in the second sputum sample. Neither further correlations between the individual numbers of fungal and bacterial OTUs nor correlations between specific fungal species and bacterial pathogens were observed.
FIG 3.
Differences in individual bacterial (right) and fungal (left) OTU counts for a subcohort of 13 CF patients with repeated sputum collection. For each patient, the first two samples were considered. Radar plots show patients A to M on angular axes. Numbers of detected species are given in radial axes. Red and gray symbols indicate values for first and second sputum samples, respectively. Symbols are accordingly combined with red and gray lines to locate the correct values for each patient.
Special emphasis was given to the dynamics of known fungal pathogens. The CF pathogen Exophiala dermatitidis was found in two samples collected within 3 months from patient C. With two and three fungal OTU counts in total, a relatively constant number of species was observed for this patient. Scedosporium apiospermum was detected in two samples collected within 3 months from patient E, in whom ABPA (allergic bronchopulmonary aspergillosis) was diagnosed. Six fungal species were observed in the first sputum sample and nine in the second sputum sample (Fig. 3). In the latter sample, several filamentous fungi were detected, such as Penicillium sp. (Eurotiomycetes), Fusarium oxysporum (Sordariomycetes), and two Cladosporium OTUs, whereas Aspergillus fumigatus (Eurotiomycetes), another major CF pathogen, was only detected in the first sample. With the exception of this case of ABPA, no relevant correlation between clinical data or other host factors and the presence of single fungal species was observed for the patients in our cohort. In this regard, an overview of host factors, clinical data, and results from microbial diagnoses is given in Table S3 in the supplemental material for all patients of the subcohort.
Prevalent bacteria in the cohort's microbiota.
Bacterial species from nine major taxonomic groups at the class level were observed in the sputum samples of the CF cohort. An overview is given in Table S4 in the supplemental material. Bacilli with 14 OTUs were the most prevalent class, including alpha-hemolytic Streptococci, such as S. salivarius and S. parasanguinis, which were both detected in 57% of the samples. Three Staphylococcaceae species were observed, including S. aureus, which, with 25% positive samples, was the most prevalent non-Streptococcus organism from the class Bacilli. Regarding the individual prevalence of all bacterial species, all other taxonomic classes, with the exception of Bacteroidia, were dominated by a single species. Gammaproteobacteria were observed in 71% of the samples, P. aeruginosa was detected in 69.4%, and Stenotrophomonas maltophilia in 4.1%. Three species of Betaproteobacteria were detected. Achromobacter xylosoxidans was the most prevalent, with 8.3% positive samples, followed by Burkholderia cepacia and Bordetella petrii. Actinobacteria with 6 detected species were dominated by Rothia mucilaginosa, which was found in 43% of the samples. Four taxonomic classes with a single OTU each were observed. Fusobacterium nucleatum (Fusobacteria) was detected in 6.9% of the samples and Veillonella sp. (Negativicutes) in 5.5%. Unique classes found in just one sample were represented by Capnocytophaga gingivalis (Flavobacteria) and an oral TM7 candidate taxon (see Fig. S3 in the supplemental material). The most diverse genera were represented by 9 OTUs each in the genus Prevotella and the genus Streptococcus. More details on the bacterial community composition and its dynamics were published previously (16).
Prevalent fungi in the cohort's microbiota.
Ten classes from the two fungal phyla, Ascomycota and Basidiomycota, were identified (Table 1). One OTU could not be taxonomically assigned, but partial 18S and 5.8S sequences of the amplicon exhibited typical fungal structures and were therefore considered. Additionally, one OTU in each division did not exhibit sufficient similarity to be further assigned to a specific class. The prevalence of each fungal class in the cohort is shown in Fig. S4in the supplemental material, including the number of OTUs per class. Saccharomycetes were the most frequently found fungi, and this class comprised the five most prevalent species. Overall, 89% of samples were positive for 19 OTUs. The polymorphic yeasts Candida albicans and C. dubliniensis were the dominant fungal species in the CF cohort, with 44.4% and 23.6% positive samples, respectively, followed by Saccharomyces cerevisiae, with 19.4%, and C. parapsilosis, with 13.8% positive samples (Table 1). Several filamentous mold species from the classes Dothiomycetes and Eurotiomycetes, including four Aspergillus-like species (A. fumigatus, A. conicus, A. versicolor, and Neosartorya pseudofischeri), were observed in the CF cohort. From Dothiomycetes, 10 species in 25% of the samples were observed. Two OTUs were dominant in this class: the Cladosporium cladosporioides complex, with 11.1%, and the Cladosporium herbarum complex, with 9.7%. The basidiomycete class Pucciniomycetes showed only two species: the yeasts Sporobolomyces roseus and Sporobolomyces ruberrimus, which were detected in 11.1% and 5.5% of sputum samples, respectively (for details on phylogeny, see Fig. S5 and S6 in the supplemental material). These two species were repeatedly detected together in the same samples (n = 3). Within the other taxonomic classes, individual prevalence of the associated OTUs was more equally distributed.
TABLE 1.
Prevalence of major fungal OTUs identified by their ITS sequences in 72 CF sputum samples from 56 patients
| Phylum | Classa (no. of incidences) | Major species | % Positive samples |
|---|---|---|---|
| Ascomycota | Saccharomycetes(101) | Candida albicans | 44.4 |
| Candida dubliniensis | 23.6 | ||
| Saccharomyces cerevisiae | 19.4 | ||
| Candida parapsilosis | 13.8 | ||
| Candida glabrata | 12.5 | ||
| Candida tropicalis | 4.1 | ||
| Cyberlindnera jadinii | 4.1 | ||
| Candida sake | 2.7 | ||
| Dothideomycetes(24) | Cladosporium cladosporioides | 11.1 | |
| Cladosporium herbarum | 9.7 | ||
| Lewia infectoria | 2.7 | ||
| Eurotiomycetes (9) | Exophiala dermatitidisb | 4.1 | |
| Aspergillus fumigatusb | 2.7 | ||
| Sordariomycetes(5) | Scedosporium apiospermumb | 4.1 | |
| Leotiomycetes(3) | Blumeria graminis | 2.7 | |
| Basidiomycota | Pucciniomycetes(12) | Sporobolomyces roseus | 11.1 |
| Sporobolomyces ruberrimus | 5.5 | ||
| Microbotryomycetes(3) | Rhodotorula glutinis | 2.7 |
Table includes only fungal OTUs detected more than once. Fungal OTUs are grouped according to taxonomic classification and ranked according to number of positive samples. The absolute number of detected incidences for each class is given in parentheses (considering all detected OTUs, including unique species). The following classes of Basidiomycetes were represented by unique OTUs only and are therefore not considered in this table: Agaricomycetes, Tremellomycetes, Exobasidiomycetes.
Emerging or acknowledged pathogenic species for CF patients.
Exposure of human airways to fungal spores.
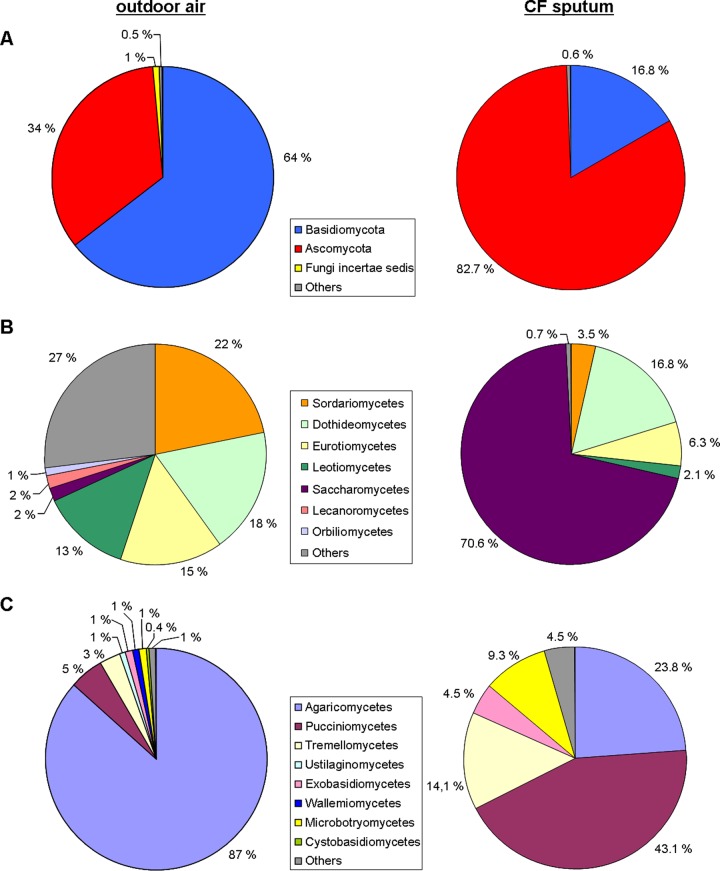
Overall, a high species diversity of fungi was observed in sputum samples from the CF cohort. Both major fungal divisions were represented by several classes and species. Yeastlike fungi showed the highest diversity. Polymorphic and dimorphic fungal species were found, as well as species limited to a single type of growth. Further comparisons with fungal diversity from urban and rural outdoor air from a comparable region in Germany revealed close similarity in terms of taxonomy in both types of samples (data for outdoor air was retrieved from Fröhlich-Nowoisky et al. [26]) (Fig. 4). Although individual numbers were different, fungi with a considerable proportion in outdoor air were also present in the sputum samples of the cohort. While yeasts such as Saccharomycetes or the Pucciniomycetes species were dominant in the sputum samples, Agaricomycetes were dominant in mixed rural and urban air. However, the same fungus classes were found in sputum and in outdoor air.
FIG 4.
Comparison of relative proportion of fungi in outdoor air and in sputum of CF patients. Pie charts for outdoor air are based on data from Fröhlich-Nowoisky et al. (26), and these data were compared with the mycobiome present in 72 samples of the cohort. Each color represents one taxonomic unit further subdivided in the figure. (A) Different phyla. (B) Different classes of Ascomycota. (C) Different classes of Basidiomycota.
DISCUSSION
In this study, we did an extensive comparative analysis of the bacterial and fungal communities of the airway microbiome in a cohort of 56 adult CF patients. In addition to the assessment of structural differences, we presented a taxonomic overview of the observed mycobiomes and correlated our results with fungi present in the air. Thus, this study was a comprehensive effort to evaluate the airway mycobiome in CF and may serve as a basis for more detailed studies on the infection ecology of fungi in the lower respiratory tract. Our findings suggest that colonization and infection of CF airways by pathogenic (filamentous) fungi are exceptional events that are triggered by unknown factors. Collectively, a more comprehensive understanding of the ecological processes during fungal infection may offer new opportunities in CF health care to prevent fungi-related diseases progression.
Our findings demonstrated that there are major differences between the structures of the fungal and bacterial communities. A considerably smaller number of bacterial species was observed in relation to the broad range of fungi. The individual bacterial microbiome is dominated by only a few species with high relative abundances (5), whereas the rare biosphere, including fungi, is known to be more diverse (27). In a simplistic model, Fuhrman (28) reasoned that abundant microorganisms are more likely to be detected, whereas rare microorganisms in one place are more likely below the detection limit in other locations. Consequently, individual communities might partly be diverse, but the more frequently samples in a cohort are analyzed, the more often previously observed species will be detected. Assuming that fungi are generally rare members of the human microbiome, the probability of detection is much lower, resulting in a higher number of observed species, each represented by only a few cells in the individual communities. This is further supported by our observation that, on average, twice as many bacterial species than fungal species were detected per sample, thereby indicating more complex bacterial microbiota in the lungs of individual CF patients. Furthermore, in the current study, bacteria were detected in every sample, whereas this was not always the case for fungi. Collectively, these findings suggest that the fungal communities are dominated by transient members and not by persistent colonizers.
Another important observation for the two communities was that the temporal dynamics of the bacteria and fungi present in CF sputum samples gave very different pictures. Relatively constant numbers of bacterial species were detected, indicating stable communities with permanent colonizers. In contrast, high fluctuations were observed for fungi among the different time points and patients, suggesting dominance of transient community members instead of stable fungal communities. Drawing attention to the dynamics of fungal pathogens, such as A. fumigatus, S. apiospermum, or E. dermatitidis, we observed a persistence of fungal species, which in turn confirmed the accuracy of our detection method. A. fumigatus was observed in the only patient of the cohort with diagnosed ABPA. In the same patient, S. apiospermum was repeatedly detected. This is in agreement with the previously reported association between S. apiospermum and A. fumigatus (13). The pathogen E. dermatitidis was also repeatedly found in sputum from the same patient within 3 months, suggesting permanent colonization by this fungus.
A major strength of this study is the comprehensive taxonomic overview of the mycobiome by a molecular approach that detects all major fungal taxa. By analyzing the phylogeny, we attempted to gain insights into the origin of the microbial community members. For bacteria, oral cavities potentially act as a reservoir and “stepping stone” for subsequent migration to the CF lung (29). The healthy bacterial microflora of the oral cavity recently presented by the Human Microbiome Project revealed the genera Streptococcus, Haemophilus, Actinomyces, and Prevotella to be the most abundant taxa and S. parasanguinis and S. salivarius to be the most dominant species (30). Likewise, the genera Prevotella and Streptococcus were observed to be the most diverse in our study, with high prevalences of S. parasanguinis and S. salivarius. These similarities suggest that the upper and lower respiratory tract microflora are closely related. For CF patients, a notably high species diversity for the families Prevotellaceae and Streptococcaceae was recently described, which matches the outcome of our study (31). CF end-stage lungs have been shown to harbor relatively low numbers of species, and 16S rRNA gene analysis of explanted lung tissue samples revealed the same four phyla of the five bacterial classes with more than one OTU found in the current study: Proteobacteria, Bacteroidia, Firmicutes, and Actinobacteria (32). Great similarity was observed between the dominant OTUs in the current and other studies; therefore, the communities in CF airways are presumably greatly influenced by these species, e.g., P. aeruginosa, S. aureus, Rothia mucilaginosa, and Streptococcus spp. (33–35). Occurrence frequencies of other bacterial pathogens, such as A. xylosoxidans, S. maltophilia, and B. cepacia, were in the ranges reported in the literature (36).
Aerial transport is considered a major mechanism of dispersal for many fungal species. Consequently, we compared our results with those of a previous study of airborne fungal presence in a comparable geographic area (26). All fungal classes identified in the current study were also observed in outdoor air and some, such as Dothideomycetes and Eurotiomycetes, even with similar prevalence (26). Therefore, air can be considered a major source for the diversity of fungal species. Despite the high exposure, the number of reported fungal lung infections is remarkably low (13). However, the real prevalence and role of fungal pathogens in CF patients remain to be elucidated. Apparently, a few species find conditions to colonize the respiratory tracts under certain circumstances. In a first study analyzing the fungal respiratory microbiome, the bronchoalveolar lavage (BAL fluid) of healthy individuals exhibited only low fungal presence, whereas high levels were observed in the BAL fluid of lung transplant recipients, with Candida and Aspergillus being the dominant genera (4). These findings support the hypothesis that fungal colonization of healthy lungs is inhibited by a host defense, including bacterial microbiome-mediated resistance (5). We propose that fungal colonization of the CF lung by environmental fungi can be considered a rare event triggered by exceptional conditions that promote infection and remain to be elucidated.
Although there is no reliable estimate of the fungal load of the human body, nobody is fungus free (5). Particularly, the oral cavity is known to harbor a mycobiome, and the genus Candida represents the most prevalent species of this microbiome (3). In comparing the proportion of fungi in outdoor air and in the sputum of CF patients, yeastlike species were found to make the largest difference. We hypothesize that if the oral cavities potentially act as a reservoir and stepping stone for bacteria (29), similar mechanisms might be involved for migration of yeasts into the airways. Saccharomycetes and particularly Candida were the most frequently detected fungi in our study and are presumably (temporary) colonizers of the CF airways. Likewise, C. albicans was previously shown to be the most isolated fungus from CF patients (37–39), followed by C. dubliniensis in the current study. C. dubliniensis is rarely found in non-CF individuals, which may be due to its ability to display cell surface hydrophobicity-associated adherence at 37°C, which enables the fungus to take advantage of the dehydrated respiratory secretion of these patients (40, 41). The other important Candida species, namely, C. parapsilosis, C. glabrata, and C. tropicalis, were observed with lower occurrence rates, but different CF studies showed a ranking of occurrence surprisingly similar to that found in our study (11, 42). This conformity in observations supports the hypothesis of similar mechanisms for the transport of Candida from oral cavities to the lower respiratory tract, although to a minor extent compared with bacteria. Besides Candida, several others of the observed classes were yeastlike species, including the most prevalent Basidiomycetes, represented by two Sporobolomyces species. Whether the same transport mechanism can likewise be assumed for these and other yeasts is unclear, since nothing is known about their prevalence in the oral microbiome. Sporobolomyces roseus together with Rhodotorula and Cryptococcus species (the latter belonging to the class Tremellomycetes represented by unique OTUs in our study) are considered the most important viable yeasts in outdoor air, although generally only ∼2% of all viable spore counts are yeast species (43). However, the pathogenicity of Candida and other yeastlike species in CF airways remains to be elucidated.
A limitation of our study might be the low number of patients colonized by known fungal pathogens. Future studies focusing on a cohort of patients in the exacerbation phase with reported fungal infections will be necessary to analyze the dynamics and to characterize the ecological changes before and during colonization by pathogenic (filamentous) fungi. This will bring insights into the unknown triggers and factors that promote fungal persistence. Additionally, BAL fluid analyses, the gold standard for respiratory specimens, may bring further insights into the infection ecology of fungi. Despite these limitations, we were able to clearly demonstrate the different community structures of bacteria and fungi in the CF airways. In summary, our study indicates a strong predominance of transient elements in the mycobiome, including pathogens, despite a high exposure of human airways to fungal contaminants. Only yeast species, particularly Candida species, were detected frequently. Collectively, circumstances favoring fungal colonization seem to be rare events, and fungal contaminants are presumably immediately cleared out of the lung. The knowledge derived from this study is crucial, since a deeper understanding of the airway microbiome and its infection ecology is critical to substantially improve CF health care.
Supplementary Material
ACKNOWLEDGMENTS
We thank the volunteers from the CF outpatient clinic of the Medical School Hannover for participating in the study.
We also thank Josefin Koch and Verena Maiberg for excellent assistance with the SSCP fingerprinting.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01094-15.
REFERENCES
- 1.Iliev ID, Underhill DM. 2013. Striking a balance: fungal commensalism versus pathogenesis. Curr Opin Microbiol 16:366–373. doi: 10.1016/j.mib.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullins J, Seaton A. 1978. Fungal spores in lung and sputum. Clin Allergy 8:525–533. doi: 10.1111/j.1365-2222.1978.tb01506.x. [DOI] [PubMed] [Google Scholar]
- 3.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. 2010. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, Bushman FD, Collman RG. 2012. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med 186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huffnagle GB, Noverr MC. 2013. The emerging world of the fungal microbiome. Trends Microbiol 21:334–341. doi: 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. 2013. The role of the lung microbiome in health and disease: a National Heart, Lung, and Blood Institute workshop report. Am J Respir Crit Care Med 187:1382–1387. doi: 10.1164/rccm.201303-0488WS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies JC, Alton EWFW, Bush A. 2007. Cystic fibrosis. BMJ 335:1255–1259. doi: 10.1136/bmj.39391.713229.AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauser AR, Jain M, Bar-Meir M, McColley SA. 2011. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin Microbiol Rev 24:29–70. doi: 10.1128/CMR.00036-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delhaes L, Monchy S, Fréalle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, Sime-Ngando T, Chabé M, Viscogliosi E. 2012. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community–implications for therapeutic management. PLoS One 7:e36313. doi: 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Müller F-MC, Seidler M. 2010. Characteristics of pathogenic fungi and antifungal therapy in cystic fibrosis. Expert Rev Anti Infect Ther 8:957–964. doi: 10.1586/eri.10.72. [DOI] [PubMed] [Google Scholar]
- 12.Giraud S, Favennec L, Bougnoux M-E, Bouchara J-P. 2013. Rasamsonia argillacea species complex: taxonomy, pathogenesis and clinical relevance. Future Microbiol 8:967–978. doi: 10.2217/fmb.13.63. [DOI] [PubMed] [Google Scholar]
- 13.Pihet M, Carrere J, Cimon B, Chabasse D, Delhaes L, Symoens F, Bouchara J-P. 2009. Occurrence and relevance of filamentous fungi in respiratory secretions of patients with cystic fibrosis–a review. Med Mycol 47:387–397. doi: 10.1080/13693780802609604. [DOI] [PubMed] [Google Scholar]
- 14.Willger SD, Grim SL, Dolben EL, Shipunova A, Hampton TH, Morrison HG, Filkins LM, O'Toole GA, Moulton LA, Ashare A, Sogin ML, Hogan DA. 2014. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome 2:40. doi: 10.1186/2049-2618-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kramer R, Sauer-Heilborn A, Welte T, Guzman CA, Höfle MG, Abraham WR. 2015. A rapid method for breath analysis in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis 34:745–751. doi: 10.1007/s10096-014-2286-5. [DOI] [PubMed] [Google Scholar]
- 16.Kramer R, Sauer-Heilborn A, Welte T, Jauregui R, Brettar I, Guzman CA, Höfle MG. 2015. High individuality of respiratory bacterial communities in a large cohort of adult cystic fibrosis patients under continuous antibiotic treatment. PLoS One 10:e0117436. doi: 10.1371/journal.pone.0117436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wos-Oxley ML, Plumeier I, von Eiff C, Taudien S, Platzer M, Vilchez-Vargas R, Becker K, Pieper DH. 2010. A poke into the diversity and associations within human anterior nare microbial communities. ISME J 4:839–851. doi: 10.1038/ismej.2010.15. [DOI] [PubMed] [Google Scholar]
- 18.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen YC, Eisner JD, Kattar MM, Rassoulian-Barrett SL, Lafe K, Bui U, Limaye AP, Cookson BT. 2001. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J Clin Microbiol 39:4042–4051. doi: 10.1128/JCM.39.11.4042-4051.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichler S, Christen R, Höltje C, Westphal P, Bötel J, Brettar I, Mehling A, Höfle MG. 2006. Composition and dynamics of bacterial communities of a drinking water supply system as assessed by RNA- and DNA-based 16S rRNA gene fingerprinting. Appl Environ Microbiol 72:1858–1872. doi: 10.1128/AEM.72.3.1858-1872.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bassam BJ, Caetano-Anollés G, Gresshoff PM. 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83. doi: 10.1016/0003-2697(91)90120-I. [DOI] [PubMed] [Google Scholar]
- 22.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Thorsten Lumbsch H, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai Y-C, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson K-H, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo J-M, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schüssler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao YJ, Zhang N. 2007. A higher-level phylogenetic classification of the fungi. Mycol Res 111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drozd P, Novotny V. 2010. AccuCurve. Version 1 http://prf.osu.cz/kbe/dokumenty/sw/AccuCurve/AccuCurve.xls. [Google Scholar]
- 26.Fröhlich-Nowoisky J, Pickersgill DA, Després VR, Pöschl U. 2009. High diversity of fungi in air particulate matter. Proc Natl Acad Sci U S A 106:12814–12819. doi: 10.1073/pnas.0811003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedrós-Alió C. 2012. The rare bacterial biosphere. Annu Rev Mar Sci 4:449–466. doi: 10.1146/annurev-marine-120710-100948. [DOI] [PubMed] [Google Scholar]
- 28.Fuhrman JA. 2009. Microbial community structure and its functional implications. Nature 459:193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- 29.Rogers GB, Carroll MP, Serisier DJ, Hockey PM, Jones G, Kehagia V, Connett GJ, Bruce KD. 2006. Use of 16S rRNA gene profiling by terminal restriction fragment length polymorphism analysis to compare bacterial communities in sputum and mouthwash samples from patients with cystic fibrosis. J Clin Microbiol 44:2601–2604. doi: 10.1128/JCM.02282-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sibley CD, Grinwis ME, Field TR, Eshaghurshan CS, Faria MM, Dowd SE, Parkins MD, Rabin HR, Surette MG. 2011. Culture enriched molecular profiling of the cystic fibrosis airway microbiome. PLoS One 6:e22702. doi: 10.1371/journal.pone.0022702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudkjøbing VB, Thomsen TR, Alhede M, Kragh KN, Nielsen PH, Johansen UR, Givskov M, Høiby N, Bjarnsholt T. 2011. True microbiota involved in chronic lung infection of cystic fibrosis patients found by culturing and 16S rRNA gene analysis. J Clin Microbiol 49:4352–4355. doi: 10.1128/JCM.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guss AM, Roeselers G, Newton ILG, Young CR, Klepac-Ceraj V, Lory S, Cavanaugh CM. 2011. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J 5:20–29. doi: 10.1038/ismej.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW, Carroll MP, Parkhill J, Bruce KD. 2011. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 5:780–791. doi: 10.1038/ismej.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J, Schloss PD, Kalikin LM, Carmody LA, Foster BK, Petrosino JF, Cavalcoli JD, VanDevanter DR, Murray S, Li JZ, Young VB, LiPuma JJ. 2012. Decade-long bacterial community dynamics in cystic fibrosis airways. Proc Natl Acad Sci U S A 109:5809–5814. doi: 10.1073/pnas.1120577109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cystic Fibrosis Foundation. 2012. Patient registry 2011 annual data report. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 37.Bakare N, Rickerts V, Bargon J, Just-Nübling G. 2003. Prevalence of Aspergillus fumigatus and other fungal species in the sputum of adult patients with cystic fibrosis. Mycoses 46:19–23. doi: 10.1046/j.1439-0507.2003.00830.x. [DOI] [PubMed] [Google Scholar]
- 38.Valenza G, Tappe D, Turnwald D, Frosch M, König C, Hebestreit H, Abele-Horn M. 2008. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros 7:123–127. doi: 10.1016/j.jcf.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Muthig M, Hebestreit A, Ziegler U, Seidler M, Müller F-MC. 2010. Persistence of Candida species in the respiratory tract of cystic fibrosis patients. Med Mycol 48:56–63. doi: 10.3109/13693780802716532. [DOI] [PubMed] [Google Scholar]
- 40.Hazen KC, Wu JG, Masuoka J. 2001. Comparison of the hydrophobic properties of Candida albicans and Candida dubliniensis. Infect Immun 69:779–786. doi: 10.1128/IAI.69.2.779-786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peltroche-Llacsahuanga H, Döhmen H, Haase G. 2002. Recovery of Candida dubliniensis from sputum of cystic fibrosis patients. Mycoses 45:15–18. doi: 10.1046/j.0933-7407.2001.00719.x. [DOI] [PubMed] [Google Scholar]
- 42.Doern GV, Brogden-Torres B. 1992. Optimum use of selective plated media in primary processing of respiratory tract specimens from patients with cystic fibrosis. J Clin Microbiol 30:2740–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rantio-Lehtimäki A. 1985. Mould spores and yeasts in outdoor air. Allergy 40(Suppl 3):S17–S20. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.