Abstract
The possibility of performing genotypic tropism testing (GTT) with proviral DNA (pvDNA) even during suppressed viremia would facilitate the use of CCR5 inhibitors as part of switching, simplification, or intensification strategies. Thus, we aimed to evaluate the tropism concordance between plasma RNA and pvDNA samples and to assess which factors could affect possible discrepancies between the two compartments. GTT was performed using both plasma RNA and pvDNA from 55 sample pairs from drug-experienced patients. Potential differences between the two compartments were evaluated by analyzing coreceptor usage and genetic variability. Paired samples were also stratified in three levels of viremia (<50, 51 to 500, and >500 copies/ml). Overall, Geno2Pheno comparisons of false-positive rates in the two compartments showed good correlation (r = 0.72). A high level of concordance in tropism predictions for the two compartments was found (46/55 sample pairs [83.6%]). Among the 9 sample pairs with discordant tropisms, a larger proportion of pvDNA samples harboring CXCR4/dual-mixed-tropic viruses was found, in comparison with plasma RNA samples (88.9% versus 11.1%; P = 0.0034). Discordant samples were characterized by greater genetic variability than were concordant samples. With stratification of the paired samples according to viremia levels, the prevalence of discordant samples decreased with increasing viremia (<50 copies/ml, 21.4%; 51 to 500 copies/ml, 15.4%; >500 copies/ml, 6.7%; P = 0.2). Our findings confirm that prediction of viral tropism using pvDNA is feasible even in low-level viremia and provides useful information for therapy optimization for patients with low or suppressed viremia.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) requires contact with secondary coreceptors (chemokine receptors) to start infection. CCR5 (R5) and CXCR4 (X4) coreceptors are the major cellular coreceptors involved in the entry of HIV-1 into host cells. Pure R5-tropic and pure X4-tropic viruses can use only CCR5 and CXCR4 coreceptors, respectively, to enter target cells (1, 2), while dual-tropic virus can use both coreceptors. In a virus population, the use of both coreceptors can be due to the presence of either dual-tropic clones or a mixture of pure R5-tropic and X4-tropic clones, or both. This is cumulatively defined as a dual/mixed phenotype (3).
Maraviroc (MVC) is the first R5 antagonist approved for the treatment of patients already experiencing virological failure because of resistance to other antiretroviral agents (4, 5). In the context of virological suppression, regimen switching to MVC is now feasible. Coreceptor usage in virologically suppressed patients can be determined from proviral DNA (pvDNA) obtained from peripheral blood mononuclear cells (PBMCs) (6).
The major viral determinant of HIV-1 tropism is the V3 region of the envelope protein gp120; therefore, analysis of the V3 loop sequence with genotypic tropism testing (GTT) is of clinical relevance (7). GTT is performed using plasma HIV-1 RNA in clinical practice but in some settings, such as cases with very low or undetectable viral loads or a lack of plasma, pvDNA may be considered a potential alternative source of viral genetic material for tropism testing (8). Plasma viral RNA represents currently replicating HIV-1, while pvDNA is intracellular viral DNA that contains the reservoir of archived viruses (9–11). It has been shown that there are high frequencies of X4-tropic or dual-tropic viruses in pvDNA that could persist for long periods (12). Therefore, the possibility of performing GTT with pvDNA would facilitate the use of R5 antagonists as part of switching, simplification, or intensification strategies, even during suppressed viremia (13–15). Recent studies have demonstrated that the performance of V3 region sequencing with both plasma RNA and pvDNA is very successful, even if the performance of pvDNA is higher than that of plasma RNA, especially in the setting of low-level viremia (93.3% and 63%, respectively) (3, 16), confirming that GTT with pvDNA is feasible and significantly relevant. Although the use of MVC after DNA tropism testing has potential, this strategy is not yet recommended in current guidelines, because of the lack of large studies on this topic (6).
Several studies showed a high level of concordance between HIV-1 genotypic coreceptor tropism predictions based on plasma RNA and pvDNA findings (2, 11, 17–19), but the factors that might be associated with the discrepancies between the two compartments have not yet been clearly determined. Moreover, little is known about the tropism concordance between the two compartments in patients with virological suppression (viremia levels of <50 copies/ml).
Thus, the aim of this study was to analyze the degree of GTT concordance between plasma HIV-1 RNA and pvDNA overall and according to different viremia levels (including low or undetectable viremia values). Moreover, the factors that may affect the discrepancies between the two compartments were evaluated.
(This work was presented in part at the 6th National Workshop of the Società Italiana Virologia Medica, Rome, Italy, 10 to 12 December 2014.)
MATERIALS AND METHODS
Patients.
This retrospective study included samples from HIV-1-drug-experienced subjects that had been tested for the V3 loop region of gp120 in both pvDNA and plasma HIV-1 RNA in 2010 to 2014, in three clinical centers in Italy, in routine clinical practice. Patients selected had paired samples with blood sampling dates that were concomitant or within a period of 24 months at most, on the basis of the observation that no evolution is found in samples within 24 months of follow-up monitoring (20). For each sample, the viremia value at the time of genotyping was known.
Viral RNA/DNA extraction.
Viral RNA was extracted from 1 ml of plasma samples, as described previously (21). Proviral DNA was obtained from lymphomonocytic cells after separation from peripheral blood from HIV-1-infected patients with a Ficoll-Hypaque gradient, as described previously (22). HIV-1 DNA was extracted from PBMCs by using a commercially available kit (QIAamp DNA Viral minikit; Qiagen), according to the product specifications.
PCR amplifications and sequencing.
The V3 region of the env gene was amplified using a homemade one-step reverse transcription (RT)-PCR system (for viral RNA samples) or PCR (for viral DNA samples). When the RT-PCR product was not visible on an agarose gel, a nested PCR was performed as described previously (3, 16).
PCR products were then subjected to full-length sequencing in the sense and antisense orientations by using the BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems) and an automated sequencer (ABI 3130). Sequences with a mixture of wild-type and mutant residues at a single position were considered to have a mutation at that position. When mixtures contained at least two different mutations, all of the mutations were considered and the sequences were reported as quasispecies.
Phylogenetic analysis.
A phylogenetic analysis of HIV-1 V3 sequences was performed for the following purposes: (i) to determine viral subtypes, (ii) to check for any cross-contamination, and (iii) to evaluate the proper clustering of the pvDNA and plasma RNA sequences from the same subjects. Briefly, the V3 sequences were aligned with HIV-1 reference sequences of all subtypes (http://www.hiv.lanl.gov). The alignment was edited using the BioEdit program, version 7.0.5.3. MEGA6 (23) was used to perform phylogenetic analysis. A first phylogenetic tree was constructed with a neighbor-joining (NJ) method. Distances were calculated using MEGA6, based on the Kimura 2-parameter (K2P) model (24). The reliability of the branching orders was assessed by bootstrap analysis of 1,000 replicates.
The robustness of the paired sample clusters was further tested using the maximum likelihood (ML) method. This was inferred with the generalized time-reversible (GTR) nucleotide substitution model, with gamma distribution of rates among site heterogeneity, a proportion of invariable sites (GTR + I + Γ5) (25), and 1,000 bootstrap replicates. Paired sample clusters were identified by bootstrap support of >70%. FigTree software was used to draw trees. HIV-1 subtypes were confirmed by analyzing HIV-1 pol (protease/reverse transcriptase) sequences obtained from the same samples, by repeating the phylogenetic analysis and by using the REGA subtype tool (http://www.bioafrica.net/rega-genotype/html/subtypinghiv.html) and the COMET subtype tool (http://comet.retrovirology.lu). To improve the accuracy of recombinant and unique forms, RDP3 software (http://web.cbio.uct.ac.za/∼darren/rdp.html) and Splits Tree software (http://en.bio-soft.net/tree/SplitsTree.html) were used.
Tropism determination.
HIV-1 coreceptor usage was inferred from the V3 nucleotide sequence by using the Geno2Pheno (G2P) algorithm (http://coreceptor.bioinf.mpi-inf.mpg.de). For plasma RNA samples, G2P was set at a false-positive rate (FPR) of 10%, according to guidelines on the clinical management of HIV-1 tropism testing (8). For pvDNA samples, G2P was set at FPR values of both 10%, as reported in several studies (2, 11, 18, 26, 27), and 20%, as recommended by guidelines (8). Thus, sequences with FPRs of <10% from both plasma RNA and pvDNA samples and <20% from pvDNA samples were considered X4/dual-mixed tropic. The concordance of genotypically inferred tropism patterns was evaluated for each sample pair.
Evaluation of V3 variability.
Pairwise genetic distance analyses were conducted using the Tajima-Nei model (28). The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.5). The analysis involved 110 nucleotide sequences. The codon positions included were first plus second plus third plus noncoding. All positions with <95% site coverage were eliminated. That is, <5% alignment gaps, missing data, and ambiguous bases were allowed at any position. Evolutionary analyses were conducted with MEGA6 (23).
Statistical analysis.
All analyses were performed using the statistical software package SPSS (version 19.0) for Windows (SPSS Inc., Chicago, IL). P values of <0.05 were considered statistically significant.
Evaluation of potential differences between pvDNA and RNA samples.
Potential differences among DNA and RNA paired samples were evaluated with the Mann-Whitney test for continuous variables (such as viral loads, CD4+ cell counts, FPRs, genetic distances, numbers of mutated positions, and numbers of quasispecies) and with Fisher's exact test for categorical variables (such as the proportions of samples with X4/dual-mixed-tropic viruses). Spearman's correlation coefficient (r) was used to study the correlation of FPRs determined from genotypic tropism testing of RNA and pvDNA samples.
Evaluation of tropism concordance between compartments in different viremia ranges.
The concordance of tropism results for the two compartments was also evaluated according to different levels of viremia. Paired samples were divided according to viremia ranges of <50 copies/ml, 51 to 500 copies/ml, or >500 copies/ml. This analysis was restricted to the 42 pairs of samples belonging to the same viremia range. The chi-squared test for trend was used to evaluate potential differences in the concordances for the different viremia ranges.
Evaluation of V3 mutation prevalence in DNA and RNA samples according to tropism.
Among the concordant paired samples, the association of V3 mutations with genotypically determined tropism was evaluated. For this analysis, paired samples were divided into two groups (X4/dual-mixed group and R5 group) according to the tropism. The prevalence of each V3 mutation was calculated for each group, and results were compared for pvDNA and RNA samples. Potential differences in the mutation prevalence between DNA and RNA samples were calculated by using Fisher's exact test. All mutations found at the 35 V3 positions with an overall prevalence of ≥10% were evaluated.
RESULTS
Study population.
Samples from a total of 55 HIV-1-drug-experienced subjects were analyzed; 37 patients had concomitant paired samples, while 18 had paired samples collected within a period of 1 to 11 months. Demographic and clinical characteristics of patients at the time of pvDNA genotyping are presented in Table 1.
TABLE 1.
Patient characteristics at the time of pvDNA V3 genotyping
| Characteristica | Value |
|---|---|
| No. of patients | 55 |
| Male (no. [%]) | 38 (69.1) |
| Age (median [IQR]) (yr) | 50 (43–55) |
| Nationality (no. [%]) | |
| Italian | 44 (80.0) |
| African | 6 (10.9) |
| Other | 5 (9.1) |
| HIV exposure (no. [%]) | |
| Heterosexual sex | 17 (30.9) |
| MSM | 12 (21.8) |
| IDU | 6 (10.9) |
| Unknown or other | 20 (36.4) |
| Subtype (no. [%]) | |
| B | 43 (78.2) |
| Non-Bb | 12 (21.8) |
| CD4+ cell count (median [IQR]) (cells/mm3) | |
| At RNA sequencing | 375 (233–615) |
| At DNA sequencing | 375 (262–714) |
| Past drug treatment (no. [%]) | |
| NRTI | 53 (96) |
| NNRTI | 29 (53) |
| PI | 51 (93) |
| INI | 17 (31) |
| Enfuvirtide | 3 (5.4) |
| Maraviroc | 7 (12.7) |
| Treatment status (no. [%]) | |
| Receiving treatment | 48 (87.3) |
| In treatment interruption | 7 (12.7) |
| Current drug treatment (no. [%])c | |
| NRTI | 42 (76.4) |
| NNRTI | 13 (24) |
| PI | 31 (56.4) |
| INI | 10 (18) |
| Enfuvirtide | 5 (9) |
| Maraviroc | 5 (9) |
MSM, men having sex with men; IDU, injection drug use; NRTI, nucleotide reverse transcriptase inhibitor; NNRTI, nonnucleoside reverse transcriptase inhibitor; PI, protease inhibitor; INI, integrase inhibitor.
C (n = 3), BF (n = 3), F1 (n = 2), G (n = 2), BC (n = 1), or CRF06_cpx (n = 1).
Six (10.9%) of 55 patients were undergoing different treatments at the time of RNA genotyping versus the time of pvDNA genotyping.
The population was mainly represented by male (69.1%) and Italian (80%) subjects. Phylogenetic analysis of the V3 sequences revealed that the B subtype was the most prevalent strain (78.2%). The remaining 21.8% of patients were infected with non-B-subtype viruses (3 C, 3 BF, 2 F1, 2 G, 1 BC, and 1 CRF06_cpx). Phylogenetic analysis of the pol sequences confirmed the subtypes determined with V3 sequences.
Overall, at the time of sample collection, median viremia levels were 3.32 log10 copies/ml (interquartile range [IQR], 2.36 to 4.8 log10 copies/ml) and 1.72 log10 copies/ml (IQR, 1.60 to 2.14 log10 copies/ml) for RNA and DNA samples, respectively. In particular, for the nonconcomitant samples, the plasma viral load was significantly higher for RNA samples (medians of 2.10 log10 copies/ml [IQR, 1.71 to 3.0 log10 copies/ml] and 1.60 log10 copies/ml [IQR, 1.37 to 1.98 log10 copies/ml] for RNA and DNA samples, respectively; P = 0.006).
Median CD4+ cells counts at the time of sample collection were 375 cells/mm3 (IQR, 233 to 615 cells/mm3) and 375 cells/mm3 (IQR, 262 to 714 cells/mm3) for RNA and DNA samples, respectively. Of note, no difference in CD4+ cell counts was found between concomitant and nonconcomitant samples (medians of 375 cells/mm3 [IQR, 267 to 590 cells/mm3] and 338 cells/mm3 [IQR, 234 to 682 cells/mm3], respectively; P = 0.92).
Seven patients had received MVC; of those patients, five continued to receive MVC at the time of pvDNA sampling. All other patients were naive to MVC.
V3 genotyping reliability.
By evaluating each cluster with the paired plasma RNA and pvDNA samples, we found that sequences belonging to the same subject showed a high level of homology (bootstrap value of >70%) in 100% of cases, denoting genetic relatedness (see Fig. S1 in the supplemental material).
Characterization of HIV-1 tropism, viremia, and genetic diversity in paired plasma RNA and pvDNA samples.
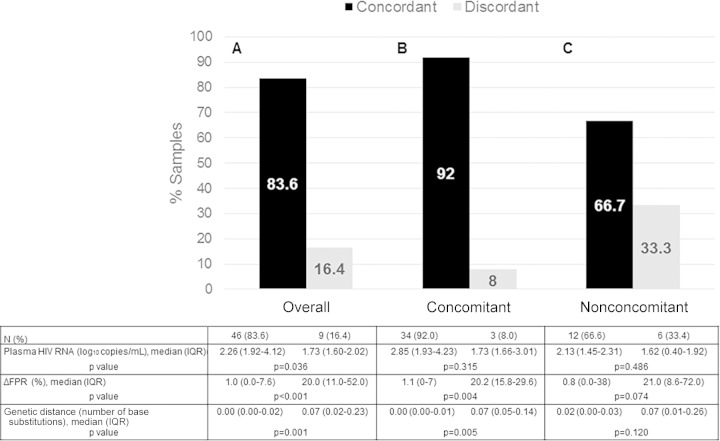
Overall, setting the FPR at 10%, a genotypically inferred tropism concordance between compartments was found for 46/55 samples (83.6%) (Fig. 1A). R5 tropism was predicted for 38/46 sample pairs (82.6%), with FPRs ranging from 13.0% to 96.1%; X4/dual-mixed tropism was predicted for 8/46 samples (17.4%), with FPRs ranging from 0.2% to 8.6%. The concordance of tropism findings was very high among concomitant samples (92.0% [34/37 samples]) (Fig. 1B) and was still considerable among nonconcomitant samples (66.7% [12/18 samples]) (Fig. 1C). The proportion of X4/dual-mixed-tropic viruses in discordant samples was significantly greater for pvDNA samples than for plasma RNA samples, as expected (8/9 samples [88.9%] versus 1/9 samples [11.1%]; P = 0.0034, by Fisher's exact test). Overall, most of the discordant samples had nonconcomitant sampling dates, in a range of 30 to 256 days, and were characterized by greater differences in FPRs between plasma RNA and pvDNA samples than were concordant samples (median ΔFPR values of 20.0% [IQR, 11.0 to 52.0%] versus 1.0% [IQR, 0.0 to 7.6%]; P < 0.001) (Fig. 1A). A higher FPR in discordant samples was also found by repeating the analysis within concomitant and nonconcomitant samples (Fig. 1B and C). Four (44.4%) of nine pairs of discordant samples belonged to patients infected with non-B-subtype viruses (1 BC, 1 G, and 2 F1). Of note, no patient treated with MVC had discordant tropisms in the two compartments.
FIG 1.
Characterization of HIV-1 tropism, viremia, and genetic diversity for paired RNA and DNA samples. Percentages of concordant and discordant samples for the overall paired samples (A), the concomitant paired samples (B), and the nonconcomitant paired samples (C) are shown. Potential differences among paired RNA and DNA samples were evaluated with the Mann-Whitney test.
Overall, among the patients analyzed, lower viremia levels were found for those with discordant samples than for those with concordant samples (medians of 1.73 log10 copies/ml [IQR, 1.60 to 2.02 log10 copies/ml] versus 2.26 log10 copies/ml [IQR, 1.92 to 4.12 log10 copies/ml]; P = 0.036) (Fig. 1A). This difference was maintained even when concomitant and nonconcomitant samples were analyzed separately (Fig. 1B and C). No difference in CD4+ cell counts was found between concordant and discordant samples (medians of 379 cells/mm3 [IQR, 264 to 637 cells/mm3] and 358 cells/mm3 [IQR, 240 to 691 cells/mm3], respectively; P = 0.98).
To evaluate the pairwise genetic variability between the two compartments, genetic distances, in terms of the number of base substitutions per site, were calculated (Fig. 1). Data showed greater genetic variability in discordant samples than in concordant samples (median genetic distances of 0.00 [IQR, 0.00 to 0.02] versus 0.07 [IQR, 0.02 to 0.23]; P = 0.001) (Fig. 1A).
Numbers of quasispecies and mutated positions were also analyzed. Substantial similarity among the numbers of mutated positions between plasma RNA and pvDNA was found (medians of 7 positions [IQR, 5 to 9 positions] and 6 positions [IQR, 5 to 8 positions], respectively; P = 0.82), while, as expected, the number of quasispecies increased in pvDNA (medians of 0 quasispecies [IQR, 0 to 4 quasispecies] for RNA sequences and 4 quasispecies [IQR, 0 to 8 quasispecies] for DNA sequences; P = 0.008).
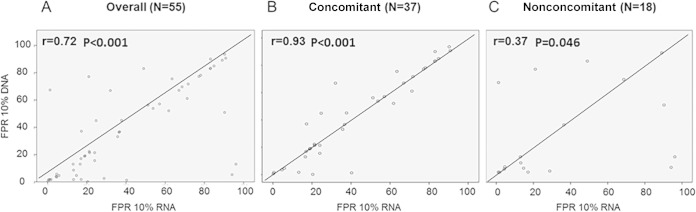
Comparison of the overall G2P FPRs in plasma RNA and pvDNA compartments showed good correlation (r = 0.72; P < 0.001) (Fig. 2A). A very good correlation was found for the 37 concomitant samples (r = 0.93; P < 0.001) (Fig. 2B), while a lower correlation was found among the 18 nonconcomitant samples (r = 0.37; P = 0.046) (Fig. 2C), which was explained by a large difference in FPRs between the 2 compartments (median of 7.3% [IQR, 0.2 to 43.5%]).
FIG 2.
Scatterplots for comparisons between plasma RNA and pvDNA V3-based coreceptor tropism predictions. Tropism predictions were performed with the Geno2Pheno (G2P) coreceptor tool, and results are expressed as the false-positive rate (FPR). Correlation coefficients are shown for the overall samples (A), for the 37 concomitant samples (B), and for the 18 nonconcomitant samples (C).
With the FPR set at 20% for pvDNA samples, as suggested by the European guidelines on the clinical management of HIV-1 tropism testing (8), the number of discordant pairs of samples increased from nine to 16 (29%). Of note, the difference between FPRs for the two compartments for the additional seven discordant couples with FPRs in the range 10 to 20% was very low (median ΔFPR of 0.0% [IQR, 0.0 to 8.4%]).
Evaluation of tropism concordance between compartments at different viremia levels.
The concordance of tropism findings for the two compartments was evaluated according to the viremia levels. Among the 42 sample pairs with the same viremia levels, 33.3%, 31%, and 35.7% had viremia levels of <50, 51 to 500, and >500 copies/ml, respectively. With stratification of the paired samples according to the viremia levels, the prevalence of discordant samples increased with decreasing viremia, although without statistical significance, probably due to the small numbers of samples analyzed for each group (<50 copies/ml, 21.4%; 51 to 500 copies/ml, 15.4%; >500 copies/ml, 6.7%; P = 0.2, by chi-squared test for trend) (Fig. 3). In considering the tropism of concordant paired samples (36/42 samples [85.7%]), the prevalence of X4/dual-mixed tropism was higher for viremia values of <50 copies/ml than for viremia values of >50 copies/ml (4/11 samples [36.4%] versus 2/25 samples [8.0%]; P = 0.052).
FIG 3.
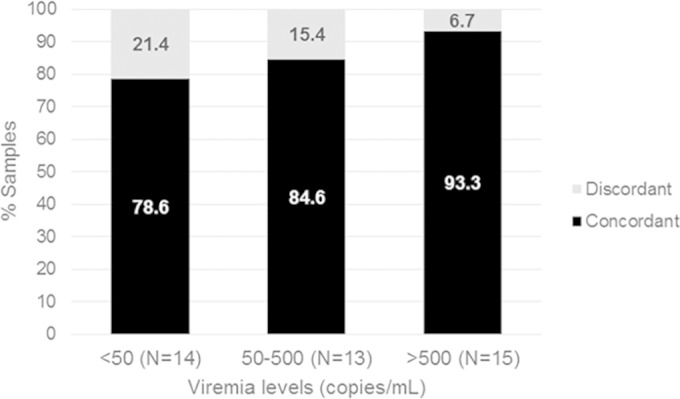
Evaluation of tropism concordance between paired plasma RNA and pvDNA samples according to the viremia level, among the 42 sample pairs with the same viremia levels. P = 0.2, by chi-squared test for trend.
Evaluation of V3 mutation prevalence in pvDNA and plasma RNA samples according to tropism.
Among the concordant paired samples (n = 46), the prevalence of V3 mutations was evaluated in X4/dual-mixed and R5 paired samples. Among the X4/dual-mixed paired samples (n = 8), the V3 mutations S11G (61.5%), H13R (37.5%), E25R (50.0%), Q32K (37.5%), and H34Y (50.0%) were found with greater prevalence in pvDNA samples. In contrast, the V3 mutations N5D (37.5%), K10R (62.5%), H13S (37.5%), T22A (75.0%), G24R (37.5%), E25K (62.5%), and I26V (62.5%) were mostly prevalent in plasma RNA, suggesting different viral evolution in the two compartments (see Fig. S2A in the supplemental material). These differences in mutation prevalence did not have any statistical significance, however, probably due to the small number of samples. No difference between the two compartments in the mutation prevalence was found in R5-tropic concordant samples (n = 38) (see Fig. S2B in the supplemental material).
DISCUSSION
In this study, we aimed at evaluating the genotypically inferred tropism in paired plasma HIV-1 RNA and pvDNA samples from 55 HIV-1-infected patients. In particular, we characterized the tropism concordance between the two compartments overall and according to viremia levels. Patients with viremia levels of <50 copies/ml in both plasma RNA and pvDNA samples were also analyzed, whereas the great majority of the previous studies did not compare the two compartments in the undetectable range of viremia but focused their attention on plasma HIV-1 RNA samples from patients with detectable viral loads (11, 18, 29, 30).
We found a high level of concordance between the two compartments (∼84%), confirming findings described previously in other studies (2, 11, 17, 18). With the FPR set at 10%, concordance was found mainly for R5-tropic viruses (83%). The overall FPR values were linearly correlated between the two compartments (r = 0.72; P < 0.001). In particular, the FPR correlation increased when we considered only the concomitant samples (r = 0.93; P < 0.001), while it decreased when we considered nonconcomitant samples (r = 0.37; P = 0.046). Discordance was attributable mainly to X4/dual-mixed prediction from pvDNA samples and R5 prediction from plasma RNA samples, as found in other studies (10–12, 26, 31). These findings indicate that, although plasma and PBMCs often provide similar tropism reports (32), tropism testing with PBMCs may not produce biological results equivalent to those from plasma, because the structures of viral populations vary between compartments (30), and can be useful in excluding slightly more individuals from treatment with a CCR5 inhibitor. In the evaluation of the genetic variability between the two compartments, a greater median number of quasispecies in pvDNA was found with either concomitant or nonconcomitant samples, with a consistent increase in quasispecies in the latter group. These findings confirm that V3 pvDNA and RNA populations can be different, suggesting that archived viruses may not always correspond to the viruses most prevalent in plasma (33). However, the presence of a larger number of quasispecies could influence the correct algorithmic interpretation, overestimating the presence of discordant paired samples.
Most of the discordant paired samples found in our study (66.7%) had nonconcomitant sampling dates (30 to 256 days apart), suggesting viral evolution in a short period. Therefore, simultaneous sample collection should be carried out for proper comparison of the two compartments. Tropism discordance between the two compartments could even be due to subtype. Indeed, 44.4% of discordant pairs were from patients infected with non-B-subtype viruses. In this regard, it should be noted that estimations of HIV-1 tropism using bioinformatics tools based on V3 sequences are worse for testing non-B strains than for testing B strains. In particular, G2P is a tool that mainly overestimates X4 predictions for non-B subtypes (34). Therefore, further optimization of genotypic methods is needed, and larger studies to determine the utility of such methods for tropism prediction for diverse HIV-1 non-B subtypes are necessary, especially in light of increased circulation of non-B strains in areas with previously homogeneous B strains, such as several European countries, including Italy (35). In general, discordant samples were found mostly at lower viremia levels, compared with concordant samples (median plasma HIV RNA levels of 1.73 log10 copies/ml [IQR, 1.60 to 2.02 log10 copies/ml] versus 2.26 log10 copies/ml [IQR, 1.92 to 4.12 log10 copies/ml]). With stratification of the paired samples according to the viremia level (<50, 51 to 500, or >500 copies/ml), the prevalence of discordant samples decreased with increasing levels of viremia (21.4%, 15.4%, and 6.7%, respectively). Similar results were found by Verhofstede and colleagues (11). These findings suggest that, in patients with low-level viremia, viral quasispecies in the plasma compartment did not fully represent the proviral reservoir.
By analyzing the most prevalent V3 mutations (overall prevalence of ≥10%), we confirmed that some mutations are associated with X4 (e.g., E25K [more prevalent in plasma RNA] and E25R, Q32K, and H34Y [more prevalent in pvDNA]) or R5 (i.e., F20L, Y21F, T22A, and E25D [similar prevalence rates in the two compartments]) coreceptor usages in both compartments (36, 37). The main limitations of our study were the small number of paired samples included and the heterogeneity of the population (such as the presence of nonconcomitant and non-B-subtype samples), which could have yielded overestimations of the differences between the two compartments.
In conclusion, our findings confirm that GTT produces a relatively good correlation between pvDNA and RNA tropism estimations, suggesting that prediction of viral tropism using PBMC DNA is feasible. Among discordant samples, the presence of X4/dual-mixed-tropic viruses in pvDNA in the setting of low viremia levels suggests that genotypic tropism determination with pvDNA provides useful information for the optimization of therapy for patients with low-level or suppressed viremia. Further studies are needed to determine whether the use of MVC for patients with virological suppression and with discordant tropisms in the two compartments (R5 in plasma RNA and X4/dual-mixed in pvDNA) could be effective over time.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the European Commission Framework 7 Programme (Collaborative HIV and Anti-HIV Drug Resistance Network, Integrated Project 223131), the Italian Ministry of Health (Progetto Ricerca Finalizzata, grant RF-2009-1539999), the Italian Ministry of Instruction, University, and Research (Bandiera InterOmics Protocollo PB05 1°), and an unrestricted grant from the AVIRALIA Foundation.
We gratefully thank Daniele Armenia, Andrea Biddittu, Massimiliano Bruni Stefania Carta, Fabio Continenza, Roberta D'Arrigo, Domenico Di Carlo, Domenico Di Pinto, Valentina Fedele, Federica Forbici, Sara Giannella, Tania Guenci, Daniele Pizzi, Marzia Romani, Valentina Serafini, and Francesca Stazi for sequencing and data management.
We have no conflicts of interest directly related to this manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00893-15.
REFERENCES
- 1.Berger EA. 1998. HIV entry and tropism: when one receptor is not enough. Adv Exp Med Biol 452:151–157. [PubMed] [Google Scholar]
- 2.Seclén E, Del Mar González M, De Mendoza C, Soriano V, Poveda E. 2010. Dynamics of HIV tropism under suppressive antiretroviral therapy: implications for tropism testing in subjects with undetectable viraemia. J Antimicrob Chemother 65:1493–1496. doi: 10.1093/jac/dkq156. [DOI] [PubMed] [Google Scholar]
- 3.Svicher V, D'Arrigo R, Alteri C, Andreoni M, Angarano G, Antinori A, Antonelli G, Bagnarelli P, Baldanti F, Bertoli A, Borderi M, Boeri E, Bonn I, Bruzzone B, Callegaro AP, Cammarota R, Canducci F, Ceccherini-Silberstein F, Clementi M, Monforte AD, De Luca A, Di Biagio A, Di Gianbenedetto S, Di Perri G, Di Pietro M, Fabeni L, Fadda G, Galli M, Gennari W, Ghisetti V, Giacometti A, Gori A, Leoncini F, Maggiolo F, Maserati R, Mazzotta F, Micheli V, Meini G, Monno L, Mussini C, Nozza S, Paolucci S, Parisi S, Pecorari M, Pizzi D, Quirino T, Re MC, Rizzardini G, Santangelo R, Soria A, Stazi F, Sterrantino G, Turriziani O, Viscoli C, Vullo V, Lazzarin A, Perno CF. 2010. Performance of genotypic tropism testing in clinical practice using the enhanced sensitivity version of Trofile as reference assay: results from the OSCAR Study Group. New Microbiol 33:195–206. [PubMed] [Google Scholar]
- 4.Sayana S, Khanlou H. 2009. Maraviroc: a new CCR5 antagonist. Expert Rev Anti Infect Ther 7:9–19. doi: 10.1586/14787210.7.1.9. [DOI] [PubMed] [Google Scholar]
- 5.MacArthur RD, Novak RM. 2008. Reviews of anti-infective agents: maraviroc: the first of a new class of antiretroviral agents. Clin Infect Dis 47:236–241. doi: 10.1086/589289. [DOI] [PubMed] [Google Scholar]
- 6.DHHS Panel on Antiretroviral Guidelines for Adults and Adolescents. 2015. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. National Institutes of Health, Bethesda, MD: http://aidsinfo.nih.gov/contentfiles/lvguidelines/adultandadolescentgl.pdf. [Google Scholar]
- 7.Harrigan PR, Geretti AM. 2011. Genotypic tropism testing: evidence-based or leap of faith? AIDS 25:257–264. doi: 10.1097/QAD.0b013e32834113f9. [DOI] [PubMed] [Google Scholar]
- 8.Vandekerckhove LP, Wensing AM, Kaiser R, Brun-Vézinet F, Clotet B, De Luca A, Dressler S, Garcia F, Geretti AM, Klimkait T, Korn K, Masquelier B, Perno CF, Schapiro JM, Soriano V, Sönnerborg A, Vandamme AM, Verhofstede C, Walter H, Zazzi M, Boucher CA. 2011. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect Dis 11:394–407. doi: 10.1016/S1473-3099(10)70319-4. [DOI] [PubMed] [Google Scholar]
- 9.Soulié C, Fourati S, Lambert-Niclot S, Malet I, Wirden M, Tubiana R, Valantin MA, Katlama C, Calvez V, Marcelin AG. 2010. Factors associated with proviral DNA HIV-1 tropism in antiretroviral therapy-treated patients with fully suppressed plasma HIV viral load: implications for the clinical use of CCR5 antagonists. J Antimicrob Chemother 65:749–751. doi: 10.1093/jac/dkq029. [DOI] [PubMed] [Google Scholar]
- 10.Frange P, Galimand J, Goujard C, Deveau C, Ghosn J, Rouzioux C, Meyer L, Chaix ML. 2009. High frequency of X4/DM-tropic viruses in PBMC samples from patients with primary HIV-1 subtype-B infection in 1996–2007: the French ANRS CO06 PRIMO Cohort Study. J Antimicrob Chemother 64:135–141. doi: 10.1093/jac/dkp151. [DOI] [PubMed] [Google Scholar]
- 11.Verhofstede C, Brudney D, Reynaerts J, Vaira D, Fransen K, De Bel A, Seguin-Devaux C, De Wit S, Vandekerckhove L, Geretti AM. 2011. Concordance between HIV-1 genotypic coreceptor tropism predictions based on plasma RNA and proviral DNA. HIV Med 12:544–552. doi: 10.1111/j.1468-1293.2011.00922.x. [DOI] [PubMed] [Google Scholar]
- 12.Verhofstede C, Vandekerckhove L, Eygen VV, Demecheleer E, Vandenbroucke I, Winters B, Plum J, Vogelaers D, Stuyver L. 2009. CXCR4-using HIV type 1 variants are more commonly found in peripheral blood mononuclear cell DNA than in plasma RNA. J Acquir Immune Defic Syndr 50:126–136. doi: 10.1097/QAI.0b013e31819118fa. [DOI] [PubMed] [Google Scholar]
- 13.Obermeier M, Symons J, Wensing AM. 2012. HIV population genotypic tropism testing and its clinical significance. Curr Opin HIV AIDS 7:470–477. doi: 10.1097/COH.0b013e328356eaa7. [DOI] [PubMed] [Google Scholar]
- 14.Vitiello P, Brudney D, MacCartney M, Garcia A, Smith C, Marshall N, Johnson M, Geretti AM. 2012. Responses to switching to maraviroc-based antiretroviral therapy in treated patients with suppressed plasma HIV-1-RNA load. Intervirology 55:172–178. doi: 10.1159/000332023. [DOI] [PubMed] [Google Scholar]
- 15.Swenson LC, Dong WW, Mo T, Demarest J, Chapman D, Ellery S, Heera J, Valdez H, Poon AF, Harrigan PR. 2013. Use of cellular HIV DNA to predict virologic response to maraviroc: performance of population-based and deep sequencing. Clin Infect Dis 56:1659–1666. doi: 10.1093/cid/cit105. [DOI] [PubMed] [Google Scholar]
- 16.Svicher V, Alteri C, Montano M, D'Arrigo R, Andreoni M, Angarano G, Antinori A, Antonelli G, Allice T, Bagnarelli P, Baldanti F, Bertoli A, Borderi M, Boeri E, Bon I, Bruzzone B, Callegaro AP, Capobianchi MR, Carosi G, Cauda R, Ceccherini-Silberstein F, Clementi M, Chirianni A, Colafigli M, D'Arminio Monforte A, De Luca A, Di Biagio A, Di Nicuolo G, Di Perri G, Di Pietro M, Di Santo F, Fabeni L, Fadda G, Galli M, Gennari W, Ghisetti V, Giacometti A, Gori C, Gori A, Gulminetti R, Leoncini F, Maffongelli G, Maggiolo F, Manca G, Gargiulo F, Martinelli C, Maserati R, Mazzotta F, Meini G, Micheli V, Monno L, Mussini C, Narciso P, Nozza S, Paolucci S, Pal G, Parisi S, Parruti G, Pignataro AR, Pollicita M, Quirino T, Re MC, Rizzardini G, Santangelo R, Scaggiante R, Sterrantino G, Turriziani O, Vatteroni ML, Vecchi L, Viscoli C, Vullo V, Zazzi M, Lazzarini A, Perno CF. 2012. Performance of genotypic tropism testing on proviral DNA in clinical practice: results from the DIVA Study Group. New Microbiol 35:17–25. [PubMed] [Google Scholar]
- 17.Paar C, Geit M, Stekel H, Berg J. 2011. Genotypic prediction of human immunodeficiency virus type 1 tropism by use of plasma and peripheral blood mononuclear cells in the routine clinical laboratory. J Clin Microbiol 49:2697–2699. doi: 10.1128/JCM.00336-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta S, Neogi U, Srinivasa H, Shet A. 2013. High concordance of genotypic coreceptor prediction in plasma-viral RNA and proviral DNA of HIV-1 subtype C: implications for use of whole blood DNA in resource-limited settings. J Antimicrob Chemother 68:2003–2006. doi: 10.1093/jac/dkt138. [DOI] [PubMed] [Google Scholar]
- 19.Saracino A, Monno L, Punzi G, Cibelli DC, Tartaglia A, Scudeller L, Brindicci G, Lagioia A, Scotto G, Angarano G. 2007. HIV-1 biological phenotype and predicted coreceptor usage based on V3 loop sequence in paired PBMC and plasma samples. Virus Res 130:34–42. doi: 10.1016/j.virusres.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Palmisano L, Giuliano M, Galluzzo CM, Amici R, Andreotti M, Weimer LE, Pirillo MF, Fragola V, Bucciardini R, Vella S. 2009. The mutational archive in proviral DNA does not change during 24 months of continuous or intermittent highly active antiretroviral therapy. HIV Med 10:477–481. doi: 10.1111/j.1468-1293.2009.00715.x. [DOI] [PubMed] [Google Scholar]
- 21.Santoro MM, Fabeni L, Armenia D, Alteri C, Di Pinto D, Forbici F, Bertoli A, Di Carlo D, Gori C, Carta S, Fedele V, D'Arrigo R, Berno G, Ammassari A, Pinnetti C, Nicastri E, Latini A, Tommasi C, Boumis E, Petrosillo N, D'Offizi G, Andreoni M, Ceccherini-Silberstein F, Antinori A, Perno CF. 2014. Reliability and clinical relevance of the HIV-1 drug resistance test in patients with low viremia levels. Clin Infect Dis 58:1156–1164. doi: 10.1093/cid/ciu020. [DOI] [PubMed] [Google Scholar]
- 22.Aquaro S, Perno CF. 2005. Assessing the relative efficacy of antiretroviral activity of different drugs on macrophages. Methods Mol Biol 304:445–453. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 25.Tavaré S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci 17:57–86. [Google Scholar]
- 26.Bon I, Turriziani O, Musumeci G, Clò A, Montagna C, Morini S, Calza L, Gibellini D, Antonelli G, Re MC. 2015. HIV-1 coreceptor usage in paired plasma RNA and proviral DNA from patients with acute and chronic infection never treated with antiretroviral therapy. J Med Virol 87:315–322. [DOI] [PubMed] [Google Scholar]
- 27.Parisi SG, Andreoni C, Sarmati L, Boldrin C, Buonomini AR, Andreis S, Scaggiante R, Cruciani M, Bosco O, Manfrin V, d'Ettorre G, Mengoli C, Vullo V, Palù G, Andreoni M. 2011. HIV coreceptor tropism in paired plasma, peripheral blood mononuclear cell, and cerebrospinal fluid isolates from antiretroviral-naïve subjects. J Clin Microbiol 49:1441–1445. doi: 10.1128/JCM.02564-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tajima F, Nei M. 1984. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol 1:269–285. [DOI] [PubMed] [Google Scholar]
- 29.Brown J, Burger H, Weiser B, Pollard RB, Li XD, Clancy LJ, Baumann RE, Rogers AA, Hamdan HB, Pesano RL, Kagan RM. 2014. A genotypic HIV-1 proviral DNA coreceptor tropism assay: characterization in viremic subjects. AIDS Res Ther 11:14. doi: 10.1186/1742-6405-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pou C, Codoñer FM, Thielen A, Bellido R, Pérez-Álvarez S, Cabrera C, Dalmau J, Curriu M, Lie Y, Noguera-Julian M, Puig J, Martínez-Picado J, Blanco J, Coakley E, Däumer M, Clotet B, Paredes R. 2013. HIV-1 tropism testing in subjects achieving undetectable HIV-1 RNA: diagnostic accuracy, viral evolution and compartmentalization. PLoS One 8:e67085. doi: 10.1371/journal.pone.0067085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumann RE, Rogers AA, Hamdan HB, Burger H, Weiser B, Gao W, Anastos K, Young M, Meyer WA III, Pesano RL, Kagan RM. 2015. Determination of HIV-1 coreceptor tropism using proviral DNA in women before and after viral suppression. AIDS Res Ther 12:11. doi: 10.1186/s12981-015-0055-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raymond S, Delobel P, Mavigner M, Cazabat M, Encinas S, Souyris C, Bruel P, Sandres-Sauné K, Marchou B, Massip P, Izopet J. 2010. CXCR4-using viruses in plasma and peripheral blood mononuclear cells during primary HIV-1 infection and impact on disease progression. AIDS 24:2305–2312. [DOI] [PubMed] [Google Scholar]
- 33.Prosperi MC, Bracciale L, Fabbiani M, Di Giambenedetto S, Razzolini F, Meini G, Colafigli M, Marzocchetti A, Cauda R, Zazzi M, De Luca A. 2010. Comparative determination of HIV-1 co-receptor tropism by Enhanced Sensitivity Trofile, gp120 V3-loop RNA and DNA genotyping. Retrovirology 7:56. doi: 10.1186/1742-4690-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrido C, Roulet V, Chueca N, Poveda E, Aguilera A, Skrabal K, Zahonero N, Carlos S, García F, Faudon JL, Soriano V, de Mendoza C. 2008. Evaluation of eight different bioinformatics tools to predict viral tropism in different human immunodeficiency virus type 1 subtypes. J Clin Microbiol 46:887–891. doi: 10.1128/JCM.01611-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santoro MM, Perno CF. 2013. HIV-1 genetic variability and clinical implications. ISRN Microbiol 2013:481314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dimonte S, Mercurio F, Svicher V, D'Arrigo R, Perno CF, Ceccherini-Silberstein F. 2011. Selected amino acid mutations in HIV-1 B subtype gp41 are associated with specific gp120v3 signatures in the regulation of co-receptor usage. Retrovirology 8:33. doi: 10.1186/1742-4690-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svicher V, Alteri C, Artese A, Zhang JM, Costa G, Mercurio F, D'Arrigo R, Alcaro S, Palù G, Clementi M, Zazzi M, Andreoni M, Antinori A, Lazzarin A, Ceccherini-Silberstein F, Perno CF. 2011. Identification and structural characterization of novel genetic elements in the HIV-1 V3 loop regulating coreceptor usage. Antivir Ther 16:1035–1045. doi: 10.3851/IMP1862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.