Abstract
Marriage rules, the community prescriptions that dictate who an individual can or cannot marry, are extremely diverse and universally present in traditional societies. A major focus of research in the early decades of modern anthropology, marriage rules impose social and economic forces that help structure societies and forge connections between them. However, in those early anthropological studies, the biological benefits or disadvantages of marriage rules could not be determined. We revisit this question by applying a novel simulation framework and genome-wide data to explore the effects of Asymmetric Prescriptive Alliance, an elaborate set of marriage rules that has been a focus of research for many anthropologists. Simulations show that strict adherence to these marriage rules reduces genetic diversity on the autosomes, X chromosome and mitochondrial DNA, but relaxed compliance produces genetic diversity similar to random mating. Genome-wide data from the Indonesian community of Rindi, one of the early study populations for Asymmetric Prescriptive Alliance, are more consistent with relaxed compliance than strict adherence. We therefore suggest that, in practice, marriage rules are treated with sufficient flexibility to allow social connectivity without significant degradation of biological diversity.
Keywords: mating systems, Asymmetric Prescriptive Alliance, genetic diversity, Indonesia, Approximate Bayesian Computation
Introduction
Human societies are characterized by a myriad of often elaborate marriage rules. Describing the diversity of these rules and their central role in the organization of human communities was once a major focus of anthropological research (Mascie-Taylor and Boyce 1988). Following the seminal works of Van Wouden (1935) and Lévi-Strauss (1949) in the 1930s and 1940s, studies of marriage systems proliferated during the 1950s and 1960s (Lévi-Strauss 1965; Gilbert and Hammel 1966; Jacquard 1967, 1970; MacCluer et al. 1971), but interest declined as anthropologists recognized that they lacked the necessary tools to formally test the hypotheses they had developed. Today, access to large genetic data sets and fast computer simulation provides a springboard to revisit many of these historically unanswered questions.
Marriage rules are universal, yet extraordinarily diverse (Lévi-Strauss 1949). Although enforcement varies, all communities, both traditional and westernized, impose at least some constraints on who individuals can or cannot marry. Many marriage rules are famously intricate, such as Asymmetric Prescriptive Alliance (APA), which is characterized by a complex, but clearly defined, intergenerational framework (fig. 1) (Needham 1964; Beatty 1990; Forth 2009). Men are required to marry their mother’s brother’s daughter (MBD), and women move from “wife-giver” to “wife-taker” communities in what Van Wouden (1935) describes as a “circulating connubium.” Consequently, although women move in one direction around the network of communities, bride wealth flows in the opposite direction. Hence marriage to the MBD’s clan creates asymmetric alliance ties between patrilines. APA is particularly common in the small islands of eastern Indonesia (Forth 1990), but found globally, it is unclear why such intricate marriage rules emerged, almost certainly independently, in multiple places at multiple times.
Fig. 1.
Kinship under APA. Each box represents a clan; each row represents a generation. The red clan acts as wife-giver to the black clan, which in turn acts as wife-giver to the blue clan. Dashed arrows represent marriage alliances with women moving from their natal clan to the community of their husband.
Marriage rules appear to be important for both social and biological reasons. The connections they create often underpin stable long-term trade and support networks (Winterhalder and Smith 2000; Marlowe 2003; Huber et al. 2011; Henrich et al. 2012), but because marriage is also the primary institution leading to offspring, marriage rules should strongly affect patterns of genetic diversity, particularly in small traditional communities. Research on this question has been surprisingly limited. Departures from expected genetic patterns have sometimes been attributed to social factors (Watkins 2004; Chaix et al. 2007; Moorad et al. 2011; Heyer et al. 2012), such as reduced Y chromosome diversity under polygyny (Lansing et al. 2008), where few men are permitted to marry and reproduce, or excessive mitochondrial DNA (mtDNA) diversity under patrilocality (Tumonggor et al. 2013), where women preferentially move between villages. Comparison of X chromosome and autosomal diversity has also revealed other social constructs, such as sex-specific patterns of postmarital dispersal (Ségurel et al. 2008; Verdu et al. 2013) and sex-biased admixture (Cox et al. 2010). Theoretical studies are even rarer. The effects of polygyny (Guillot and Cox 2014) and marriage alliances around ring communities (Billari et al. 2007) have been modeled. Both studies required special methods, as most marriage rules cannot be studied using standard population genetic theory, such as the coalescent, which assumes random mating, asexual populations, and the crucial trait of exchangeability (Kingman 1982). In consequence, the biological effects of marriage rules remain largely unexplored.
Here, we ask whether marriage rules affect patterns of genetic diversity. We determine whether marriage systems, such as APA, produce biological benefits or disadvantages; whether asymmetry is an important biological feature of such marriage systems; and whether the effects of marriage rules can be meaningfully detected in real-world genetic data. To address these questions, we employ new modeling software, SMARTPOP, which we designed specifically to simulate the intricate marriage rules observed in human communities (Guillot and Cox 2014). Finally, we also use genome-wide single nucleotide polymorphism (SNP) data from Rindi, a community in eastern Indonesia that practices APA (Forth 1981), to estimate the historic degree of compliance with this mating system.
Results
The first major question is whether and how marriage rules affect patterns of genetic diversity. We explored this for APA by simulating data across a grid of values for πMBD, the probability that the MBD rule is followed, and πmig, the probability that a woman migrates according to the wife-giver/wife-taker scheme. In the anthropological literature, APA typically follows a classificatory rule that defines all female siblings of the same generation within a clan as equal (Maybury-Lewis 1965), thus obliging men to marry a woman from the wife-giving group, but not specifically their cousin (). However, unlike this more general case, in Rindi the genealogical cousin is explicitly preferred (Forth 1981). Our framework, which separates the migration and cousin marriage aspects of the APA rule system, allows us to represent the entire gamut of APA-type systems.
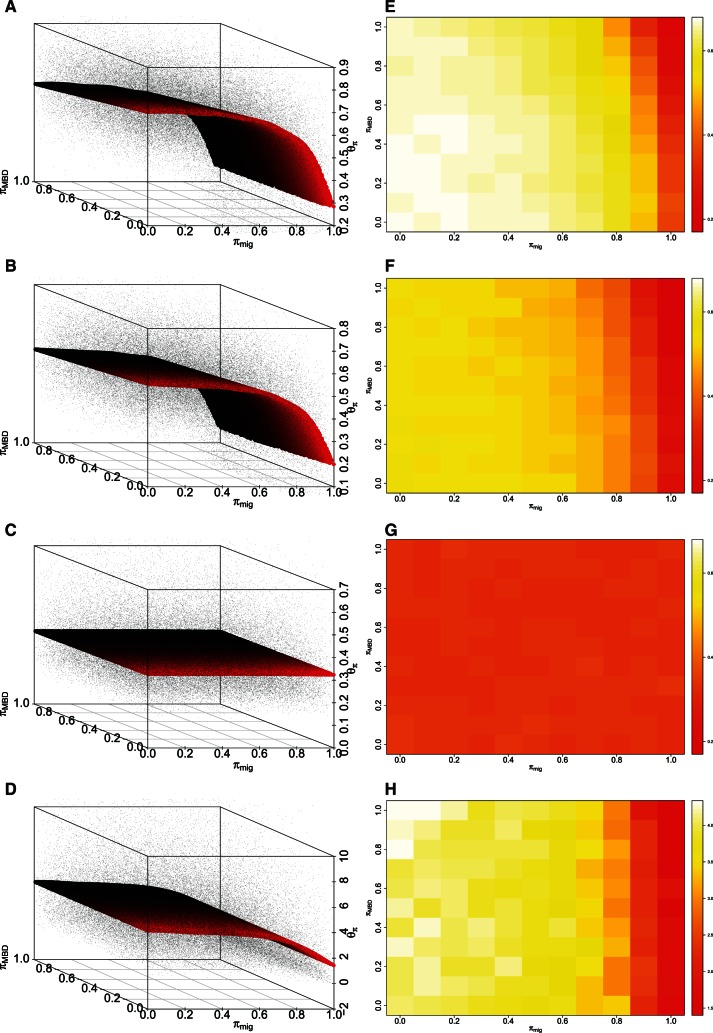
Under a strict interpretation of APA (), genetic diversity shows reproducible reductions in genetic diversity compared with random mating () (fig. 2). Nonlinear regressions show that APA influences genetic diversity on the autosomes, X chromosome and mtDNA, but not the paternally inherited Y chromosome (table 1). This is expected because the system modeled here is patrilocal—women move between communities to marry, whereas men remain in their natal clan (fig. 1). This holds true whether women follow the APA rules or not. However, following the MBD rule negatively affects genetic diversity on the autosomes and X chromosome, and following the wife-giver/wife-taker migration rule negatively affects diversity on the autosomes, X chromosome, and mtDNA. As shown by the regression coefficients, this loss of genetic diversity is driven more by constraints on migration than cousin mating. We therefore conclude that strong adherence to APA is detrimental to the maintenance of genetic diversity.
Fig. 2.
Genetic diversity () across a grid of random values for πMBD and πmig under APA for (A,E) autosomes, (B,F) X chromosome, (C,G) Y chromosome, and (D,H) mtDNA over 50,000 simulations (3,000 individuals, 20 demes). (A)–(D) present fitted values (sheet with black to red color range for better visualization in 3D) from the GAMs, together with simulated values (dots). (E)–(H) are projections of simulated mean diversity values across the grid of parameters.
Table 1.
Local GAM to Fit Mean Pairwise Diversity as a Function of πMBD and πmig.
| Genomic Region | R2 | P( | P( |
|---|---|---|---|
| Autosomes | 0.43 | 0 | 0 |
| X chromosome | 0.35 | 0 | 0 |
| Y chromosome | 7.0 | 0.10 | 0.24 |
| mtDNA | 0.091 | 0.35 | 0 |
Note.—Significant values for the correlation with πMBD and πmig are shown in italics (50,000 simulations).
The equivalent symmetric migration scheme (Symmetric Prescriptive Alliance [SPA]) decreases genetic diversity in a similar manner to APA across the parameter space. We therefore suggest that symmetry versus asymmetry in the movements of women has little biological effect.
The second question is the extent to which marriage rules can be followed stringently. Within a strict APA system, not all men can follow the rule to marry their MBD, as an appropriate individual of the right sex is not always available. The effects of deviating from the MBD rule have been discussed previously (Kunstadter et al. 1963; Ackerman 1964; McFarland 1970; MacCluer et al. 1971; Fredlund 1976; Mascie-Taylor and Boyce 1988), but not determined objectively due to the unavailability of computer simulation as a readily accessible tool in the 1950s–1970s. In our simulations, we can measure the rate at which MBD marriage actually occurs, as a function of πMBD and πmig (fig. 3). In the extreme case where rules are followed strictly, on average only 30% of marriages can be made to a MBD due to the unavailability of an appropriate partner. As πMBD decreases (x axis), more individuals marry a random partner rather than following the rule, leading to lower rates of MBD marriages. When there is compliance with the migration rule, but not the cousin alliance rule, MBD marriages still occur at moderate frequency (∼12%) due to the random chance of marrying the right cousin. However, when the migration rule is not followed, women move to a nonprescribed clan, where no appropriate cousin is present for them to marry. Therefore, as πmig decreases (colored lines), the actual rate of MBD marriage decreases rapidly as well.
Fig. 3.

Actual rate of MBD marriage observed in simulations as a function of πMBD (x axis) and πmig (color scale) (25,000 observations during 50 simulations for each data point). The dashed line represents the rate of MBD marriage observed by Forth (1981) in Rindi (10%); the observed migration rate to the prescribed clan is 26%.
The third question is whether the effects of marriage rules can be inferred in practice. We applied Approximate Bayesian Computation (ABC) to genome-wide data from Rindi, a population on the eastern Indonesian island of Sumba in which APA has been well studied. Figure 4 shows posterior distributions for N, πMBD, and πmig. The population size is estimated at nearly 7,000 individuals (mode = 6,608, 95% credible region 5,086–10,630). We estimate that the migration rule is followed in just over half of cases (mode, 95% credible region ). The mating parameter also shows moderate compliance with the rule (mode), but again has a wide 95% credible region (). Despite considerable uncertainty in these values, the modes of the posterior densities suggest that the Rindi community has not followed either of the extreme cases—strict APA or random mating. In practice, therefore, this community would fall near the center of the graphs in figure 2, where genetic diversity does not differ markedly from random mating.
Fig. 4.
ABC for Rindi, a population practicing APA on Sumba in eastern Indonesia. Posterior distributions are shown for N, πMBD, and πmig.
Cross-validation reveals the accuracy of parameter inference by testing simulated cases with known values (see supplementary material, Supplementary Material online, for details). Population size (Epred = 0.94) and the migration parameter (Epred = 1.24) show moderate linear relationships between estimated and real values, indicating that the inference procedure has reasonable statistical power to infer these parameters. Less power is available to infer the mating parameter (Epred = 1.53).
Finally, using simulated data sets, we asked how genomic sequence data, without the ascertainment bias of SNP genotyping chips, would improve statistical inference. Although predictions for population size (Epred = 0.54) are improved, values for the migration (Epred = 0.94) and mating parameters () suggest that it will always prove challenging to infer mating systems from genomic data, even when they are unbiased. Larger sample sizes and targeted clan sampling designs may help ameliorate these issues.
Discussion
Marriage rules are a ubiquitous feature of all traditional societies. From early in the twentieth century, an extensive body of anthropological literature has attempted to determine their purpose. Although little consensus was reached on the details, anthropologists developed a broadly held view that marriage rules help structure connections within and between communities, and that they therefore play a fundamental role in social cohesion. However, any social rules that affect marriage also have a direct impact on offspring, and hence, the genetics of communities. In small communities, which were the only type that existed throughout most of human history, genetic diversity is easily lost through genetic drift, which in turn can lead to reduced individual fitness, lower reproductive success, increased levels of genetic disorders, and ultimately, community extinction (Ober et al. 1999; Winata et al. 1995).
Before this study, it was not known whether marriage rules help or hinder the maintenance of genetic diversity. We are able to address this question through two recent advances. First, the availability of new simulation tools that can simultaneously model marriage rules and population genetics. Most population genetic methods do not explicitly model two sexes (men and women), let alone specific marriage rules. New computer programs, such as SMARTPOP (Guillot and Cox 2014), now make extensive modeling of social rules and population genetics possible. Second, marriage rules act at very small scales: Either within communities or across a small cluster of communities. Community-level sampling (as opposed to regional collections from schools or medical clinics) is now starting to become more common.
We apply these advances to address the role of APA in mediating patterns of genetic variation in Rindi, the well-studied APA community on Sumba, a small island in eastern Indonesia. APA describes a specific form of cousin marriage, which also implies a regular intergenerational movement of women between clans. Our theoretical simulations show that APA reduces genetic diversity, but only when it is followed strictly. In particular, the migration component of the APA rules elicits a dramatic decline in diversity if followed by more than 80% of women. More variable compliance with the marriage rules leads to genetic diversity that does not differ markedly from random mating.
Here, we make the necessary simplification that the alliance model has been relatively stable through time. Although these simulations assume adherence to APA during the recent past, other scenarios are of course possible. For instance, a recent shift to APA would weaken the effect of the rules on genetic diversity. Occasional reassortment of clans, a process known as fusion–fission (Smouse et al. 1981; Chaix et al. 2007), could also variously increase or decrease genetic diversity among the groups. As genomic data sets improve, it should become feasible to model such complex social dynamics. The simpler scenario modeled here provides an obligatory first step.
The two parameters that underpin APA, the MBD rule and wife-giver/wife-taker migration, together with the population size, were inferred for Rindi from genome-wide SNP chip data. The results show that the size of the Rindi population is large, which is consistent with female immigration due to patrilocal postmarriage residence patterns (Guillot et al. 2013). Patterns of genetic diversity in Rindi seem most consistent with intermediate values of πMBD and πmig, thus arguing against random mating or a strict adoption of the APA rules.
Forth (1981) undertook a detailed ethnographic study of Rindi in the 1970s. He observed the actual proportion of MBD marriages (10%), as well as the proportion of marriages to the prescribed clan (26%). There is no simple relationship between the modeled parameters and the observed rate of cousin marriage, as an appropriate spouse may not be available even if the rules are followed strictly by the community. Using the simulated relationships in figure 3, we deduce that Forth’s observed rates imply a theoretical compliance with the MBD rule of approximately 90% for those individuals who do marry into the prescribed clan. Our overall estimates of πMBD and πmig from the genomic data, 56% and 59%, respectively, differ from those Forth observed. However, we note that our values represent long-term averages, perhaps suggesting that adherence to the APA rules once varied from Forth’s observations in the late twentieth century. Our estimates are most consistent with only moderate long-term compliance with the migration and marriage rules, which in turn would help the community to maintain genetic diversity.
Marriage rules are therefore perhaps best viewed as convenient ideologies: Revered more in theory than in practice. Nevertheless, these results show that marriage rules have important biological outcomes for communities, and that strict adherence can be biologically disadvantageous. We argue that the flexibility with which marriage rules are implemented in practice is therefore not so much a problem as the key point. Although small human communities almost certainly do not think in genetic terms, there are both social and biological reasons to overlook violations of marriage rules. The moderate observance rate in Rindi suggests relatively weak enforcement of marriage rules in this community. Elsewhere, strict compliance can be driven by strong sanctions against transgressors, often mediated through belief systems, and not uncommonly leading to the ultimate sanction, death (Lansing 2006).
This study shows how modern computer simulations can provide new insight into old anthropological questions. The stochastic behavior of individuals, such as instances where the required spouse is unavailable or a different spouse is chosen for an alternative social reason, appears to be a dominant feature of traditional marriage systems. The effects of deviating from a strict interpretation of marriage rules can now be modeled, as can other community choices, such as symmetric versus asymmetric migration. The addition of a genetic element to these models further allows exploration of the effects of marriage systems on biological diversity. Symmetric migration between communities in an SPA setting produces much the same biological outcome as APA. Hence, the preference for APA over SPA in eastern Indonesia (Forth 1990) may be better explained by socioeconomic factors, such as the long-term stability of asymmetric wife-giver/wife-taker exchange, which creates enduring networks of relationships between patrilocal kin groups (Van Wouden 1935; Lévi-Strauss 1965).
The addition of a statistical inference framework to our theoretical work allows us to estimate the long-term biological effects of marriage rules on specific communities such as Rindi. Although the statistical power of the analyses presented here is relatively low, this partly reflects the need for summary statistics that are able to circumvent a small sample size and the ascertainment bias found in current genotyping chips. Unbiased data from whole-genome sequencing will become increasingly common in coming years and the approach presented here is ready to take full advantage of these new data. However, power analyses show that reconstructing mating systems from any sort of genetic data will always be a challenging undertaking.
We do, however, show that genetic evidence has the potential to reconstruct aspects of the social systems by which communities historically lived. Marriage rules are ubiquitous, but we suggest that it is unlikely they were followed strictly. The majority of these violations probably had prosaic local causes. In many cases, the individual required by the marriage rule may not have been available to marry. Alternately, reduced genetic diversity in small communities quickly leads to the accumulation of genetic disorders. Although communities presumably had little understanding of genetic inheritance, they may have linked social behaviors, such as adherence to marriage rules, to unfavorable biological outcomes. Certainly, reduced genetic diversity under a strict interpretation of the APA marriage rules suggests that there was little biological incentive for communities to enforce marriage rules strongly, at least for long periods of time. Whether this holds true across the wide gamut of marriage rules recorded globally by anthropologists is now a question that can feasibly be revisited.
Materials and Methods
Ethics
Biological samples were collected by J.S.L., H.S., and a team from the Eijkman Institute for Molecular Biology, with the assistance of Indonesian Public Health clinic staff, following protocols for the protection of human subjects established by both the Eijkman Institute and the University of Arizona institutional review boards. Permission to conduct research in Indonesia was granted by the State Ministry of Research and Technology.
Sampling and Genetic Screening
Genetic markers were screened in 28 consenting, closely unrelated and apparently healthy individuals from Rindi, a community on the eastern Indonesian island of Sumba. Apart from excluding immediate relatives, individuals were approached randomly during the course of a medical visit. Participant interviews confirmed ethnic, linguistic, and geographic affiliations with Rindi for at least two generations into the past. mtDNA markers are as described elsewhere (Tumonggor et al. 2013). Autosomal (n = 664,475), X chromosome (n = 16,034), and Y chromosome (n = 266) SNPs were screened in 24 individuals using the Illumina HumanOmniExpress-24 BeadChip (GeneByGene, Houston, TX). SNP chip genotype data for Rindi are available from the authors on request.
Computer Simulations
Simulations were run with SMARTPOP v.2.0 (Guillot and Cox 2014), a free open-source C++ forward-in-time simulator, which was purpose-built to model the effects of marriage rules on human communities. Twenty demes of equal size (150 individuals in the regression, varying population sizes in the ABC study) were modeled, thus approximating the number of clans in the APA system recorded for Rindi (Forth 1981). Genetic data were simulated across four genomic regions: 200 unlinked loci on the autosomes (n = 200; 32 bp), 10 unlinked loci on the X chromosome (n = 10; 1,000 bp), a fully linked locus on the Y chromosome (n = 1; 10,000 bp), and a fully linked mtDNA locus (n = 1; 544 bp). This data structure was selected to mimic key features of the real data set as closely as possible, while still meeting nontrivial constraints imposed by runtime speed in the obligatory forward-in-time simulation setting. Slightly more individuals were sampled for mtDNA (n = 28) than nuclear loci (n = 24) to match the observed data.
Demes evolved through phases of migration, mating, and mutation at each generation. Due to considerable uncertainty surrounding human mutation rates (Scally and Durbin 2012) and relative insensitivity to exact values in this component of the analysis, average mutation rates were employed for the autosomes ( mutations/site/generation), X chromosome ( mutations/site/generation), Y chromosome ( mutations/site/generation), and mtDNA ( mutations/site/generation) (Soares et al. 2009; Lynch 2010) using a generation interval of 25 years (Fenner 2005). To simplify the computation, generations did not overlap, thus not allowing us to model intergenerational marriages. Migration and mating were implemented according to the marriage rules of the given model (see details below). For each simulation, the system was allowed to reach equilibrium within a single large randomly mating population, before dispersal of structured demes that followed a particular set of marriage rules for 1,000 generations. Simulated data were strongly robust to these initialization parameters (see also Guillot and Cox 2014).
Model System
Although APA has been described by anthropologists in different ways (Needham and Elkin 1973), two integral components are 1) cousin mate prescription and 2) structured migration. Migration was implemented as a wife-giver/wife-taker system, in which a deme always takes wives from the same set of source populations and gives wives to a different set of sink populations (fig. 1). Each deme was permitted up to three wife-giver and three wife-taker clans, although for any given family, the mother’s brother’s deme is always the wife-giver clan. Mate choice is the prescription for a male to marry his MBD.
APA can be envisaged as a two-parameter system: πMBD, the probability that the MBD rule is followed, and πmig, the probability that a women migrates according to the wife-giver/wife-taker scheme. In its most stringent form (), women always move to their prescribed partner clan and marry their paternal cousin (if one exists). The opposite situation () represents random patrilocal migration and random mating. Because a suitable cousin may not always be available to marry (e.g., in a family with no children of the required sex), we track the effective (i.e., actual) rate of MBD marriage in addition to πMBD. As Rindi practices polygyny (Lansing et al. 2011), as do most other APA communities, simulations allow up to three wives per male.
For comparison, we also simulate SPA, where any two demes exchange wives at each generation (i.e., each clan acts simultaneously as wife-giver and wife-taker). This model is simulated as for APA, with changes only to the migration scheme.
Summary Statistics
As almost all traditional summary statistics ultimately reflect aspects of the folded site frequency spectrum (Achaz 2009), we use the site frequency spectrum itself as a summary statistic. For historical reasons, we also report several commonly used summaries, such as the site homozygosity H (Nei 1978), Watterson’s nucleotide diversity θW, Tajima’s mean pairwise diversity , and the observed number of singleton polymorphisms η1. Summaries were calculated separately for the autosomes, X chromosome, Y chromosome and mtDNA, as mating systems are expected to affect each of these genetic regions in different ways. The known ascertainment bias of existing SNP chips (an inherent feature of their design; Clark et al. 2005) overrepresents polymorphic sites and underrepresents invariant sites. This bias was addressed by developing unbiased summaries that capture the relative frequencies of polymorphic sites on the autosomes and X chromosome (see supplementary material, Supplementary Material online, for details).
Summary statistics from the simulated data were compared with published observations of unbiased autosomal and X chromosome sequence data to confirm comparability in summary values (Hammer et al. 2008). Length normalized values of the mean pairwise divergence for southern Han Chinese () and Melanesian populations () are broadly consistent with simulated values (). We note that the effective population sizes of southern Han Chinese and Melanesians (the geographically and historically closest populations to Rindi for which unbiased autosomal and X chromosome sequence data are available) are likely to be considerably greater than for the small communities that our simulations are intended to mimic. We suggest that the reduced levels of genetic diversity seen in the simulations can be attributed to this lower population size.
General Additive Model Regression
The effects of πMBD and πmig on and H were modeled for the autosomes, X chromosome, Y chromosome, and mtDNA using a general additive model (GAM), which accommodates local and global nonlinear effects (Hastie and Tibshirani 1990). Regressions were fitted to simulated values using the formula , where is a smoothing spline function for the migration parameter. GAM regressions were calculated using the R package MGCV (Wood 2011).
Approximate Bayesian Computation
The fit between genomic data from Rindi and simulations was determined using ABC (Beaumont et al. 2010; Sunnåker et al. 2013). This likelihood-free statistical inference method estimates model parameters by comparing outcomes from simulations with real data. Three parameters were inferred: The population size N, πMBD, and πmig. All priors were drawn from continuous uniform distributions with , and . From simulations, 0.1% were accepted using a rejection algorithm (Beaumont et al. 2002). ABC was performed using the R packages abc and abctools (Csillery et al. 2012; Nunes and Prangle 2015).
Different sets of summary statistics were explored and the optimal set selected that returned the lowest prediction error (a measure of distance between estimated and true values for each parameter) (Csillery et al. 2012). The Y chromosome data were discarded due to the limited number of SNPs screened by the HumanOmniExpress chip (n = 266) and insensitivity of the Y chromosome to πMBD and πmig observed in initial simulations. Cross-validation was used to confirm the accuracy of the inference method.
Finally, to determine the potential role of larger data sets available in the future, the power of the ABC framework was determined for simulated full sequence data without the ascertainment bias inherent with SNP chips. ABC was performed using the same parameters as described above and cross-validated over 1,000 simulations to generate prediction errors for N, πMBD, and πmig.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Royal Society of New Zealand through a Rutherford Fellowship (RDF-10-MAU-001) and Marsden Grant (11-MAU-007) to M.P.C. E.G.G. was funded by a doctoral scholarship from the Institute of Fundamental Sciences, Massey University. Computational resources were provided by Massey University and the New Zealand eScience Infrastructure (NeSI). The authors thank Gregory Forth (University of Alberta) and three anonymous reviewers for their constructive comments.
References
- Achaz G. 2009. Frequency spectrum neutrality tests: one for all and all for one. Genetics 183(1):249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman C. 1964. Structure and statistics: the Purum case. Am Anthropol. 66(1):53–65. [Google Scholar]
- Beatty A. 1990. Asymmetric alliance in Nias, Indonesia. Man 25(3):454–471. [Google Scholar]
- Beaumont MA, Nielsen R, Robert C, Hey J, Knowles L, Hickerson M, Scott A. 2010. In defence of model-based inference in phylogeography. Mol Ecol. 19:436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Zhang W, Balding DJ. 2002. Approximate Bayesian computation in population genetics. Genetics 162:2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billari F, Fent T, Prskawetz A, Aparicio Diaz B. 2007. The “Wedding-Ring”: an agent based model based on social interaction. Demogr Res. 17:59–82. [Google Scholar]
- Chaix R, Quintana-Murci L, Hegay T, Hammer MF, Mobasher Z, Austerlitz F, Heyer E. 2007. From social to genetic structures in central Asia. Curr Biol. 17(1):43–48. [DOI] [PubMed] [Google Scholar]
- Clark AG, Hubisz MJ, Bustamante CD, Williamson SH. 2005. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 15:1496–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MP, Karafet TM, Lansing JS, Sudoyo H, Hammer MF. 2010. Autosomal and X-linked single nucleotide polymorphisms reveal a steep Asian-Melanesian ancestry cline in eastern Indonesia and a sex bias in admixture rates. Proc R Soc Lond B Biol Sci. 277(1687):1589–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csillery K, Francois O, Blum MGB. 2012. abc: an R package for approximate Bayesian computation (ABC). Methods Ecol Evol. 3:475–479. [Google Scholar]
- Fenner JN. 2005. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 128:415–423. [DOI] [PubMed] [Google Scholar]
- Forth GL. 1990. From symmetry to asymmetry. An evolutionary interpretation of eastern Sumbanese relationship terminology. Anthropos 85(4):373–392. [Google Scholar]
- Forth GL. 2009. Human beings and other people. Bijdr Taal Land Volkenkd. 165(4):493–514. [Google Scholar]
- Forth GL. 1981. Rindi: an ethnographic study of a traditional domain in eastern Sumba. The Hague (The Netherlands): Kininklijk Instituut voor Taal-, Land- en Volkenkude. [Google Scholar]
- Fredlund EV. 1976. Measuring marriage preference. Ethnology 15(1):35–45. [Google Scholar]
- Gilbert JP, Hammel EA. 1966. Computer simulation and analysis of problems in kinship and social structure. Am Anthropol. 68:71–93. [Google Scholar]
- Guillot EG, Cox MP. 2014. SMARTPOP: inferring the impact of social dynamics on genetic diversity through high speed simulations. BMC Bioinformatics 15:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot EG, Tumonggor MK, Lansing JS, Sudoyo H, Cox MP. 2013. Climate change influenced female population sizes through time across the Indonesian archipelago. Hum Biol. 85(1–3):135–152. [DOI] [PubMed] [Google Scholar]
- Hammer MF, Mendez FL, Cox MP, Woerner AE, Wall JD. 2008. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 4(9):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie TJ, Tibshirani RJ. 1990. Generalized additive models. London: CRC Press. [Google Scholar]
- Henrich J, Boyd R, Richerson PJ. 2012. The puzzle of monogamous marriage. Philos Trans R Soc Lond B Biol Sci. 367:657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer E, Chaix R, Pavard S, Austerlitz F. 2012. Sex-specific demographic behaviours that shape human genomic variation. Mol Ecol. 21(3):597–612. [DOI] [PubMed] [Google Scholar]
- Huber BR, Danaher WF, Breedlove WL. 2011. New cross-cultural perspectives on marriage transactions. Cross Cult Res. 45(4):339–375. [Google Scholar]
- Jacquard A. 1967. La reproduction humaine en régime malthusien. Un modèle de simulation par la méthode de Monte-Carlo. Population 22(5):897–920. [Google Scholar]
- Jacquard A. 1970. Panmixie et structure des familles. Population 25(1):69–76. [Google Scholar]
- Kingman JFC. 1982. The coalescent. Stoch Process Appl. 13(3):235–248. [Google Scholar]
- Kunstadter P, Buhler R, Stephan FF, Westoff CF. 1963. Demographic variability and preferential marriage patterns. Am J Phys Anthropol. 21(4):511–519. [DOI] [PubMed] [Google Scholar]
- Lansing JS. 2006. Perfect order: recognizing complexity in Bali. Princeton (NJ): Princeton University Press. [Google Scholar]
- Lansing JS, Cox MP, De Vet TA, Downey S, Hallmark B, Sudoyo H. 2011. An ongoing Austronesian expansion in Island Southeast Asia. J Anthropol Archaeol. 30(3):262–272. [Google Scholar]
- Lansing JS, Watkins JC, Hallmark B, Cox MP, Karafet TM, Sudoyo H, Hammer MF. 2008. Male dominance rarely skews the frequency distribution of Y chromosome haplotypes in human populations. Proc Natl Acad Sci U S A. 105(33):11645–11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévi-Strauss C. 1949. Les Structures Élémentaires de la Parenté. Paris: PUF. [Google Scholar]
- Lévi-Strauss C. 1965. The future of kinship studies. Proceedings of the Royal Anthropological Institute of Great Britain and Ireland, 1965: 13–22. [Google Scholar]
- Lynch M. 2010. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 107(3):961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCluer JW, Neel JV, Chagnon NA. 1971. Demographic structure of a primitive population: a simulation. Am J Phys Anthropol. 35(2):193–207. [DOI] [PubMed] [Google Scholar]
- Marlowe FW. 2003. The mating system of foragers in the standard cross-cultural sample. Cross Cult Res. 37(3):282–306. [Google Scholar]
- Mascie-Taylor N, Boyce AJ. 1988. Human mating patterns. Cambridge: Cambridge University Press. [Google Scholar]
- Maybury-Lewis DHP. 1965. Prescriptive marriage systems. Southwest J Anthropol. 21(3):207–230. [Google Scholar]
- McFarland DD. 1970. Effects of group size on the availability of marriage partners. Demography 7(4):475–476. [PubMed] [Google Scholar]
- Moorad JA, Promislow DE, Smith KR, Wade MJ. 2011. Mating system change reduces the strength of sexual selection in an American frontier population of the 19th century. Evol Hum Behav. 32(2):147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham R. 1964. Descent, category, and alliance in Sirionó society. Southwest J Anthropol. 20(3):229–240. [Google Scholar]
- Needham R, Elkin AP. 1973. Prescription. Oceania 43(3):166–181. [Google Scholar]
- Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89(3):583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes M, Prangle D. 2015. abctools: tools for ABC analyses. Available from: http://cran.r-project.org/web/packages/abctools/index.html [Google Scholar]
- Ober C, Hyslop T, Hauck WW. 1999. Inbreeding effects on fertility in humans: evidence for reproductive compensation. Am J Hum Genet. 64(1):225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A, Durbin R. 2012. Revising the human mutation rate: implications for understanding human evolution. Genetics 13:745–753. [DOI] [PubMed] [Google Scholar]
- Ségurel L, Martínez-Cruz B, Quintana-Murci L, Balaresque P, Georges M, Hegay T, Aldashev A, Nasyrova F, Jobling MA, Heyer E, et al. 2008. Sex-specific genetic structure and social organization in Central Asia: insights from a multi-locus study. PLoS Genet. 4(9):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smouse PE, Vitzthum VJ, Neel JV. 1981. The impact of random and lineal fission on the genetic divergence of small human groups: a case study among the Yanomama. Genetics 98:179–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, Ermini L, Thomson N, Mormina M, Rito T, Röhl A, Salas A, Oppenheimer S, Macaulay V, Richards MB. 2009. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 84(6):740–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnåker M, Busetto AG, Numminen E, Corander J, Foll M, Dessimoz C. 2013. Approximate Bayesian computation. PLoS Comput Biol. 9(1):e1002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumonggor MK, Karafet TM, Hallmark B, Lansing JS, Sudoyo H, Hammer MF, Cox MP. 2013. The Indonesian archipelago: an ancient genetic highway linking Asia and the Pacific. J Hum Genet. 58(3):165–173. [DOI] [PubMed] [Google Scholar]
- Van Wouden FAE. 1935. Sociale Structuurtypen in de Groote Oost. Leiden (The Netherlands): Ginsberg. [Google Scholar]
- Verdu P, Becker NSA, Froment A, Georges M, Grugni V, Quintana-Murci L, Hombert J-M, Van der Veen L, Le Bomin S, Bahuchet S, et al. 2013. Sociocultural behavior, sex-biased admixture, and effective population sizes in Central African Pygmies and non-Pygmies. Mol Biol Evol. 30(4):918–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JC. 2004. The role of marriage rules in the structure of genetic relatedness. Theor Popul Biol. 66(1):13–24. [DOI] [PubMed] [Google Scholar]
- Winata S, Arhya IN, Moeljopawiro S, Hinnant JT, Liang Y, Friedman TB, Asher JH., Jr 1995. Congenital non-syndromal autosomal recessive deafness in Bengkala, an isolated Balinese village. J Med Genet. 32:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterhalder B, Smith EA. 2000. Analyzing adaptive strategies: human behavioral ecology at twenty-five. Evol Anthropol. 9(2):51–72. [Google Scholar]
- Wood SN. 2011. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J R Stat Soc B. 73(1):3–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.