Abstract
Environmental control of flowering allows plant reproduction to occur under optimal conditions and facilitates adaptation to different locations. At high latitude, flowering of many plants is controlled by seasonal changes in day length. The photoperiodic flowering pathway confers this response in the Brassicaceae, which colonized temperate latitudes after divergence from the Cleomaceae, their subtropical sister family. The CONSTANS (CO) transcription factor of Arabidopsis thaliana, a member of the Brassicaceae, is central to the photoperiodic flowering response and shows characteristic patterns of transcription required for day-length sensing. CO is believed to be widely conserved among flowering plants; however, we show that it arose after gene duplication at the root of the Brassicaceae followed by divergence of transcriptional regulation and protein function. CO has two close homologs, CONSTANS-LIKE1 (COL1) and COL2, which are related to CO by tandem duplication and whole-genome duplication, respectively. The single CO homolog present in the Cleomaceae shows transcriptional and functional features similar to those of COL1 and COL2, suggesting that these were ancestral. We detect cis-regulatory and codon changes characteristic of CO and use transgenic assays to demonstrate their significance in the day-length-dependent activation of the CO target gene FLOWERING LOCUS T. Thus, the function of CO as a potent photoperiodic flowering switch evolved in the Brassicaceae after gene duplication. The origin of CO may have contributed to the range expansion of the Brassicaceae and suggests that in other families CO genes involved in photoperiodic flowering arose by convergent evolution.
Keywords: tandem gene duplication, Brassicaceae, flowering, CONSTANS
Introduction
Gene duplication is one of the mechanisms by which genes conferring novel functions arise during evolution (Ohno 1970; Soltis et al. 2009; Levin 2011; Ramsey 2011). Such duplications occur at the level of single genes through tandem or transposition duplication or as part of whole-genome duplications (WGDs) (Freeling 2009). Although one of the duplicates is usually lost, in some cases both are retained allowing divergence in their function and evolution of novel activities. Different models for retention of duplicates have been proposed including subfunctionalization, where each duplicate takes on part of the function of the original gene, and neofunctionalization in which one of the duplicates exhibits a novel advantageous activity (Lynch and Conery 2000; Freeling et al. 2008; Flagel and Wendel 2009; Freeling 2009). In plants, WGDs have occurred many times and are implicated in providing the genetic flexibility required for species diversification (Soltis et al. 2009; Schranz et al. 2012). During the evolutionary history of the model species Arabidopsis thaliana at least five WGDs have occurred. The oldest (At-ζ) is shared with all seed plants (Jiao et al. 2011), whereas the most recent (At-α) is specific to the Brassicaceae. At-α is detected in the genomes of species in the core Brassicaceae and their sister tribe Aethionemae (Vision et al. 2000; Bowers et al. 2003; Jiao et al. 2011; Haudry et al. 2013), but is not present in the Cleomaceae, the sister family of the Brassicaceae. Rather the Cleomaceae underwent an independent genome triplication (Th-α) as revealed in the genome of Tarenaya hassleriana (Cheng et al. 2013; Haudry et al. 2013). Thus At-α occurred in the progenitor of the core Brassicaceae and their sister tribe Aethionemae around 30–40 Ma, and this duplication may have provided the flexibility required for the evolutionary success of the core Brassicaceae (Couvreur et al. 2010; Schranz et al. 2012).
Tandem duplications are also commonly found in plant genomes (Leister 2004; Casneuf et al. 2006; Rizzon et al. 2006). These are gene family members located adjacent or close to one another on the same chromosome. They are likely to mainly arise through recombination-based mechanisms, particularly unequal crossing over, because they are present at highest frequency in regions of the genome where recombination occurs most frequently and genes in tandem arrays are most often present in the same orientation (Zhang and Gaut 2003; Rizzon et al. 2006). In the well-characterized genome of A. thaliana, such tandemly arrayed genes comprise only a slightly lower proportion of all genes (18%) than those retained after WGDs (25%) (Lockton and Gaut 2005; Rizzon et al. 2006). As discussed above for genes involved in WGD, genes that arise by tandem duplication can also diverge in function and expression pattern (Zhang et al. 2000; Kliebenstein et al. 2001; Vlad et al. 2014). Indeed, tandemly duplicated genes are reported to be more divergent in their expression patterns than those that arise by WGD (Casneuf et al. 2006). Also, WGDs occur rarely within a phylogeny, whereas tandem duplications can occur frequently and thus can be restricted to closely related species or even only be present within a species (Leister 2004; Hanikenne et al. 2008; Vlad et al. 2014). In this way, they can contribute to adaptation to particular environments or to species specific traits (Zhang et al. 2000; Hanikenne et al. 2008; Vlad et al. 2014). Consistent with this conclusion, analysis of proteins encoded by tandemly duplicated genes indicated that those involved in environmental responses to biotic or abiotic stimuli are overrepresented (Leister 2004; Rizzon et al. 2006).
Environmental control of the transition to flowering, the first step in plant reproduction, is an important adaptive trait in many species. These responses allow flowering to be induced at the optimal time to maximize opportunities for cross-fertilization, to minimize floral damage and to ensure completion of seed development before the onset of adverse conditions (Anderson et al. 2011). Seasonal cues such as changes in photoperiod (day length) and response to vernalization (prolonged periods of cold) are critical in the adaptation of flowering time to environment in many plant families (Hayama et al. 2003; Yan et al. 2003; Turck et al. 2008; Wang et al. 2009; Blackman et al. 2011). Consistent with this, latitudinal or altitudinal clines for flowering responses to seasonal cues and allelic variation in some of the genes conferring them have been reported (Ray and Alexander 1966; Stinchcombe et al. 2004; Mendez-Vigo et al. 2011) as have selection for flowering in response to other environmental pressures (Hall and Willis 2006; Toomajian et al. 2006; Franks et al. 2007). The molecular-genetic control of seasonal flowering is best understood in the model species A. thaliana (Andrés and Coupland 2012).
The vernalization response to winter cold is widely dispersed in the Brassicaceae. The FLOWERING LOCUS C (FLC) gene encodes a MADS box transcription factor that plays a critical role in vernalization response of A. thaliana and other Brassicaceae species by repressing flowering until plants are exposed to winter temperatures (Michaels and Amasino 1999; Sheldon et al. 1999; Schranz et al. 2002; Okazaki et al. 2007; Wang et al. 2009). Allelic variation at FLC and its upstream regulator FRIGIDA is widespread in natural populations of different species (Johanson et al. 2000; Michaels et al. 2003; Lempe et al. 2005; Shindo et al. 2005), and some of this shows patterns of variation consistent with selection (Stinchcombe et al. 2004; Toomajian et al. 2006; Mendez-Vigo et al. 2011).
Flowering of A. thaliana is also regulated by seasonal changes in day length, being accelerated by long days (LDs) typical of spring and summer. The CONSTANS (CO) gene is a key regulatory component of the photoperiodic flowering time pathway (Putterill et al. 1995), and natural genetic variation has been described at this locus in A. thaliana (Rosas et al. 2014). In the genetic cascade that triggers flowering in response to photoperiod, a class of redundant repressive transcription factor called CYCLING DOF FACTOR (CDF), which are regulated by both light and the circadian clock, contains the major regulators of CO transcription (Imaizumi et al. 2005; Sawa et al. 2007; Fornara et al. 2009). This regulation ensures that CO mRNA levels are low when plants are exposed to light under short days (SDs), but high during the evening under LDs, when exposure to light stabilizes CO protein (Suarez-Lopez et al. 2001; Valverde et al. 2004; Sawa et al. 2007; Fornara et al. 2009). Consequently CO protein only accumulates to promote flowering under inductive LDs, and does so by activating the transcription of FLOWERING LOCUS T (FT) and TWIN SISTER OF FT (TSF) in the leaf vasculature (Samach et al. 2000; Corbesier et al. 2007; Jaeger and Wigge 2007; Mathieu et al. 2007; Song et al. 2012). This regulatory cascade confers photoperiodic flowering even in distantly related monocotyledons (Yano et al. 2000; Hayama et al. 2003; Campoli et al. 2012), and CO homologs are also associated with other day-length-dependent developmental processes such as tuberization in potato or bud dormancy in poplar trees (Bohlenius et al. 2006; González-Schain et al. 2012). Moreover, CO homologs confer diverse responses to photoperiod, functioning as photoperiodic sensors in plants that respond to long photoperiods, such as A. thaliana, or to short photoperiods, such as Oryza sativa and Solanum tuberosum (Yano et al. 2000; González-Schain et al. 2012). Thus CO-related genes have been utilized during evolution to confer different photoperiodic responses on diverse developmental processes.
CO was the first member of a small family of CONSTANS-LIKE (COL) genes to be identified (Putterill et al. 1995). In A. thaliana, the family contains 17 members (Robson et al. 2001; Khanna et al. 2009). All COL proteins contain one or two zinc finger B-Boxes that are assumed to be protein–protein interaction domains and a CO, CONSTANS-LIKE, TOC1 (CCT) domain important for mediating interaction with DNA (Griffiths et al. 2003; Tiwari et al. 2010; Song et al. 2012). Although CO is a master regulator of photoperiodic flowering in A. thaliana, its closest homologs COL1 and COL2 were implicated in circadian clock function but not reproductive development (Putterill et al. 1995; Ledger et al. 2001; Kim et al. 2013). CO, COL1, and COL2 represent a single clade within the family referred to as subgroup Ia COL genes (Griffiths et al. 2003). COL1 exists as a tandem duplication with CO on chromosome 5, a structure also present in Brassica napus (Robert et al. 1998), whereas COL2 is located on chromosome 3. These two chromosomal regions of A. thaliana arose in the most recent WGD (At-α) early in the evolution of the Brassicaceae (Bowers et al. 2003; Khanna et al. 2009; Cheng et al. 2013; Haudry et al. 2013). Interestingly COL1 and COL2 are differently expressed than CO with a high amplitude peak of transcription only in the morning (Ledger et al. 2001). Similar diurnal expression patterns to COL1/2 have been identified in close homologs of CO from many dicot plants including Pharbitis nil, S. tuberosum, and Vitis vinifera (Ledger et al. 2001; Liu et al. 2001; Almada et al. 2009; González-Schain et al. 2012). In contrast, CO homologs in the monocot models rice and barley show transcriptional patterns similar to CO of A. thaliana (Hayama et al. 2003; Turner et al. 2005; Campoli et al. 2012). How the photoperiodic role of CO genes and their characteristic patterns of expression evolved has not been investigated.
In this study, we investigate the evolution of the photoperiodic flowering pathway in the Brassicaceae. We examine the evolutionary history of subgroup Ia COL genes, which includes both WGD and tandem duplication. We analyze the expression patterns of the single CO homolog ThCOL from T. hassleriana of the Cleomaceae family (Schranz and Mitchell-Olds 2006) as well as those of subgroup Ia COL genes of the basal Brassicaceae species Aethionema arabicum, the sister of the core Brassicaceae, and demonstrate that the CO transcriptional pattern is shown by CO genes in the Brassicaceae but not by COL genes in the Brassicaceae or Cleomaceae. This novel transcriptional pattern seems to have originated with the tandem duplication of COL1 and CO in the Brassicaceae, which led to a significant and conserved increase in DOF binding sites in the promoters of CO genes. In addition, functional tests in A. thaliana showed that the Brassicaceae CO proteins are more effective at promoting flowering than subgroup Ia COL proteins from the Brassicaceae or of T. hassleriana. We conclude that the change in expression pattern together with enhancement of the functionality of CO protein in promoting FT transcription compared with its ancestral gene generated a potent photoperiodic switch early in the evolution of the Brassicaceae and this was then conserved throughout the family. These responses are likely to have been important as Brassicaceae, in contrast to the Cleomaceae, colonized northern latitudes where responses to seasonal fluctuations of day length are particularly significant. The origin of CO at the root of the Brassicaceae suggests that similarly expressed CO-like genes involved in photoperiodic responses in monocotyledonous plants or in potato arose by convergent evolution.
Results
Aethionema arabicum, a Member of a Basal Brassicaceae Lineage, Contains a Functional CO Homolog
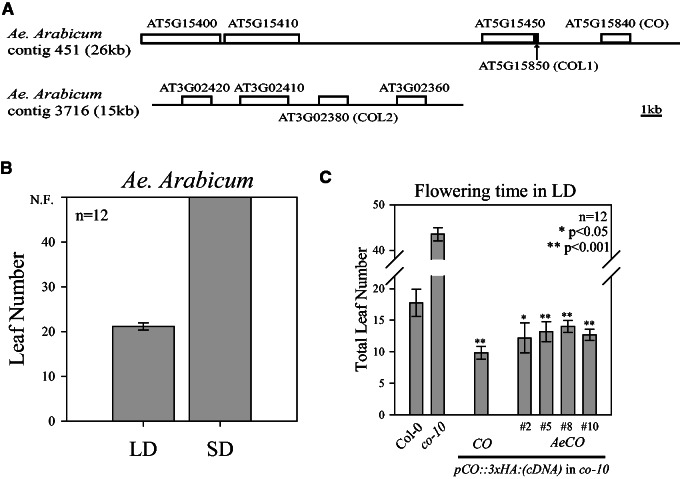
CO orthologs are found in many Brassicaceae species but the conservation of their expression patterns and functions has not been analyzed across distantly related family members. To this end, we identified subgroup Ia orthologs of CO in the Brassicaceae species Ae. arabicum, which belongs to a basal sister lineage of the core Brassicaceae (Bailey et al. 2006; Haudry et al. 2013; supplementary fig. S1, Supplementary Material online). Orthologs of CO (AeCO) and COL2 (AeCOL2) were identified in Ae. arabicum based on their sequence homology and synteny with A. thaliana (fig. 1A), but the ortholog of COL1 was not present due to an apparently recent rearrangement (fig. 1A).
Fig. 1.
Identification of a functional CO homolog in the LD plant Aethionema arabicum. (A) Two contigs containing subgroup Ia COL genes in Ae. arabicum. AGI locus identifiers indicate homologous genes in Arabidopsis thaliana (Tair 10). A chromosomal rearrangement removed most of the COL1 ortholog in Ae. arabicum of which only the last 59 bp were retained (black arrow). (B) Flowering time of Ae. arabicum as total number of leaves at appearance of first inflorescence in LD (16 h light/8 h dark) and SD conditions (8 h light/16 h dark). Mean ± SD; n = 12; N.F., not flowering. (C) Flowering time of A. thaliana as mean (±SD) of total leaf number of Col-0, co-10, pCO::3xHA:CO (co-10) and pCO::HA:AeCO (co-10) in LD conditions (Ae, Aethionema arabicum). Transgenic plants are independent homozygous T3 lines with n = 12 plants per genotype. Asterisks (*P < 0.05; **P < 0.001) indicate statistically significantly earlier flowering compared with Col-0 (wt).
The flowering time of Ae. arabicum was tested under controlled conditions to determine whether the presence of AeCO was associated with a photoperiodic flowering response. Aethionema arabicum plants flowered with approximately 20 leaves in LDs (fig. 1B) but did not flower under SDs, indicating that these plants exhibit an obligate photoperiodic flowering response. To test the functionality of AeCO its cDNA was fused to the A. thaliana CO promoter (pCO) to confer a similar transcriptional pattern as CO. This construct was introduced into A. thaliana co-10 null mutants. Plants carrying the transgene flowered at a comparable time to the control line carrying an CO transgene, and significantly earlier than the co-10 progenitor or Col-0 wild-type (wt) (fig. 1C). Thus Ae. arabicum contains a CO ortholog that shares protein activity with CO of A. thaliana, suggesting that CO was already present before diversification of the major lineages of the Brassicaceae.
Distinct Transcriptional Patterns of CO and COL2 Are Conserved in Ae. arabicum
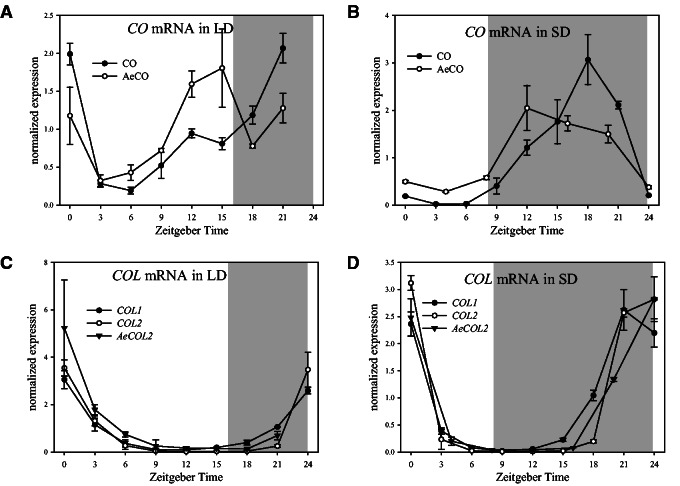
CO exhibits a temporal transcriptional pattern distinct from COL1 and COL2 in A. thaliana. In order to determine whether this difference is conserved in Ae. arabicum, the mRNA levels of AeCO and AeCOL2 were examined every 3 h for 24 h in seedlings growing under LDs or SDs (fig. 2). Under LDs, AeCO showed a similar pattern of mRNA accumulation to CO, including high mRNA levels at lights on (dawn) that fall in the middle of the day before rising again to high amplitude in the evening and night (fig. 2A). Under SDs, AeCO mRNA levels were low during day light but increased during the night, again similar to CO (fig. 2B). AeCOL2 mRNA levels exhibited a single peak in the morning (lights on, Zeitgeber Time (ZT)0) and trough levels in the evening (ZT9 and ZT15) under both LDs and SDs (fig. 2C and D). AeCOL2 expression levels rose earlier in the night in SD grown seedlings compared with those grown in LD. This pattern of transcription is highly consistent with the expression patterns of COL2 and COL1 (fig. 2C and D). Thus both CO and COL2 have conserved expression patterns in A. thaliana and Ae. arabicum, suggesting that these differences arose very early in the diversification of the Brassicaceae and have been conserved in distinct lineages.
Fig. 2.
Diurnal patterns of CO and COL1/2 mRNA abundance in Arabidopsis thaliana and Aethionema arabicum. (A, B) Diurnal pattern of CO mRNA was measured in LD and SD grown seedlings of A. thaliana (CO) and Ae. arabicum (AeCO). (C, D) Diurnal expression patterns of COL1 and COL2 from A. thaliana and Ae. arabicum (AeCOL2) in LD or SD conditions. Values are shown as mean ± SD of two biological replicates after normalization to PP2A (A. thaliana) or ACTIN (Ae. arabicum). Gray areas indicate darkness.
Tarenaya hassleriana COL Exhibits the Transcriptional Pattern and Protein Activity of COL1/2
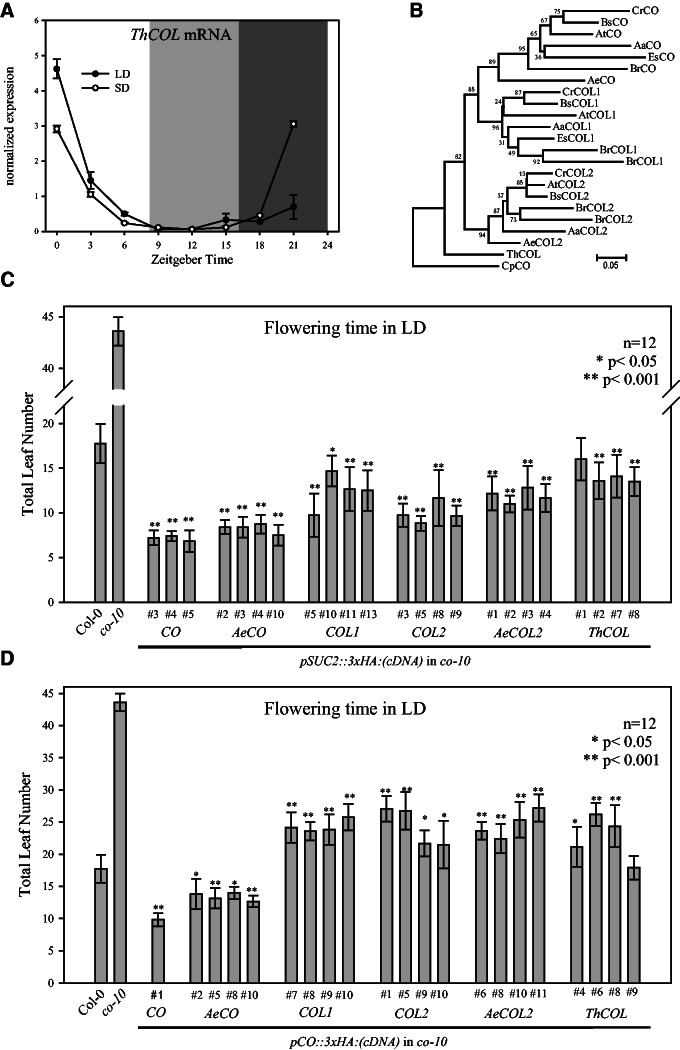
CO, COL1, and COL2 represent a monophyletic group that diversified in the Brassicaceae, whereas T. hassleriana of the Cleomaceae family, which is sister to the Brassicaceae, contains only a single gene (ThCOL) in this group. ThCOL is therefore believed to be homologous to CO, COL1, and COL2 (Schranz and Mitchell-Olds 2006; supplementary fig. S3C, Supplementary Material online). ThCOL expression pattern and protein function were tested to determine whether they are more similar to CO or COL1/2. The expression pattern of ThCOL in T. hassleriana seedlings grown under LD and SD (fig. 3A) was highly similar to the temporal expression patterns of COL1 and COL2 genes in the Brassicaceae but clearly distinct from CO and AeCO (fig. 2). This result suggests that the last common ancestor of the Brassicaceae and Cleomaceae families carried a single subgroup Ia COL gene that was expressed similarly to COL1/2 and that the mRNA expression pattern of CO arose early in the diversification of the Brassicaceae so that it was conserved in each lineage.
Fig. 3.
Properties of ThCOL in comparison to Brassicaceaen subgroup Ia COL genes. (A) Diurnal expression of ThCOL mRNA in Tarenaya hassleriana seedlings grown under LD or SD conditions. Values are shown as mean ± SD for two biological replicates after normalization to ACTIN. Light gray area indicates darkness in SD, whereas dark gray indicates darkness in both light conditions. (B) Neighbor-joining tree of amino acid sequence from subgroup Ia COL proteins from Arabidopsis thaliana (At), Arabis alpina (Aa), Aethionema arabicum (Ae), Boechera stricta (Bs), Brassica rapa (Br), Capsella rubella (Cr), Carica papaya (Cp), Eutrema salsugineum (Es), and T. hassleriana (Th). Numbers indicate bootstrap values of 10,000 iterations. Arabidopsis thaliana co-10 null mutant was complemented with cDNA of COL1, COL2, AeCO, AeCOL2, and ThCOL fused to 3xHA-tag expressed from pSUC2 (C) or pCO (D). Flowering time (mean total leaf number ± SD) was examined in independent homozygous transgenic lines (T3) in LD conditions. Col-0, co-10, and homozygous pCO::3xHA:CO, pAeCO::3xHA:CO (from fig. 1C) and pSUC2::3xHA:CO lines are shown as controls. Asterisks indicate statistically significant differences in leaf number (*P < 0.05; **P < 0.001) compared with Col-0 (wt).
The amino acid sequences of Brassicaceae subgroup Ia COL proteins were compared with the sequence of ThCOL and the amino acid sequence of a COL protein from Carica papaya was used to root the phylogenetic tree (fig. 3B). CO, COL1, and COL2 proteins from the Brassicaceae family form monophyletic groups and ThCOL acts as an outgroup to all Brassicaceae subgroup Ia COL proteins (fig. 3B). The ratio of nonsynonymous to synonymous substitution rates (dn/ds ratio) between A. thaliana or Ae. arabicum Ia COL genes and ThCOL was lower than 1.0, suggesting stabilizing selection (supplementary fig. S3B, Supplementary Material online). Based on this analysis ThCOL is equally distantly related to all three subgroup Ia COL genes (CO, COL1, and COL2).
In order to identify functional differences among them, subgroup Ia COL proteins were tested in a transgenic complementation assay. To examine the capacity of subgroup Ia COL proteins to accelerate flowering in A. thaliana, cDNAs encoding these proteins in A. thaliana, Ae. arabicum and T. hassleriana were fused to the strong, phloem-specific SUCROSE TRANSPORTER 2 (SUC2) promoter (pSUC2, Imlau et al. 1999). This promoter was chosen because CO and its downstream target FT are expressed in the phloem companion cells in A. thaliana and overexpression of CO from pSUC2 greatly accelerates flowering (An et al. 2004; Corbesier et al. 2007). All constructs were introduced into the co-10 null mutant of A. thaliana to test their functionality. The co-10 plants carrying each of the pSUC2 driven constructs flowered much earlier than the co-10 progenitor and at least as early as wt (Col-0) (fig. 3C and supplementary fig. S2A, Supplementary Material online). Nonetheless, the lines carrying either CO or AeCO were among the earliest flowering, with a mean number of approximately seven to approximately eight total leaves compared with lines carrying COL genes that had a mean number of approximately 9 to approximately 16 total leaves. In all lines tested early flowering was associated with elevated FT mRNA levels compared with co-10 control, suggesting that each of these proteins is capable of activating FT transcription when expressed from pSUC2 (supplementary fig. S2C and D, Supplementary Material online). Thus, expression from the SUC2 promoter demonstrates that all subgroup Ia COL proteins as well as the Cleomaceae ortholog of these proteins are able to promote flowering when overexpressed in the appropriate cells in A. thaliana.
The SUC2 promoter causes high expression levels in phloem companion cells (Imlau et al. 1999) that might reduce the specificity of these proteins. Thus, similar experiments were performed using the native A. thaliana pCO that were expected to more reliably reveal the capacity with which these proteins can promote flowering. CO and AeCO expressed from pCO were judged to fully complement the late-flowering phenotype of co-10 (figs. 1C and 3D), because transgenic lines carrying these constructs flowered with a mean of approximately 10 to approximately 14 total leaves, which was much earlier than the co-10 mutant (∼44 leaves) and even earlier than the wt control (∼18 total leaves). The other proteins (COL1, COL2, AeCOL2, ThCOL) were not able to fully replace CO function leading to flowering times later than wt but earlier than co-10 (fig. 3D and supplementary fig. S2B, Supplementary Material online). The level of mRNA expressed from the transgenes was tested in each line (supplementary fig. S2E, Supplementary Material online). Those lines that promoted earliest flowering (expressing CO and AeCO) did not express higher levels of transgene mRNA than later flowering lines (expressing COL1, COL2, AeCOL2, ThCOL) (supplementary fig. S2E, Supplementary Material online). Furthermore, transgenic lines expressing COL1, COL2, AeCOL2, and ThCOL from pCO consistently expressed lower levels of FT mRNA than those expressing CO or AeCO (supplementary fig. S2F, Supplementary Material online). Collectively, these experiments show that CO and AeCO are more potent in promoting FT transcription and flowering than the COL1/2 homologs and the Cleomaceae ortholog.
To identify residues potentially responsible for these differences in protein activity, the COL and CO proteins from different species were aligned. Amino acid differences in the B-Boxes and CCT domain were searched for because these regions are functionally important based on the analysis of co mutant alleles (Robson et al. 2001). Our analysis revealed four amino acid positions that are conserved in COL1/2 homologs (including ThCOL and CpCO) and different in all CO homologs (supplementary fig. S3A, Supplementary Material online). Three are located in the B-Boxes (S49A; D79A; I90V) and the remaining residue is in the CCT domain (N342A). These four residues are candidates for conferring functional differences between COL and CO proteins.
These experiments indicate that COL1 and COL2 are able to accelerate flowering and activate FT transcription when expressed from pSUC2 or pCO, but the late-flowering phenotype and very low FT mRNA level of the co-10 mutant suggest that COL1 and COL2 do not carry out these functions when expressed from their native promoters (Jang et al. 2009). We thus also examined the spatial expression patterns of COL1 and COL2 by using promoter β-glucuronidase (GUS) fusion constructs in transgenic A. thaliana plants. pCO::GUS, which serves as a control, is expressed mainly in leaf vascular tissue, where CO activates FT transcription (supplementary fig. S3D, Supplementary Material online; Takada and Goto 2003; An et al. 2004). GUS activity driven by pCOL1, on the other hand, is more evenly distributed in the leaf and therefore differs from the pCO expression pattern. More precisely, pCOL1::GUS activity is less pronounced in the vascular tissue compared with pCO::GUS and it is also detectable in the lower part of young leaves (supplementary fig. S3D, Supplementary Material online). In pCOL2::GUS plants, enzyme activity is detected in the stem below or overlapping with the shoot apical meristem but it is very weak in other tissues (supplementary fig. S3D, Supplementary Material online). Thus, in addition to differences between CO and COL1/2 protein activity, cis-regulatory differences in the temporal and spatial expression of COL1/2 in comparison to CO likely contribute to subfunctionalization (Ledger et al. 2001; Kim et al. 2013) of the three genes following duplication.
The Promoter of CO Is Conserved in the Brassicaceae Family and Differs from COL Gene Promoters in Cis-Element Composition
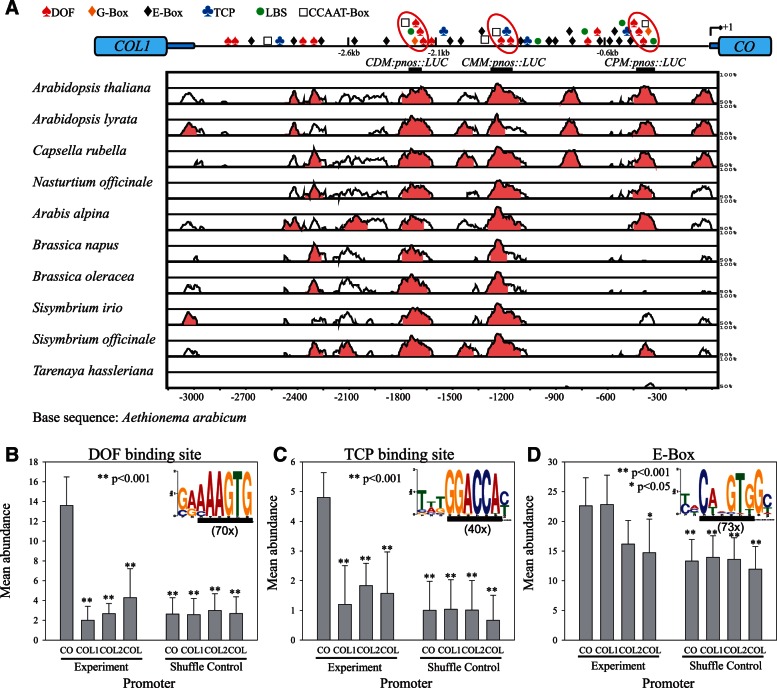
The CO transcriptional pattern is a novelty that arose in the Brassicaceae and is conserved in the different lineages represented by A. thaliana and Ae. arabicum (fig. 2). Therefore, the extent of promoter sequence conservation among these genes was examined. The intergenic region between CO and COL1 was identified and aligned with those from ten Brassicaceae species. The comparison using Ae. arabicum as a base genome revealed conserved regions (>70% identity, fig. 4A). Three conserved blocks, about − 200 bp, −1.4 kb and − 2.2 kb upstream of the ATG of CO (supplementary fig. S4A, Supplementary Material online), were identified (about − 300 bp, − 1.2 kb, and − 1.7 kb in Ae. arabicum; fig. 4A), which were named Conserved Proximal Motif (CPM), Conserved Middle Motif (CMM), and Conserved Distal Motif (CDM). In agreement with the differential expression patterns of ThCOL and CO, the promoter of ThCOL does not exhibit conservation of more than 50% of nucleotides (in a 100-bp sliding window) to Brassicaceae CO promoters (fig. 4A and supplementary fig. S4A, Supplementary Material online).
Fig. 4.
Conservation and cis-element composition of CO promoter in the Brassicaceae. (A) Pairwise alignment of the Aethionema arabicum CO promoter to orthologous sequences from ten Brassicaceae CO promoters as well as Tarenaya hassleriana COL promoter displayed as VISTA plots. Sequence similarity (%) was calculated in 100-bp sliding window while light-red color indicates greater than 75% identity. For clarity a diagram of Arabidopsis thaliana CO promoter is displayed above, indicating position of cis-elements (DOF = AAAGTG, TCP = GGACCA, E-Box = CANNTG, G-Box = CACGTG, LBS = GATWCG, and CCAAT-Box = CCAAT). Approximate positions of CO promoter sequences used for LUC reporter (CDM:pnos:LUC, CMM:pnos::LUC, CPM:pnos::LUC) and GFP:CO (2.6, 2.1, and 0.6 kb) complementation constructs in this study are indicated in relation to the vista plot. (B–D) Short motif search (MEME, 6–10 bp) in CO promoters from ten Brassicaceae species (see A) resulted in three elements that were found 70 (B), 40 (C), and 73 times (D), respectively (displayed as WEBLOGO). Identified motifs (underlined) contain the extended DOF binding site (B, AAAGTG), the TCP binding site (C, GGACCA), and the E-Box (D, CANNTG). Mean Abundance of these three cis-elements was determined in promoters (−4 kb) of Brassicaceae CO, COL1, and COL2 as well as non-Brassicaceae COL orthologs (COL) (B–D, Experiment, ±SEM). The same analysis was performed 100 times with shuffled data sets as control for base composition (Shuffle Control). Brassicaceae sequences include homologs from A. thaliana, A. alpina, Eutrema parvulum, Brassica rapa (1 × CO, 2 × COL1, and 2 × COL2), and Ae. arabicum (CO and COL2). Promoters from non-Brassicaceae species include T. hassleriana (ThCOL), Medicago truncatula (MtCOLa), Populus Trichocarpa (PtCO1; PtCO2), Solanum tuberosum (StCO1; StCO2), and Vitis vinifera (VvCO). Statistical analysis indicates significant differences (*P < 0.05; **P < 0.001) in mean abundance compared with CO.
Next, we investigated the cis-element composition of the three conserved motifs (fig. 4A and supplementary fig. S6, Supplementary Material online). We found two or three DOF binding sites in all three conserved motifs. The extended DOF binding site (AAAGTG) occurs in clusters in the CO promoter and was shown to be recognized by the CO repressor CDF1 (Imaizumi et al. 2005; Sawa et al. 2007; Rosas et al. 2014). Additionally all three conserved elements contain a CCAAT Box (CCAAT), which has not previously been described to be important in CO transcriptional regulation. However, CCAAT-Boxes are linked to vascular cell-specific transcriptional regulation through Nuclear Factor Y transcription factors and in promotion of transcription of FT, a downstream target of CO (Wenkel et al. 2006; Siefers et al. 2008; Cao et al. 2014). The CPM and CDM motifs additionally contain two elements not so far linked to CO transcriptional regulation: A G-Box (CACGTG) and a LUX Binding site (LBS) (GATWCG). G-Boxes are cis-elements implicated in control of transcription by light (Terzaghi and Cashmore 1995), whereas LBS motifs are bound by the Evening Complex through the LUX TF and are found in the promoters of circadian clock regulated genes (Helfer et al. 2011; Nusinow et al. 2011). CMM, but not CPM and CDM, contains a cis-element (GGACCA) that was associated with regulation by TCP4, a member of the TEOSINTE BRANCHED/CYCLOIDEA/PCF (TCP) transcription factor family, that has not been associated with regulation of CO (Schommer et al. 2008). In summary, CO promoter has two conserved motifs, CPM and CDM containing the same combination of cis-elements (3 × DOF, CCAAT-Box, G-Box, LBS) and one motif, the CMM with a partially overlapping combination (2 × DOF, CCAAT-Box, TCP) (fig. 4A and supplementary fig. S6, Supplementary Material online).
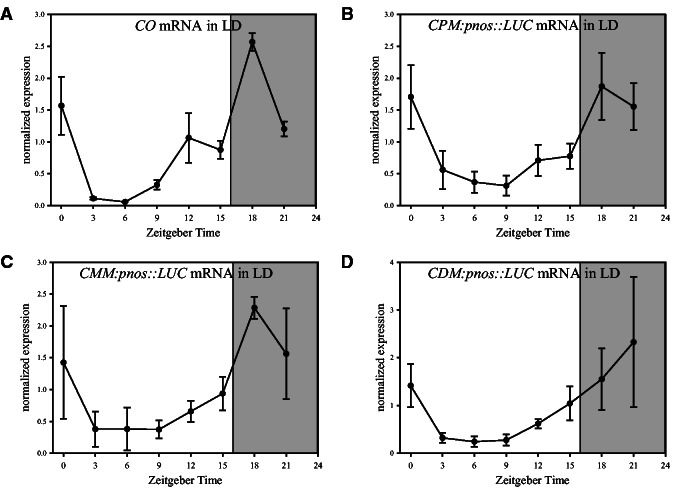
To investigate the functional importance of the three conserved motifs, the short promoter regions (fig. 4A and supplementary fig. S6, Supplementary Material online) were fused to the NOPALINE SYNTHASE (NOS) minimal promoter (pnos; Shaw et al. 1984; Puente et al. 1996) to drive LUCIFERASE (LUC) expression in transgenic A. thaliana plants. Luminescence due to LUC expression was detected in the vasculature of CPM:pnos::LUC, CMM:pnos::LUC and CDM:pnos::LUC plants but not in the pnos empty vector control line (supplementary fig. S5E, Supplementary Material online). The temporal expression of LUC mRNA was then tested using reverse transcriptase polymerase chain reaction (PCR), which was more precise than following luminescence due to the low expression level of each transgene. Compared with the full-length CO promoter (2.6 kb) driving LUC cDNA, CPM (135 bp), CMM (171 bp), and CDM (93 bp) confer much lower overall expression levels (supplementary fig. S5B, Supplementary Material online). Nevertheless, normalization of the expression patterns and comparison to endogenous CO transcription revealed that these short motifs confer diurnal transcriptional patterns similar to those of native CO (fig. 5), whereas the pnos empty vector control does not (supplementary fig. S5A, Supplementary Material online). In CPM:pnos::LUC and CMM:pnos::LUC lines, a shoulder was present in the peak of expression of LUC mRNA (ZT12-15) and a peak was present during the night (ZT18), which are characteristic of the CO mRNA temporal pattern, whereas CDM:pnos::LUC lines showed a more gradual upregulation in the evening and night (fig. 5). In summary, three conserved parts of CO promoter are independently sufficient to confer a diurnal and spatial pattern of expression similar to that of CO but at lower amplitude.
Fig. 5.
Functional analysis of conserved sequences in the Arabidopsis thaliana CO promoter. (A) Diurnal expression pattern of CO mRNA in LD conditions in 10-day-old A. thaliana Col-0 seedlings. CO expression values were determined in the material used in (B) to (D) and represent the mean data from four biological replicates. Diurnal expression of LUC driven by Conserved Proximal Motif (CPM:pnos::LUC, #4, #7, B), Conserved Middle Motif (CMM:pnos::LUC, #1, #2, C), or Conserved Distal Motif (CDM:pnos::LUC, #2, #4, D) in LD conditions. Expression values were normalized to PP2A and the average of each experiment. Each data point represents the mean ± SD of two independent transgenic lines from two biological replicates. Position and composition of CPM, CMM, and CDM are explained in figure 4A and supplementary figure S6, Supplementary Material online.
The functional impact on flowering-time regulation of the three conserved elements was tested in a promoter complementation assay. The elements are distributed along the full length of the region previously defined as the CO promoter (fig. 4A; An et al. 2004). The pCO::GFP:CO fusion construct was used to complement the co-2 mutation in A. thaliana Landsberg erecta (Ler) accession. Complementation with this 2.6-kb promoter caused early flowering in homozygous transgenic lines (supplementary fig. S5C and D, Supplementary Material online). Although co-2 mutant plants flowered with 20–30 leaves in LD conditions, the pCO::GFP:CO (2.6 kb) co-2 lines flowered with approximately six total leaves. The CDM motif was deleted from the CO promoter in the 2.1 kb pCO fragment (fig. 4A), and pCO::GFP:CO (2.1 kb) co-2 plants flowered significantly later (9–15 total leaves) than those carrying the full-length 2.6-kb promoter construct in three of four independent transgenic lines (supplementary fig. S5C, Supplementary Material online). This result indicates that the distal conserved motif (CDM) is located in a functionally important part of the CO promoter and that this segment is required for full acceleration of flowering in LDs. Whether a fusion of the proximal 0.6 kb of CO promoter to GFP:CO is sufficient to cause earlier flowering of co-2 mutants was then examined. However, no significant acceleration of flowering of pCO::GFP:CO (0.6 kb) co-2 was detected compared with co-2, although this proximal promoter contains the CPM motif (supplementary fig. S5D, Supplementary Material online). This promoter complementation assay indicates that the activity of pCO gradually decreases with progressive deletions from 2.6, 2.1, and 0.6 kb and only the fragment containing all three conserved regions provides full promoter activity.
The CO promoter evolved in the Brassicaceae and contains functionally important conserved sequences that harbor distinct combinations of cis-elements (figs. 4A and 5 and supplementary fig. S5 and S6, Supplementary Material online). To identify cis-element motifs associated with CO expression pattern, 6–10 bp motifs overrepresented in the ten Brassicaceae CO promoters were sought using the Multiple Em for Motif Elicitation (MEME) tool (Bailey et al. 2009). The most enriched sequences (present 73 × in the ten promoters) contain the E-Box (CANNTG, fig. 4D), which is the binding site for FLOWERING BHLH (FBH) transcription factors that activate CO transcription in A. thaliana (Ito et al. 2012). The second most abundant motif (70×) contains the extended DOF binding site (AAAGTG, fig. 4B) described earlier. The third motif identified (40×) carries a core sequence (GGACCA) that is bound by TCP transcription factors (Schommer et al. 2008; fig. 4C) and is part of the CMM. In order to investigate whether the identified cis-elements represent a novelty in CO promoters, the abundance of these elements was determined in CO, COL1 and COL2 promoters (−4 kb) from five Brassicaceae as well as subgroup Ia COL genes from six non-Brassicaceae species (fig. 4B–D). Available mRNA data for these COL genes indicated that they showed an expression pattern similar to COL1/2 of A. thaliana (Hecht et al. 2005, 2007; Bohlenius et al. 2006; Almada et al. 2009; González-Schain et al. 2012; Hsu et al. 2012; Kloosterman et al. 2013). CO promoters contained significantly more DOF and TCP binding sites compared with the COL gene promoters of Brassicaceae COL genes as well as those from other species and significantly more than the CO promoters shuffle control (fig. 4B and C). Additionally, the CO promoters contained more E-Boxes compared with the shuffle control and non-Brassicaceae COL promoters (fig. 4D) but were not significantly different in this respect to COL1 and COL2 genes from the Brassicaceae. A significant enrichment of G-Boxes was detected in Brassicaceae CO promoters compared with the shuffle control (supplementary fig. S4C, Supplementary Material online), but no enrichment for LBSs and CCAAT Boxes (supplementary fig. S4B and D, Supplementary Material online). In conclusion, a significant increase in cis-element number between CO and COL promoters was always observed for DOF binding sites, TCP binding sites, and G-Boxes.
Model for Evolution of CO Regulation by CDF Transcription Factors in the Brassicaceae Family
DOF binding sites are present in CPM, CMM, and CDM that confer a CO expression pattern and CDFs regulate the transcription of CO in A. thaliana (Imaizumi et al. 2005; Sawa et al. 2007; Fornara et al. 2009; fig. 5B–D). The cdf1-R, cdf2-1, cdf3-1, and cdf5-1 (cdf1235) quadruple mutant exhibits higher levels of CO mRNA (Fornara et al. 2009; supplementary fig. S7A and B, Supplementary Material online). To test how important CDFs and DOF binding sites are for discriminating CO from COL1/2 expression patterns, COL1 and COL2 transcript levels were tested in the cdf1235 quadruple mutant (supplementary fig. S7C–F, Supplementary Material online). No significant difference in the expression levels of these mRNAs was detected, indicating that, in contrast to CO, the COL1 and COL2 genes are not regulated by CDFs. These data suggest that CDF regulation of CO is either a novelty that arose in the Brassicaceae or that it was present in the last common ancestor of CO, COL1 and COL2 but was lost in COL1 and COL2 during the process of subfunctionalization.
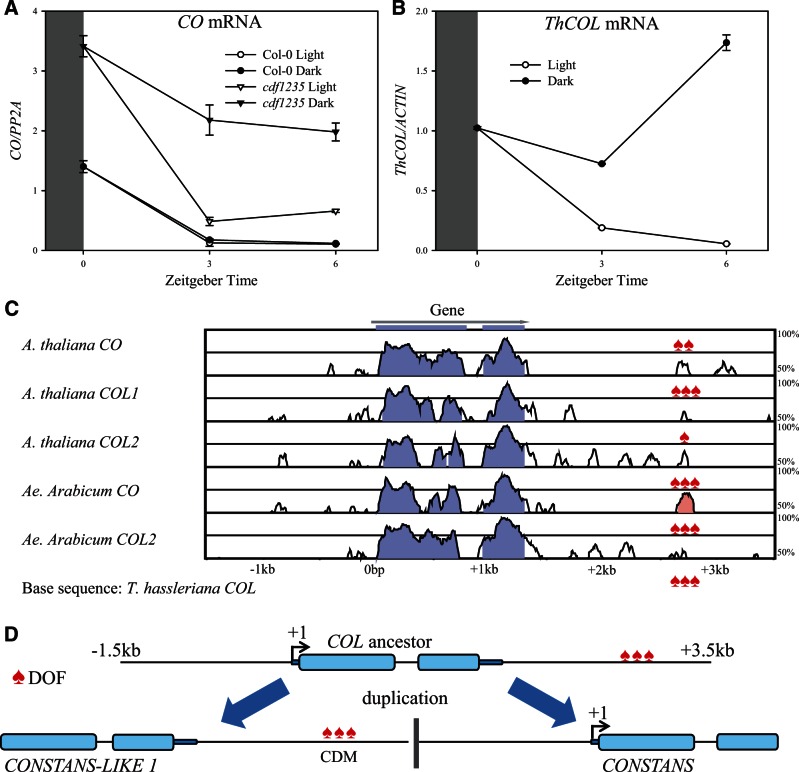
Another indication that DOF regulation is a novelty associated with the CO gene came from further comparison of the cdf1235 mutant and wt (Col-0). In wt A. thaliana, CO mRNA levels are efficiently downregulated in the morning to ensure the gene is not transcribed when plants are exposed to light early in the day. The CDFs contribute to this repression of CO, because in the cdf1235 mutant CO mRNA levels are generally higher (Fornara et al. 2009). However, if the cdf1235 mutant seedlings grown in LDs remain in darkness at subjective dawn, even higher CO mRNA levels are observed than if the plants are transferred to light (fig. 6A). This suggests that CO transcription is also repressed at dawn by a second mechanism that is independent of CDFs but dependent on light. This light-dependent mechanism is not detected in wt plants because it is overridden by CDF activity that strongly represses CO transcription. A similar effect of light is detected for ThCOL mRNA (fig. 6B) as well as COL1 and COL2 (supplementary fig. S8, Supplementary Material online; Ledger et al. 2001). These data indicate that ThCOL, such as COL1 and COL2, is not repressed by CDF transcription factors at dawn but by exposure to light, and that this regulation can also be detected for the CO promoter if CDF activity is removed in the cdf1235 mutant.
Fig. 6.
Model for evolution of CO regulation by CDF transcription factors in the Brassicaceae family. CO (A) or ThCOL (B) mRNA levels in LD grown seedlings (Arabidopsis thaliana Col-0 compared with cdf1235, A) (Tarenaya hassleriana, B) either kept in light or moved to dark before lights on. Mean ± SD after normalization to PP2A for technical triplicates and a biological replicate gave similar results. (C) Pairwise alignment of subgroup Ia COL gene loci (−1.5 to 3.5 kb) from A. thaliana (CO, COL1, and COL2), Aethionema arabicum (AeCO, AeCOL2), and T. hassleriana (ThCOL) displayed as VISTA plot. Sequence similarity (%) was calculated in 100-bp sliding window while color indicates greater than 75% base identity. Blue color indicates protein coding region of CO as annotated in TAIR10. ♠ indicates position of DOF binding sites (AAAGTG) in the conserved downstream region. (D) Model for the tandem duplication that led to COL1 and CO in the Brassicaceae. CO/COL1 gene body (−1.5 to +3.5 kb) became duplicated including conserved DOF binding sites in the downstream sequence of the ancestor (see C). In Brassicaceae CO promoters, this region overlaps with CDM (fig. 4A).
CO but not COL1/2 transcription is regulated by CDFs, raising the question of how this novelty arose. The number of DOF binding sites, which were shown to be bound by at least CDF1 (Sawa et al. 2007), increased during the evolution of the CO promoter based on the comparison with the promoters of ThCOL and Brassicaceae COL1/2 genes (fig. 4B). However, a conserved region was identified 3′ of the coding sequence of subgroup Ia COL genes, and specifically in the case of COL1 this sequence represents CDM found in the CO promoter (fig. 6C). Indeed, the 2 kb downstream of subgroup Ia COL genes is enriched in DOF binding sites, so that all of these loci contain between 1 and 3 such sites in the homologous region (CDM; fig. 6C). Thus, the tandem duplication that led to COL1 and CO in the Brassicaceae introduced this sequence motif distally upstream of CO to form the CDM (fig. 6D). This event placed DOF binding sites in the promoter of CO which probably influenced its mRNA expression pattern based on the functional analysis of deleted versions. Taken together the tandem duplication leading to CO in the Brassicaceae probably changed the transcriptional pattern of the gene by including sequences 3′ of the COL ancestor in the CO promoter.
Discussion
Gene duplications are often associated with the evolution of novelties or range expansions (Ohno 1970; Soltis et al. 2009; Levin 2011; Ramsey 2011). Such duplications can occur at the level of single genes or of whole genomes (Freeling 2009). In plants, WGDs have occurred many times and have been implicated in species diversification (Soltis et al. 2009; Schranz et al. 2012). The WGD At-α occurred prior to diversification of the core Brassicaceae (Vision et al. 2000; Bowers et al. 2003; Jiao et al. 2011; Haudry et al. 2013), around 30–40 Ma, and may have contributed to the evolutionary success of these plants in a cooling climate during the Late Eocene (Bowers et al. 2003; Couvreur et al. 2010). This capacity to survive in cool conditions likely also contributed to the range expansion of the core Brassicaceae into temperate and cold regions of central Asia as well as the Northern Hemisphere, where they are now predominately found (Couvreur et al. 2010). Brassicaceae species are rare in the tropics, in contrast to their sister family, the Cleomaceae. Most plant species in temperate regions show responses to photoperiod, synchronizing key stages of their life cycle with the changing seasons and thereby adapting to extreme seasonal fluctuations in their environment (Thomas and Vince-Prue 1997). Photoperiodic control of flowering or of bud dormancy is proposed to be adaptive (Ray and Alexander 1966; Bohlenius et al. 2006), and the expansion of the Brassicaceae into temperate regions extending to high latitudes likely required the evolution of photoperiodic responses. Here we demonstrate that one of the key genes in the photoperiodic flowering response of the Brassicaceae, CO, evolved by tandem duplication in a similar time interval to At-α. Our data indicate that CO arose by tandem duplication of COL1 in the ancestor of the Brassicaceae and is thus, like the At-α, shared by Aethionemae but absent from the Cleomaceae species T. hassleriana. We show that the characteristic spatial and temporal transcriptional patterns of CO as well as the capacity of its protein product to efficiently activate transcription of the downstream gene FT evolved soon after the tandem duplication and prior to diversification of the core Brassicaceae and Aethionemae but after divergence from the Cleomaceae. Therefore, efficient promotion of flowering upon exposure to inductive long-day stimuli likely arose in the Brassicaceae around the time they were exposed to a cooling climate. This conclusion is supported by the finding that a strong photoperiodic response is conserved between A. thaliana and Ae. arabicum but weak in the non-Brassicaceae species T. hassleriana that does not contain CO (Koevenig 1973; fig. 1B). Several models of gene retention after duplication have been proposed (Freeling 2009) and the history of several duplicate genes has been followed in the Brassicaceae (Hanikenne et al. 2008; Hofberger et al. 2013; Vlad et al. 2014). The duplication of CO was initially present in all Brassicaceae lineages after duplication, similar to what has been described for some genes involved in glucosinolate biosynthesis and regulation (Hofberger et al. 2013). In the case of CO this is likely due to positive selection after neofunctionalization, because after duplication and divergence of promoter and amino acid sequences, it appears to have provided a strong photoperiodic flowering response that was then frequently retained throughout the family.
Evolution of CO Protein Function
CO is a potent activator of flowering in A. thaliana (Rédei 1962; Putterill et al. 1995) and was the first identified member of a family of proteins that arose early during plant evolution, as indicated by their presence in algae and mosses (Zobell et al. 2005; Serrano et al. 2009). To compare the function of CO from A. thaliana with its closest homologs COL1 and COL2 as well as with orthologs from related species, all proteins were expressed in A. thaliana co null mutants from the same promoters. Overexpression of these proteins from a phloem specific promoter (pSUC2) demonstrated that Brassicaceae COL1 and COL2 proteins as well as their putative ortholog from T. hassleriana (ThCOL) induced co-10 plants to flower earlier than wt, although slightly later than plants carrying pSUC2::3xHA:CO or pSUC2::3xHA:AeCO (fig. 3C). Therefore, when overexpressed in the appropriate cells all subgroup Ia COL genes tested can activate transcription of FT and induce flowering of A. thaliana, although CO proteins are the most effective. When the native CO promoter was used instead of pSUC2, the difference in activity between CO (CO and AeCO) and COL proteins (COL1, COL2, AeCOL2, ThCOL) was more pronounced, and most of the pCO::3xHA:COL transformants flowered later than wt plants (fig. 3D). Taken together these data demonstrate that when expressed in A. thaliana from the same promoters, CO proteins are more effective at activating FT transcription and flowering than the most closely related COL proteins.
Four amino acids were identified that were altered in the B-Boxes and CCT domains of all Brassicaceae CO proteins compared with the Brassicaceae COLs. These are functionally important domains of the protein based on analysis of available mutant alleles (Putterill et al. 1995; Robson et al. 2001). After divergence these amino acids are highly conserved in the CO and COL proteins of species in different Brassicaceae lineages, and are candidates for explaining the functional differences between these proteins. Interestingly, in three of four cases (AAs 49, 79 and 342; supplementary fig. S3A, Supplementary Material online) CO is the derived state when compared with B-Box and CCT domain proteins from other plant families (Griffiths et al. 2003; Khanna et al. 2009; Gendron et al. 2012), suggesting that these changes contributed to a novel Brassicaceae-specific function of CO proteins compared with COLs. No effect of the S49A substitution on CO protein function was found in a previous report, but the interpretation of this experiment was complicated by overexpression of the mutant form using the 35 S promoter (Kim et al. 2013) and changing all four amino acids might have a synergistic effect that is required to detect functional differences.
CO Promoter Conservation and Evolution
The temporal and spatial patterns of transcription of CO differ from those of COL1 and COL2. The temporal differences are maintained in all species tested in the core Brassicaceae and Aethionemae, whereas the gene of T. hassleriana shows the temporal expression pattern of COL1/2. These results suggest that the ancestral gene at the root of the Brassicaceae family, prior to the diversification of the core Brassicaceae and Aethionemeae but after separation from Cleomaceae, showed a similar temporal pattern of expression to COL1/2. Thus evolutionary changes, particularly the CO/COL1 tandem duplication (fig. 6D), which occurred before radiation of the core Brassicaceae and the Aethionemae lineage, generated a novel transcriptional pattern characteristic of CO. Cis-regulatory regions responsible for the novel expression pattern of CO are conserved among all Brassicaceae species investigated and are not found in COL gene promoters (fig. 4A).
The temporal expression pattern of CO is strongly shaped by CDFs (Imaizumi et al. 2005; Sawa et al. 2007; Fornara et al. 2009). More DOF binding sites were found in the promoters of CO genes than in those of COL genes, and many of these sites are organized in clusters (CPM, CMM, and CDM). Interestingly, two conserved and functionally important clusters have the same organization and combination of cis-elements but appear to have differing origins (fig. 4 and supplementary fig. S6, Supplementary Material online). The distal conserved cluster (CDM) shows homology with a conserved region downstream of subgroup Ia COL genes and that we propose was introduced into the CO promoter as part of the tandem duplication that produced CO and COL1 (fig. 6C and D). A similar scenario in which 3′-sequences of an ancestral gene contributed to the promoter of a tandemly duplicated copy was previously described for tandemly duplicated MYB transcription factors in maize (Zhang et al. 2000), and may be a more general mechanism by which the transcriptional patterns of tandemly duplicated copies diverge (Casneuf et al. 2006). The proximal and middle clusters (CPM and CMM) may have subsequently arisen to reinforce the transcriptional pattern of CO. How these evolved is unknown, but the G-Box element in CPM is also present in the proximal promoters of COL1 and COL2, indicating that the proximal promoter of the COL ancestral gene was modified to give rise to the CPM. The number of DOF binding sites in CPM varies among Brassicaceae species and even among A. thaliana accessions (supplementary fig. S6, Supplementary Material online; Rosas et al. 2014). The DOF binding sites in this region are multiples of the same sequence (ACACTTT) which may have originated from a single site by defective DNA replication or also by unequal crossing over. These variations in the number of DOF binding sites in the proximal promoter probably affected its regulation (Rosas et al. 2014). Further examination of the role of classes of cis-elements in CO regulation such as the DOF binding sites or TCP motifs requires systematic site directed mutagenesis of all members of specific classes of motifs and the mutant versions could then be functionally tested in complementation assays.
Evolution of CO as a Photoperiod Sensor in Flowering Plants
We propose that the promoter of CO and the activity of its protein product coevolved to generate a photoperiodic switch in the Brassicaceae. Tandem duplication of a COL gene produced the progenitors of COL1 and CO early in the diversification of the Brassicaceae family (fig. 6C and D). On the protein level, changes in the coding sequence of CO occurred after the duplication, causing the protein to be more potent in the promotion of FT transcription and acceleration of flowering. Furthermore, the tandem duplication created a novel CO promoter by bringing the gene under the control of sequences present at the 3′-end of COL1. This arrangement is suggested to have initiated evolution of the characteristic temporal and spatial patterns of CO expression. These changes to CO transcription and protein function are proposed to have produced a potent photoperiodic flowering switch that conferred an adaptive advantage contributing to the expansion of Brassicaceae species into cooler, temperate environments.
The function of COL1 and COL2 as well as of the ancestral COL gene that gave rise to CO remains unclear. The high sequence conservation of CO, COL1, and COL2 suggested that these genes might be functionally redundant (Ledger et al. 2001). However, in contrast to CO, overexpression of COL1/2 from the ubiquitous 35 S promoter had no effect on flowering time, but altered circadian clock period length (Ledger et al. 2001; Kim et al. 2013). The failure of COL1 or COL2 to promote flowering when expressed from 35 S is probably caused by this promoter not being highly active in companion cells, because we detected clear acceleration of flowering when either protein was expressed from the SUC2 or CO promoter. In wt plants, a strong role for COL1 and COL2 in the promotion of flowering in response to LD under standard light conditions seems unlikely because the co-10 mutant is almost unresponsive to LD, showing a similar phenotype to plants carrying null mutations of FT and the related gene TSF (Jang et al. 2009). Nevertheless, COL1/2 might promote flowering under specific environmental conditions such as particular wavelengths of light, different light intensities or under nonstandard growth temperatures. No loss of function mutants for COL1 or COL2 has been studied in detail preventing thorough analysis of their function in wt plants. Furthermore, the similarity of their temporal expression patterns and protein sequences suggests that double mutants might be required to overcome functional redundancy. We are in the process of analyzing existing insertion alleles to determine whether they abolish activity of COL1 and COL2, and we are using CRISPR/Cas technology to generate true null alleles (Hyun et al. 2015). Nevertheless, at present whether these genes contribute to flowering-time control in wt plants remains unclear. Similarly, whether the ancestral COL gene that gave rise to CO had any effect on flowering time is unknown, although the observation that T. hassleriana is almost insensitive to photoperiod with respect to flowering time makes it less likely that the ancestral COL gene controlled photoperiodic responses. Reverse genetic analysis of the evolutionarily related COL gene in T. hassleriana would allow analysis of whether it plays a role in flowering-time control.
Genes related to CO regulate photoperiodic flowering in other plant families, and these are probably examples of convergent evolution. In potato, COL genes have been implicated in controlling the photoperiodic response of tuberization (González-Schain et al. 2012). In particular, a subgroup Ia COL gene, S. tuberosum CO1 (StCO1), that acts in this process is also under control of a potato CDF gene (StCDF1.2) and exhibits a similar expression pattern to CO in A. thaliana (Kloosterman et al. 2013; Morris et al. 2013). Interestingly StCO1 is found in the context of a recent tandem duplication where it is located directly downstream of its closest ortholog (StCO2). Furthermore, as in the tandem duplication in A. thaliana, StCO2 is not regulated by StCDFs and shows a clear morning pattern of expression similar to COL1 and COL2 (Kloosterman et al. 2013; Morris et al. 2013). The comparison of potato StCO1/2 and A. thaliana CO/COL1 indicates that changes in expression pattern are tightly linked to tandem duplication events and suggests that these two occurrences arose independently. In the cereals rice and barley, OsHd1 and HvCO1 (the closest homologs of CO) are also expressed in a similar temporal pattern to A. thaliana CO and contribute to photoperiodic flowering (Yano et al. 2000; Hayama et al. 2003; Turner et al. 2005; Li et al. 2009; Campoli et al. 2012; Huang et al. 2012). There are no reports on regulation of these genes by DOF transcription factors. Our results indicate that the transcriptional patterns of CO, OsHd1, and StCO1 are the result of convergent evolution and that these evolved independently to confer strong photoperiodic responses. Such a conclusion might also explain why in some families, such as legumes, CO-related genes do not appear to be involved in controlling photoperiodic flowering (Wong et al. 2014). CO genes might be particularly suited to confer photoperiodic flowering leading to their independent recruitment in different families. Their transcriptional and posttranslational regulation by light as well as their transcriptional control by the circadian clock is highly appropriate for conferring responses to day length (Andrés and Coupland 2012). In addition they encode transcription factors that can activate expression of FT gene homologs, which seem to transduce the photoperiodic signal in all plants tested. This combination of attributes might result in the independent recruitment of CO-related genes and proteins as sensitive photoperiodic sensors in different families of flowering plants.
Materials and Methods
Plant Material and Growth Conditions
Arabidopsis thaliana plant material from the Col-0 accession includes co-10 (Laubinger et al. 2006) and the quadruple cdf1-R cdf2-1 cdf3-1 cdf5-1 mutant (Fornara et al. 2009) as well as the co-2 mutant in the Ler accession (Koornneef et al. 1991). Experiments were performed on T. hassleriana (ES1100; Cheng et al. 2013) and Ae. arabicum (Bioproject Accession code PRJNA202984; Haudry et al. 2013). Flowering time measurements were done on soil grown plants in controlled environment rooms under LD (16 h light/8 h dark) or SD (8 h light/16 h dark).
Plasmid Construction and Plant Transformation
Full length coding sequences (CDS) from COL1 (At5g15850), COL2 (At3g02380), AeCO (AA40G00516, v2.5, id23428), AeCOL2 (AA10G00370, v2.5, id23428), and ThCOL (gb|ABD96940.1|) were amplified from cDNA made from A. thaliana (Col-0), Ae. arabicum or T. hassleriana and introduced into SUC2::3xHA:GW and pCO::3xHA:GW (2.5 kb CO promoter) using GATEWAY (GW) technology. Both vectors were derived from pAlligator2 (Bensmihen et al. 2004; Jang et al. 2009). The NOS gene minimal promoter (pnos 105 bp; Shaw et al. 1984) was inserted into GW::LUC using HINDIII restriction sites to produce GW:pnos::LUC. Short motifs from CO promoter (Ler, CDM, −2,308 to –2,214 bp; CMM, −1,599 to −1,428 bp; and CPM, −237 to −102 bp) were introduced into GW:pnos::LUC using GW technology. For complementation of co-2 mutation in A. thaliana, CO promoter fragments of 2.6 kb (−2,617 to −5 bp), 2.1 kb ( −2,075 to −5 bp), or 0.6 kb (−621 to −5 bp) were introduced by GW technology into GW::GFP:CO (An et al. 2004) or GW::LUC (both pGreenII). For GUS expression analysis, promoters of COL1 (1,102 bp from ATG) and COL2 (1,105 bp from ATG) were introduced by GW recombination into pGreenII (GW::GUS). All plasmids for plant transformation were introduced into Agrobacterium strain GV3101 (pMP90RK) and transformed into Col-0, co-10 or co-2 plants by the floral dip method (Clough and Bent 1998). All pGreenII transgenic lines were selected using BASTA (Bayer CropScience). For pAlligator2 transgenic lines, GFP-positive seeds were selected using a Leica MZFLIII stereomicroscope equipped with GFP filters.
DNA and Protein Sequences Used for Phylogenetic Reconstruction
Sequences for protein alignment were either identified by Basic Local Alignment Search Tool (BLAST) search or predicted from genomic and cDNA sequencing data (Arabis alpina, Ae. arabicum, and C. papaya) and accession numbers can be found in supplementary table S2, Supplementary Material online. For the phylogenetic analysis of CO promoter, intergenic region of CO (including its start codon) and the upstream gene COL1 were used for analysis (except for ThCOL where no COL1 is present). Arabis alpina CO promoter was assembled from short reads identified by BLAST search of the preliminary genome assembly (MPIPZ, Cologne). Sequences were assembled using SeqMan Pro (DNASTAR Lasergene Version 8.0.2). CO Promoters from Capsella rubella, Nasturtium officinale, and Sisymbrium officinale were amplified from genomic DNA (kindly provided by Markus Berns, MPIPZ, Cologne) using degenerate primers that anchor in COL1 and CO, respectively (COL1 Fw: TGACACMGGATATGGAATTG; CO Rv: TGGCAGAGTGRACTTGAGCA). The intergenic region between COL1 and CO orthologs was subsequently sequenced by primer walk. The CO promoter of Ae. arabicum was identified by BAC (bacterial artificial chromosome) screen using an CO probe (BAC probe Fw: ACTGGTGGTGGATCAAGAGG; Rv: TCTTGGGTGTGAAGCTGTTG) and sequenced by primer walk from two independent BACs. Accession numbers for CO promoters used in the alignment can be found in supplementary table S3, Supplementary Material online. For the comparison of cis-element composition, 5′-sequences (4,000 bp including the start codon) were identified in publicly available genomes. Accession numbers are given in supplementary table S4, Supplementary Material online. The accession numbers for the comparison of whole loci can be found in supplementary table S5, Supplementary Material online. To obtain Ae. arabicum, whole loci sequences paired-end next generation sequencing reads of Ae. arabicum genomic sequence (http://mustang.biol.mcgill.ca:8885/cgi-bin/hgGateway?org=A.+arabicum&db=aa_4&hgsid=1696, last accessed May 19, 2015) were de novo assembled using CLC Genomics Workbench (CLC Bio) with standard parameters. Desired genes (AeCO, AeCOL2) were subsequently identified in the Ae. arabicum genome running BLAST on a local database.
Bioinformatics Data Analysis Tools
Phylogenetic tree for relationship of subgroup Ia COL protein sequences was inferred using the Neighbor-Joining method in MEGA5 with bootstrap values from 10,000 replicates (Saitou and Nei 1987; Tamura et al. 2007). Substitution rates were calculated from a codon alignment of the respective CDS followed by calculation of substitution rates corrected by Jukes–Cantor method using SNAP v1.1.1 (Rodrigo and Learn 2001, Chapter 4, p. 55–72). CO promoter and COL locus alignments were performed using LAGAN algorithm on mVISTA (Mayor et al. 2000; Brudno et al. 2003). Output is displayed as conservation in per cent in 100-bp sliding windows to the corresponding reference sequence. Alignments are displayed using ClustalX 2.0.11 (Larkin et al. 2007). For motif identification in CO promoters, data sets were submitted to MEME motif identification tool (Version 4.6, Bailey and Elkan 1994) and searched for 6–10 bp motifs that can occur in any number. WebLogo was used to present conserved motifs (Version 2.8.2; Crooks et al. 2004). For statistical comparison of cis-element number, the − 4 kb sequence beginning with the start codon was used from the indicated genes. Cis-elements were counted using customized Perl script (supplementary material, Supplementary Material online, Geo Velikkakam James, MPIPZ, Cologne). Shuffle control data sets were generated with the algorithm “shuffle” (100 times, Version 1.02, Press et al. 2007, p. 281). To test for statistical differences in cis-element analysis and flowering time, one-way analysis of variance and Tukey test were performed using SigmaStat 3.5 (Systat Software).
GUS Assay
GUS staining was performed as described earlier (Sieburth and Meyerowitz 1997). In brief, 14–day-old A. thaliana seedlings were vacuum infiltrated (10 min) and incubated in X-Gluc solution at 37 °C for 4 h (pCO, pCOL1) or 12 h (pCOL2) and fixed in FAA (formaldehyde [3.7%], acetic acid [5%], ethanol [50%]) for 30 min. Plant material was destained using an ethanol series of 30%, 50%, and 70%. pCO control line was previously described (An et al. 2004). Images were taken with Leica MZFLIII stereomicroscope.
Spatial Expression of LUC
Ten-day-old, LD grown seedlings of pnos::LUC transgenic lines (Col-0) were sprayed with Luciferin (2.5 mM) and chemiluminescence was imaged with the Image Quant LAS4000 biomolecular imager (GE Healthcare Life Sciences) for 15 min at high resolution.
Quantification of mRNA Expression
RNA was extracted from 10-day-old seedlings of A. thaliana (∼25 seedlings per sampling), Ae. arabicum (∼25 seedlings per sampling), and T. hassleriana (2–5 seedlings per sampling) grown in indicated light conditions (LD, SD, and LD + dark shift) using RNeasy Plant Mini Kit (Qiagen). Samples were treated with Ambion DNase (life technologies) and 1.5–3 µg of RNA was used for cDNA synthesis (SuperScript II Reverse Transcriptase; Invitrogen). Real time PCR was performed using IQ SYBR Green Supermix (Bio-Rad) on LightCycler480 (Roche) for the genes indicated (supplementary table S1, Supplementary Material online) using standard protocol (95 °C 5 min; [95 °C 20 s; 60 °C 20 s; 72 °C 20 s] 45×) followed by melting curve. Data were normalized against arbitrary dilution series to check PCR efficiency. Concentrations were calculated as 2(cycle number). Values are normalized to PROTEIN PHOSPHATASE 2 A (PP2A) (AT1G69960) for A. thaliana and ACTIN for Ae. arabicum and T. hassleriana. For comparison of expression pattern, samples were additionally normalized to the mean of each data set (8 ZT values). Mean and standard deviation (SD) of two biological replicates were calculated. For dark shift experiments and comparison of pCO::LUC to CDM:pnos::LUC, CMM:pnos::LUC and CPM:pnos::LUC, single biological replicates are displayed as three technical replicates ± SD.
Supplementary Material
Supplementary tables S1–S5 and figures S1–S8 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Eric Schranz, Geo Velikkakam James, and Markus Berns for providing material; Rafael Martinez-Gallegos and Tobias Gabriel for technical support; and Christiane Kiefer as well as Federico Valverde for critical comments on the manuscript. This work was supported by the International Graduate School for Development Health and Disease, Cologne, Germany, as well as by the Deutsche Forschungsgemeinschaft through SFB680 and the Cluster of Excellence in Plant Sciences (CEPLAS). The laboratory of G.C. receives a core grant from the Max Planck Society. Accession numbers can be found in supplementary tables S2–S5, Supplementary Material online.
References
- Almada R, Cabrera N, Casaretto JA, Ruiz-Lara S, González Villanueva E. 2009. VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Rep. 28:1193–1203. [DOI] [PubMed] [Google Scholar]
- An H, Roussot C, Suarez-Lopez P, Corbesier L, Vincent C, Pinero M, Hepworth SR, Mouradov A, Justin S, Turnbull C, et al. 2004. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131:3615–3626. [DOI] [PubMed] [Google Scholar]
- Anderson JT, Willis JH, Mitchell-Olds T. 2011. Evolutionary genetics of plant adaptation. Trends Genet. 27:258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F, Coupland G. 2012. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 13:627–639. [DOI] [PubMed] [Google Scholar]
- Bailey CD, Koch MA, Mayer M, Mummenhoff K, O’Kane SL, Jr, Warwick SI, Windham MD, Al-Shehbaz IA. 2006. Toward a global phylogeny of the Brassicaceae. Mol Biol Evol. 23:2142–2160. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 2:28–36. [PubMed] [Google Scholar]
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F. 2004. Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561:127–131. [DOI] [PubMed] [Google Scholar]
- Blackman BK, Michaels SD, Rieseberg LH. 2011. Connecting the sun to flowering in sunflower adaptation. Mol Ecol. 20:3503–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. 2006. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312:1040–1043. [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:428–433. [DOI] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. 2003. LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13:721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli C, Drosse B, Searle I, Coupland G, von Korff M. 2012. Functional characterisation of HvCO1, the barley (Hordeum vulgare) flowering time ortholog of CONSTANS: functional characterisation of HvCO1 in barley. Plant J. 69:868–880. [DOI] [PubMed] [Google Scholar]
- Cao S, Kumimoto RW, Gnesutta N, Calogero AM, Mantovani R, Holt BF. 2014. A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 26:1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y. 2006. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biol. 7:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, van den Bergh E, Zeng P, Zhong X, Xu J, Liu X, Hofberger J, de Bruijn S, Bhide AS, Kuelahoglu C, et al. 2013. The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell 25:2813–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735–743. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316:1030–1033. [DOI] [PubMed] [Google Scholar]
- Couvreur TL, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K. 2010. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol Biol Evol. 27:55–71. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. 2009. Gene duplication and evolutionary novelty in plants. New Phytol. 183:557–564. [DOI] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KCS, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G. 2009. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 17:75–86. [DOI] [PubMed] [Google Scholar]
- Franks SJ, Sim S, Weis AE. 2007. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc Natl Acad Sci U S A. 104:1278–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. 2009. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 60:433–453. [DOI] [PubMed] [Google Scholar]
- Freeling M, Lyons E, Pedersen B, Alam M, Ming R, Lisch D. 2008. Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 18:1924–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA. 2012. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci U S A. 109:3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Schain ND, Díaz-Mendoza M, Żurczak M, Suárez-López P. 2012. Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner: regulation of tuberization by potato CONSTANS. Plant J. 70:678–690. [DOI] [PubMed] [Google Scholar]
- Griffiths S, Dunford RP, Coupland G, Laurie DA. 2003. The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol. 131:1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Willis JH. 2006. Divergent selection on flowering time contributes to local adaptation in Mimulus guttatus populations. Evol Int J Org Evol. 60:2466–2477. [PubMed] [Google Scholar]
- Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, Motte P, Kroymann J, Weigel D, Krämer U. 2008. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453:391–395. [DOI] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly-Lopez Z, Steffen JG, Hazzouri KM, et al. 2013. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet. 45:891–898. [DOI] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. 2003. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422:719–722. [DOI] [PubMed] [Google Scholar]
- Hecht V, Foucher F, Ferrandiz C, Macknight R, Navarro C, Morin J, Vardy ME, Ellis N, Beltran JP, Rameau C, et al. 2005. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137:1420–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V, Knowles CL, Vander Schoor JK, Liew LC, Jones SE, Lambert MJM, Weller JL. 2007. Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 144:648–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA. 2011. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol. 21:126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofberger JA, Lyons E, Edger PP, Chris Pires J, Eric Schranz M. 2013. Whole genome and tandem duplicate retention facilitated glucosinolate pathway diversification in the mustard family. Genome Biol Evol. 5:2155–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C-Y, Adams JP, No K, Liang H, Meilan R, Pechanova O, Barakat A, Carlson JE, Page GP, Yuceer C. 2012. Overexpression of Constans homologs CO1 and CO2 fails to alter normal reproductive onset and fall bud set in woody perennial poplar. PLoS One 7:e45448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhao X, Weng X, Wang L, Xie W. 2012. The rice B-box zinc finger gene family: genomic identification, characterization, expression profiling and diurnal analysis. PLoS One 7:e48242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Kim J, Cho SW, Choi Y, Kim J-S, Coupland G. 2015. Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 241:271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. 2005. FKF1 F-Box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309:293–297. [DOI] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N. 1999. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11:309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T. 2012. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci U S A. 109:3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. 2007. FT protein acts as a long-range signal in Arabidopsis. Curr Biol. 17:1050–1054. [DOI] [PubMed] [Google Scholar]
- Jang S, Torti S, Coupland G. 2009. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 60:614–625. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100. [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290:344–347. [DOI] [PubMed] [Google Scholar]
- Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu S-H. 2009. The Arabidopsis B-Box zinc finger family. Plant Cell 21:3416–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-K, Park H-Y, Jang YH, Lee JH, Kim J-K. 2013. The sequence variation responsible for the functional difference between the CONSTANS protein, and the CONSTANS-like (COL) 1 and COL2 proteins, resides mostly in the region encoded by their first exons. Plant Sci. 199–200:71–78. [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T. 2001. Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13:681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman B, Abelenda JA, Gomez MdMC, Oortwijn M, de Boer JM, Kowitwanich K, Horvath BM, van Eck HJ, Smaczniak C, Prat S, et al. 2013. Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495:246–250. [DOI] [PubMed] [Google Scholar]
- Koevenig JL. 1973. Floral development and stamen filament elongation in Cleome hassleriana. Am J Bot. 60:122. [Google Scholar]
- Koornneef M, Hanhart CJ, Van der Veen JH. 1991. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 229:57–66. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- Laubinger S, Marchal M, Le Gourrierec J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoecker U. 2006. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133:3213–3222. [DOI] [PubMed] [Google Scholar]
- Ledger S, Strayer C, Ashton F, Kay SA, Putterill J. 2001. Analysis of the function of two circadian-regulated CONSTANS-LIKE genes. Plant J. 26:15–22. [DOI] [PubMed] [Google Scholar]
- Leister D. 2004. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet. 20:116–122. [DOI] [PubMed] [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D. 2005. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 1:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. 2011. Polyploidy and ecological transfiguration in Achillea. Proc Natl Acad Sci U S A. 108:6697–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Yang C, Li X, Gan Q, Zhao X, Zhu L. 2009. Functional characterization of rice OsDof12. Planta 229:1159–1169. [DOI] [PubMed] [Google Scholar]
- Liu J, Yu J, McIntosh L, Kende H, Zeevaart JA. 2001. Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol. 125:1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockton S, Gaut BS. 2005. Plant conserved non-coding sequences and paralogue evolution. Trends Genet. 21:60–65. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. 2007. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol. 17:1055–1060. [DOI] [PubMed] [Google Scholar]
- Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. 2000. VISTA : visualizing global DNA sequence alignments of arbitrary length. Bioinforma Oxf Engl. 16:1046–1047. [DOI] [PubMed] [Google Scholar]
- Mendez-Vigo B, Pico FX, Ramiro M, Martinez-Zapater JM, Alonso-Blanco C. 2011. Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol. 157:1942–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM. 2003. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci US A. 100:10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris WL, Hancock RD, Ducreux LJM, Morris JA, Usman M, Verrall SR, Sharma SK, Bryan G, Mcnicol JW, Hedley PE, et al. 2013. Day length dependent re-structuring of the leaf transcriptome and metabolome in potato genotypes with contrasting tuberisation phenotypes. Plant Cell Environ. 37:1351–1363. [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA. 2011. The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475:398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Berlin-Heidelberg-New York: Springer-Verlag. [Google Scholar]
- Okazaki K, Sakamoto K, Kikuchi R, Saito A, Togashi E, Kuginuki Y, Matsumoto S, Hirai M. 2007. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor Appl Genet. 114:595–608. [DOI] [PubMed] [Google Scholar]
- Press WH, Teukolsky SA, Vetterling WT, Flannery BP. 2007. Numerical recipes: the art of scientific computing. 3 rd ed. Cambridge: Cambridge University Press. [Google Scholar]
- Puente P, Wei N, Deng XW. 1996. Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 15:3732. [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. 1995. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80:847–857. [DOI] [PubMed] [Google Scholar]
- Ramsey J. 2011. Polyploidy and ecological adaptation in wild yarrow. Proc Natl Acad Sci U S A. 108:7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PM, Alexander WE. 1966. Photoperiodic adaptation to latitude in Xanthium strumarium. Am J Bot. 53:806. [Google Scholar]
- Rédei GP. 1962. Supervital mutants of Arabidopsis. Genetics 47:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS. 2006. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol. 2:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert LS, Robson F, Sharpe A, Lydiate D, Coupland G. 1998. Conserved structure and function of the Arabidopsis flowering time gene CONSTANS in Brassica napus. Plant Mol Biol. 37:763–772. [DOI] [PubMed] [Google Scholar]
- Robson F, Costa MMR, Hepworth SR, Vizir I, Reeves PH, Putterill J, Coupland G. 2001. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28:619–631. [DOI] [PubMed] [Google Scholar]
- Rodrigo AG, Learn GH. 2001. Computational and evolutionary analysis of HIV molecular sequences. Boston: Kluwer Academic Publishers. [Google Scholar]
- Rosas U, Mei Y, Xie Q, Banta JA, Zhou RW, Seufferheld G, Gerard S, Chou L, Bhambhra N, Parks JD, et al. 2014. Variation in Arabidopsis flowering time associated with cis-regulatory variation in CONSTANS. Nat Commun. 5:3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288:1613–1616. [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T. 2007. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318:261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. 2008. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz ME, Mitchell-Olds T. 2006. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell 18:1152–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz ME, Mohammadin S, Edger PP. 2012. Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr Opin Plant Biol. 15:147–153. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Quijada P, Sung S-B, Lukens L, Amasino R, Osborn TC. 2002. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162:1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano G, Herrera-Palau R, Romero JM, Serrano A, Coupland G, Valverde F. 2009. Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr Biol. 19:359–368. [DOI] [PubMed] [Google Scholar]
- Shaw CH, Carter GH, Watson MD. 1984. A functional map of the nopaline synthase promoter. Nucleic Acids Res. 12:7831–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. 1999. The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11:445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C. 2005. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138:1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. 1997. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. Plant Cell 9:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefers N, Dang KK, Kumimoto RW, Bynum WE, Tayrose G, Holt BF. 2008. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149:625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, dePamphilis CW, Wall PK, Soltis PS. 2009. Polyploidy and angiosperm diversification. Am J Bot. 96:336–348. [DOI] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. 2012. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336:1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J. 2004. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc Natl Acad Sci U S A. 101:4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. 2001. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410:1116–1120. [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K. 2003. TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15:2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 24:1596–1599. [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. 1995. Light-regulated transcription. Annu Rev Plant Biol. 46:445–474. [Google Scholar]
- Thomas B, Vince-Prue D. 1997. Photoperiodism in plants. San Diego (CA): Academic Press. [Google Scholar]
- Tiwari SB, Shen Y, Chang H-C, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, et al. 2010. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187:57–66. [DOI] [PubMed] [Google Scholar]
- Toomajian C, Hu TT, Aranzana MJ, Lister C, Tang C, Zheng H, Zhao K, Calabrese P, Dean C, Nordborg M. 2006. A nonparametric test reveals selection for rapid flowering in the Arabidopsis genome. PLoS Biol. 4:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. 2008. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 59:573–594. [DOI] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. 2005. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310:1031–1034. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. 2004. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303:1003–1006. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. 2000. The origins of genomic duplications in Arabidopsis. Science 290:2114–2117. [DOI] [PubMed] [Google Scholar]
- Vlad D, Kierzkowski D, Rast MI, Vuolo F, Dello Ioio R, Galinha C, Gan X, Hajheidari M, Hay A, Smith RS, et al. 2014. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343:780–783. [DOI] [PubMed] [Google Scholar]
- Wang R, Farrona S, Vincent C, Joecker A, Schoof H, Turck F, Alonso-Blanco C, Coupland G, Albani MC. 2009. PEP1 regulates perennial flowering in Arabis alpina. Nature 459:423–427. [DOI] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. 2006. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18:2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A, Hecht VF, Picard K, Diwadkar P, Laurie RE, Wen J, Mysore K, Macknight RC, Weller JL. 2014. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Plant Genet Genomics. 5:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J. 2003. Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci U S A. 100:6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna M, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12:2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gaut BS. 2003. Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res. 13:2533–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Chopra S, Peterson T. 2000. A segmental gene duplication generated differentially expressed myb-homologous genes in maize. Plant Cell 12:2311–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobell O, Coupland G, Reiss B. 2005. The family of CONSTANS-like genes in Physcomitrella patens. Plant Biol. 7:266–275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.