Abstract
Long noncoding RNAs (lncRNAs) are important developmental regulators in bilaterian animals. A correlation has been claimed between the lncRNA repertoire expansion and morphological complexity in vertebrate evolution. However, this claim has not been tested by examining morphologically simple animals. Here, we undertake a systematic investigation of lncRNAs in the demosponge Amphimedon queenslandica, a morphologically simple, early-branching metazoan. We combine RNA-Seq data across multiple developmental stages of Amphimedon with a filtering pipeline to conservatively predict 2,935 lncRNAs. These include intronic overlapping lncRNAs, exonic antisense overlapping lncRNAs, long intergenic nonprotein coding RNAs, and precursors for small RNAs. Sponge lncRNAs are remarkably similar to their bilaterian counterparts in being relatively short with few exons and having low primary sequence conservation relative to protein-coding genes. As in bilaterians, a majority of sponge lncRNAs exhibit typical hallmarks of regulatory molecules, including high temporal specificity and dynamic developmental expression. Specific lncRNA expression profiles correlate tightly with conserved protein-coding genes likely involved in a range of developmental and physiological processes, such as the Wnt signaling pathway. Although the majority of Amphimedon lncRNAs appears to be taxonomically restricted with no identifiable orthologs, we find a few cases of conservation between demosponges in lncRNAs that are antisense to coding sequences. Based on the high similarity in the structure, organization, and dynamic expression of sponge lncRNAs to their bilaterian counterparts, we propose that these noncoding RNAs are an ancient feature of the metazoan genome. These results are consistent with lncRNAs regulating the development of animals, regardless of their level of morphological complexity.
Keywords: long noncoding RNAs, evolution, gene expression, complexity, basal metazoans
Introduction
Bilaterian animal genomes (vertebrates, insects, worms, and their allies) encode a vast range of nonprotein coding RNAs (ncRNAs) that differ in size and level of conservation (Eddy 2001; Storz 2002; Amaral et al. 2008; Dinger et al. 2009; Mattick 2009b). ncRNAs are comprised a raft of different small RNA (sRNA) types, including microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs) and small interfering RNAs (siRNAs), and long noncoding RNAs (lncRNAs) that have been implicated in transcriptional and posttranscriptional regulation of gene expression or in guiding DNA modification (Eddy 2001). Although many of these broad classes of ncRNAs can be found in other kingdoms of eukaryotes, including plants, it remains unclear if these are conserved features of the ancestral eukaryotic genome or if they evolved independently multiple times (Pang et al. 2006; Ponting et al. 2009; Qu and Adelson 2012). To understand the origin and evolution of metazoan ncRNAs, the genomes of early-branching metazoan lineages need to be analyzed for regulatory RNA content and function (Grimson et al. 2008; Moran et al. 2013, 2014; Moroz et al. 2014).
lncRNAs are a case in point. They have been characterized in only a limited number of bilaterians (vertebrates, insects, and worms), budding yeast and plants, where they have emerged as an important class of regulators of gene expression (Carninci et al. 2005; Ravasi et al. 2006; Guttman et al. 2009; Cabili et al. 2011; Ulitsky et al. 2011; Boerner and McGinnis 2012; Derrien et al. 2012; Geisler et al. 2012; Liu et al. 2012; Nam and Bartel 2012; Pauli et al. 2012; Rinn and Chang 2012; Young et al. 2012; Cloutier et al. 2013; Sauvageau et al. 2013; Brown et al. 2014; Li et al. 2014; Zhang, Liao, et al. 2014; Zhou et al. 2014). lncRNAs are endogenous RNAs that resemble mRNAs in terms of CpG islands, complex splicing patterns, 5′ terminal methylguanosine cap and poly(A) 3′ tails (Carninci et al. 2005; Birney et al. 2007; Guttman et al. 2009, 2010; Derrien et al. 2012; Guttman and Rinn 2012). However, they are not translated in a similar manner to mRNAs (Guttman et al. 2013; Ingolia et al. 2014). Although some lncRNAs are transcribed by RNA polymerase III (Dieci et al. 2007; Kapranov et al. 2007) or produced by partial processing by the snoRNA machinery (Yin et al. 2012; Zhang, Yin, et al. 2014), the majority of lncRNAs shows a clear signature of RNA polymerase II transcription, with the promoters marked by histone H3 lysine 4 trimethylation (H3K4me3) and the transcribed gene bodies marked by histone H3 lysine 36 trimethylation (H3K36me3) (Guttman et al. 2009; Khalil et al. 2009).
Although most lncRNAs have not been functionally characterized, those that have been suggest lncRNAs are versatile molecules that can interact with DNA, other RNAs and proteins, either through nucleotide base pairing or through formation of structural domains generated by RNA folding (Wilusz et al. 2009; Poliseno et al. 2010; Salmena et al. 2011; Wang and Chang 2011). As expected for regulatory molecules, lncRNAs display specific spatiotemporal expression patterns, high tissue specificity (Cabili et al. 2011; Djebali et al. 2012; Pauli et al. 2012; Li et al. 2014; Necsulea et al. 2014; Washietl et al. 2014) and can regulate expression of genes in close genomic proximity (cis-acting) or at distance (trans-acting) (Mercer et al. 2009; Ponting et al. 2009; Rinn and Chang 2012; Ulitsky and Bartel 2013). Several lncRNAs have been shown to act as decoys that titrate away miRNAs or regulatory proteins, such as transcription factors and chromatin modifiers (Wang and Chang 2011). Other lncRNAs may act as scaffolds to bring two or more proteins into a complex or in physical proximity (Wang and Chang 2011). An example of a scaffold lncRNA is HOTAIR, which can epigenetically silence gene expression at many sites across the human genome by recruitment of both the Polycomb Repressive Complex 2 (PRC2) and the Lysine (K)-specific demethylase 1A/RE1-silencing transcription factor/REST corepressor 1 (LSD1/REST/CoREST) repressive chromatin modifying complexes (Rinn et al. 2007; Tsai et al. 2010). Also, many lncRNAs act as guides to recruit chromatin-modifying enzymes and are individually required for proper localization of these ribonucleoprotein complexes to specific targets (Wang and Chang 2011).
Although some lncRNAs are highly conserved within vertebrates (Feng et al. 2006; Chodroff et al. 2010; Ulitsky et al. 2011; Necsulea et al. 2014; Washietl et al. 2014; Zhou et al. 2014), previous studies established that genomic sequence and gene structure conservation are rare at putative orthologous lncRNA loci, and that lncRNAs are subjected to rapid turnover during evolution (Wang et al. 2004; Pang et al. 2006; Guttman et al. 2009; Mercer et al. 2009; Ponting et al. 2009; Kelley and Rinn 2012; Kutter et al. 2012; Ulitsky and Bartel 2013; Kapusta and Feschotte 2014; Washietl et al. 2014). Although conservation indicates functionality, lack of sequence conservation does not imply lack of function (Pang et al. 2006). Because of the flexible relationship between lncRNA primary sequence and function, lncRNA primary sequences may be more pliable to evolutionary pressures than protein-coding genes, as evidenced by the existence of many lineage-specific lncRNAs (Pang et al. 2006). However, the question of what fraction of lncRNAs act as functional transcripts remains controversial.
It has been proposed that there is a positive correlation between ncRNAs number and diversity, and developmental and cognitive complexity (Mattick and Makunin 2006; Taft et al. 2007; Mattick 2009a; Liu et al. 2013), and that lncRNAs have contributed to the evolution of complex metazoan features, in particular the mammalian brain (Mattick 2011; Sauvageau et al. 2013). However, there currently is a paucity of comparative data from morphologically simple, early-branching metazoans (Srivastava et al. 2008, 2010; Ryan et al. 2013; Moroz et al. 2014). Here, we report the systematic identification and characterization of developmentally regulated lncRNAs in the marine demosponge Amphimedon queenslandica (Hooper and Van Soest 2006). Amphimedon belongs to the phylum Porifera (fig. 1A), an ancient phyletic lineage of morphologically simple animals that diverged from other metazoans at least 700 Ma, well before the Cambrian explosion (Erwin et al. 2011). We combine developmental RNA-Seq data for Amphimedon with a stringent computational filtering pipeline to predict a high-confidence set of lncRNA transcripts. Notably, sponge lncRNAs are remarkably similar to their bilaterian counterparts, showing features typical of regulatory molecules, including dynamic and stage-specific developmental expression profiles. lncRNA features shared between bilaterians and a sponge are likely to have been present in their last common ancestor. Our analysis, the first systematic identification of lncRNAs in a basal metazoan, therefore suggests antiquity of complex metazoan genome regulation by lncRNAs, and we propose that lncRNAs may be essential regulatory elements that fulfill a wide range of functions in development, regardless of the level of morphological complexity.
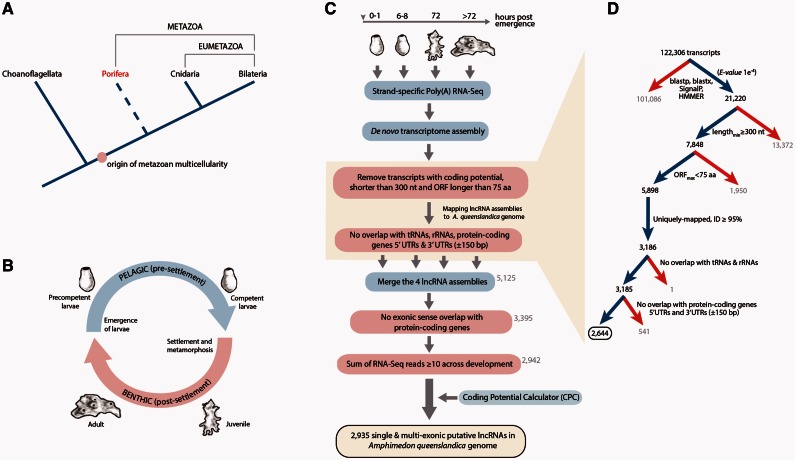
Fig. 1.
Identification of Amphimedon lncRNAs. (A) Animal phylogeny. The phylogenetic relationship of Porifera, Cnidaria, Bilateria, and the sister group to metazoans, Choanoflagellata, is shown here, along with the evolutionary origin of metazoan multicellularity. Monophyly of Porifera (sponges; in red) remains controversial, indicated by dashed line. (B) Schematic representation of the demosponge Amphimedon queenslandica life cycle. Larvae emerge from maternal brood chambers and then swim in the water column as precompetent larvae before they develop competence to settle and initiate metamorphosis. Upon settling, the larva adopts a flattened morphology as it metamorphoses into a juvenile, which displays the hallmarks of the adult body plan, including an aquiferous system with canals, choanocytes chambers, and oscula (Leys and Degnan 2002). This juvenile will grow and mature into a benthic adult. (C) Overview of the computational filtering pipeline used for the identification of sponge lncRNAs. See main text and Materials and Methods for details. Red boxes highlight the major filtering steps. Yellow box highlights the final number of transcripts that passed all filters and were considered high-confidence Amphimedon lncRNAs. (D) Details of the filtering pipeline used for the identification of putative lncRNAs in competent larvae. At each step, a blue arrow denotes the transcripts that passed the filter; a red arrow, those that did not pass the filter. Black bold numbers indicate the number of transcripts that passed the filter.
Results
A Comprehensive Yet Conservative Catalog of 2,935 Sponge lncRNAs
To identify lncRNA transcripts expressed during sponge development, we performed RNA-Seq experiments across four time points that span the pelagobenthic life cycle of A. queenslandica: 1) 0–1 h after emergence of the swimming larvae from the brood chambers of the adult (precompetent larva), 2) 6–8 h after emergence when larvae become competent to respond to environmental cues and initiate settlement and metamorphosis (competent larva), 3) 72 h after settlement when a functional water-filtering system is established (juvenile), and 4) adult (fig. 1B) (Fernandez-Valverde et al. 2015). Approximately 84 million raw 100-bp paired-end sequence reads were obtained from poly(A) RNA from each of the four stages and about 78 million sequence reads per stage passed initial quality thresholds (supplementary table S1, Supplementary Material online). We assembled transcripts expressed in each developmental stage in a genome-independent manner using the de novo assembler Trinity (Haas et al. 2013). This approach performs well in genomes with high gene density such as in Amphimedon (Srivastava et al. 2010; Fernandez-Valverde et al. 2015). This resulted in a comprehensive de novo assembly of a total number of 443,650 transcripts across the whole developmental time course, from precompetent larva to adult (supplementary table S1, Supplementary Material online).
We developed a highly stringent filtering pipeline designed to remove transcripts with evidence for protein-coding potential based on current approaches (Boerner and McGinnis 2012; Nam and Bartel 2012; Pauli et al. 2012; Kaushik et al. 2013; Li et al. 2014; Zhou et al. 2014). We used four core filtering criteria: 1) Homology with known proteins and protein domains, 2) presence of signal peptides, 3) transcript length, and 4) open reading frame (ORF) size (fig. 1C and D). First, we removed transcripts with similarity to known proteins based on BLASTp and BLASTx (NCBI nr database [db]) (Altschul et al. 1990). Second, we removed transcripts with similarity to Amphimedon-specific predicted peptides (local db) and subsequently to known protein domains and signal peptides based on HMMER (Finn et al. 2011) (Pfam domains) and SignalP (Petersen et al. 2011), respectively (fig. 1D). These filters retained 15,400 ncRNAs in the precompetent larva, 21,220 ncRNAs in the competent larva, 12,926 ncRNAs in the juvenile, and 14,207 ncRNAs in the adult. We filtered these remaining transcripts based on their length, removing those shorter than 300 nt, a stricter cutoff than the 200 nt commonly used to identify lncRNAs (Orom et al. 2010; Cabili et al. 2011; Boerner and McGinnis 2012; Derrien et al. 2012; Young et al. 2012; Zhou et al. 2014) (fig. 1D). We subjected the residual putative lncRNAs to an ORF prediction and, subsequently, removed any remaining transcripts of uncertain protein-coding potential by applying a strict ORF size cutoff (fig. 1D). The complete discrimination of a functional ORF from a nonfunctional one is challenging without experimentally assessing for the presence of predicted peptides. However, it is expected that a large, complete ORF is more likely to be translated into a protein (Boerner and McGinnis 2012). To examine the effect of varying the ORF size cutoff, we analyzed all putative lncRNAs longer than 300 nt in each developmental stage, selecting for specific ORF size cutoffs (50, 75, 100, 150, 200, 250, 300, and >300 amino acids). When the ORF size selection is increased, the number of lncRNAs in each developmental stage that passes through selection gradually decreases, displaying a unimodal distribution centered on a median ORF size of approximately 50 amino acids (supplementary fig. S1, Supplementary Material online). Thus, to retain a significant level of strictness without losing an excessive number of potential lncRNAs, we selected an ORF size cutoff of 75 amino acids.
We then mapped the putative lncRNAs to Amphimedon genome (Srivastava et al. 2010) using UCSC’s BLAT software (Kent 2002), retaining only those that uniquely mapped to the genome with at least 95% identity (fig. 1D). These mapped transcripts were filtered to remove those that overlapped on the same strand of annotated transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), protein-coding gene 5′-untranslated regions (UTRs) plus 150 bp upstream, and protein-coding gene 3′-UTRs plus 150 bp downstream (fig. 1C and D). This approach retained 2,596 lncRNAs in the precompetent larva, 2,644 lncRNAs in the competent larva, 1,702 lncRNAs in the juvenile, and 1,964 lncRNAs in the adult. We then merged the putative lncRNAs from these four time points with Cuffmerge (Trapnell et al. 2010) and, to exclude peptide-encoding transcripts resulting from potentially incomplete transcript structures, we removed any transcript that had a sense exonic overlap with a protein-coding gene. The resulting set contained 3,395 candidate lncRNAs (fig. 1C). Finally, to reduce noise without losing low-abundance transcripts, we retained lncRNAs with an overall expression of at least ten raw read counts in total across the developmental stages. This step retained a set of 2,942 lncRNAs expressed in Amphimedon larvae, juveniles, and adults (fig. 1C). To corroborate our custom-filtering pipeline, we evaluated the coding potential of these sponge lncRNA candidates using the coding potential calculator (CPC) software (Kong et al. 2007) (fig. 1C), which assesses the quality, completeness, and sequence similarity of potential ORFs to proteins in the NCBI protein db. Only 7 of the 2,942 (0.2%) lncRNAs showed either homology to known proteins and/or protein domains or were defined as “coding” by CPC, and were subsequently removed from the sponge lncRNAs repertoire (supplementary table S2, Supplementary Material online).
Thus, with our comprehensive yet conservative pipeline we identified a final set of 2,935 high-confidence lncRNAs expressed throughout A. queenslandica development (fig. 1C) (supplementary table S3, Supplementary Material online).
Sponge lncRNAs Share Many of the Features of Their Bilaterian Counterparts
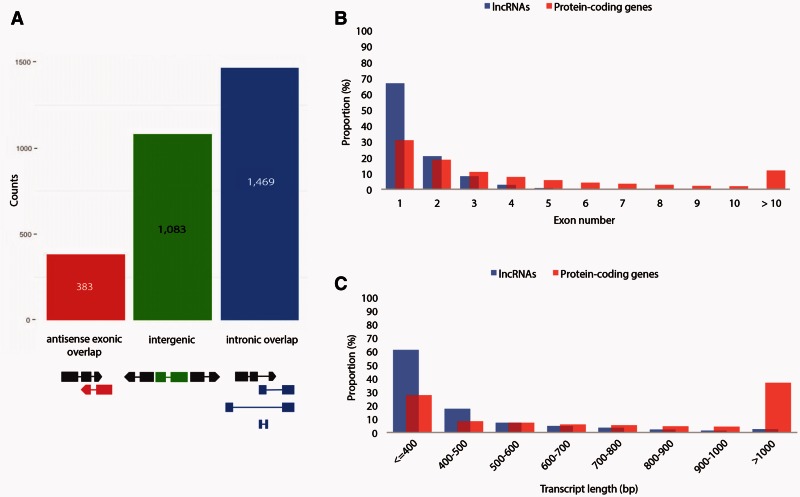
According to their genomic location, the 2,935 lncRNAs are further divided into 1,083 long intergenic ncRNAs (lincRNAs) that do not overlap with any protein-coding genes, 1,469 intronic lncRNAs, and 383 antisense exonic overlapping lncRNAs. Intronic lncRNAs are defined as lncRNAs that overlap with a coding gene in either sense or antisense orientation but have no exon–exon overlap (fig. 2A). This categorization is consistent with previous studies in vertebrates (e.g., Derrien et al. 2012; Pauli et al. 2012).
Fig. 2.
Classification and characterization of Amphimedon lncRNAs. (A) Number of lncRNAs in each of the three main classes defined by their genomic location relative to protein-coding genes. A schematic representation of lncRNAs (color) position relative to protein-coding genes (black) is shown at the bottom. lncRNAs with “antisense exonic overlap” (red) have at least one exon that overlaps with an exon of a protein-coding gene on the opposite strand. “Intergenic” lncRNAs (green) have no overlap with any protein-coding gene. lncRNAs with “intronic overlap” (light blue) are defined as transcripts that have overlap with another protein-coding gene but no exon–exon overlap (no overlap with exons of the overlapping genes). (B) Number of exons of Amphimedon lncRNAs in comparison to protein-coding genes. lncRNAs have fewer exons per transcript (median 1; average 1.5) than protein-coding genes (median 2; average 4.9). (C) Length of Amphimedon lncRNAs in comparison to protein-coding genes (mean length of 424 nt for lncRNAs vs. 1,118 nt for protein-coding genes) based on the current genome assembly.
One expected role of these sponge high-confidence lncRNAs would be to act as precursor molecules that are further processed into sRNAs (Birney et al. 2007; Wilusz et al. 2008). To identify putative sRNA-precursor lncRNAs, we compared our sponge lncRNAs catalogue with data sets of sRNAs (Grimson et al. 2008; Calcino AD, Degnan BM, et al., unpublished data) expressed at the same major life cycle transitions previously described. We identified 69 (2.4%) lncRNAs that appeared to be precursors for the production of piRNAs, endogenous small-interfering RNAs (endo-siRNAs), or sRNAs of unknown categories (supplementary table S4, Supplementary Material online). This analysis indicates that the majority of sponge lncRNAs is not processed into sRNAs, consistent with previous findings in vertebrates (Pauli et al. 2012) and plants (Zhang, Liao, et al. 2014).
From a reference catalog of transposable elements (TEs) in Amphimedon, which was established using RepeatMasker (Smit et al. 1996–2010), we determined the TE content of lncRNAs by calculating the percentage of lncRNA transcripts with at least one exon overlapping with a TE by at least 10 bp. We found that 46% of sponge lncRNAs (1,341 of 2,935) contain exonic sequences of at least partial TE origin, which is lower than protein-coding genes (50%; 22,568 of 44,719) (Fernandez-Valverde et al. 2015). Class II DNA transposons, long interspersed elements, and long terminal repeats were the three most abundant known repetitive elements to overlap with sponge lncRNA exons (supplementary tables S5 and S6, Supplementary Material online). To determine the total coverage of TE-derived sequences in sponge lncRNA exons in comparison to protein-coding genes, we intersected the reference catalog of TEs in Amphimedon with the genomic coordinates of all lncRNA exons and protein-coding exons. This approach, which is similar to that employed for vertebrates (Kapusta et al. 2013), revealed that TE coverage in this sponge is considerably lower for lncRNA exons (22%; 0.27 Mb) than for protein-coding exons (31%; 14.93 Mb) and the whole genome (34%; 56.33 Mb). Thus, although little is known about repetitive elements in Amphimedon, our findings are consistent with TEs being more likely to contribute to the origin of protein-coding genes than sponge lncRNAs in Amphimedon.
To determine whether sponge lncRNAs have comparable features to their bilaterian counterparts, we analyzed the primary structure of these lncRNAs, and their developmental expression profiles and sequence conservation (see below). We found that sponge lncRNAs were on average shorter (mean length of 424 nt for lncRNAs vs. 1,118 nt for protein-coding genes) and had fewer exons per transcript (median 1; average 1.5) than protein-coding genes (median 2; average 4.9) (fig. 2B and C). These properties are in agreement with the finding that lncRNAs are generally shorter and have fewer exons than protein-coding genes as previously shown for both bilaterian and plant lncRNAs (Guttman et al. 2010; Cabili et al. 2011; Nam and Bartel 2012; Pauli et al. 2012; Young et al. 2012; Li et al. 2014; Zhang, Liao, et al. 2014; Zhou et al. 2014).
Sponge lncRNAs Are Dynamically Expressed during Development
To examine whether significant changes in the level of lncRNA expression occur during development, we combined triplicate 100-bp single-end directional RNA-Seq data sets (supplementary table S1, Supplementary Material online) with our paired-end directional data sets across the main four developmental stages previously described (four biological replicates for each stage of development). This time-series of RNA-Seq experiments allowed us to follow the expression dynamics of lncRNAs and protein-coding genes as development proceeds. The differential expression pattern of lncRNAs at the main developmental stage transitions was analyzed using the Bioconductor package DESeq2 (Love et al. 2014).
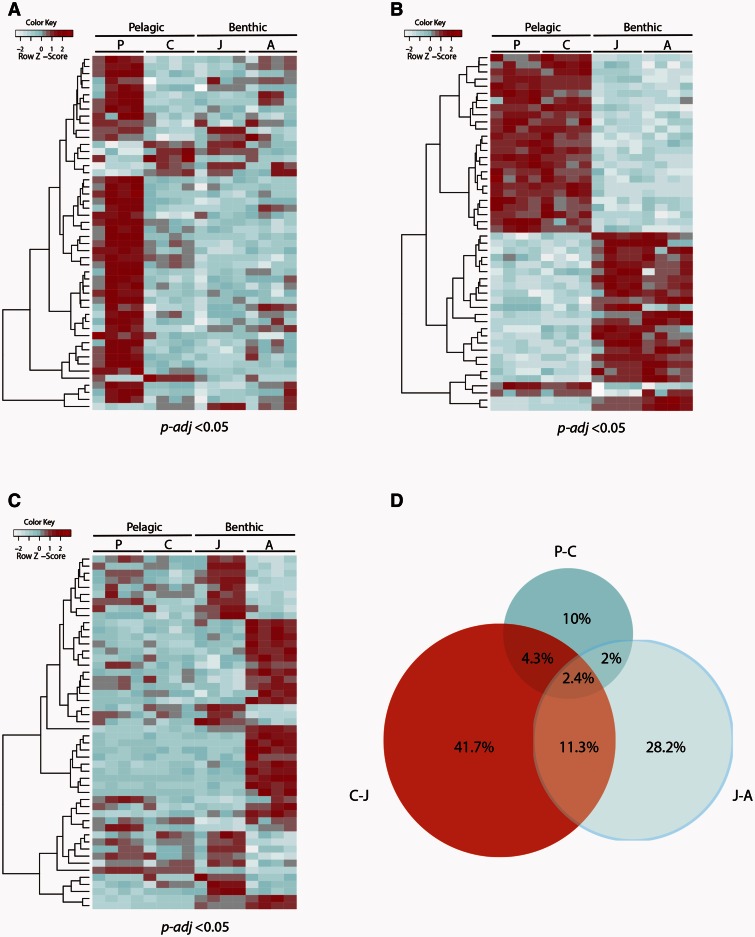
From this analysis, we identified 900 (30.7%) lncRNAs that exhibited significant changes in expression between any two successive developmental stages (P-adj < 0.05) (supplementary tables S7–S9, Supplementary Material online). Precompetent and competent larval stages showed similar lncRNA transcription profiles, with only 169 differentially expressed lncRNAs detected between the two stages (table 1 and fig. 3A). In contrast, a significant change in expression profile was evident at the pelagobenthic transition, when competent free-swimming larvae settled on the benthos and metamorphosed into the juvenile (table 1 and fig. 3B). This pelagobenthic transition was accompanied by the differential expression of 538 lncRNAs (P-adj < 0.05), accounting for approximately 60% of all differentially expressed lncRNAs detected by our analysis. Maturation from the juvenile to adult was accompanied by the differential expression of 396 lncRNAs (P-adj < 0.05) (table 1 and fig. 3C). A Venn diagram representing the proportion of differentially expressed lncRNAs detected at each of the main developmental stage transitions is shown in figure 3D. Together these results suggest that sponge lncRNAs are dynamically expressed during development.
Table 1.
Number of Differentially Expressed lncRNAs at Each of the Main Amphimedon Developmental Stage Transitions (P-adj < 0.05).
| Developmental Stage Transition | Number of Differentially Expressed lncRNAs (%) |
|---|---|
| Precompetent–competent larva | 169 (18.7) |
| Competent larva–juvenile | 538 (59.7) |
| Juvenile–adult | 396 (44) |
Fig. 3.
Developmental expression profiles of Amphimedon lncRNAs. (A) Expression profiles of the top 50 differentially expressed lncRNAs during the development from precompetent to competent larva (P-adj <0.05). (B) Expression profiles of the top 50 differentially expressed lncRNAs during the pelagobenthic transition from pelagic swimming competent larva to benthic juvenile (P-adj < 0.05). (C) Expression profiles of the top 50 differentially expressed lncRNAs during maturation from juvenile to adult (P-adj < 0.05). Expression levels were measured by RNA-Seq (four replicates per stage) and rescaled by row. Each row represents data for one lncRNA. lncRNAs were clustered by hierarchical clustering. Pelagic stages include precompetent (P) and competent (C) larva; benthic stages include juvenile (J) and adult (A). Red indicates high expression level, light blue low expression (see supplementary tables S7–S9, Supplementary Material online, for the IDs of these differentially expressed lncRNAs). (D) Venn diagram representing the proportion of differentially expressed lncRNAs detected at each of the main developmental stage transitions; P-C, precompetent–competent larva; C-J, competent larva–juvenile; J-A, juvenile–adult.
Sponge lncRNAs Are Regulated Independently of Their Neighboring Coding Genes
Recent studies suggested that bilaterian lncRNAs are preferentially located next to protein-coding genes involved in development and transcriptional regulation (Dinger et al. 2008; Guttman et al. 2009; Ponjavic et al. 2009; Orom et al. 2010; Cabili et al. 2011), raising the possibility that a relationship may exist between some lncRNAs and the regulation of gene transcription.
We therefore analyzed the Gene Ontology (GO) terms of protein-coding genes that are neighbors of or that overlap with the differentially expressed lncRNAs. These closest neighbors of the differentially expressed lncRNAs are enriched for GO terms associated with transcription factor activity, protein binding, and sequence-specific DNA binding (Fisher’s exact test, P-adj lt; 0.05) (supplementary table S10, Supplementary Material online). However, the mere physical proximity of lncRNAs and genes with regulatory functions does not necessarily imply a functional relationship between the protein-coding gene and the lncRNA (Pauli et al. 2012). Indeed, previous studies in plants, worm, zebrafish, mouse, and human lncRNAs established that the expression levels of lncRNAs are not more correlated to their protein-coding gene neighbors than expected for a pair of neighboring protein-coding gene loci (Cabili et al. 2011; Guttman et al. 2011; Ulitsky et al. 2011; Guttman and Rinn 2012; Nam and Bartel 2012; Pauli et al. 2012; Zhang, Liao, et al. 2014).
To assess whether this is the case for the sponge lncRNAs, we used CEL-Seq data (Hashimshony et al. 2012; Anavy et al. 2014) comprising 82 Amphimedon developmental samples from early cleavage to adult compressed into 17 stages. In line with the previous studies, our analysis indicates that the developmental expression of Amphimedon lncRNAs generally are not correlated with neighboring or overlapping protein-coding genes, and thus appear not to be coregulated or part of a common regulatory network. Importantly, this lack of correlation in expression is consistent with intronic lncRNAs in Amphimedon being independently regulated transcripts that are not the side-product of the pre-mRNA processing of overlapping protein-coding genes (Mercer et al. 2008; St Laurent et al. 2012).
Sponge lncRNAs Show Temporally Restricted Expression Patterns
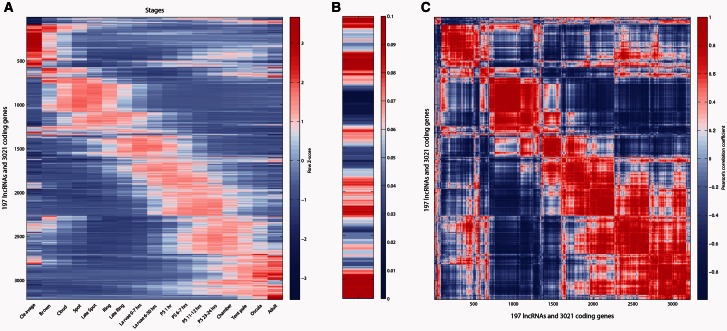
In bilaterians, lncRNAs tend to be expressed in a tissue- and stage-specific manner (Cabili et al. 2011; Ulitsky et al. 2011; Nam and Bartel 2012; Pauli et al. 2012). To assess whether this is the case for the sponge lncRNAs, we interrogated CEL-Seq data (Hashimshony et al. 2012; Anavy et al. 2014) (see above) for developmentally restricted lncRNA expression. Highly expressed and dynamic lncRNAs (>50 transcripts per million; tpm) were clustered with highly expressed protein-coding genes (>1,000 tpm) based on similarity of expression profiles. In total, 197 lncRNAs and 3,021 correlated protein-coding genes exhibited highly restricted temporal expression profiles (fig. 4A). On average, 15 protein-coding genes correlated with a given lncRNA (lncRNA to protein-coding gene ratio of 0.065), although some clusters showed lncRNA to protein-coding gene ratio as high as 0.25. Although lncRNAs were detected throughout development and present in embryos, larvae, postlarvae, juveniles, and adults, there were three stages that had a greater predominance of lncRNA transcripts (fig. 4B). Early embryos (i.e., cleavage), where there is a strong maternal influence, displayed the greatest number of dynamically expressed lncRNAs. A high diversity of transiently expressed lncRNAs also was present during the first 24 h of metamorphosis, when the larval body plan is being resculpted into the juvenile/adult body plan. Finally, the number of expressed lncRNAs increased at the establishment of the juvenile body plan and in the adult. Pairwise comparison of the combined lncRNA and protein-coding gene sets confirms that the expression of these genes, in general, is tightly correlated and restricted to specific developmental periods (fig. 4C). Together, these analyses indicate that lncRNAs have restricted developmental expression profiles that tightly match a subset of highly expressed protein-coding genes, consistent with these genes being coregulated.
Fig. 4.
Temporal expression patterns of Amphimedon lncRNAs and protein-coding genes. (A) Hierarchical clustering of lncRNA and protein-coding gene (rows) expression profiles across Amphimedon development (columns), from early cleavage to adult. Red indicates high expression level, blue low expression. Expression levels were measured by CEL-Seq and rescaled by row (Z score). Only lncRNAs (197) with an overall expression of at least 50 tpm in total across the stages and only protein-coding genes (3,021) with an overall expression of at least 1,000 tpm in total across the stages were used. PS, postsettlement postlarva. (B) Fraction of lncRNAs in a window of 200 genes (both lncRNAs and protein-coding genes), showing that lncRNAs are more popular in some clusters than in others. Red indicates high fraction of lncRNAs per window, blue low fraction. (C) Hierarchical clustering of expression correlations, for lncRNAs (197) with protein-coding genes (3,021). The average lncRNA to protein-coding gene ratio is 0.065. Red indicates positive Pearson’s correlation, blue negative Pearson’s correlation.
A “Guilt-by-Association” Analysis Suggests Developmental Regulatory Functions for Specific Sponge lncRNAs
Given the high number of correlated lncRNA and protein-coding gene developmental expression profiles, we employed the so-called “guilt-by-association” method to predict lncRNAs function. This method, which has been applied in a number of bilaterians (Dinger et al. 2008; Guttman et al. 2009; Cabili et al. 2011; Pauli et al. 2012), assigns a putative function to a specific lncRNA based on the known functions of the coexpressed, and thus presumably coregulated, protein-coding genes. Perturbation experiments are then essential to test the presumed function of specific lncRNAs.
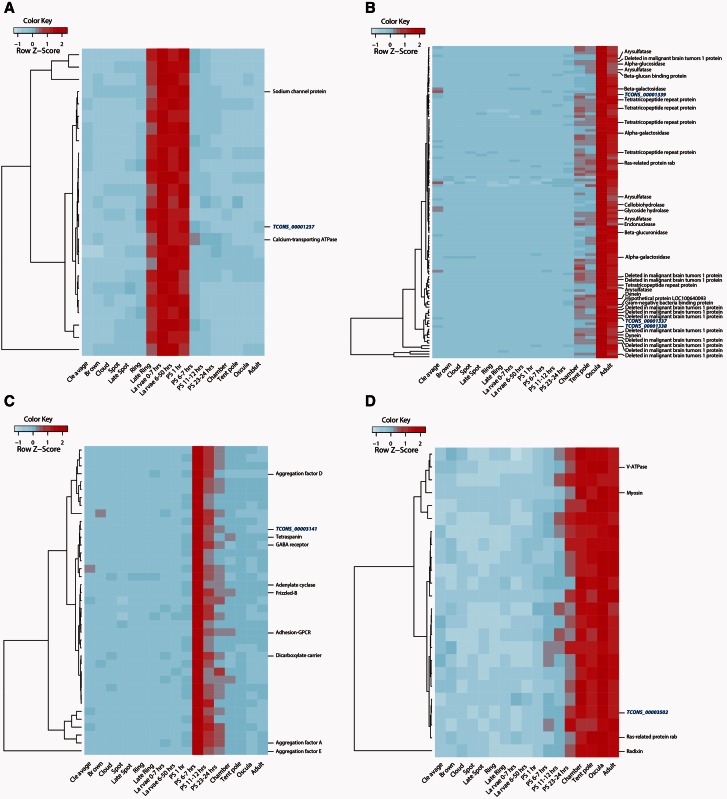
Here, we identified 17 differentially expressed lncRNAs that strongly correlated with the expression profiles of sets of protein-coding genes (Pearson’s correlation r2 > 0.95; Fisher’s exact test, P value < 0.05) (supplementary fig. S2, Supplementary Material online). GO enrichment analysis (Al-Shahrour et al. 2004) of the coexpressed protein-coding genes revealed six Amphimedon lncRNAs that were coexpressed with protein-coding genes involved in key metazoan developmental processes, such as cell adhesion, morphogenesis, and signal transduction. The latter also includes the G protein-coupled receptor (GPCR) Frizzled B (UniProt:I1G9T3_AMPQE), a key component of the Wnt signaling pathway (Adamska et al. 2007) (Fisher’s exact test, P-adj < 0.05) (fig. 5A–D; for a complete list of enriched GO terms and corresponding protein-coding genes, see supplementary table S11 and fig. S3, Supplementary Material online). These results suggest putative developmental regulatory functions for a subset of the sponge lncRNAs.
Fig. 5.
Putative developmental regulatory functions for specific Amphimedon lncRNAs. Developmental expression profiles of four distinct coexpression groups, each of which includes one lncRNA and protein-coding genes involved in key metazoan physiological and developmental processes. Expression levels were measured by CEL-Seq and rescaled by row. Red indicates high expression level, light blue low expression. Rows corresponding to protein-coding genes with an enriched GO term (Fisher’s exact test, P-adj < 0.05) are shown on the right. For a complete list of enriched GO terms and relative protein-coding genes, see supplementary table S11, Supplementary Material online. lncRNAs are shown in blue. (A) TCONS_00001237 is coexpressed with ion channels and genes enriched for calcium-transporting ATPase activity. In line with this lncRNA expression pattern, ion channels are highly expressed right before settlement and calcium signaling is a gene functional group upregulated during metamorphosis (Conaco et al. 2012). (B) TCONS_00001338 is expressed late in development and is coexpressed with protein-coding genes enriched for scavenger receptor activity, carbohydrate metabolic processes, and hydrolase activity. Consistent with this, an increase in the expression of scavenger receptors, multiple sulfatases, and polysaccharide-binding molecules is observed in the adult transcriptome (Conaco et al. 2012). (C) TCONS_00003141 is precisely activated 6–7 h after settlement and is coexpressed with protein-coding genes involved in key intercellular signaling pathways that might regulate morphogenetic events during metamorphosis, including the GPCR Frizzled-B (UniProt:I1G9T3_AMPQE), a key component of the Wnt signaling pathway. Extensive cellular transdifferentiation, proliferation, and rearrangement are observed during this stage of metamorphosis (Nakanishi et al. 2014). (D) TCONS_00003502 is coexpressed with protein-coding genes involved in cellular component organization processes. In agreement with increased expression of this lncRNA from late postlarva to adult, genes involved in tissue morphogenesis and cell proliferation are enriched in the adult transcriptome (Conaco et al. 2012).
Sponge lncRNAs Exhibit Low Sequence Conservation
Although several conserved lncRNAs are known within vertebrates, lncRNAs generally have low levels of sequence conservation (Guttman et al. 2009; Marques and Ponting 2009; Chodroff et al. 2010; Cabili et al. 2011; Ulitsky et al. 2011; Kapusta and Feschotte 2014; Necsulea et al. 2014; Washietl et al. 2014).
To assess the level of conservation of the sponge lncRNAs, we first searched each lncRNA against genomic sequences from Drosophila melanogaster (Berkeley Drosophila Genome Project Release 5/dm3), Caenorhabditis elegans (WS242), Nematostella vectensis (Nemve1), Trichoplax adhaerens (Triad1), Mnemiopsis leidyi (Mnemiopsis Genome Project Portal; Ryan et al. 2013; Moreland et al. 2014), Pleurobrachia bachei (Moroz et al. 2014), Monosiga brevicollis (Monbr1), Saccharomyces cerevisiae (sacCer3), Dictyostelium discoideum (dictybase.01), Arabidopsis thaliana (TAIR10) and Zea mays (AGPv3), and then searched each lncRNA against the transcriptome of 12 sponge species, spanning over 650 My of evolution across the four classes of Porifera (supplementary fig. S4, Supplementary Material online). These include the demosponges Crella elegans (Perez-Porro et al. 2013), Chondrilla nucula (Riesgo et al. 2014), Ircinia fasciculate (Riesgo et al. 2014), Petrosia ficiformis (Riesgo et al. 2014), Spongilla lacustris (Riesgo et al. 2014), Ephydatia muelleri (Hemmrich and Bosch 2008), Microciona prolifera (Fernandez-Valverde SL, Degnan BM, et al., unpublished data) and Pseudospongosorites suberitoides (Riesgo et al. 2014), the homoscleromorphs Oscarella carmela (Hemmrich and Bosch 2008) and Corticium candelabrum (Riesgo et al. 2014), the hexactinellid Aphrocallistes vastus (Riesgo et al. 2014), and the calcisponge Sycon coactum (Riesgo et al. 2014).
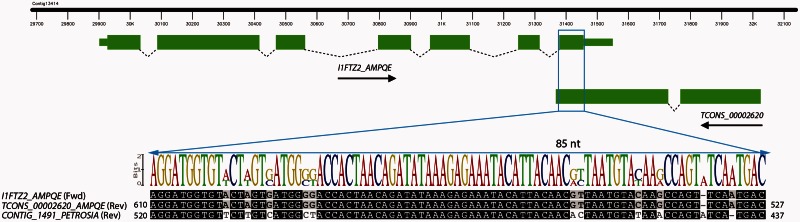
With our sequence similarity analysis we found that Amphimedon lncRNAs had no detectable orthologs outside demosponges, which diverged from other animals at least 700 Ma, well before eumetazoan cladogenesis (Erwin et al. 2011). Interestingly, two antisense exonic lncRNA transcripts (TCONS_00001844 and TCONS_00002620) had detectable orthologs with Pe. ficiformis (order Haplosclerida; family Petrosiidae), the closest related species to Amphimedon among the sponges surveyed (supplementary fig. S4, Supplementary Material online), which diverged from each other at least 450 Ma (Erwin et al. 2011). Our BLASTn search identified a conserved 156 nt match between TCONS_00001844 and the Petrosia transcript contig_13053 (E value 2e-28). Both transcripts show significant complementarity to the sponge hypothetical protein BRAFLDRAFT_78705. The other conserved lncRNA, TCONS_00002620, is a 661-nt transcript encoded by two exons, located in antisense orientation to the sponge 5′-AMP-activated protein kinase subunit beta-1-like gene (UniProt:I1FTZ2_AMPQE), and differentially expressed at metamorphosis. Our BLASTn search identified a conserved 85 nt match between this lncRNA and the Petrosia transcript contig_1491 (E value 3e-19) (fig. 6). Contig_1491 shows significant complementarity to this sponge’s 5′-AMP-activated protein kinase subunit beta-1-like gene (E value 9e-19), as also found for TCONS_00002620 (fig. 6). Finally, we evaluated the coding potential of the putative Petrosia lncRNA orthologs using CPC software (Kong et al. 2007). Contig_13053 and contig_1491 had a coding potential score of −0.94 and −0.63, respectively, and were therefore defined as “noncoding.” However, in both cases, the presence of a highly conserved ortholog gene in antisense orientation presumably contributes to the high level of lncRNA sequence conservation.
Fig. 6.
A syntenic sponge lncRNA. The blue box shows the region with sequence conservation. Alignment tracks show an 85-nt syntenic segment between Amphimedon and the demosponge Petrosia ficiformis, which diverged from each other at least 450 Ma (Erwin et al. 2011). This segment has complementarity to a predicted Amphimedon 5′-AMP-activated protein kinase subunit beta-1-like gene (Uniprot:I1FTZ2_AMPQE). A gray scale indicates sequence similarity: White, less than 60% similar; light gray, 60–80% similar; dark gray, 80–100% similar; black, 100% similar. The consensus logo highlights the 85-nt conserved sequence, which was identified from a BLASTn search (E value 3e-19). A score of 2 bits indicates that these bases are perfectly conserved between these two sponge species.
Discussion
Although an increasing number of lncRNAs have been identified in a range of multicellular and unicellular eukaryotes, these have been largely restricted to established model organisms (Carninci et al. 2005; Ravasi et al. 2006; Guttman et al. 2009; Cabili et al. 2011; Ulitsky et al. 2011; Boerner and McGinnis 2012; Derrien et al. 2012; Geisler et al. 2012; Liu et al. 2012; Nam and Bartel 2012; Pauli et al. 2012; Young et al. 2012; Cloutier et al. 2013; Sauvageau et al. 2013; Brown et al. 2014; Li et al. 2014; Liao et al. 2014; Zhang, Liao, et al. 2014). The lack of lncRNA annotations in early-branching metazoans has thus precluded detailed comparative analyses. By characterizing here lncRNAs in a morphologically simple representative of one of the oldest phyletic lineages of animals, the poriferans, it can be determined 1) whether the commonalities shared between vertebrate, insect and nematode lncRNAs originated early in metazoan evolution, as has been shown to be the case for many other gene families and genomic features, including miRNAs and piRNAs (e.g., Grimson et al. 2008; Srivastava et al. 2008, 2010), and 2) what role lncRNAs may play in the evolution of animal complexity.
Deep sequencing of the transcriptome of the marine demosponge A. queenslandica, as it develops from a pelagic larva to a benthic adult, and the subsequent comprehensive de novo transcripts reconstruction have allowed us to generate a high-confidence catalog of lncRNAs expressed across the key developmental stages. We defined a comprehensive yet conservative set of 2,935 single and multiexonic noncoding RNA transcripts, which includes lincRNAs, intronic lncRNAs, antisense overlapping lncRNAs, and precursors for sRNAs. This conservative estimate of A. queenslandica lncRNAs—the first lncRNAs catalog in an early-branching metazoan—shares many of the characteristics of their bilaterian counterparts (Guttman et al. 2009, 2010, 2011; Cabili et al. 2011; Nam and Bartel 2012; Pauli et al. 2012; Young et al. 2012; Brown et al. 2014; Zhou et al. 2014). Specifically, they are relatively short in length, have a low number of exons, display temporally restricted expression profiles throughout development, and have low sequence conservation in comparison to protein-coding genes. These observations are consistent with the characteristics of bilaterian lncRNAs originating very early in evolution, before the divergence of poriferan and eumetazoan lineages.
The dynamic expression of lncRNAs during A. queenslandica embryogenesis, larval development, and metamorphosis not only indicates that these genes must be developmentally regulated but is also consistent with their regulatory function throughout development, as has been observed for specific bilaterian lncRNAs with gene regulatory function (Dinger et al. 2008; Cabili et al. 2011; Guttman et al. 2011; Derrien et al. 2012; Nam and Bartel 2012; Pauli et al. 2012; Sauvageau et al. 2013; Brown et al. 2014). Analysis of the 197 A. queenslandica lncRNAs that display the highest and most dynamic developmental expression profiles reveals that lncRNA abundances correlate with morphogenetic and developmental events and milestones. These expression profiles match closely with those observed for subsets of coding genes (Conaco et al. 2012; Anavy et al. 2014), with on average 15 protein-coding genes that tightly correlate in expression with a given sponge lncRNA. As in vertebrates, where lncRNAs have been shown to be highly interconnected with multiple protein-coding genes (Necsulea et al. 2014), our findings are consistent with these genes being part of a common regulatory network.
Amphimedon queenslandica metamorphosis, which takes approximately 3 days, is the transition from the pelagic larval to the benthic juvenile/adult body plan and entails extensive but localized programmed cell death, transdifferentiation, cell proliferation, and movement (Leys and Degnan 2002; Nakanishi et al. 2014). The dynamic activation and repression of lncRNAs through this dramatic developmental period are perhaps expected. A detailed view of expression profiles through metamorphosis reveals specific lncRNA levels fluctuate rapidly, with notable differences between postlarval stages that are only a few hours apart. This is consistent with lncRNA expression in A. queenslandica being tightly regulated to a specific developmental context.
A large number of lncRNAs are highly expressed exclusively in cleavage-stage embryos. In early A. queenslandica embryos, as is the case in other demosponge embryos (reviewed in Ereskovsky 2010), cleavage is accompanied by embryo growth by the fusion of nutritive maternal nurse cells that, once embedded in the embryo, undergo programmed cell death. The complexity of the morphogenetic events during A. queenslandica cleavage appears to be reflected in the high diversity of the lncRNAs expressed at this stage. By the next stage of development—the brown stage—the maternal input of lncRNAs appears to have diminished. This and subsequent embryonic stages have markedly fewer lncRNAs expressed at high levels. This observation is similar to previous findings in bilaterians (Pauli et al. 2012), where parentally supplied lncRNAs are specifically enriched in cleavage-stage embryos and rapidly decay after the first few hours of embryogenesis (Pauli et al. 2012). These lncRNAs might have a role in the regulation of maternal transcripts or transcription of cell-cycle genes (Hung et al. 2011; Pauli et al. 2011).
Different lncRNA expression profiles in A. queenslandica correspond closely with specific morphogenetic stages, when specific protein-coding gene classes are known to be activated (Conaco et al. 2012). For instance, the transition from the planktonic competent larval stage to the benthic juvenile stage is accompanied by the activation of genes involved in secondary metabolism, immune system, cell proliferation, and tissue morphogenesis (Conaco et al. 2012). Consistent with sponge lncRNAs being important developmental regulators, “guilt-by-association” analyses reveal that specific lncRNA expression profiles correlate with the expression profiles of conserved metazoan developmental genes, such as the Wnt signaling pathway components. This is consistent with sponge lncRNAs also playing a role as regulators of gene activity during differentiation and development, although the exact regulatory mechanisms still need to be elucidated. Thus, we conclude that sponges, despite having a simple morphology, possess an lncRNA repertoire akin to their bilaterian counterparts. In addition to developmental transcription factors and signaling pathway genes (Srivastava et al. 2008, 2010), lncRNAs may regulate the development of multicellular animals, regardless of the level of morphological complexity.
Our understanding of other ncRNAs has been improved by examining their evolutionary conservation (Bartel 2009). Although Amphimedon lncRNAs are similar to bilaterian lncRNAs in terms of composition, structure, and expression, they have no significant sequence similarity to known bilaterian lncRNAs. This supports the hypothesis that lncRNA sequences largely evolve more rapidly than protein-coding sequences (Pang et al. 2006; Kapusta and Feschotte 2014). lncRNAs appear to be able to accept primary sequence changes, additions, and deletions over evolutionary time without detrimental effects on functionality (Smith et al. 2013; Johnson et al. 2014; Washietl et al. 2014). This suggests that negative selection is acting on only portions of lncRNAs or on their higher-order structure (Washietl et al. 2014). Highly conserved elements within lncRNA sequences, interspersed with longer and less conserved stretches of nucleotide sequences, have been reported (Guttman and Rinn 2012; Ulitsky and Bartel 2013). Well-known examples of such elements, which could have evolved for specific protein and/or RNA interactions, include the PRC2-binding elements in the lncRNA Xist (Maenner et al. 2010), the 26 nt miR-7 binding site in the zebrafish lncRNA Cyrano (Ulitsky et al. 2011), and the Splicing factor 1 (Sf1) binding site in the mammalian lncRNA Miat (Rapicavoli et al. 2010; Tsuiji et al. 2011). Consistent with this proposition, we have identified two syntenic demosponge-specific lncRNAs, between Pe. ficiformis and Amphimedon, where only a small portion of these lncRNAs is conserved; these regions overlap with conserved protein-coding sequences on the opposite strand. The presence of a highly conserved coding gene in antisense orientation may be the reason for the high level of lncRNA sequence conservation. As the majority of Amphimedon lncRNAs does not have identifiable orthologs in other sponges, it appears that sponge lncRNAs are evolving in a similar manner to bilaterian lncRNAs. Possibly, with our sequence-homology-based search we have underestimated lncRNA conservation (Ulitsky and Bartel 2013). Computational approaches that rely on structural, rather than sequence, similarity may identify additional sponge lncRNA orthologs. Our analysis was also limited by the quality of available transcript data sets in other species and other poriferan genomes and will be improved as more comprehensive lncRNA catalogs are released in other early-branching metazoans.
The discrimination of protein-coding and noncoding transcripts remains a challenge, particularly in determining whether a hypothetical ORF truly encodes a protein (Ingolia et al. 2011; Chew et al. 2013; Guttman et al. 2013; Magny et al. 2013; Slavoff et al. 2013; Bazzini et al. 2014; Cohen 2014; Pauli et al. 2014; Ruiz-Orera et al. 2014; Smith et al. 2014; Anderson et al. 2015). Although we identified thousands of lncRNAs in A. queenslandica, we predict that more lncRNAs will be annotated in this species. These will comprise many genes that are not polyadenylated, and thus, have been missed in the poly(A)-based RNA-Seq and CEL-Seq data sets used in this study. Other genes that remain unannotated might comprise those that are not expressed in the developmental stages surveyed, those that did not map to the genome under our mapping criteria, and those filtered out by our stringent filtering pipeline. Nonetheless, the differential expression of lncRNAs in A. queenslandica is consistent with them having a developmental role akin to bilaterian lncRNAs. In addition, our finding of lncRNAs in a sponge strongly suggests that lncRNAs are an ancient feature of the metazoan regulatory system, and evolved before the divergence of sponge and eumetazoan lineages.
Materials and Methods
Animal Collection and Sequencing
Amphimedon queenslandica adults were collected from Heron Island Reef, Great Barrier Reef, Queensland, Australia, and larvae and juveniles were reared as previously described (Leys et al. 2008). Total RNA was isolated using the standard TRIzol (Invitrogen) protocol and genomic DNA removed by DNase treatment. The quality of the RNA was confirmed using the Agilent 2100 Bioanalyzer. Strand-specific libraries for both 100-bp paired-end and single-end sequencing were prepared and sequenced on the Illumina HiSeq 2000 (Illumina, San Diego) (supplementary table S1, Supplementary Material online) (Fernandez-Valverde et al. 2015).
De Novo Transcriptome Assembly
Raw sequencing data were quality filtered using Trimmomatic (HEADCROP: 7, SLIDINGWINDOW: 4:15) (Bolger et al. 2014). Unpaired reads and reads smaller than 60 nt were discarded. Quality-filtered paired-end reads were assembled de novo using Trinity (Haas et al. 2013) (supplementary table S1, Supplementary Material online). Each developmental stage was assembled independently with default parameters, with the exception of a lower transcript size of 200 nt (Fernandez-Valverde et al. 2015).
Bioinformatics Pipeline for the Identification of lncRNAs
For each of the four main developmental stages, classification of each transcript as either coding or noncoding was determined using a stringent stepwise filtering pipeline. First, all transcript candidates were subjected to BLASTp (Altschul et al. 1990) (both NCBI nr and Amphimedon-specific dbs), BLASTx (Altschul et al. 1990) (both NCBI nr and Amphimedon-specific dbs), HMMER (Finn et al. 2011) (both Pfam-A and Pfam-B), and SignalP (Petersen et al. 2011). For BLASTp, HMMER, and SignalP analyses, the transcripts were translated (stop-to-stop codon) using Getorf tool (Rice et al. 2000) and the longest unique ORF for each transcript was retained. Any transcript with an E value lower than 1e-4 in any of the search algorithms was considered as protein-coding (for SignalP we used D-cutoff: 0.45). To reduce the number of potential spurious transcripts found in the transcriptome assemblies, transcripts shorter than 300 nt were removed. Any remaining transcripts of uncertain coding potential were removed by applying a strict ORF size cutoff of 75 amino acids. lncRNAs were then mapped to Amphimedon genome assembly using BLAT (Kent 2002) (parameters: fine, minIdentity = 95). Next, lncRNAs were removed that had sense exonic overlap with annotated tRNAs, rRNAs, protein-coding gene 5′-UTRs plus 150 bp upstream, and protein-coding gene 3′-UTRs plus 150 bp downstream, using overlapSelect. Cuffmerge (Trapnell et al. 2010) was used to merge the lncRNA assemblies corresponding to each of the four developmental stages. lncRNA transcripts that had sense exonic overlap with a protein-coding gene were removed. Only lncRNA transcripts with an overall expression of at least ten raw read counts across the whole developmental time course were retained. Finally, the CPC (Kong et al. 2007) was used to evaluate the sensitivity of our discrimination pipeline. Only transcripts that were classified as noncoding by CPC and did not show homology to any known proteins (E-value threshold of 1e-10) were finally classified as high-confidence Amphimedon lncRNAs.
Classification of lncRNAs
The high-confidence set of lncRNAs was subdivided into three main categories using overlapSelect: 1) Intergenic lncRNAs that do not overlap with other genes, 2) intronic lncRNAs that overlap with a coding gene in either sense or antisense orientation but have no exon–exon overlap, and 3) exonic antisense lncRNAs that overlap with an exon of a protein-coding gene but are transcribed in the opposite direction.
sRNA Analysis
sRNAs expressed at the same four developmental stages as lncRNAs were used (Grimson et al. 2008; Calcino AD, Degnan BM, et al., unpublished data). The number of sRNAs overlapping lncRNAs was counted using overlapSelect with a 95% threshold. Transcripts with at least ten uniquely mapped overlapping sRNAs were considered as potential sRNA-precursors. These potential sRNA-precursors were then classified according to A. queenslandica sRNA categories, that is, piRNAs (26–30 nt), endo-siRNAs (20–22 nt), and unknown sRNA types (23–25 nt).
TE Content Analysis
Detailed annotation of repeats in the A. queenslandica genome was generated using RepeatMasker (Smit et al. 1996–2010). The annotation was then parsed to exclude low complexity DNA sequences and non-TE repeats, and to retain only known and unknown classes of TEs. The percentage of transcripts with at least one exon overlapping with a TE was determined by intersecting the genomic coordinates of both known and unknown classes of TEs with genomic coordinates of lncRNA and protein-coding gene exons, respectively, using BEDTools v.2.17.0 (Quinlan and Hall 2010). Coverage of TEs in the genome and the amount of overlap in base pairs that is observed between the lncRNA exons or protein-coding exons and the TEs was determined using BEDTools v.2.17.0 (Quinlan and Hall 2010). For the genome, total nucleotide amount (100%) corresponds to nucleotide amount of assembly without gaps (167 Mb). For lncRNAs and protein-coding genes, total nucleotide amount of genomic projection of all exons is considered (1.21 and 47.80 Mb, respectively). Only overlaps of minimum 10 bp were kept.
Differential Gene Expression Analysis
Quality-filtered reads from the 4 stranded paired-end libraries and the 12 stranded single-end libraries (three biological replicates per stage) were mapped to A. queenslandica genome (Srivastava et al. 2010) using TopHat2 (‐i 30 –g 30 –p 8) (Kim et al. 2013). The counts of reads mapping to each protein-coding gene and our lncRNAs catalog were calculated using HTSeq (version 0.6.0) (Anders et al. 2014). These raw gene counts were analyzed using DEseq2 1.4.1 (Love et al. 2014) and tested for differential expression, using a multifactor design, at a 5% False Discovery Rate (adjusted P value for multiple testing using the Benjamini–Hochberg correction).
CEL-Seq Data Expression Analysis
CEL-Seq reads were processed and mapped back to the A. queenslandica genome using Bowtie (Langmead et al. 2009). We then compressed the 82 Amphimedon developmental samples, from early cleavage to adult, into 17 stages averaging the biological replicates for each developmental stage across them. Larval stages have been combined in two different groups (Larvae 0–7 h and Larvae 6–50 h), as these developmental time points only have one replicate per time point. The mean and standard deviation were then calculated for each protein-coding gene and lncRNA in the CEL-Seq data set. Only lncRNAs (197) with an overall expression of at least 50 tpm in total across the developmental stages were retained. Only protein-coding genes (3,021) with an overall expression of at least 1,000 tpm in total across the developmental stages were retained. lncRNA and protein-coding gene expression levels were rescaled by row (Z score) and clustered by hierarchical clustering. The fraction of lncRNAs in a window of 200 genes (both lncRNAs and protein-coding genes) was calculated. These profiles were then smoothed by computing the average over windows of 200 genes. Pearson’s correlation was used to correlate the expression level of lncRNAs with the protein-coding genes. All analyses were performed using MATLAB (2012).
“Guilt-by-Association” Analysis
CEL-Seq reads were processed and mapped back to the A. queenslandica genome using Bowtie (Langmead et al. 2009). We then compressed the 82 Amphimedon developmental samples, from early cleavage to adult, into 17 stages as described above. To reduce noise, the differentially expressed lncRNAs and protein-coding genes with an overall expression of less than 100 CEL-Seq normalized counts throughout the whole developmental time course were discarded. Pearson’s correlation and a Fisher’s exact test were then used to correlate the expression level of each differentially expressed lncRNA with the protein-coding genes, using R (R Development Core Team 2010). Only genes that showed more than 0.95 correlation (positive and negative) and a P-value < 0.05 were considered to be coexpressed. Only lncRNAs that were coexpressed with at least ten protein-coding genes were used for the subsequent GO term enrichment analysis. Single GO term enrichments were then performed using fatiGO at a 5% FDR (Al-Shahrour et al. 2004) with custom annotation.
Nearest Neighbor Analysis
For each of the differentially expressed lncRNA, the nearest protein-coding neighbor was identified using BEDTools v.2.17.0 (Quinlan and Hall 2010). For antisense and intronic differentially expressed lncRNAs, the overlapping protein-coding gene(s) were identified using BEDTools v.2.17.0 (Quinlan and Hall 2010). The list of lncRNA/protein-coding gene pairs was tested for GO term enrichment using fatiGO (Al-Shahrour et al. 2004) at a 5% FDR with custom annotation. As described above, CEL-Seq reads were processed and mapped back to the A. queenslandica genome using Bowtie (Langmead et al. 2009). To reduce noise, differentially expressed lncRNAs and protein-coding genes with an overall expression of less than 100 CEL-Seq normalized counts throughout the whole developmental time course were discarded. Pearson’s correlation and a Fisher’s exact test were then used to correlate the expression level of each differentially expressed lncRNA with the nearest protein-coding neighbor, using R (R Development Core Team 2010). Only genes that showed more than 0.95 correlation (positive and negative) and a P-value < 0.05 were considered to be coexpressed.
Sequence Similarity Analysis
To assess the level of conservation of sponge lncRNAs, we searched each lncRNA against genome/transcriptome sequences of D. melanogaster (Berkeley Drosophila Genome Project Release 5/dm3), C. elegans (WS242), N. vectensis (Nemve1), T. adhaerens (Triad1), M. leidyi (Mnemiopsis Genome Project Portal; Ryan et al. 2013; Moreland et al. 2014), P. bachei (Moroz et al. 2014), Mo. brevicollis (Monbr1), S. cerevisiae (sacCer3), Di. discoideum (dictybase.01), Ar. thaliana (TAIR10), Z. mays (AGPv3), Cr. elegans (Perez-Porro et al. 2013), Ch. nucula (Riesgo et al. 2014), I. fasciculate (Riesgo et al. 2014), Pe. ficiformis (Riesgo et al. 2014), Sp. lacustris (Riesgo et al. 2014), E. muelleri (Hemmrich and Bosch 2008), Mi. prolifera (Fernandez-Valverde SL, Degnan BM, et al., unpublished data), Ps. suberitoides (Riesgo et al. 2014), O. carmela (Hemmrich and Bosch 2008), C. candelabrum (Riesgo et al. 2014), Ap. vastus (Riesgo et al. 2014), and Sy. coactum (Riesgo et al. 2014) using BLASTn (Altschul et al. 1990) with an E-value cutoff of 1e-6 and requiring a match coverage of at least 50 bases.
Data Access
Amphimedon queenslandica genome assembly ampQue1 (http://metazoa.ensembl.org/Amphimedon_queenslandica/Info/Index, last accessed May 25, 2015) was used throughout the study. The transcriptome sequencing data have been submitted to NCBI’s Sequence Read Archive (SRA) under the accession Number PRJNA255066. The replicated directional single-end RNA-Seq data set has been submitted to NCBI’s SRA under the accession Number SRP055403. All lncRNA sequences have been submitted to NCBI’s Transcriptome Shotgun Assembly (TSA). The TSA project has been deposited at DDBJ/EMBL/GenBank under the accession GBXN00000000. The version described in this article is the first version, GBXN01000000 (see supplementary table S3, Supplementary Material online, for lists of contig accession numbers-to-contigIDs for all the lncRNA sequences deposited at DDBJ/EMBL/GenBank). CEL-Seq data sets used can be obtained from NCBI GEO (GSE54364) (Anavy et al. 2014). The complete set of lncRNAs and protein-coding genes, along with the Trinity assembled transcriptomes, can be accessed and visualized in our website: http://amphimedon.qcloud.qcif.edu.au/lncRNAs/, last accessed May 25, 2015. The codes used for the gene coexpression analysis are available for download at: https://bitbucket.org/selene_fernandez/amphimedon-lncrnas (last accessed May 25, 2015).
Supplementary Material
Supplementary tables S1–S11 and figures S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Sandie Degnan for her critical comments on the manuscript and Degnan lab members for constructive discussions. This research was supported by an Australian Research Council grant to B.M.D. and the University of Queensland Early Career Researcher grant to N.N. The authors declare that they have no competing interests. F.G., S.L.F.V., M.T., and B.M.D. conceived and designed the study. F.G. and S.L.F.V. carried out all the computational analysis. N.N. and A.D.C. contributed with sample collection, library preparation, and sequencing of the replicated directional single-end RNA-Seq and small RNA-Seq data sets, respectively. I.Y. contributed with the library preparation, sequencing, and analysis of the CEL-Seq data sets. F.G. and B.M.D. wrote the manuscript, which was edited and approved by all authors.
References
- Adamska M, Degnan SM, Green KM, Adamski M, Craigie A, Larroux C, Degnan BM. 2007. Wnt and TGF-beta expression in the sponge Amphimedon queenslandica and the origin of metazoan embryonic patterning. PLoS One 2:e1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Diaz-Uriarte R, Dopazo J. 2004. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20:578-580. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. 2008. The eukaryotic genome as an RNA machine. Science 319:1787-1789. [DOI] [PubMed] [Google Scholar]
- Anavy L, Levin M, Khair S, Nakanishi N, Fernandez-Valverde SL, Degnan BM, Yanai I. 2014. BLIND ordering of large-scale transcriptomic developmental timecourses. Development 141:1161-1166. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2014. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, et al. 2015. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160:595-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, et al. 2014. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 33:981-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerner S, McGinnis KM. 2012. Computational identification and functional predictions of long noncoding RNA in Zea mays. PLoS One 7:e43047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, et al. 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512:393-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25:1915-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309:1559-1563. [DOI] [PubMed] [Google Scholar]
- Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. 2013. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development 140:2828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnar Z, Ponting CP. 2010. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 11:R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier SC, Wang S, Ma WK, Petell CJ, Tran EJ. 2013. Long noncoding RNAs promote transcriptional poising of inducible genes. PLoS Biol. 11:e1001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM. 2014. Everything old is new again: (linc)RNAs make proteins! EMBO J. 33:937-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Neveu P, Zhou N, Arcila M, Degnan SM, Degnan BM, Kosik KS. 2012. Transcriptome profiling of the demosponge Amphimedon queenslandica reveals genome-wide events that accompany major life cycle transitions. BMC Genomics 13:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22:1775-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. 2007. The expanding RNA polymerase III transcriptome. Trends Genet. 23:614-622. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Mattick JS. 2009. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct Genomic Proteomic. 8:407-423. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, RU K, Solda G, Simons C, et al. 2008. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 18:1433-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489:101-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2001. Non-coding genes and the modern RNA world. Nat Rev Genet. 2:919-929. [DOI] [PubMed] [Google Scholar]
- Ereskovsky AV. 2010. The comparative embryology of sponges. Dordrecht (The Netherlands): Springer-Verlag. [Google Scholar]
- Erwin DH, Laflamme M, Tweedt SM, Sperling EA, Pisani D, Peterson KJ. 2011. The Cambrian conundrum: early divergence and later ecological success in the early history of animals. Science 334:1091-1097. [DOI] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. 2006. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 20:1470-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Valverde SL, Calcino AD, Degnan BM. 2015. Deep developmental transcriptome sequencing uncovers numerous new genes and enhances gene annotation in the sponge Amphimedon queenslandica. BMC Genomics 16:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Lojek L, Khalil AM, Baker KE, Coller J. 2012. Decapping of long noncoding RNAs regulates inducible genes. Mol Cell. 45:279-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, Degnan BM, Rokhsar DS, Bartel DP. 2008. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455:1193-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477:295-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 28:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. 2012. Modular regulatory principles of large non-coding RNAs. Nature 482:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. 2013. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154:240-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8:1494-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T, Wagner F, Sher N, Yanai I. 2012. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2:666-673. [DOI] [PubMed] [Google Scholar]
- Hemmrich G, Bosch TC. 2008. Compagen, a comparative genomics platform for early branching metazoan animals, reveals early origins of genes regulating stem-cell differentiation. BioEssays 30:1010-1018. [DOI] [PubMed] [Google Scholar]
- Hooper JNA, Van Soest RWM. 2006. A new species of Amphimedon (Porifera, Demospongiae, Haplosclerida, Niphatidae) from the Capricorn-Bunker Group of Islands, Great Barrier Reef, Australia: target species for the “sponge genome project.” Zootaxa 1314:31-39. [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. 2011. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 43:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris HS, Talhouarne GJS, Jackson SE, Wills MR, Weissman JS. 2014. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 8:1365-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147:789-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Guigo R. 2014. The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs. RNA 20:959-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316:1484-1488. [DOI] [PubMed] [Google Scholar]
- Kapusta A, Feschotte C. 2014. Volatile evolution of long noncoding RNA repertoires: mechanisms and biological implications. Trends Genet. 30:439-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. 2013. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 9:e1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik K, Leonard VE, Kv S, Lalwani MK, Jalali S, Patowary A, Joshi A, Scaria V, Sivasubbu S. 2013. Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. PLoS One 8:e83616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D, Rinn J. 2012. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 13:R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 106:11667-11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. 2007. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35:W345–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, Odom DT, Marques AC. 2012. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 8:e1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leys SP, Degnan BM. 2002. Embryogenesis and metamorphosis in a haplosclerid demosponge: gastrulation and transdifferentiation of larval ciliated cells to choanocytes. Invertebr Biol. 121:171-189. [Google Scholar]
- Leys SP, Larroux C, Gauthier M, Adamska M, Fahey B, Richards GS, Degnan SM, Degnan BM. 2008. Isolation of amphimedon developmental material. CSH Protoc. 2008:pdb prot5095. [DOI] [PubMed] [Google Scholar]
- Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, et al. 2014. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 15:R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, Shen J, Liu J, Sun X, Zhao G, Chang Y, Xu L, Li X, Zhao Y, Zheng H, et al. 2014. Genome-wide identification and functional annotation of Plasmodium falciparum long noncoding RNAs from RNA-seq data. Parasitol Res. 113:1269-1281. [DOI] [PubMed] [Google Scholar]
- Liu G, Mattick JS, Taft RJ. 2013. A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH. 2012. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24:4333-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2010. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 8:e1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magny EG, Pueyo JI, Pearl FM, Cespedes MA, Niven JE, Bishop SA, Couso JP. 2013. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 341:1116-1120. [DOI] [PubMed] [Google Scholar]
- Marques AC, Ponting CP. 2009. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 10:R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATLAB. 2012. MATLAB and Statistics Toolbox Release. Natick (MA): The MathWorks, Inc. [Google Scholar]
- Mattick JS. 2009a. Deconstructing the dogma: a new view of the evolution and genetic programming of complex organisms. Ann N Y Acad Sci. 1178:29-46. [DOI] [PubMed] [Google Scholar]
- Mattick JS. 2009b. The genetic signatures of noncoding RNAs. PLoS Genet. 5:e1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. 2011. The central role of RNA in human development and cognition. FEBS Lett. 585:1600-1616. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. 2006. Non-coding RNA. Hum Mol Genet. 15:R17–R29. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. 2009. Long non-coding RNAs: insights into functions. Nat Rev Genet. 10:155-159. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. 2008. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 105:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, Fredman D, Praher D, Li XZ, Wee LM, Rentzsch F, Zamore PD, Technau U, Seitz H. 2014. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. 24:651-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran Y, Praher D, Fredman D, Technau U. 2013. The evolution of microRNA pathway protein components in Cnidaria. Mol Biol Evol. 30:2541-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland RT, Nguyen AD, Ryan JF, Schnitzler CE, Koch BJ, Siewert K, Wolfsberg TG, Baxevanis AD. 2014. A customized Web portal for the genome of the ctenophore Mnemiopsis leidyi . BMC Genomics 15:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, Povolotskaya IS, Grigorenko AP, Dailey C, Berezikov E, Buckley KM, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510:109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Sogabe S, Degnan BM. 2014. Evolutionary origin of gastrulation: insights from sponge development. BMC Biol. 12:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JW, Bartel DP. 2012. Long noncoding RNAs in C. elegans. Genome Res. 22:2529-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grutzner F, Kaessmann H. 2014. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505:635-640. [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143:46-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS. 2006. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 22:1-5. [DOI] [PubMed] [Google Scholar]
- Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S, Mitchell A, MA J, Dubrulle J, Reyon D, et al. 2014. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 343:1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Rinn JL, Schier AF. 2011. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 12:136-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. 2012. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res. 22:577-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Porro AR, Navarro-Gomez D, Uriz MJ, Giribet G. 2013. A NGS approach to the encrusting Mediterranean sponge Crella elegans (Porifera, Demospongiae, Poecilosclerida): transcriptome sequencing, characterization and overview of the gene expression along three life cycle stages. Mol Ecol Resour. 13:494-509. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 8:785-786. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465:1033-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP. 2009. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 5:e1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. 2009. Evolution and functions of long noncoding RNAs. Cell 136:629-641. [DOI] [PubMed] [Google Scholar]
- Qu Z, Adelson DL. 2012. Evolutionary conservation and functional roles of ncRNA. Front Genet. 3:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2010. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. [Google Scholar]
- Rapicavoli NA, Poth EM, Blackshaw S. 2010. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol. 10:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. 2006. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 16:11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP. 2014. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Mol Biol Evol. 31:1102-1120. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 81:145-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129:1311-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. 2014. Long non-coding RNAs as a source of new peptides. eLife 3:e03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF, Pang K, Schnitzler CE, Nguyen AD, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, NISC Comparative Sequencing Program, et al. 2013. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342:1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language?. Cell 146:353-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. 2013. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife 2:e01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. 2013. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 9:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 1996–2010 RepeatMasker Open-3.0. Available from: http://www.repeatmasker.org.
- Smith JE, Alvarez-Dominguez JR, Kline N, Huynh NJ, Geisler S, Hu W, Coller J, Baker KE. 2014. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep. 7:1858-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Gesell T, Stadler PF, Mattick JS. 2013. Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res. 41:8220-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature 454:955-960. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G, Shtokalo D, Tackett M, Yang Z, Eremina T, Wahlestedt C, Urcuqui-Inchima S, Seilheimer B, McCaffrey T, Kapranov P, et al. 2012. Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells. BMC Genomics 13:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G. 2002. An expanding universe of noncoding RNAs. Science 296:1260-1263. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS. 2007. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays 29:288-299. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28:511-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329:689-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuiji H, Yoshimoto R, Hasegawa Y, Furuno M, Yoshida M, Nakagawa S. 2011. Competition between a noncoding exon and introns: Gomafu contains tandem UACUAAC repeats and associates with splicing factor-1. Genes Cells 16:479-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. 2013. lincRNAs: genomics, evolution, and mechanisms. Cell 154:26-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147:1537-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, Samudrala R, Yu J, Wong G. 2004. Mouse transcriptome: neutral evolution of “non-coding” complementary DNAs. Nature 431:1. [PubMed] [Google Scholar]
- Wang KC, Chang HY. 2011. Molecular mechanisms of long noncoding RNAs. Mol Cell. 43:904-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Kellis M, Garber M. 2014. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 24:616-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 2008. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135:919-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 23:1494-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL. 2012. Long noncoding RNAs with snoRNA ends. Mol Cell. 48:219-230. [DOI] [PubMed] [Google Scholar]
- Young RS, Marques AC, Tibbit C, Haerty W, Bassett AR, Liu JL, Ponting CP. 2012. Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol Evol. 4:427-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XO, Yin QF, Wang HB, Zhang Y, Chen T, Zheng P, Lu X, Chen LL, Yang L. 2014. Species-specific alternative splicing leads to unique expression of sno-lncRNAs. BMC Genomics 15:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-C, Liao J-Y, Li Z-Y, Yu Y, Zhang J-P, Li Q-F, Qu L-H, Shu W-S, Chen Y-Q. 2014. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 15:512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZY, Li AM, Adeola AC, Liu YH, Irwin DM, Xie HB, Zhang YP. 2014. Genome-wide identification of long intergenic noncoding RNA genes and their potential association with domestication in pigs. Genome Biol Evol. 6:1387-1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.